Abstract
The emergence of influenza A viruses resistant to the two existing classes of antiviral drugs highlights the need for additional antiviral drugs, particularly considering the potential threat of a pandemic of H5N1 influenza A viruses. Here, we determine whether influenza A virus replication can be selectively inhibited by blocking the ability of its NS1A protein to inhibit the 3′-end processing of cellular pre-mRNAs, including beta interferon (IFN-β) pre-mRNA. Pre-mRNA processing is inhibited via the binding of the NS1A protein to the cellular CPSF30 protein, and mutational inactivation of this NS1A binding site causes severe attenuation of the virus. We demonstrate that binding of CPSF30 is mediated by two of its zinc fingers, F2F3, and that the CPSF30/F2F3 binding site on the NS1A protein extends from amino acid 144 to amino acid 186. We generated MDCK cells that constitutively express epitope-tagged F2F3 in the nucleus, although at only approximately one-eighth the level of the NS1A protein produced during virus infection. Influenza A virus replication was inhibited in this cell line, whereas no inhibition was observed with influenza B virus, whose NS1B protein lacks a binding site for CPSF30. Influenza A virus, but not influenza B virus, induced increased production of IFN-β mRNA in the F2F3-expressing cells. These results, which indicate that F2F3 inhibits influenza A virus replication by blocking the binding of endogenous CPSF30 to the NS1A protein, point to this NS1A binding site as a potential target for the development of antivirals directed against influenza A virus.
Influenza A viruses cause a highly contagious respiratory disease in humans that results in a significant loss of life each year and are responsible for human pandemics that have resulted in higher mortality rates (2). Three pandemics occurred in the 20th century, in 1918, 1957, and 1968 (23). The 1918 pandemic (“Spanish flu”) was the most devastating, causing at least 20 to 40 million deaths worldwide (19). H5N1 avian influenza A viruses, which have had a human mortality rate of approximately 50% since 1997 (22), are prime candidates for the next pandemic influenza A virus. At present, H5N1 viruses are not readily transmissible between humans, but it is quite possible that they can acquire such transmissibility via mutations and/or reassortment of genes with circulating human influenza A viruses.
The primary means for controlling influenza virus epidemics is vaccination, but antivirals provide an important additional line of defense, particularly for a rapidly spreading pandemic (5, 11). Only two classes of influenza virus antivirals are currently available: inhibitors of the viral M2 ion channel protein (amantadine and rimantadine) and inhibitors of the viral neuraminidase (zanamivir and oseltamivir) (reviewed in reference 23). The emergence of influenza A viruses resistant to the M2 inhibitors occurs at high frequency in treated patients (4, 21). Many of the human isolates of H5N1 viruses are already resistant to these inhibitors (17). In addition, a recent study has shown that influenza A viruses resistant to the neuraminidase inhibitor oseltamivir occurred in 20% of the children treated with this drug (8). In fact, H5N1 viruses that are partially resistant to oseltamivir have recently been reported (9). The emergence of influenza A viruses resistant to these two classes of antiviral drugs highlights the need for additional antiviral drugs against influenza A virus.
We undertook the present study to determine whether one of the functions of the influenza A virus-encoded nonstructural, or NS1A, protein can be targeted for the development of antiviral drugs. We focused on the NS1A protein-mediated inhibition of the 3′-end processing of cellular pre-mRNAs, which results in the inhibition of the production of functional cellular mRNAs during infection (3, 14, 16, 20). As a consequence, the production of alpha/beta interferon (IFN-α/β)-independent antiviral mRNAs [e.g., ISG15, p56, and 2′-5′-oligo(A) synthetase mRNAs] is essentially eliminated, and the production of functional IFN-β mRNA is substantially reduced, although not eliminated (16). The NS1A protein inhibits the 3′-end processing of cellular pre-mRNAs by binding two cellular proteins: the 30-kDa subunit of CPSF (cleavage and polyadenylation specificity factor) (CPSF30) and PABII [poly(A)-binding protein II] (3, 14). The NS1A sequence centered at amino acid 186 is required for the binding of CPSF30, and mutation of this binding site renders the NS1A protein largely inactive in the inhibition of the 3′-end processing of cellular pre-mRNAs (10, 16). This binding site is also required for efficient virus replication, because a recombinant influenza A virus encoding an NS1A protein with a mutated 186 sequence (M186 mutant virus) is highly attenuated (16). This attenuation is most likely due to the enhanced production of functional cellular antiviral mRNAs, particularly IFN-β mRNA, that occurs in M186 virus-infected cells (16).
Based on these results, it is reasonable to propose that the CPSF30 binding site of the NS1A protein is a potential target for the development of antivirals directed against influenza A virus. Specifically, molecules which block the binding of CPSF30 to this region of the NS1A protein might be expected to be effective inhibitors of virus replication. However, it is essential that such inhibitory molecules do not inhibit the function that CPSF30 carries out in the 3′-end processing of cellular pre-mRNAs. In other words, these molecules would need to specifically block this viral NS1A function without affecting cellular pre-mRNA processing.
In the present study, we demonstrate that the function of the CPSF30 binding site of the NS1A protein can be inhibited during influenza A virus infection in vivo, resulting in the inhibition of influenza A virus replication, without detectable effects on cellular functions. For these experiments, we utilized a fragment of CPSF30, specifically, a 61-amino-acid sequence comprising the second and third zinc fingers (F2F3) of this protein. We show that the F2F3 fragment binds specifically and efficiently to the CPSF30 binding site of the NS1A protein and does not inhibit the 3′-end processing of cellular pre-mRNAs as measured in transient-transfection experiments. Most significantly, we generated MDCK cells that constitutively express epitope-tagged F2F3 in the nucleus and demonstrate that influenza A virus replication is inhibited in these cells. In contrast, the replication of influenza B virus, whose NS1 protein (NS1B protein) lacks a CPSF30 binding site (16, 24), is not inhibited. Influenza A virus, but not influenza B virus, induced increased production of IFN-β mRNA in the F2F3-expressing cells compared to control cells, which is most likely responsible for the selective inhibition of influenza A virus replication. The F2F3-expressing cells have been maintained in tissue culture for 2 years, and we have not observed any effect on their growth. These results indicate that the CPSF30 binding site of the NS1A protein is a potential target for the development of small-molecule antiviral drugs directed against influenza A virus.
MATERIALS AND METHODS
Virus infections.
For multiple-cycle growth, MDCK cells were infected at a multiplicity of infection (MOI) of 0.001 PFU/cell with either influenza A/Udorn/72, A/WSN/33, or B/Lee/40 virus and were incubated in serum-free Dulbecco's modified Eagle's medium (DMEM) supplemented with 2.5 μg/ml of N-acetylated trypsin (NAT). Incubation was at 37°C for the two influenza A viruses and at 34°C for influenza B/Lee/40 virus. Maximal yields were obtained after 30 to 36 h for the two influenza A viruses and after 50 to 60 h for influenza B virus. Plaque assays were carried out in MDCK cells. For the plaque reduction assays, monolayers of MDCK cells were infected with approximately 100 PFU of either influenza A/Udorn/72, influenza A/WSN/33, or influenza B/Lee/40 virus. After 1 h of incubation at 37°C or 34°C, the inoculum was removed, and the cells were overlaid with 1% agarose containing DMEM plus 2.5 μg/ml NAT. The cells were incubated for 3 days at 37°C for the development of influenza A virus plaques and for 4 days at 34°C for influenza B/Lee/40 virus plaques. For single-cycle infections, MDCK cells were infected with 5 PFU/cell of either influenza A/Udorn/72 or influenza B/Lee/40 virus. After 1 hour of incubation, the inoculum was removed and the cells were washed twice with DMEM and then overlaid with DMEM.
Glutathione-Sepharose affinity selection.
The DNAs encoding the following fragments of CPSF30 were generated by PCR using appropriate primers: 1-F4 (amino acids 1 to 145), 1-F3 (amino acids 1 to 121), 1-F2 (amino acids 1 to 92), F1-F3 (amino acids 34 to 121), and F2F3 (amino acids 61 to 121). These DNAs were then fused in frame (using PCR) into glutathione S-transferase (GST) in the pGEX3X vector. Each GST fusion was expressed in Escherichia coli BL21 and purified as previously described (18). The indicated GST fusion protein was mixed with 35S-labeled NS1A protein (wild type, M186 mutant, or M144 mutant) and subjected to glutathione-Sepharose affinity selection as previously described (15). To prepare the labeled NS1A protein, the DNA encoding the appropriate NS1A protein was subcloned into pcDNA3 and translated using a Promega TnT Coupled Transcription/Translation kit in the presence of [35S]methionine.
Assay for 3′-end cleavage of pre-mRNAs in vivo.
293 cells were cotransfected with a pBC12 plasmid containing a human β-globin gene and a pcDNA3 plasmid encoding the appropriate protein (see Fig. 2A and 4) using FuGENE 6 transfection reagent. The transfected cells were collected at 40 h posttransfection, and RNA was extracted using Trizol reagent (Invitrogen). An aliquot of the total RNA was analyzed by RNase protection using a uniformly labeled RNA probe (see Fig. 2A), which was prepared as previously described (10). After this labeled RNA probe was annealed to the cellular RNA sample, followed by digestion with RNase A and phenol extraction, the protected RNA fragments were resolved by electrophoresis on a urea-polyacrylamide (5%) gel.
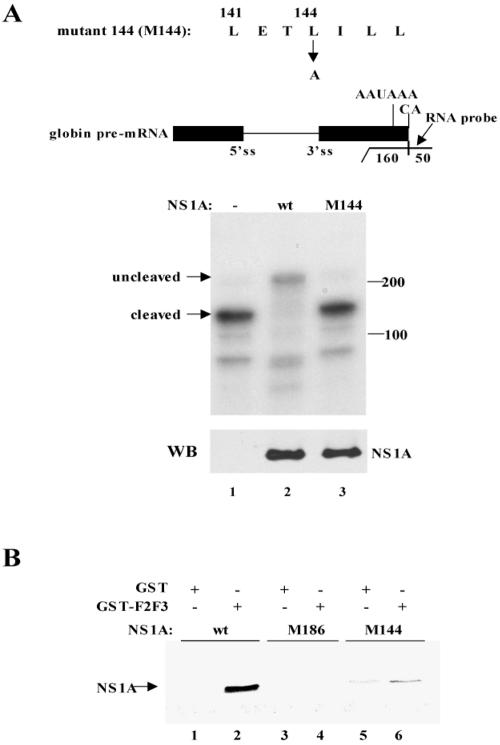
FIG. 2.
The NS1A protein containing an L-to-A substitution at position 144 did not inhibit the 3′-end processing of β-globin pre-mRNA and did not bind the F2F3 fragment of CPSF30. (A) 3′-end processing assay. 293 cells were cotransfected with a pBC12 plasmid containing a human β-globin gene and either an empty pcDNA3 plasmid (lane 1) or a pcDNA3 plasmid encoding wt NS1A protein (lane 2) or the M144 mutant NS1A protein (lane 3). The M144 sequence, which is diagrammed above the blot, was generated by RT-PCR using appropriate primers. RNA was analyzed by RNase protection using the indicated uniformly labeled RNA probe (270 nucleotides long). The protected RNA fragments were resolved by electrophoresis on a urea-polyacrylamide (5%) gel. The positions of the RNA fragments corresponding to the uncleaved and 3′-end-cleaved pre-mRNAs are indicated. No residual probe containing 270 nucleotides was detected. The amount of the wt and M144 NS1A proteins was determined by a Western blot (WB) using NS1A antibody. (B) GST pull-down assay. GST-F2F3 or GST was mixed with 35S-labeled wt, M186 mutant, or M144 mutant NS1A protein as indicated, followed by affinity chromatography on glutathione-Sepharose.
FIG. 4.
Transient expression of the F2F3 protein fragment did not inhibit the 3′-end processing of cellular pre-mRNAs. 293 cells were cotransfected with a pBC12 plasmid containing a human β-globin gene and either an empty pcDNA3 plasmid (lane 1) or a pcDNA3 plasmid encoding wt NS1A protein (lane 2) or the F2F3 fragment (lane 3). The sequence of the F2F3 protein fragment is diagrammed above. Cells were collected 40 h posttransfection, and RNA was analyzed by RNase protection as described in the legend to Fig. 2A. The positions of the RNA fragments corresponding to the uncleaved and 3′-end-cleaved pre-mRNAs are indicated. No residual probe containing 270 nucleotides was detected.
Generation of a recombinant influenza A/Udorn/72 virus encoding mutant 144 NS1A protein.
Position 144 in the NS1A protein of influenza A/Udorn/72 virus was changed from L to A by PCR mutagenesis, and the resulting DNA was cloned into pHH21. This plasmid, plus the seven pHH21 plasmids carrying the other Udorn genomic RNAs, was cotransfected into 293T cells, along with the four plasmids encoding the PB1, PB2, PA, and NP proteins. At 12 h posttransfection, the medium was changed to Opti-MEM supplemented with 3 μg/ml of NAT. After an additional 24 to 30 h, the 293T cells were overlaid onto MDCK cells for virus amplification. Culture supernatants were collected when a positive hemagglutinin assay titer was observed. Viruses were titered by plaque assay on MDCK cells, and individual plaques were amplified in 10-day-old embryonic chicken eggs at 34°C. Amplified virus was titered by plaque assay.
Measurement of IFN-β mRNA by real-time quantitative reverse transcription (RT)-PCR.
RNA was isolated from infected cells by using the Trizol reagent at the appropriate times after infection of MDCK cells. For each sample, 1 μg of total RNA, which corresponds to equal cell equivalents, was reverse transcribed using an oligo(dT) primer to generate the DNA complementary to all mRNAs. The amount of IFN-β mRNA was determined using the TaqMan Gene Expression Assay (Applied Biosystems) with 5′ and 3′ primers specific for canine IFN-β mRNA and a 6-carboxyfluorescein dye-labeled TaqMan MGB (minor groove binder) internal probe. Real-time PCR analysis was carried out using the Perkin-Elmer/Applied Biosystems 7900HT sequence detector.
Indirect immunofluorescence and confocal microscopy.
Cells were fixed with 4% paraformaldehyde for 20 min and with 0.5% Triton X-100 for another 10 min and then incubated with the appropriate rabbit or mouse antibody at 37°C for 1 h. Following three washes in phosphate-buffered saline, the cells were incubated for 45 min with the secondary antibody, either fluorescein isothiocyanate-conjugated goat anti-rabbit antibody or rhodamine-conjugated goat anti-mouse antibody. The cells were examined by confocal microscopy as described previously (3).
Generation of an MDCK cell stably expressing the F2F3 protein fragment.
The F2F3 protein fragment that was expressed in MDCK cells contained an N-terminal nuclear localization signal (NLS) from the simian virus 40 T antigen (DPKKKKRKV) linked to the 61-amino-acid F2F3 sequence from CPSF30, which in turn was linked at its C terminus to 13 myc epitopes. The DNA sequence encoding this F2F3 fragment was produced using the pAJ1026 plasmid, which contains 13 myc epitopes (13x EQKLISEEDL) (12). The NLS-F2F3 sequence containing a 5′ EcoRI site was inserted into the N terminus of the 13xmyc sequence by PCR, which also generated a 3′ EcoRI site. The fused sequence was excised using EcoRI and inserted into the EcoRI site of the pcDNA3 plasmid. In addition, as a control, we generated an F2F3 fragment containing a C-to-A mutation in the F2 (amino acid 76) and F3 (amino acid 105) zinc fingers. Another control was a pcDNA3 plasmid lacking an insert. MDCK cells were transfected by electroporation with 10 μg of ScaI-linearized plasmid. Forty-eight hours after transfection, DMEM containing 1.0 mg/ml neomycin sulfate was added, and the cells were incubated for approximately 2 weeks, at which time the mock-transfected cells were all dead. Individual stable clones were picked and incubated with 25 μl trypsin-EDTA, followed by being plated onto 96-well tissue culture dishes. Each clone was grown and maintained in DMEM containing neomycin sulfate. For the cells transfected with the pcDNA3 containing an F2F3 insert, each cell clone was analyzed by immunoblotting using myc antibody to identify the highest-expressing cell clone.
RESULTS
Identification of the region of CPSF30 that binds to the influenza A virus NS1A protein.
The influenza A virus NS1A protein binds CPSF30, a key component of the mammalian 3′-end processing machinery (10, 14). CPSF30 contains five C3H zinc finger repeats (1). To identify the region of CPSF30 that mediates its binding to the NS1A protein, we expressed GST fusions of N-terminal fragments of CPSF30 in bacteria and used these GST fusions in pull-down assays with labeled NS1A protein (Fig. 1A, lanes 3 to 5). These assays showed that the N-terminal fragment containing zinc fingers 1 to 3 (1-F3) is the shortest N-terminal fragment that binds the NS1A protein. Deletion of the region N-terminal to zinc fingers 1 to 3, thereby generating the F1-F3 sequence, did not affect binding to the NS1A protein (lane 6), demonstrating that these three zinc fingers alone are sufficient for efficient binding to NS1A. In fact, the F1 zinc finger is not required, because F2F3, a 61-amino-acid sequence, was sufficient for such binding (lane 7). The binding of F2F3 to NS1A requires the zinc finger structure, because a C-to-A mutation in either F2 (amino acid 76) or F3 (amino acid 105) greatly reduced binding (data not shown).
FIG. 1.
Identification of the region of CPSF30 that binds to the 186-amino-acid region of the NS1A protein. (A) GST fusions containing the indicated regions of CPSF30 were mixed with 35S-labeled wt NS1A protein, followed by affinity chromatography on glutathione-Sepharose. (B) GST-F2F3 or GST was mixed with 35S-labeled wt or M186 mutant NS1A protein as indicated, followed by affinity chromatography on glutathione-Sepharose (lanes 3 to 5). Lanes 1 and 2 show the input wt and M186 NS1A proteins. RBO, RNA-binding domain.
The NS1A sequence centered at amino acid 186 is required for the binding of CPSF30 (10, 16). As shown in Fig. 1B, the NS1A sequence centered at amino acid 186 is also required for binding F2F3. Whereas GST-F2F3 efficiently bound labeled wild-type (wt) NS1A (lane 3), no detectable M186 NS1A protein bound to GST-F2F3 (lane 4).
Identification of the extent of the NS1A sequence that mediates inhibition of 3′-end processing via F2F3 binding.
Because the F2F3 fragment of CPSF30 is 61 amino acids long, it was reasonable to expect that its binding site on the NS1A protein would include amino acids in addition to those in the M186 region. To determine whether this was the case, we made L-to-A substitutions at positions upstream (N terminal) of position 186. The mutated NS1A proteins were assayed by expressing them in transient-transfection experiments to determine whether they retained the wild-type NS1A protein activity of inhibiting the 3′-end processing of cellular pre-mRNAs. A plasmid expressing β-globin pre-mRNA provided the target pre-mRNA in these assays. The mutant NS1A protein containing an L-to-A substitution at position 141 retained wild-type activity (data not shown). In contrast, as shown in Fig. 2A, the mutant NS1A protein containing an L-to-A substitution at position 144 (M144) did not inhibit the 3′-end processing of β-globin pre-mRNA, unlike the wt NS1A protein (compare lanes 2 and 3). In addition, this amino acid substitution eliminates most of the binding of the NS1A protein to the F2F3 fragment of CPSF30 (Fig. 2B). Only approximately 5 to 10% of wild-type binding to GST-F2F3 was observed with the M144 mutant protein (compare lanes 2 and 6). We conclude that binding to CPSF30 (as measured by F2F3 binding) and the resulting inhibition of 3′-end processing of cellular pre-mRNAs is mediated by amino acid 144 of the NS1A protein, as well as by amino acids 184 to 188 (the 186 region).
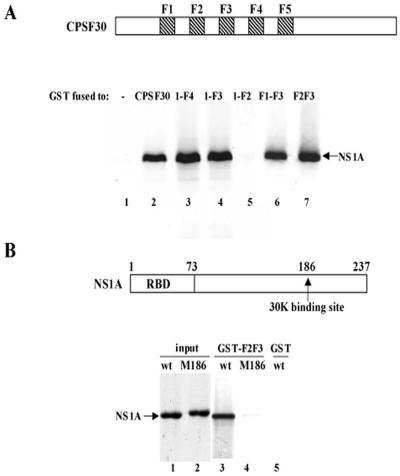
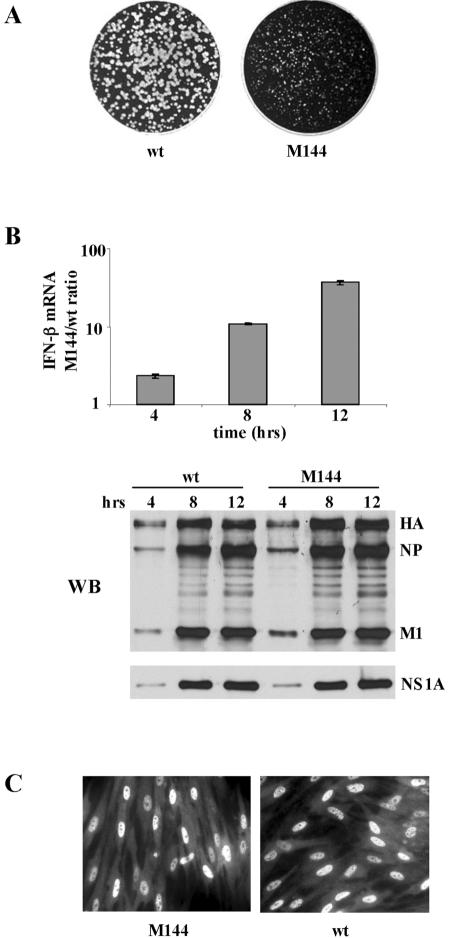
Based on the above-mentioned results, we generated a recombinant influenza A/Udorn virus encoding an NS1A protein with an L-to-A substitution at position 144. The M144 mutant virus is attenuated: it forms pinpoint plaques (Fig. 3A), and the rate of replication and virus yield at a low multiplicity of infection (MOI, 0.001 PFU/cell) was approximately 1,000-fold lower than with wild-type virus (data not shown). This attenuation is attributable to the enhanced production of IFN-β mRNA in M144 virus-infected cells relative to wt virus-infected cells, as measured by quantitative RT-PCR (Fig. 3B). During single-cycle virus growth (MOI, 5), the amounts of IFN-β mRNA produced in M144 virus-infected cells at 8 and 12 h postinfection were 12 and 40 times more, respectively, than that produced in wt virus-infected cells. We used high-MOI conditions to ensure that we were measuring the amount of IFN-β mRNA produced per infected cell. Under these high-MOI conditions, equal amounts of all viral proteins, including the NS1A protein, were synthesized in wt virus- and M144 virus-infected cells (Fig. 3B). The M144 mutant NS1A protein, like the wt NS1A protein, is localized in the nuclei of infected cells (Fig. 3C), demonstrating that the L-to-A substitution at position 144 does not affect the nuclear localization of the NS1A protein.
FIG. 3.
Characterization of the M144 mutant virus. (A) Plaques formed by wt and M144 mutant viruses on MDCK cells. (B) Relative amounts of IFN-β mRNA produced during single-cycle growth by M144 and wt virus. MDCK cells were infected with either M144 or wt virus at an MOI of 5, and at the indicated times after infection the relative amounts of IFN-β mRNA produced were determined by quantitative RT-PCR. The error bars indicate standard deviations for three experiments. WB, Western blot analysis of the viral proteins synthesized in wt and M144 virus-infected cells using either an antibody against virion proteins (top) or NS1A antibody (bottom). HA, hemagglutinin. (C) Localization of the NS1A protein in cells infected by M144 and wt virus at 8 h postinfection was determined by indirect immunofluorescence. The primary antibody was a rabbit polyclonal against the NS1A protein.
Inhibition of influenza A virus replication by F2F3.
Because F2F3 binds strongly to the 144-to-186 region of the NS1A protein, we hypothesized that it would block the access of full-length endogenous CPSF30 to the NS1A protein and hence inhibit the replication of influenza A virus. To test this hypothesis, we constructed a plasmid expressing an F2F3 molecule containing an N-terminal NLS to ensure that the F2F3 was localized in the nucleus. In addition, we added 13 myc tags at the C terminus of F2F3 to increase its size to 249 amino acids and to provide an epitope to analyze its production and localization. We first carried out transient-transfection experiments to determine whether high expression of this F2F3 protein construct inhibited endogenous CPSF30 function in the 3′-end processing of cellular pre-mRNAs. As shown in Fig. 4, the F2F3 protein construct did not inhibit the 3′-end processing of β-globin pre-mRNA, in contrast to the inhibition observed with the NS1A protein (compare lanes 2 and 3).
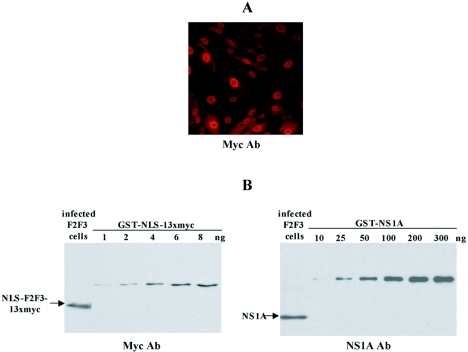
Based on the above-mentioned results, it might be expected that a cell line that constitutively expresses the F2F3 protein construct would effectively carry out the 3′-end processing of cellular pre-mRNAs and hence would be viable. Consequently, we generated such a cell line to determine whether the F2F3 protein construct inhibits influenza A virus replication. MDCK cells were transfected with a pcDNA3 plasmid expressing the F2F3 protein construct under the control of a cytomegalovirus promoter. Forty G418-resistant cell colonies were selected and screened by immunoblotting using myc antiserum to identify the cells expressing the highest level of the F2F3 protein construct. The highest-expressing MDCK cell line has been maintained in tissue culture for 2 years and has not exhibited any discernible growth impediment. As shown in Fig. 5A, the expressed F2F3 protein construct is localized in the nuclei of these cells, demonstrating that the NLS at the N terminus of the F2F3 construct is functional. In contrast, in cells expressing F2F3 constructs containing shorter epitope tags, e.g., the 3 FLAG epitope, the F2F3 construct was diffusely distributed throughout the cell and was not localized in the nucleus (data not shown), which led us to use the 13xmyc epitope. Because the putative target of the F2F3 protein construct is the NS1A protein, we determined whether the F2F3 protein is expressed at a level comparable to that of the NS1A protein synthesized during influenza A virus infection. The F2F3-expressing cells were infected with influenza A virus at an MOI of 5 to infect all the cells, and at 6 h postinfection, the amounts of the F2F3 protein construct and the NS1A protein were estimated by immunoblotting using either anti-myc or anti-NS1A antibody (Fig. 5B). Based on the protein standards, we estimated that on a molar basis the F2F3 construct was present in approximately one-eighth the amount of the NS1A protein (see the legend to Fig. 5B).
FIG. 5.
Characterization of F2F3-expressing MDCK cells. (A) Localization of the F2F3 fragment was determined by indirect immunofluorescence using anti-myc antibody (Ab). (B) Determination of the relative amounts of the F2F3 protein fragment and the NS1A protein in influenza A virus-infected F2F3-expressing MDCK cells. An aliquot of the infected cells was analyzed by immunoblotting using either anti-myc (left) or anti-NS1A (right) antibody. To estimate the amount of the F2F3 protein fragment, increasing amounts of GST-NLS-13xmyc were applied to the anti-myc immunoblot (left). The GST-NLS-13xmyc was generated using the pAJ1026 plasmid, which contains 13 myc epitopes (12). Based on this immunoblot, as well as another immunoblot containing 10 to 30 ng of GST-NLS-13xmyc, we estimated that the aliquot from the virus-infected cells contained 10 ng of the F2F3 protein fragment. To estimate the amount of the NS1A protein, increasing amounts of GST-NS1A protein were applied to the anti-NS1A immunoblot (right). Based on this immunoblot, we estimated that the aliquot from the virus-infected cells contained approximately 80 ng. Because the molecular weights of the F2F3 protein fragment and the NS1A protein were approximately the same, the molar ratio of the F2F3 fragment/NS1A protein was approximately 1/8.
Although this is not an optimal F2F3/NS1A ratio, we determined whether the replication of influenza A virus was inhibited in these cells. To generate control cell lines, MDCK cells were transfected with either an empty pcDNA3 plasmid or a pcDNA3 plasmid expressing an NLS-F2F3-13xmyc construct containing a C-to-A mutation in the F2 and F3 zinc fingers, thereby eliminating the ability to bind to the NS1A protein. G418-resistant cells were then selected. However, the highest level of the mutant F2F3 construct that was expressed was only approximately 10% of the level of the wild-type F2F3 construct in the cell line analyzed in Fig. 5. The two control cell lines yielded identical results in the subsequent assays employed in this study.
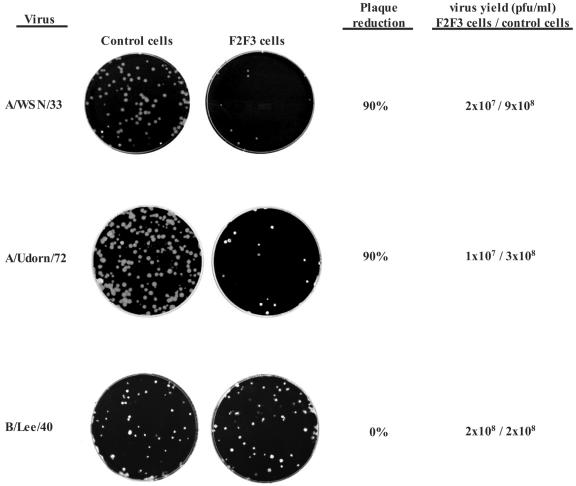
As the first approach to monitor virus replication, we employed a plaque reduction assay. Monolayer cultures of the control and F2F3-expressing cells were infected with approximately 100 PFU of either influenza A/WSN/33 virus or influenza A/Udorn/72 virus per 60-mm culture dish and the viruses were allowed to form plaques under soft agar. The number of plaques on the F2F3-expressing cells was only approximately 10% of the number on the control cells (Fig. 6). To determine whether this 90% plaque reduction was specific for influenza A virus, we carried out the same assay with an influenza B virus (B/Lee/40) whose NS1B protein does not bind CPSF30 (16, 24). No reduction in plaque number of the influenza B virus was observed in the F2F3-expressing cells compared to the control cells. The same selectivity was observed when virus yields were measured after low-MOI (0.001 PFU/cell) infections. The maximal yields of the two influenza A virus strains (36 h at 37°C) in the F2F3-expressing cells were 35- to 60-fold lower than in the control cells (Fig. 6). In contrast, the maximal virus yields of influenza B/Lee/40 virus (60 h at 34°C) were the same in both the control and the F2F3-expressing cells. We conclude that the replication of influenza A virus, but not influenza B virus, is inhibited in the F2F3-expressing cells.
FIG. 6.
Plaque reduction assays and virus yields after low-MOI infections in control and F2F3-expressing MDCK cells. The viruses used in these assays were influenza A/WSN/33, influenza A/Udorn/72, and influenza B/Lee/40 viruses.
The influenza A virus plaques on the F2F3-expressing cells were approximately the same size as those on the control cells (Fig. 6), suggesting two possibilities: (i) a small fraction (approximately 10%) of the F2F3-expressing cells contained suboptimal levels of the F2F3 fragment that did not block virus replication or (ii) the plaques on the F2F3-expressing cells were formed by preexisting virus mutants that were resistant to inhibition by the F2F3 fragment. To distinguish between these two possibilities, several individual plaques from the F2F3-expressing cells were reassayed for plaque formation on control and F2F3-expressing cells. The virus from these plaques behaved identically to the original influenza A virus stock shown in Fig. 6: the number of plaques on the F2F3-expressing cells was only approximately 10% of the number on the control cells (data not shown). We conclude that these viruses were still sensitive to F2F3 inhibition. The most likely interpretation is that approximately 10% of the F2F3-expressing cells lack sufficient amounts of the F2F3 fragment to inhibit virus replication. It is quite possible that most of the influenza A virus replication detected in the F2F3-expressing cells actually occurs in the small fraction of cells containing suboptimal levels of the F2F3 fragment.
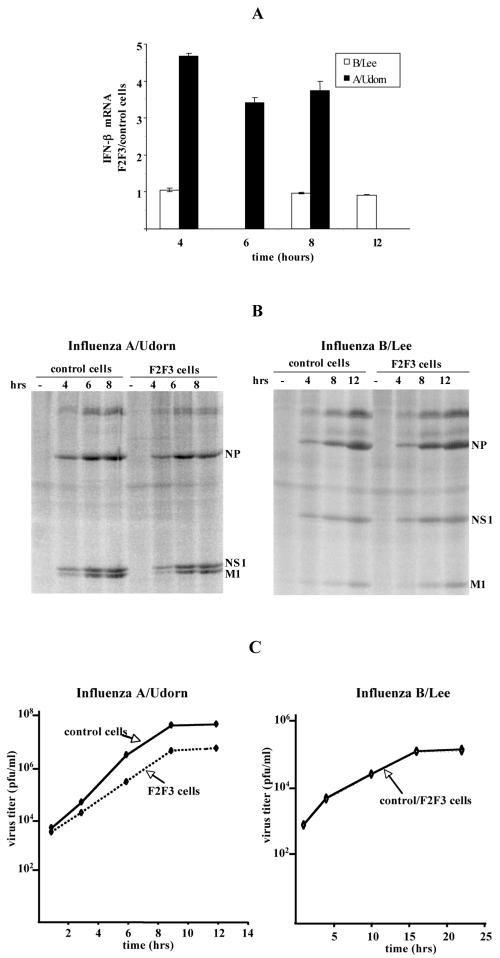
The selective inhibition of influenza A virus replication in the F2F3-expressing cells is consistent with our hypothesis that the expressed F2F3 protein fragment selectively relieves the inhibition of the 3′-end processing of IFN-β pre-mRNA mediated by the CPSF30 binding site on the NS1A protein. To determine whether this is the case, we measured the amount of IFN-β mRNA produced per cell during single-cycle growth after high-MOI infection with influenza A or influenza B virus (Fig. 7A). The amount of IFN-β mRNA produced after influenza A virus infection was three- to fivefold higher in the F2F3-expressing cells than in the control cells. Equal amounts of the NS1A protein were synthesized after infection of the two cell lines (Fig. 7B), whereas virus replication was inhibited approximately 10-fold in the F2F3-expressing cells compared to the control cells (Fig. 7C). This level of inhibition is consistent with the 35- to 60-fold inhibition observed in the low-MOI multiple-cycle infection. In contrast, the amounts of IFN-β mRNA produced by influenza B virus were the same in the F2F3-expressing and control cells (Fig. 7A), and the replication of influenza B virus was not inhibited in the F2F3-expressing cells (Fig. 7B and C). We conclude that enhanced production of IFN-β mRNA is induced by influenza A virus in the F2F3-expressing cells and that this enhanced production is most likely responsible for the inhibition of influenza A virus replication during single-cycle as well as multiple-cycle infections.
FIG. 7.
Production of IFN-β mRNA and infectious virus during single-cycle growth in control and F2F3-expressing cells. (A) Relative amounts of IFN-β mRNA produced in F2F3-expressing and control cells after high-MOI infection (5 PFU/cell) with either influenza A/Udorn/72 or influenza B/Lee/40 virus. The error bars indicate standard deviations for three experiments. (B) Viral-protein synthesis in F2F3-expressing and control cells after high-MOI infection with either influenza A/Udorn/72 virus (left) or influenza B/Lee/40 virus (right). At the indicated times after infection, the cells were washed twice with methionine-free DMEM, 5 μl of a mixture of [35S]methionine and [35S]cysteine (Promix; Amersham) was added in a final volume of 1 ml of serum-free DMEM, followed by incubation for 30 min. After incubation, the cells were washed twice with phosphate-buffered saline and lysed in 200 μl of Laemmli sample buffer. An aliquot was loaded onto SDS-polyacrylamide gels (12 to 15%) for analysis by autoradiography. (C) Replication of influenza A/Udorn/72 and influenza B/Lee/40 after high-MOI infection of F2F3-expressing and control cells.
DISCUSSION
The goal of this study was to determine whether the site on the influenza A virus NS1A protein that binds CPSF30 can be targeted to inhibit influenza A virus replication. The binding of CPSF30 to the NS1A protein results in the inhibition of the 3′-end processing of IFN-β pre-mRNA, as well as other cellular pre-mRNAs, in influenza A virus-infected cells (reference 16 and the present study). This inhibition is crucial, because influenza A virus, like several other RNA viruses, efficiently activates the RIG-I RNA helicase (13) to trigger the activation of IRF-3 and NF-κB and hence the synthesis of IFN-β pre-mRNA (6, 7, 13, 16). Because 3′-end processing of the newly synthesized IFN-β pre-mRNA is inhibited by the NS1A protein, only a small amount of mature IFN-β mRNA is produced (16). Mutational inactivation of the NS1A site for binding CPSF30 results in increased IFN-β mRNA production and substantial attenuation of the virus (reference 16 and the present study). For example, as shown here, the L-to-A mutation at position 144 results in a 40-fold increase in the amount of IFN-β mRNA produced during single-cycle virus infection. Because mutations at position 144 and in the 186 region of the NS1A protein result in this phenotype, the CPSF30 binding site on the NS1A protein likely includes the region between 144 and 186 (reference 16 and the present study). Based on our demonstration that the region of CPSF30 comprising two of its zinc fingers, F2F3, mediates its binding to this region of the NS1A protein, we proposed that a protein fragment containing these two zinc fingers might occupy this NS1A binding site, thereby blocking the binding of full-length endogenous CPSF30 and inhibiting virus replication.
To test this hypothesis, we generated a cell line expressing an F2F3 protein fragment. The highest-expressing cell line produced an amount of the F2F3 fragment that was only one-eighth that of the NS1A protein produced during virus infection. Despite this relatively low-level expression, influenza A virus replication was selectively inhibited in the F2F3-expressing cell line. The most dramatic evidence for this selective inhibition was obtained using plaque reduction assays. The number of influenza A virus plaques on the F2F3-expressing cells was only 10% of that on the control cells, whereas no plaque reduction was observed with influenza B virus. In addition, the virus yield after low-MOI infection with influenza A virus was reduced 35- to 60-fold in the F2F3-expressing cells compared to the control cells, whereas the virus yield of an influenza B virus was not reduced in the F2F3-expressing cells. Finally, during single-cycle growth at a high MOI, influenza A virus induced the synthesis of 3- to 5-fold more IFN-β mRNA in the F2F3-expressing cells than in the control cells, and virus replication was inhibited by 10-fold in the F2F3-expressing cells. In contrast, influenza B virus did not induce more IFN-β mRNA in the F2F3-expressing cells, nor was its replication inhibited. These results provide strong support for the model shown in Fig. 8. Because the F2F3 fragment binds to the 144-to-186 region of the NS1A protein, it blocks the binding of full-length endogenous CPSF30. As a consequence, more IFN-β mRNA is produced, resulting in the inhibition of virus replication. The replication of influenza B virus is not inhibited because its NS1B protein lacks a binding site for CPSF30 and hence for its F2F3 fragment (16, 24).
FIG. 8.
Proposed mechanism for the selective inhibition of influenza A virus replication by the F2F3 fragment of CPSF30.
The ability of the F2F3 fragment to inhibit influenza A virus replication even though it is expressed at one-eighth the level of the NS1A protein indicates that targeting the F2F3 (and CPSF30) binding site on the NS1A protein is a promising approach for the development of antivirals directed against influenza A virus. In fact, our results suggest that influenza A virus replication may occur primarily in a small fraction of the cells that express suboptimal levels of the F2F3 fragment and that the majority of the F2F3-expressing cells are largely resistant to influenza A virus replication. We propose that small chemical compounds that bind strongly and specifically to the NS1A protein at its CPSF30 binding site will be effective inhibitors of influenza A virus replication. It is expected that the concentration of such small chemical compounds that can be achieved in all cells will greatly exceed the concentration of the F2F3 fragment achieved in the present study, resulting in a reduction of virus yield similar to that observed with mutational inactivation of the CPSF30 binding site on the NS1A protein. Further, the lack of any apparent growth impediment of the F2F3-expressing cells during 2 years in tissue culture bodes well for the identification of small chemical compounds that bind with high specificity to the CPSF30 binding site on the NS1A protein without affecting the 3′-end processing of host cell pre-mRNAs. It should be pointed out that the present study has already suggested an assay for the identification of such small-molecule inhibitors of influenza A virus replication, specifically, a high-throughput assay to identify small chemical compounds that inhibit the binding of the F2F3 fragment to the NS1A protein. Small chemical compounds directed against the CPSF30 binding site of the NS1A protein would be expected to inhibit the replication of all strains of influenza A virus.
Acknowledgments
This work was supported by Public Health Service grant AI-11772 from the National Institutes of Health.
REFERENCES
- 1.Barabino, S. M., W. Hubner, A. Jenny, L. Minvielle-Sebastia, and W. Keller. 1997. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 11:1703-1716. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 28 September 2005. Key facts about influenza and the influenza vaccine. http://www.cdc.gov/flu/keyfacts.htm. [Online.]
- 3.Chen, Z., Y. Li, and R. M. Krug. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox, N. J., and K. Subbarao. 1999. Influenza. Lancet 354:1277-1282. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson, N. M., D. A. Cummings, S. Cauchemez, C. Fraser, S. Riley, A. Meeyai, S. Iamsirithaworn, and D. S. Burke. 2005. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature 437:209-214. [DOI] [PubMed] [Google Scholar]
- 6.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. Garcia-Sastre. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99:10736-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim, M. J., A. G. Latham, and R. M. Krug. 2002. Human influenza viruses activate an interferon-independent transcription of cellular antiviral genes: outcome with influenza A virus is unique. Proc. Natl. Acad. Sci. USA 99:10096-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiso, M., K. Mitamura, Y. Sakai-Tagawa, K. Shiraishi, C. Kawakami, K. Kimura, F. G. Hayden, N. Sugaya, and Y. Kawaoka. 2004. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 364:759-765. [DOI] [PubMed] [Google Scholar]
- 9.Le, Q. M., M. Kiso, K. Someya, Y. T. Sakai, T. H. Nguyen, K. H. Nguyen, N. D. Pham, H. H. Ngyen, S. Yamada, Y. Muramoto, T. Horimoto, A. Takada, H. Goto, T. Suzuki, Y. Suzuki, and Y. Kawaoka. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437:1108. [DOI] [PubMed] [Google Scholar]
- 10.Li, Y., Z. Y. Chen, W. Wang, C. C. Baker, and R. M. Krug. 2001. The 3′-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA 7:920-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Longini, I. M., Jr., A. Nizam, S. Xu, K. Ungchusak, W. Hanshaoworakul, D. A. Cummings, and M. E. Halloran. 2005. Containing pandemic influenza at the source. Science 309:1083-1087. [DOI] [PubMed] [Google Scholar]
- 12.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 13.Loo, Y.-M., B. Fredericksen, and M. Gale, Jr. Submitted for publication.
- 14.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 15.Nemeroff, M. E., X. Y. Qian, and R. M. Krug. 1995. The influenza virus NS1 protein forms multimers in vitro and in vivo. Virology 212:422-428. [DOI] [PubMed] [Google Scholar]
- 16.Noah, D. L., K. Y. Twu, and R. M. Krug. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAs. Virology 307:386-395. [DOI] [PubMed] [Google Scholar]
- 17.Puthavathana, P., P. Auewarakul, P. C. Charoenying, K. Sangsiriwut, P. Pooruk, K. Boonnak, R. Khanyok, P. Thawachsupa, R. Kijphati, and P. Sawanpanyalert. 2005. Molecular characterization of the complete genome of human influenza H5N1 virus isolates from Thailand. J. Gen. Virol. 86:423-433. [DOI] [PubMed] [Google Scholar]
- 18.Qiu, Y., and R. M. Krug. 1994. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J. Virol. 68:2425-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid, A. H., J. K. Taubenberger, and T. G. Fanning. 2001. The 1918 Spanish influenza: integrating history and biology. Microbes Infect. 3:81-87. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu, K., A. Iguchi, R. Gomyou, and Y. Ono. 1999. Influenza virus inhibits cleavage of the HSP70 pre-mRNAs at the polyadenylation site. Virology 254:213-219. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki, H., R. Saito, H. Masuda, H. Oshitani, M. Sato, and I. Sato. 2003. Emergence of amantadine-resistant influenza A viruses: epidemiological study. J. Infect. Chemother. 9:195-200. [DOI] [PubMed] [Google Scholar]
- 22.World Health Organization. 2006. Avian influenza. http://www.who.int/csr/disease/avian_influenza/en/. [Online.]
- 23.Wright, P. F., and R. G. Webster. 2001. Orthomyxoviruses, p. 1533-1579. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 24.Yuan, W., and R. M. Krug. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]