Abstract
Many eukaryotic and viral regulatory proteins are known to undergo posttranslational modifications including phosphorylation, which plays a critical role in many aspects of cell function. Previous studies from our and other laboratories indicated that the JC virus (JCV) late regulatory protein, agnoprotein, plays an important role in the JCV life cycle. Agnoprotein contains several potential phosphorylation sites, including Ser7, Ser11, and Thr21, which are potential targets for the serine/threonine-specific protein kinase C (PKC). In this study, we investigated the functional significance of these phosphorylation sites for the activity of agnoprotein. In vitro and in vivo kinase assays demonstrated that agnoprotein is a target for phosphorylation by PKC. In addition, each of the PKC phosphorylation sites was mutated to Ala singly and in combination, and the effects of these mutations on the JCV life cycle were analyzed. Although the expression of each mutant agnoprotein was detectable during the infection cycle, virus containing each of these mutations failed to propagate. These results contrast with those obtained with an agnoprotein start codon point (Pt) mutant where agnoprotein expression was completely inhibited. The Pt mutant was viable but replicates less efficiently than the wild type (WT). Moreover, conservative substitutions at PKC phosphorylation sites (Ser7, Ser11, and Thr21 to Asp) resulted in a viable virus, which further demonstrate the importance of these sites on agnoprotein function. Further analysis of the mutants by viral release assay and electron microscopy studies revealed that viral particles were efficiently released from infected cells and morphologically indistinguishable from those of WT but were deficient in DNA content. This may account for the defective propagation of the mutants. These results imply that phosphorylated forms of agnoprotein may have essential functions in the viral life cycle and serve as potential targets for therapeutic interventions to limit JCV propagation and JCV-induced diseases.
Posttranslational modifications including phosphorylation are critical for the activity of many regulatory proteins (40). Perhaps the most extensively characterized viral protein in this manner is large T antigen (LT-Ag) of simian virus 40 (SV40), which is a multifunctional nuclear phosphoprotein and regulates key events in the viral life cycle including initiation of viral DNA replication and activation of viral late genes. The binding activity of SV40 LT-Ag to the viral origin of replication is stimulated when it is phosphorylated by p34cdc2 kinase (21). Cooperative hexamer assembly is facilitated when SV40 LT-Ag is dephosphorylated by phosphatase 2A (26). Similarly, phosphorylation has also been shown to affect the activities of many other viral regulatory proteins that are involved in DNA replication, including mouse polyomavirus LT-Ag (43), the papillomavirus E1 protein (45), JCV LT-Ag (38), human immunodeficiency virus type 1 vpr (1), and herpes simplex virus type 1 origin binding protein (15).
JCV is a human polyomavirus with a double-stranded covalently linked circular genome and is the etiological agent of a fatal demyelinating disease, progressive multifocal leukoencephalopathy (PML) (5, 28, 32). Although JCV shows a high degree of sequence homology to the human BK virus (BKV) and simian SV40 polyomaviruses in coding regions (∼70%), the noncoding regulatory region of each virus is highly divergent from one another. This divergence appears to play a determining role in the unique pattern of expression of each virus in different cell types and tissues (11, 19). In addition, JCV exhibits an unusually narrow tissue tropism (10, 17, 39) in that it lytically infects oligodendrocytes in the central nervous system and causes PML. JCV is a slow-growing virus compared to SV40. It takes several weeks to grow an equivalent amount virus in tissue culture compared to 2 to 3 days for SV40. JCV also exhibits marked divergence from SV40 with respect to the mode of viral entry into cells. SV40 enters cells through a caveola-dependent endocytosis pathway (2, 36), whereas JCV enters by a clathrin-dependent receptor-mediated endocytosis pathway (25).
In addition to encoding viral capsid proteins, the late coding region of JCV encodes a small and basic regulatory protein, agnoprotein, whose functions in the JCV life cycle remain elusive. Agnoprotein localizes to the cytoplasmic and perinuclear regions when expressed in infected cells (34). We have previously demonstrated that agnoprotein functionally interacts with JCV LT-Ag and the cellular factor, YB-1 (31, 34). In addition, we showed that agnoprotein dysregulates cell cycle progression in that the cells stably expressing agnoprotein largely accumulate at the G2/M phase of the cell cycle (8). Recent observations also indicate that JCV agnoprotein may be involved in negative regulation of DNA repair by modulating the activity of Ku70 and Ku80 (9). Furthermore, it was recently reported that agnoprotein interacts with cellular proteins, FEZ1 and HP1α, and may promote the transport of virions from the nucleus to the cytoplasm (23) and thereby facilitate the propagation of JCV (37).
Computer-aided motif search analysis of agnoprotein has revealed that it contains potential phosphorylation sites for several kinases, including protein kinase C (PKC). PKC belongs to a group of kinases that phosphorylate serine or threonine residues of target proteins. This kinase plays a crucial role in the initial events of signal transduction pathways (4, 13) downstream of growth factor receptors. It relies on the stimulation by the products of phospholipid hydrolysis for its activation. A membrane bound enzyme called phospholipase C mediates the hydrolysis of inositol phospholipids into diacylglycerol (DAG) and inositol 1,4,5-triohosphate [Ins(1,4,5)P3] products. Ins(1,4,5)P3 in turn triggers the release of calcium (Ca2+) from intracellular stores. Both release Ca2+ and DAG and then activate PKC, which subsequently phosphorylates serine and threonine residues on a variety of intracellular proteins, including proteins that are components of the transcriptional machinery, regulatory proteins of cytoskeleton, growth factor receptors, and proteins that regulate ion channels (4, 13).
We have analyzed here the importance of the potential PKC phosphorylation sites of agnoprotein for the JCV life cycle. Our results demonstrated that agnoprotein was a target for phosphorylation by PKC in vitro and in vivo. Agnoprotein phosphorylation mutants were expressed during the infection cycle, but mutant viruses were unable to sustain replication cycle. In addition, we showed that the mutant viral particles were successfully released from the infected cells but were deficient in viral DNA content compared to the wild type (WT), which may account for their defective propagation in cell culture.
MATERIALS AND METHODS
Cell lines.
SVG-A is a subclonal population of a human glial cell line that was established by transformation of human fetal glial cell line with an origin-defective SV40 mutant and has been described previously (20). These cells do not express either SV40 agnoprotein or viral capsid proteins (VPs). SVG-A cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and antibiotics (penicillin-streptomycin, 100 μg/ml). They were maintained at 37°C in a humidified atmosphere with 7% CO2.
Plasmid constructs and mutant viruses.
The BstXI site of Bluescript KS vector was inactivated by cutting and filling in with Klenow, followed by religation. Then, the 4556- to 980-bp region of JCV Mad-1 strain (4556-980) was PCR amplified by using Mad-1 forward (4556-4588) primer (5′-TCTTGGGTTAAGTCACACCCAAACCATTGTCT-3′) and Mad-1 reverse (980-960) primer (5′-GTCTAGGATCTAAATAGTGAATATTG-3′) and subcloned into the SmaI site of the modified Bluescript KS vector. This new plasmid was designated Bluescript KS (BstXI−) Mad-1 (4556-980) and used for site-directed mutagenesis to convert Agno-Ser7, Agno-Ser11, and Agno-Thr21 into Ala. Three different mutants were created. In mutant 1 (Mut1), only Thr21 was substituted for Ala. In mutant 2 (Mut2), Ser7 and Ser11 were both substituted for Ala. In mutant 3 (Mut3), Ser7, Ser11, and Thr21 were altogether substituted for Ala. Ser7, Ser11, and Thr21 altogether were also substituted for Asp in the viral background (Mut-Asp). Site-directed mutagenesis was performed by using the QuikChange site-directed mutagenesis kit from Stratagene (catalog no. 200519). Mutated nucleotides were verified by DNA sequencing. In addition, JCV Mad-1 WT genome was subcloned into Bluescript KS (BstXI−) vector at BamHI site and the new plasmid was designated Bluescript KS (BstXI−) Mad-1 WT. The BstXI/HpaI fragment of Mad-1 WT was then replaced with BstX I/HpaI fragment of each mutant to create mutant viruses. The new plasmids were called Bluescript KS (BstXI−) Mad-1 Mut1(Agno Thr21 to Ala), Bluescript KS (BstXI−) Mad-1 Mut2(Agno Ser7 and Ser11 to Ala), Bluescript KS (BstXI−) Mad-1 Mut3(Agno Ser7, Ser11 and Thr21 to Ala), and Bluescript KS (BstXI−) Mad-1 Mut-Asp(Agno Ser7, Ser11, and Thr21 to Asp). JCV Mad-1 point (Pt) mutant virus genome was kindly provided by K. Nagashima of Japan. In the Pt mutant, the NcoI site (CCATGG) of Mad-1 WT, containing the translation initiation codon (ATG) of agnoprotein, was converted into XhoI site (CTCGAG) by base substitution. Thus, the point mutant virus is defective in producing agnoprotein. Pt mutant was also cloned into BamHI site of Bluescript KS, and the new plasmid was designated as Bluescript KS Mad-1 Pt.
In vitro kinase assay.
GST and GST-agnoprotein were expressed in bacteria and were purified as described previously (31, 34). One microgram of bacterially expressed GST alone or GST-agnoprotein was incubated with 10 U of PKC (Promega) in a reaction mixture (50 μl) containing 25 mM Tris-HCl (pH 7.4), 20 mM MgCl2, 5 mM dithiothreitol, 2.5 μCi of [γ-32P]ATP, and 10% glycerol at 30°C for 1 h. After the kinase reaction, phosphorylated proteins (GST alone or GST-agnoprotein) were extensively washed with reaction buffer containing no [γ-32P]ATP, fractionated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE), and visualized by autoradiography.
Phosphatase treatment and in vivo kinase assay.
To avoid the use of high level of [32P]orthophosphate in our biocontainment facility, we used an indirect approach to demonstrate that agnoprotein is phosphorylated in vivo. Agnoprotein was immunoprecipitated from whole-cell extracts (200 μg) prepared in an extraction buffer supplemented with a PhosphoSafe extraction reagent (Novagen, catalog no. 71296-3), at 5 or 10 days posttransfection from SVG-A cells transfected with JCV Mad-1 genome and subsequently treated or not treated with alkaline phosphatase (Roche, catalog no. 11097075001) in a 100-μl reaction volume using 80 U of enzyme to remove the phosphate groups from agnoprotein (30). Phosphatase-treated or nontreated agnoprotein was washed and subjected to a kinase reaction in the presence of phosphatase inhibitor (phosphatase inhibitor cocktail I; Sigma, catalog no. P2850), using 10 U of PKC (Promega) in a reaction mixture (50 μl) containing 25 mM Tris-HCl (pH 7.4), 20 mM MgCl2, 5 mM dithiothreitol, 2.5 μCi of [γ-32P]ATP, and 10% glycerol at 30°C for 1 h. Kinase-treated agnoproteins were then extensively washed with reaction buffer containing no [γ-32P]ATP, fractionated by SDS-15% PAGE, and visualized by autoradiography. Agnoprotein phosphorylation mutants were also immunoprecipitated from extracts prepared at 5 days posttransfection, subjected to the phosphorylation-rephosphorylation process, and analyzed as described above.
Transfection and/or infection.
SVG-A cells (2 × 106 cell/75-cm2 flask) were transfected either with Mad-1 WT or with Mad-1 Mut1, Mad-1 Mut2, Mad-1 Mut3, or Mad-1 Mut-Asp viral DNA (8 μg each) by using Lipofectin according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA). At 5 h posttransfection, cells were washed twice with phosphate-buffered saline (PBS) and fed with DMEM supplemented with 2% FBS and antibiotics (penicillin-streptomycin, 100 μg/ml). Cells were then maintained at 37°C in a humidified atmosphere with 7% CO2 until they were processed for extract preparation or DNA isolation.
Indirect immunofluorescence microscopy.
Indirect immunofluorescence microscopy studies were performed as previously described (3, 30, 33, 34) using an anti-agnoprotein polyclonal antibody.
Whole-cell and nuclear extract preparation.
Whole-cell and nuclear extracts were prepared as described previously (18, 30, 35).
Western blotting.
Whole-cell extracts, prepared from SVG-A cells untransfected or transfected with respective plasmid constructs, were resolved by SDS-PAGE and blotted onto nitrocellulose membranes. Blotted membranes were probed either with an anti-agnoprotein polyclonal antibody as described previously (30, 34) and detected by an ECL kit according to the manufacturer's recommendations (Amersham, England). In parallel, nuclear extracts, prepared from either transfected or untransfected cells, were analyzed for VP1 by Western blotting utilizing a monoclonal anti-VP1 antibody (pAB597, kindly provided by W. Atwood, Brown University, Rhode Island). In addition, the released viral particles were immunoprecipitated by an anti-VP1 antibody (pAB597), and immunocomplexes were probed with either anti-VP1 or an anti-SV40 antibody (Lee Biomolecular Research) that detects viral capsid proteins, which is also cross-reactive with JCV capsid proteins.
Replication assay.
Replication assays were carried out as previously described (31). Briefly, SVG-A cells (2 × 106 cell/75-cm2 flask) were transfected either with Mad-1 WT or with Mad-1 Mut1, Mad-1 Mut2, Mad-1 Mut3, Mad-1 Pt, or Mad-1 Mut-Asp viral DNA (8 μg each) using Lipofectin according to manufacturer's recommendations (Invitrogen, Carlsbad, CA). At 5 h posttransfection, cells were washed twice with PBS. SVG-A cells were fed with regular DMEM containing 2% FBS. At the indicated points posttransfection, low-molecular-weight DNA containing both input and replicated viral DNA was isolated by using QIAGEN spin columns (46), digested with BamHI and DpnI enzymes, resolved on a 1% agarose gel, and analyzed by Southern blotting using probe prepared from whole Mad-1 genome.
Viral particle release assay.
Mad-1 WT genome or its agnoprotein phosphorylation mutants (Mut1, Mut2, and Mut3) were separately transfected into SVG-A cells (8 μg of DNA/2 × 106 cells/75-cm2 flask). Whole media (12 ml) were collected at indicated time points, spun at 10,000 rpm to clear cell debris and was subjected to immunoprecipitation with an anti-VP1 antibody (10 μg). Half of the samples were analyzed by Western blotting with anti-VP1 antibody (pAB597). The other half was analyzed by Southern blotting for detection of encapsidated viral DNA. The viral DNA from capsids was purified by using QIAGEN spin columns (46), digested with BamHI and DpnI enzymes, resolved on a 1% agarose gel, and analyzed by Southern blotting with a probe prepared from whole Mad-1 genome.
Electron microscopy.
Released viral particles (WT and Mut3) were immunoprecipitated with an anti-VP1 antibody (pAB597) and processed by the electron microscopy (EM) Biomedical Imaging Core, Department of Pathology, University of Pennsylvania.
RESULTS
Agnoprotein is phosphorylated by PKC in vitro.
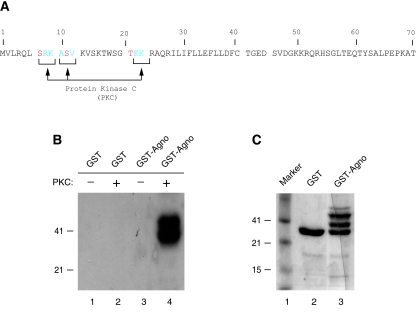
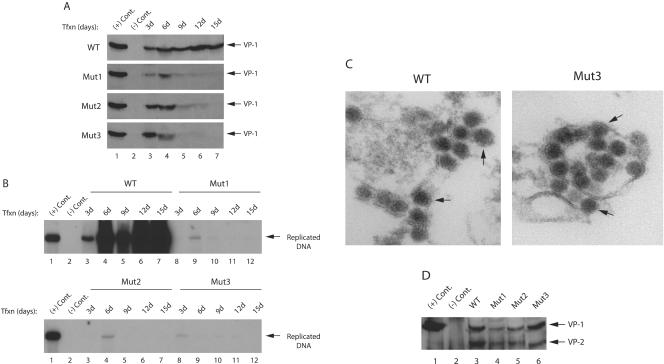
Phosphorylation plays critical roles in the function of many regulatory viral and cellular proteins (1, 41-43, 45). Agnoprotein, encoded by the leader sequences of the late transcripts of JCV, contains potential phosphorylation sites for a serine and a threonine kinase, PKC (Fig. 1A). Therefore, we sought to demonstrate whether this enzyme is able to phosphorylate agnoprotein in vitro. To do this, bacterially produced GST or GST-agnoprotein, both of which were immobilized on glutathione S-transferase (GST)-Sepharose beads, was incubated with PKC as described in Materials and Methods. Proteins were then fractionated on a SDS-PAGE and visualized by autoradiography. As shown in Fig. 1B, GST-Agno fusion protein is phosphorylated by PKC (lane 4) but GST alone is not (lane 2). This demonstrates that agnoprotein is a specific substrate for PKC (compare lane 4 to lane 2). In Fig. 1C, we analyzed the purified GST protein and GST-agnoprotein by SDS-PAGE, followed by Coomassie staining. Of note, GST-agnoprotein is not produced as a single band in bacteria as previously reported (31).
FIG. 1.
Agnoprotein is phosphorylated by PKC in vitro. (A) Deduced amino acid sequence of agnoprotein. Potential phosphorylation sites on agnoprotein for PKC (Ser7, Ser11, and Thr21) are depicted by arrows. (B) PKC phosphorylates agnoprotein in vitro. One microgram of bacterially produced GST alone or GST-agnoprotein (GST-Agno) fusion protein, immobilized on GST-Sepharose beads, was incubated with PKC as described in Materials and Methods. Phosphorylated proteins, which bound to GST-Sepharose beads, were extensively washed with reaction buffer, fractionated by SDS-12% PAGE, and visualized by autoradiography. (C) Analysis of bacterially produced GST and GST-agnoprotein by a SDS-12% PAGE, followed by Coomassie staining. Due to rapid degradation of the agnoprotein, it is difficult to produce it as a single-banded protein in bacteria (34).
Agnoprotein is phosphorylated during the infection cycle.
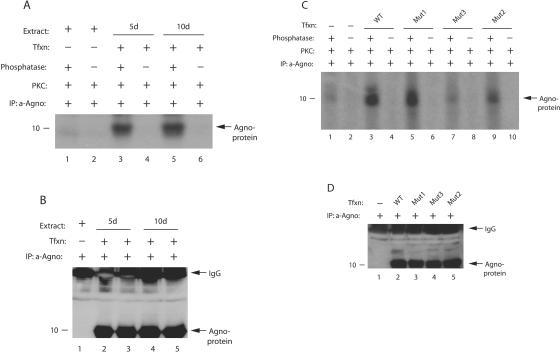
We also wanted to investigate whether agnoprotein is phosphorylated in vivo during the infection cycle. Whole-cell extracts were prepared at 5 and 10 days posttransfection from SVG-A cells either untransfected or transfected with JCV Mad-1 genome in the presence of phosphatase inhibitors. Whole-cell lysates were then subjected to immunoprecipitation with an anti-agnoprotein antibody. A small portion of the immunocomplexes was treated with phosphatase or left untreated, and a kinase reaction was performed on the samples as described in Materials and Methods. The assumption in the treatment of the samples with phosphatase was the following. If agnoprotein was phosphorylated in vivo during the infection cycle, the phosphate groups would be removed upon treatment with phosphatase, and we should obtain a relatively higher signal from the samples that were treated with phosphatase after the kinase reaction compared to the untreated ones. This in turn provides information about the phosphorylation status of agnoprotein in vivo. As shown in Fig. 2A, agnoprotein immunoprecipitated from the extracts prepared at 5 and 10 days posttransfection exhibited a strong signal upon dephosphorylation, followed by phosphorylation by PKC, compared to untreated samples (compare lanes 3 and 5 with lanes 4 and 6, respectively), suggesting that agnoprotein is phosphorylated in vivo during the infection cycle. As a negative control, we used immunocomplexes from untransfected cells for the kinase reaction but detected a faint signal at background levels after the dephosphorylation/rephosphorylation process (compare lane 1 to lanes 3 and 5), which supports the specificity of our in vivo phosphorylation reactions for agnoprotein. In Fig. 2B, we demonstrated by Western blotting that a relatively equal amount of agnoprotein was used in kinase reactions.
FIG. 2.
Agnoprotein is phosphorylated in vivo. (A) Phosphatase-treated agnoprotein exhibits higher signal upon phosphorylation. Whole-cell lysates were prepared from SVG-A cells, transfected with JCV Mad-1 genome, in an extraction buffer supplemented with a PhosphoSafe extraction reagent (Novagen, catalog no. 71296-3), at 5 and 10 days posttransfection. Extract (200 μg) was immunoprecipitated with an anti-agnoprotein antibody (α-Agno, 4 μl) as indicated. Whole-cell lysates were also prepared from untransfected cells and processed as described for the transfected cells. A small portion of the samples (one-sixth portion) was then either treated (lanes 1, 3, and 5) or not treated (lanes 2, 4, and 6) with phosphatase and rephosphorylated with [γ-32P]ATP using PKC enzyme as described in Materials and Methods. Samples were then washed extensively with reaction buffer and fractionated by SDS-15% PAGE, followed by autoradiography. Representative data are shown here. (B) A small portion (one-sixth portion) of immunoprecipitated samples was directly analyzed prior to phosphatase treatment by Western blotting with an α-Agno antibody. In lane 1, whole-cell lysate prepared from untransfected cells was immunoprecipitated by an anti-Agno antibody and loaded as a negative control. (C) Phosphorylation of Mut1, Mut2, and Mut3 by PKC. Mad-1 WT genome and its agnoprotein phosphorylation mutants (Mut1, where Thr21 was substituted for Ala; Mut2, where Ser7 and Ser11 were substituted for Ala; and Mut3, where Ser7, Ser11, and Thr21 were substituted for Ala) were transfected into SVG-A cells, and whole-cell lysates were prepared in an extraction buffer supplemented with a PhosphoSafe extraction reagent at day 5 posttransfection. Whole-cell lysate was also prepared from untransfected cells. WT and mutant agnoproteins were immunoprecipitated (200 μg of protein) with an anti-Agno antibody (4 μl). A one-sixth portion of the immunoprecipitated proteins was subjected to dephosphorylation, followed by phosphorylation by PKC, fractionation by SDS-PAGE, and analysis by autoradiography as described for panel A. The signal intensities for the bands at lanes 1, 3, 5, 7, and 9 were determined by scintillation counting. (D) A small portion (one-sixth portion) of immunoprecipitated samples was analyzed by Western blotting prior to phosphatase treatment with an α-Agno antibody. In lane 1, whole-cell lysate prepared from untransfected cells was immunoprecipitated by a anti-Agno antibody and loaded as a negative control. Tfxn, transfection; d, day; IP, immunoprecipitation.
In order to further confirm whether suspected phosphorylation sites of agnoprotein are in fact a target for PKC in vivo, we individually or combinatorially mutated these sites into Ala and the repeated kinase assays. SVG-A cells were transfected with either WT or Mut1, Mut2, or Mut3, and whole-cell lysates, prepared at 5th day posttransfection, were subjected to immunoprecipitation with an anti-agnoprotein antibody followed by dephosphorylation/rephosphosphorylation reactions as described for panel A. Of note, we were unable to produce each phosphorylation mutant of agnoprotein as a GST fusion protein in bacteria because of the rapid degradation, and therefore we did not use them in kinase assays. As shown in Fig. 2C, WT agnoprotein was phosphorylated by PKC after dephosphorylation (lane 3), which is consistent with our results from Fig. 2A. We also detected a strong signal for Mut1 but the efficiency of phosphorylation was ∼26% less than for WT because only Thr21 was mutated in this mutant and the other two potential phosphorylation sites for PKC (Ser7 and Ser11) remained intact. In addition, Mut2 also exhibited a relatively strong signal, where only Thr21 is intact, but it was significantly less than WT (∼78% less, compare lane 9 to lane 3). However, as expected, the level of signal for Mut3, where all three phosphorylation sites (Ser7, Ser11, and Thr21) were substituted for Ala, dropped to the background levels after the dephosphorylation/rephosphorylation process (compare lane 7 to lane 1). Figure 2D demonstrates the use of a relatively equal amount of agnoprotein in kinase reactions by Western blotting. Taken together, these results showed that agnoprotein is phosphorylated in vivo and that Ser7, Ser11, and Thr21 are targets for phosphorylation by PKC.
Agnoprotein phosphorylation mutants were unable to sustain infection cycle.
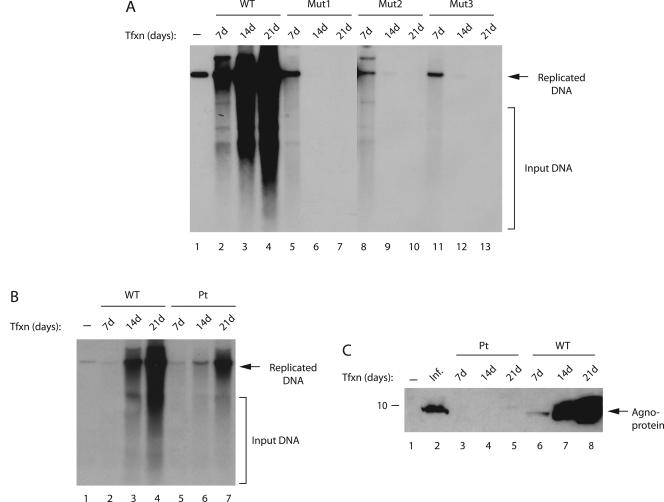
We next examined the effect of phosphorylation mutants of agnoprotein on JCV DNA replication by using a DpnI replication assay. For this purpose, JCV Mad-1 WT or its phosphorylation mutant genomes (Mut1, Mut2, and Mut3) were transfected into SVG-A cells, and low-molecular weight DNA was isolated by a column purification method (46) at 7, 14, and 21 days posttransfection. Subsequently, newly replicated DNA was digested with BamHI and DpnI enzymes and analyzed by Southern blotting. As shown in Fig. 3A, as expected, WT virus replicated efficiently during the infection cycle, and its level of replication increased gradually but significantly toward 21 day posttransfection (lanes 2 to 4). However, it was surprising that all three phosphorylation mutants of agnoprotein showed replication only during the early phases of infection cycle (lanes 5, 8, and 11) and were unable to sustain replication cycles at later time points. It was also evident from our results that mutation at Thr21 alone or those at Ser7 and Ser11 were sufficient to cause this effect.
FIG. 3.
Replication properties of agnoprotein phosphorylation and point mutants. (A) Effect of agnoprotein phosphorylation mutants on viral replication. SVG-A cells were transfected with either Mad-1 WT genome or its agnoprotein mutant genomes (Mut1, Mut2, and Mut3) (8 μg/2 × 106 cells/75-cm2 flask) using Lipofectin as described in Materials and Methods. At 7, 14, and 21 days posttransfection, low-molecular-weight DNA was isolated by using a QIAGEN spin column (46), digested with BamHI and DpnI enzymes, resolved on a 1% agarose gel, and analyzed by Southern blotting. (B) Replication efficiency of an agnoprotein point mutant (Pt) virus compared to WT. SVG-A cells were transfected with either WT or Pt mutant virus, and replicated DNA was analyzed by a DpnI assay as described for panel A. In lane 1 in each panel, Mad-1 WT genome (2 ng) digested with BamHI was loaded as a positive control. Representative data are shown here. Input DNA (transfected) is indicated by a bracket, and replicated DNA is indicated by an arrow. (C) In parallel to the studies described for panel B, whole-cell lysates were also prepared at the indicated time points and analyzed by Western blotting with an anti-agnoprotein antibody. In lane 1, whole-cell extract from untransfected cells was loaded as a negative control. In lane 2, whole-cell extract from infected cells was loaded as a positive control. Tfxn, transfection; Inf, infection. The migration pattern of a molecular mass marker is shown on the left side of the panel in kilodaltons.
Next, we compared the replication properties of an agnoprotein point (Pt) mutant with those of the WT virus. In Pt mutant, the translation initiation codon (ATG) was altered, and therefore it does not produce agnoprotein. As shown in Fig. 3B, the agnoprotein Pt mutant was capable of replicating in the absence of agnoprotein but substantially less efficiently than WT (compare lanes 3 and 4 to lanes 6 and 7, respectively), a finding which correlates with previously published reports (22, 27). In Fig. 3C, the absence of the expression of agnoprotein from Pt mutant genome was demonstrated by Western blotting (lanes 3 to 5). Altogether, these observations indicate that agnoprotein phosphorylation mutants and Pt mutant behave differently during the infection cycle.
Agnoprotein phosphorylation mutants are expressed during the infection cycle.
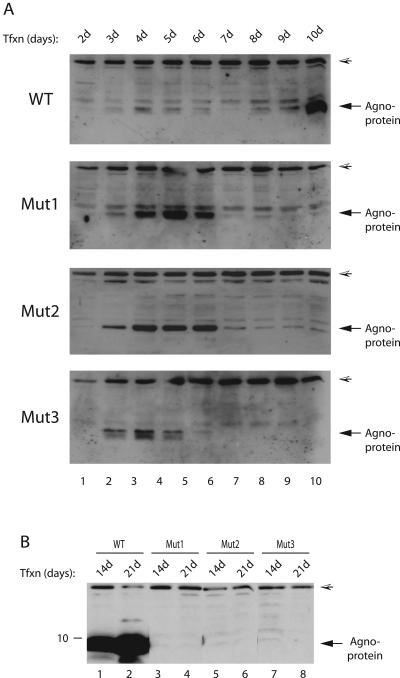
It was surprising to observe that agnoprotein phosphorylation mutants were unable to sustain viral replication cycle but displayed the ability to replicate during the early phases of infection. We then sought to determine whether each mutant agnoprotein was expressed during the infection cycle and whether the cells infected with the mutants show morphological differences with respect to WT. SVG-A cells were transfected with either Mad-1 WT or with Mut1, Mut2, or Mut3 separately, and whole-cell extracts were prepared at different time points and analyzed by Western blotting with an anti-agnoprotein antibody. As demonstrated in Fig. 4A, agnoprotein expression for both WT and mutants (lane 2) was detectable as early as day 3 posttransfection; the expression levels of mutant proteins (for all three mutants) were noticeably higher than that of WT for certain time points (lanes 2 to 6). In order to explain this observed phenomenon, we investigated a possibility that this might be due to a case, where mutant proteins are more stable than WT, by using a pulse-chase labeling assay. The results from these assays demonstrated that the turnover rates of the mutant agnoproteins are similar to that of WT, suggesting that some other mechanism(s), other than protein stability, is involved in the higher expression of mutants compared to WT (data not shown). In this regard, however, it is possible that there might exist a negative feedback mechanism, which is disrupted in mutant cases and may contribute to this observed phenomenon. In addition, further analysis of the expression levels of WT and mutant agnoproteins for later days showed that their expression decreased to minimal levels by day 7 posttransfection. This decrease was seen even earlier in the case of Mut3 (day 6, lane 6). One explanation for the observed decrease is the following. It is likely that the first round of infection cycle was complete by day 7 posttransfection, and newly infected cells did not have chance to express viral proteins at higher levels. It was also surprising that whereas WT gradually recovered from this decrease by day 10 posttransfection (lanes 8 to 10), neither of the mutants was able to do so as the infection cycle proceeded forward, suggesting that the phosphorylation mutant agnoprotein had a dramatic effect on JCV life cycle. In addition, we also examined the level of agnoprotein phosphorylation mutants beyond day 10 posttransfection. We were unable to detect mutant agnoproteins for later days of infection cycle compared to WT, which is consistent with the results from replication studies where mutants were defective for replication beyond day 7 posttransfection (Fig. 3A).
FIG. 4.
Mutant agnoproteins are expressed during the infection cycle. (A) Analysis of WT and mutant agnoproteins during the early phases of infection cycle. The Mad-1 WT genome or its phosphorylation mutants of agnoprotein (Mut1, Mut2, and Mut3) were separately transfected into SVG-A cells (8 μg of DNA/2 × 106 cells/75-cm2 flask). Whole-cell lysates were prepared at the indicated time points and analyzed by Western blotting (40 μg/lane) with an anti-agnoprotein antibody. A hatched arrowhead in each panel points to a nonspecific band, which was detected by using an anti-agnoprotein antibody. (B) In parallel, whole-cell lysates were also analyzed for agnoprotein expression for the later days of infection cycle by Western blotting as described for panel A.
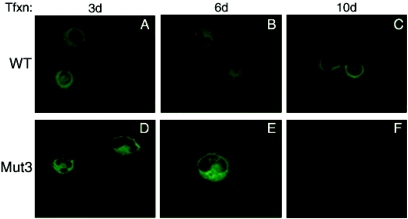
Next, we examined the effect of one of the agnoprotein phosphorylation mutants (Mut3) on the morphology of the infected cells by immunocytochemistry (Fig. 5). SVG-A cells were transfected either with WT or Mut3, fixed at 3, 6, and 10 days posttransfection; probed with an anti-agnoprotein antibody; and analyzed under a fluorescence microscope for morphological alterations. At early phases of infection (day 3), the cells transfected with WT genome had a sphere-shaped nuclear morphology and WT agnoprotein spread throughout the cytoplasm, although the highest concentrations were localized to the perinuclear area (Fig. 5A). In contrast, the cells transfected with Mut3 had a kidney-shaped nuclear morphology at the same day posttransfection (day 3), and mutant agnoprotein was more perinuclear but accumulated mostly at the hilus region of a kidney-shaped nucleus. We also observed a kidney-shaped nuclear morphology for WT, but this occurred at the later datum points (Fig. 5B). It was also evident that the effect of WT agnoprotein on cell morphology was not as dramatic as the one that was observed for Mut3 (Fig. 5D and E).
FIG. 5.
Analysis of WT agnoprotein and its phosphorylation mutant (Mut3) by immunocytochemistry. SVG-A cells, transfected with either Mad-1 WT or Mad-1 Mut3 viral DNA, were fixed with cold acetone at the indicated time points and incubated with a polyclonal anti-agnoprotein primary antibody overnight as described in Materials and Methods. Cells were then washed four times with PBS-0.01% Tween 20 for 10-min intervals and incubated with an anti-rabbit fluorescein isothiocyanate-conjugated goat secondary antibody for 45 min. Finally, the slides were washed three times with PBS, mounted, and examined with a fluorescence microscope to detect agnoprotein.
Analysis of virion release and genomic content of the viral particles.
The inability of agnoprotein phosphorylation mutants to sustain viral replication cycle suggested the possibility that the mutants were either defective in the release of the infectious viral particles or that the released particles were deficient in DNA content. To distinguish between these possibilities, the released viral particles were immunoprecipitated from the culture media and analyzed by both Western and Southern blotting. The culture media were collected from the cells transfected either with WT or mutants (Mut1, Mut2, and Mut3) at different time points, and viral particles were subjected to immunoprecipitation with an anti-VP1 antibody. The samples were then split into two equal portions. One portion was analyzed by Western blotting with anti-VP1 antibody. The other portion was analyzed by Southern blotting for the detection of encapsidated viral DNA. As shown in Fig. 6A, VP1 was readily detectable for WT at every time point evaluated posttransfection (lanes 3 to 7) by Western blotting. We also observed comparable levels of virion release for each mutant virus at days 3 and 6 posttransfection (lanes 3 and 4). An even a higher level of signal was observed for Mut2 and Mut3 at day 3 posttransfection compared to the WT. However, the observed signal declined to minimal levels after the day 6 datum point for mutants compared to the WT (lanes 5 and 6). The observed signal was not detectable for Mut1 and Mut2 beyond day 12 posttransfection. This occurred even earlier for Mut3 (day 9, lane 5). These results demonstrated that mutant viruses were capable of releasing the viral particles at least as efficiently as WT. In light of these findings, we then reasoned that perhaps the released viral particles were deficient in viral DNA and were therefore unable to successfully propagate. To investigate this possibility, we analyzed the viral DNA content of the other half of the immunoprecipitated samples by Southern blotting. As demonstrated in Fig. 6B, we detected, as expected, a relatively strong signal for encapsidated WT viral DNA, isolated at day 3 posttransfection (lane 3), and the intensity of the signal gradually increased for the later time points (lanes 4, 6, and 7) except for the day 9 (lane 5). One of the possible explanations for this discrepancy is that, since JCV completes its first round of infection cycle in about 6 to 7 days, the newly infected cells did not appear to have sufficient time to release comparable amount of virions into the culture media. In contrast to our findings for WT, we detected only a very weak signal for Mut1 and Mut2 at day 6 and for Mut3 at day 3 posttransfection and did not observe a detectable level of signal for the rest of the time points, a finding that which strongly supports the hypothesis that the viral particles, released from the cells transfected with the mutants, were deficient in DNA content. This then appears to result in a defective propagation of the mutant viruses at the subsequent rounds of infection cycles.
FIG. 6.
Analysis of virus release for phosphorylation mutants of agnoprotein. (A) Mad-1 WT genome or its agnoprotein phosphorylation mutants (Mut1, Mut2, and Mut3) were separately transfected into SVG-A cells (8 μg of DNA/2 × 106 cells/75-cm2 flask). Whole media (12 ml) were collected at the indicated time points, spun at 10,000 rpm to clear cell debris, and subjected to immunoprecipitation with an anti-VP1 antibody (10 μg). Half of the samples were analyzed by Western blotting with anti-VP1 antibody. The other half were analyzed by Southern blotting to detect encapsidated viral DNA. In lane 1, nuclear extract, prepared from infected cells, was loaded as a positive control [(+) Cont.]. In lane 2, nuclear extract, prepared from uninfected SVG-A cells, was loaded as a negative control [(−) Cont.)]. (B) Analysis of encapsidated viral DNA by Southern blotting. Encapsidated viral DNA from the released viral particles was purified by using QIAGEN spin columns (46), digested with BamHI and DpnI enzymes, resolved on 1% agarose gel, and analyzed by Southern blotting with probes prepared from whole Mad-1 genome. In lane 1, Mad-1 WT genome (2 ng) digested with BamHI was loaded as a positive control [(+) Cont.]. In lane 2, DNA isolated from untransfected cells was loaded as a negative control [(−) Cont.]. (C) Analysis of released viral particles by EM. Mad-1 WT genome or its agnoprotein mutant (Mut3) were separately transfected into SVG-A cells (8 μg of DNA/2 × 106 cells/75-cm2 flask). Released viral particles were immunoprecipitated by using an anti-VP-1 (pAB597) antibody on day 6 posttransfection and analyzed by EM. Arrows in both panels point to the JCV particles. (D) Mad-1 WT genome or its agnoprotein phosphorylation mutants (Mut1, Mut2, and Mut3) were separately transfected into SVG-A cells (8 μg of DNA/2 × 106 cells/75-cm2 flask). The released viral particles were immunoprecipitated by an anti-VP-1 antibody (pAB597) and analyzed by Western blotting with an antibody (a rabbit polyclonal antibody raised against SV40 capsid proteins, which is also cross-reactive with JCV viral capsid proteins) that detects viral capsid proteins. In lane 1, nuclear extract, prepared from infected cells, was loaded as a positive control [(+) Cont.]. In lane 2, nuclear extract, prepared from uninfected SVG-A cells, was loaded as a negative control [(−) Cont.].
Released mutant viral particles are morphologically similar to WT.
We further analyzed the nature of the released mutant viral particles by EM and Western blotting. For this purpose, the culture media were collected from the cells transfected either with WT or Mut3 on day 6 posttransfection, and viral particles were subjected to immunoprecipitation with an anti-VP1 antibody and processed for EM analysis. As shown in Fig. 6C, the mutant viral particles are morphologically indistinguishable from those of WT, which convincingly suggested that mutant virus is intact and therefore further suggested the possibility that they may also contain other viral structural capsid proteins. This was investigated by Western blot analysis of the released particles. As demonstrated in Fig. 6D, all three mutant viral particles (Mut1, Mut2, and Mut3), in addition to VP1, also contain VP2 capsid protein. Altogether, the results from the EM and Western blotting studies confidently demonstrate that the phosphorylation mutants of agnoprotein are capable of releasing mature viral capsids which are morphologically similar to WT but are defective in encapsidation of the viral DNA during the infection cycle.
Substitution of Ser7, Ser11, and Thr21 for Asp resulted in a viable virus.
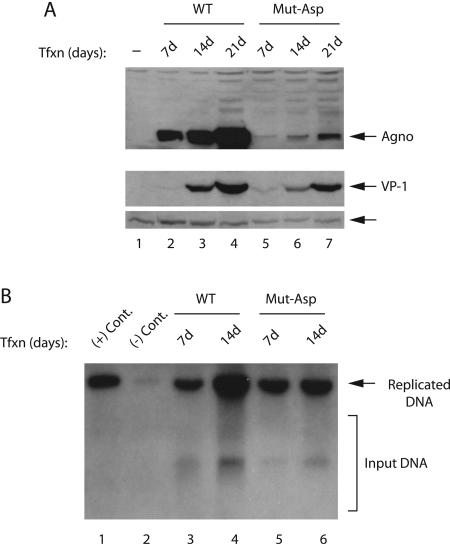
In order to further investigate the functional significance of the phosphorylation sites of agnoprotein, we made conservative substitutions at the potential PKC phosphorylation sites and changed them to Asp (i.e., Ser7, Ser11, and Thr21 to Asp). It was expected that these changes might mimic a constitutive phosphorylation of agnoprotein but still exhibit a defect in the viral life cycle. For this purpose, JCV Mad-1 WT or its Asp mutant genome (Mut-Asp) were transfected into SVG-A cells and, at indicated time points, whole-cell and nuclear extracts were prepared and analyzed by Western blotting for detection of agnoprotein and VP1. As demonstrated in Fig. 7A, conservative changes at Ser7, Ser11, and Thr21 resulted in a viable virus, where we observed the expression of agnoprotein and VP1 at detectable levels, although significantly less compared to WT (compare lanes 2 to 4 to lanes 5 to 7, respectively). Furthermore, the newly generated mutant (Mut-Asp) virus was also able to sustain its replication beyond 7 days posttransfection compared to previously analyzed mutants (Mut1, Mut2, and Mut3). These findings suggest that Asp substitutions mimic a constitutive phosphorylation of agnoprotein and further signifies the importance of phosphorylation in agnoprotein function. In parallel, we also investigated the efficiency of replication of Mut-Asp by DpnI assay. Low-molecular weight DNA was isolated by a column purification method (46) at days 7 and 14 posttransfection. Subsequently, newly replicated DNA was digested with BamHI and DpnI enzymes and analyzed by Southern blotting. These results indicate that Mut-Asp is capable of replicating at later points of infection cycle (Fig. 7B), which is consistent with our findings from the protein expression studies (Fig. 7A).
FIG. 7.
JCV is viable when changed Ser7, Ser11, and Thr21 of agnoprotein to Asp. (A) Mad-1 WT genome or its agnoprotein mutant (Mut-Asp) were separately transfected into SVG-A cells (8 μg of DNA/2 × 106 cells/75-cm2 flask). Whole-cell and nuclear extracts were prepared at indicated time points and were analyzed for the detection of agnoprotein and VP1 expression by Western blotting with both anti-agnoprotein and VP-1 (pAB597) antibodies. An arrow points to a nonspecific band detected by an anti-VP1 antibody, which is to serve as a loading control for VP1 detection. (B) In parallel, low-molecular-weight DNA was isolated at indicated time points from transfectants and analyzed by a DpnI assay as described in Fig. 3A.
DISCUSSION
Phosphorylation is a posttranslational modification that plays crucial roles in regulating the activity of many proteins. This modification can affect the stability (12), subcellular localization (6), protein-protein interaction (44), DNA binding (44), and transcriptional activity (7, 14) of various proteins. Target proteins often have many phosphorylation sites that provide them with a dynamic mechanism for executing their regulatory roles at multiple levels.
Sequence alignment of agnoproteins of polyomaviruses (JCV, BKV, and SV40) demonstrated that these three proteins show a high degree of sequence homology (∼70%). In addition, comparison of these three agnoproteins with respect to conservation of the potential PKC phosphorylation sites (Ser7, Ser11, and Thr21) showed that all three sites are completely conserved between BKV and JCV agnoprotein. Although Ser7 and Ser11 are also conserved between JCV and SV40, Thr21 on JCV agnoprotein is substituted for Ser on SV40 agnoprotein. Previous studies have demonstrated that SV40 (16) and BKV (29) agnoproteins have been reported to be phosphorylated in vivo. Although there are no data indicating a function associated with the phosphorylated form of agnoprotein of SV40 and BKV, Okada et al. reported that JCV agnoprotein may shuttle between cytoplasm and nucleus in a phosphorylation-dependent manner during the infection cycle (22). In the present study, we investigated the functional importance of phosphorylation sites of agnoprotein (Ser7, Ser11, and Thr21) by genetic and biochemical approaches and demonstrated that the phosphorylation of respective target sites on agnoprotein by PKC (Ser7, Ser11, and Thr21) is critical for virus propagation.
It was surprising to observe that neither of the phosphorylation mutants was able to successfully propagate in a tissue culture system. This finding contradicts results that we obtained from an agnoprotein point mutant, where the ATG initiation codon of agnoprotein was altered, thereby ablating agnoprotein expression. This point mutant was viable but propagated less efficiently than WT (Fig. 3B). Protein expression studies showed that each of the phosphorylation site mutant proteins expressed at comparable levels during the first round of viral infection cycle. However, it was interesting to observe that the expression level of each mutant protein declined to minimal levels toward day 7 posttransfection. A similar observation was also made for WT during the same period. However, neither of the mutants recovered from this loss, unlike WT (Fig. 4A). These results suggest that perhaps the mutant viruses are defective in the process of viral release. We investigated this possibility using a viral release assay and found that mutant viral particles were efficiently released from the infected cells compared to WT (Fig. 6A). We then investigated whether the released mutant viral particles were in fact deficient in viral DNA content and therefore defective in propagation. Analysis of such viral particles demonstrated a significant deficiency in the viral DNA content. We further investigated the morphology and nature of the released particles by EM and Western blotting, respectively (Fig. 6A and B). Our results confidently showed that the released mutant viral particles are morphologically indistinguishable from WT and also contain other viral capsid proteins, which strongly indicate that mutant viruses have indeed a deficiency in encapsidation of viral DNA. We also reasoned that conservative changes at the PKC phosphorylation sites of agnoprotein might result in a viable virus, which would then sustain the replication cycle. The substitution of Ser7, Ser11, and Thr21 for Asp indeed resulted in a virus that is viable and is capable of replicating at later points of infection cycle (Fig. 7), which demonstrate the importance of the phosphorylation sites of agnoprotein in the viral replication cycle. These findings explain why the mutants were defective in propagation but do not explain the mechanism(s) that render these mutants defective in viral maturation. A closer inspection of the agnoprotein amino acid sequence revealed that it has an excess of positive charge in the absence of phosphorylation. It is known that phosphate groups provide negative charges to a target protein which in turn perhaps affect the conformation and thereby the function of the target protein. Since neither of the phosphorylation mutants was able to propagate beyond the first round of infection cycle, it is possible that the mutant agnoproteins target specific cellular or viral proteins and restrict their functions by not releasing them at the required time of the infection cycle, thus affecting the process of the viral maturation. WT agnoprotein, on the other hand, might bind to the same factors as its unphosphorylated form, but it releases its binding partners, when it is required, thus permitting the infection cycle to proceed. An alternative mechanism is also likely. That is, while phosphorylated WT agnoprotein prevents viral capsids from forming too soon, and upon dephosphorylation, it permits a normal viral maturation process to take place. In contrast, the mutant agnoproteins may never be able to prevent viral capsids from oligomerization, and therefore the empty capsids form too early, resulting in an abortive infection. Another possibility is that the highly positive mutant agnoprotein may bind to viral DNA and prevent it from being packaged into the capsids. Such alterations may result in morphological changes in infected cells and perhaps in the process of capsid maturation. In fact, morphological comparison of the cells infected with Mut3 and with those infected with WT showed clear differences (Fig. 5). The cells infected with Mut3 displayed a kidney-shaped nuclear morphology during the early periods of infection cycle compared to WT (Fig. 5D). The cells infected with WT showed a more rounded nuclear structure during the same period (Fig. 5A). We also observed a kidney-shaped nuclear structure for WT at the late phases of infection cycle, but it was not as pronounced as the one that was observed for Mut3 (Fig. 5D). Therefore, it is possible that such morphological alterations may have a considerable effect on viral maturation and immature release of the mutant virions from the infected cells.
Based on the impact of agnoprotein phosphorylation mutants on viral propagation, it is reasonable to speculate that agnoprotein phosphorylation may be differentially regulated at the different stages of the infection cycle, and this may have functional consequences. It is possible that at the initial stages of the infection cycle agnoprotein is targeted by PKC to counterbalance the negative effect of nonphosphorylated agnoprotein. However, toward the late stages of the infection cycle, the phosphate groups are partially or completely removed from agnoprotein by certain phosphatases, which in turn contribute to the facilitated release of maturated virions. There are many examples from the literature showing that differentially phosphorylated sites of a protein can regulate its activity. A prime example of this is the regulation of Cdc25C activity during the meiotic G2/M transition. Phosphorylation of Cdc25C at Ser287 by cyclic AMP-dependent kinase A leads to the inactivation of the protein and binding to protein 4-3-2. Subsequent phosphorylation of this protein at Thr138 and Ser205 by CDK2 and p38Y, respectively, and simultaneous dephosphosphorylation of Ser287 by PP2A result in the activation of Cdc25C again. Activated Cdc25C then dephosphorylates and activates mitosis promoting factor, which in turn triggers a higher activity of Cdc2/cyclin B complex. As a result of these differential phosphorylation and dephosphorylation events of the respective proteins, the oocytes go through meiosis (24).
In addition, it was also interesting to observe that the expression level of each mutant agnoprotein was significantly higher than that of WT (Fig. 4A). This suggested the possibility that mutant proteins may have a much longer half-life of than WT. This was investigated by pulse-chase experiments, and we found no significant difference between mutant and WT with respect to stability (data not shown). The mechanism(s) that contribute to the differential expression of each mutant protein compared to WT remains unknown.
In conclusion, we have demonstrated that agnoprotein is phosphorylated by PKC and viruses containing phosphorylation mutants of agnoprotein do not propagate after the first round of the infection cycle. Given the fact that JCV is the etiologic agent of PML and may be involved in the induction of some of human malignancies, such findings make agnoprotein an attractive target for therapeutic interventions to control JCV-induced diseases. Therefore, understanding the molecular mechanisms associated with the regulatory function(s) of agnoprotein is important for unraveling the molecular secrets of the unique biology of JCV and JCV-induced diseases.
Acknowledgments
We thank past and present members of the Center for Neurovirology for sharing of ideas and reagents, particularly M. White for insightful discussions. We are thankful to W. Atwood for the kind gift of anti-VP1 monoclonal antibody. We also thank K. Nagashima for the kind gift of JCV Mad-1 agnoprotein Pt mutant viral DNA.
This study was made possible by grants awarded by the NIH to K.K. and M.S.
REFERENCES
- 1.Agostini, I., S. Popov, T. Hao, J. H. Li, L. Dubrovsky, O. Chaika, N. Chaika, R. Lewis, and M. Bukrinsky. 2002. Phosphorylation of Vpr regulates HIV type 1 nuclear import and macrophage infection. AIDS Res. Hum. Retrovir. 18:283-288. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, H. A., Y. Chen, and L. C. Norkin. 1996. Bound simian virus 40 translocates to caveolin-enriched membrane domains, and its entry is inhibited by drugs that selectively disrupt caveolae. Mol. Biol. Cell 7:1825-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari, S. A., M. Safak, L. Del Valle, S. Enam, S. Amini, and K. Khalili. 2001. Cell cycle regulation of NF-κB-binding activity in cells from human glioblastomas. Exp. Cell Res. 265:221-233. [DOI] [PubMed] [Google Scholar]
- 4.Azzi, A., D. Boscoboinik, and C. Hensey. 1992. The protein kinase C family. Eur. J. Biochem. 208:547-557. [DOI] [PubMed] [Google Scholar]
- 5.Berger, J. R., and M. Concha. 1995. Progressive multifocal leukoencephalopathy: the evolution of a disease once considered rare. J. Neurovirol. 1:5-18. [DOI] [PubMed] [Google Scholar]
- 6.Brunet, A., J. Park, H. Tran, L. S. Hu, B. A. Hemmings, and M. E. Greenberg. 2001. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a). Mol. Cell. Biol. 21:952-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi, Y., M. J. Huddleston, X. Zhang, R. A. Young, R. S. Annan, S. A. Carr, and R. J. Deshaies. 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15:1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darbinyan, A., N. Darbinian, M. Safak, S. Radhakrishnan, A. Giordano, and K. Khalili. 2002. Evidence for dysregulation of cell cycle by human polyomavirus, JCV, late auxiliary protein. Oncogene 21:5574-5581. [DOI] [PubMed] [Google Scholar]
- 9.Darbinyan, A., K. M. Siddiqui, D. Slonina, N. Darbinian, S. Amini, M. K. White, and K. Khalili. 2004. Role of JC virus agnoprotein in DNA repair. J. Virol. 78:8593-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feigenbaum, L., K. Khalili, E. Major, and G. Khoury. 1987. Regulation of the host range of human papovavirus JCV. Proc. Natl. Acad. Sci. USA 84:3695-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frisque, R. J., and F. A. White. 1992. The molecular biology of JC virus, causative agent of progressive multifocal leukoencephalopathy. Humana Press, Inc., Totowa, N.J.
- 12.Fuchs, S. Y., I. Tappin, and Z. Ronai. 2000. Stability of the ATF2 transcription factor is regulated by phosphorylation and dephosphorylation. J. Biol. Chem. 275:12560-12564. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Fernandez, J. C., A. Torrecillas, and S. Corbalan-Garcia. 2004. Diacylglycerols as activators of protein kinase C. Mol. Membr. Biol. 21:339-349. [DOI] [PubMed] [Google Scholar]
- 14.Holmberg, C. I., V. Hietakangas, A. Mikhailov, J. O. Rantanen, M. Kallio, A. Meinander, J. Hellman, N. Morrice, C. MacKintosh, R. I. Morimoto, J. E. Eriksson, and L. Sistonen. 2001. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 20:3800-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isler, J. A., and P. A. Schaffer. 2001. Phosphorylation of the herpes simplex virus type 1 origin binding protein. J. Virol. 75:628-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson, V., and R. Chalkley. 1981. Use of whole-cell fixation to visualize replicating and maturing simian virus 40: identification of new viral gene product. Proc. Natl. Acad. Sci. USA 78:6081-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenney, S., V. Natarajan, D. Strike, G. Khoury, and N. P. Salzman. 1984. JC virus enhancer-promoter active in human brain cells. Science 226:1337-1339. [DOI] [PubMed] [Google Scholar]
- 18.Kim, J., S. Woolridge, R. Biffi, E. Borghi, A. Lassak, P. Ferrante, S. Amini, K. Khalili, and M. Safak. 2003. Members of the AP-1 family, c-Jun and c-Fos, functionally interact with JC virus early regulatory protein large T antigen. J. Virol. 77:5241-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Major, E. O., and G. S. Ault. 1995. Progressive multifocal leukoencephalopathy: clinical and laboratory observations on a viral induced demyelinating disease in the immunodeficient patient. Curr. Opin. Neurol. 8:184-190. [PubMed] [Google Scholar]
- 20.Major, E. O., A. E. Miller, P. Mourrain, R. G. Traub, E. de Widt, and J. Sever. 1985. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc. Natl. Acad. Sci. USA 82:1257-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McVey, D., L. Brizuela, I. Mohr, D. R. Marshak, Y. Gluzman, and D. Beach. 1989. Phosphorylation of large tumour antigen by cdc2 stimulates SV40 DNA replication. Nature 341:503-507. [DOI] [PubMed] [Google Scholar]
- 22.Okada, Y., S. Endo, H. Takahashi, H. Sawa, T. Umemura, and K. Nagashima. 2001. Distribution and function of JCV agnoprotein. J. Neurovirol. 7:302-306. [DOI] [PubMed] [Google Scholar]
- 23.Okada, Y., T. Suzuki, Y. Sunden, Y. Orba, S. Kose, N. Imamoto, H. Takahashi, S. Tanaka, W. W. Hall, K. Nagashima, and H. Sawa. 2005. Dissociation of heterochromatin protein 1 from lamin B receptor induced by human polyomavirus agnoprotein: role in nuclear egress of viral particles. EMBO Rep. 6:452-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perdiguero, E., and A. R. Nebreda. 2004. Regulation of Cdc25C activity during the meiotic G2/M transition. Cell Cycle 3:733-737. [PubMed] [Google Scholar]
- 25.Pho, M. T., A. Ashok, and W. J. Atwood. 2000. JC virus enters human glial cells by clathrin-dependent receptor-mediated endocytosis. J. Virol. 74:2288-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prives, C. 1990. The replication functions of SV40 T antigen are regulated by phosphorylation. Cell 61:735-738. [DOI] [PubMed] [Google Scholar]
- 27.Radhakrishnan, S., J. Gordon, L. Del Valle, J. Cui, and K. Khalili. 2004. Intracellular approach for blocking JC virus gene expression by using RNA interference during viral infection. J. Virol. 78:7264-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raj, G. V., and K. Khalili. 1995. Transcriptional regulation: lessons from the human neurotropic polyomavirus, JCV. Virology 213:283-291. [DOI] [PubMed] [Google Scholar]
- 29.Rinaldo, C. H., T. Traavik, and A. Hey. 1998. The agnogene of the human polyomavirus BK is expressed. J. Virol. 72:6233-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadowska, B., R. Barrucco, K. Khalili, and M. Safak. 2003. Regulation of human polyomavirus JC virus gene transcription by AP-1 in glial cells. J. Virol. 77:665-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Safak, M., R. Barrucco, A. Darbinyan, Y. Okada, K. Nagashima, and K. Khalili. 2001. Interaction of JC virus agno protein with T antigen modulates transcription and replication of the viral genome in glial cells. J. Virol. 75:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Safak, M., E. Major, and K. Khalili. 2005. Human polyomavirus, JC virus, and progressive multifocal encephalopathy, p. 461-474. In I. G. Howard E. Gendelman, Ian Paul Everall, Stuart A. Lipton, and Susan Swindells (ed.), The neurology of AIDS. Oxford University Press, New York, N.Y.
- 33.Safak, M., G. L. Gallia, S. A. Ansari, and K. Khalili. 1999. Physical and functional interaction between the Y-box binding protein YB-1 and human polyomavirus JC virus large T antigen. J. Virol. 73:10146-10157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Safak, M., B. Sadowska, R. Barrucco, and K. Khalili. 2002. Functional interaction between JC virus late regulatory agnoprotein and cellular Y-box binding transcription factor, YB-1. J. Virol. 76:3828-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stang, E., J. Kartenbeck, and R. G. Parton. 1997. Major histocompatibility complex class I molecules mediate association of SV40 with caveolae. Mol. Biol. Cell 8:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, T., Y. Okada, S. Semba, Y. Orba, S. Yamanouchi, S. Endo, S. Tanaka, T. Fujita, S. Kuroda, K. Nagashima, and H. Sawa. 2005. Identification of FEZ1 as a protein that interacts with JC virus agnoprotein and microtubules: role of agnoprotein-induced dissociation of FEZ1 from microtubules in viral propagation. J. Biol. Chem. 280:24948-24956. [DOI] [PubMed] [Google Scholar]
- 38.Swenson, J. J., and R. J. Frisque. 1995. Biochemical characterization and localization of JC virus large T antigen phosphorylation domains. Virology 212:295-308. [DOI] [PubMed] [Google Scholar]
- 39.Tada, H., M. Lashgari, J. Rappaport, and K. Khalili. 1989. Cell type-specific expression of JC virus early promoter is determined by positive and negative regulation. J. Virol. 63:463-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tootle, T. L., and I. Rebay. 2005. Post-translational modifications influence transcription factor activity: a view from the ETS superfamily. Bioessays 27:285-298. [DOI] [PubMed] [Google Scholar]
- 41.Tzivion, G., and J. Avruch. 2002. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 277:3061-3064. [DOI] [PubMed] [Google Scholar]
- 42.Viatour, P., M. P. Merville, V. Bours, and A. Chariot. 2005. Phosphorylation of NF-κB and IκB proteins: implications in cancer and inflammation. Trends Biochem. Sci. 30:43-52. [DOI] [PubMed] [Google Scholar]
- 43.Wang, E. H., S. Bhattacharyya, and C. Prives. 1993. The replication functions of polyomavirus large tumor antigen are regulated by phosphorylation. J. Virol. 67:6788-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitmarsh, A. J., and R. J. Davis. 2000. Regulation of transcription factor function by phosphorylation. Cell Mol. Life Sci. 57:1172-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zanardi, T. A., C. M. Stanley, B. M. Saville, S. M. Spacek, and M. R. Lentz. 1997. Modulation of bovine papillomavirus DNA replication by phosphorylation of the viral E1 protein. Virology 228:1-10. [DOI] [PubMed] [Google Scholar]
- 46.Ziegler, K., T. Bui, R. J. Frisque, A. Grandinetti, and V. R. Nerurkar. 2004. A rapid in vitro polyomavirus DNA replication assay. J. Virol. Methods 122:123-127. [DOI] [PubMed] [Google Scholar]