Abstract
A pathogenic isolate of rhesus cytomegalovirus (rhCMV 180.92) was cloned, sequenced, and annotated. Comparisons with the published rhCMV 68.1 genome revealed 8 open reading frames (ORFs) in isolate 180.92 that are absent in 68.1, 10 ORFs in 68.1 that are absent in 180.92, and 34 additional ORFs that were not previously annotated. Most of the differences appear to be due to genetic rearrangements in both isolates from a region that is frequently altered in human CMV (hCMV) during in vitro passage. These results indicate that the rhCMV ORF repertoire is larger than previously recognized. Like hCMV, understanding of the complete coding capacity of rhCMV is complicated by genomic instability and may require comparisons with additional isolates in vitro and in vivo.
Rhesus cytomegalovirus (rhCMV) infection is widely prevalent in rhesus macaques and can cause disease in immunosuppressed simian immunodeficiency virus-infected macaques, indicating that it may provide a useful animal model for human CMV (hCMV) infection (2, 10-13, 19). The complete sequence of the ATCC strain rhCMV 68.1 was previously reported to contain 230 open reading frames (ORFs) that begin with an ATG, are greater than 100 amino acids (aa), and could potentially overlap larger ORFs. One hundred thirty-eight of these ORFs have homologues in hCMV (9). Defining the full coding capacity and genomic organization of wild-type hCMV has been an ongoing process due to the complexity of the hCMV genome and the genomic instability of in vitro-passaged isolates. Comparisons of hCMV strains after in vitro passage through fibroblasts versus in vivo clinical isolates have demonstrated that the hCMV genome is almost always mutated in vitro, a phenomenon highlighted by the complete sequence of the chimpanzee CMV (chCMV) genome (1, 3-7, 14, 15, 17). Thus, we cloned and sequenced a second independent rhCMV isolate for comparison and a better understanding of the rhCMV genome.
rhCMV 180.92 was isolated from diseased lung tissue of a simian immunodeficiency virus-infected macaque at the New England Primate Research Center and passaged in vitro six times on human fibroblasts (MRC-5) and seven times on primary rhesus macaque fibroblasts (10). The genome was cloned as seven overlapping cosmids and fully sequenced as previously described (18). The genome sequence was determined using 3,277 contiguous sequences (average of 500 bp) with an average 8.8-fold coverage. The first nucleotide of the genome was identified by homology to the rhCMV 68.1 genome (9).
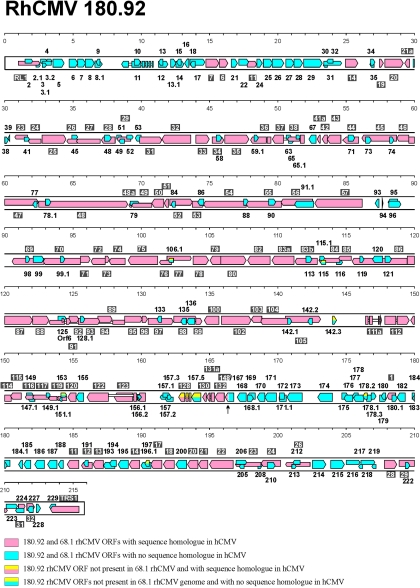
The rhCMV 180.92 genome is 215,678 nucleotides long with an overall 49% GC content. No perfect repeat regions were identified at the left end, middle, or right end of the genome. The rhCMV 180.92 genome contains 258 ORFs of 21 to 2,178 aa, with an average of 284 aa (Fig. 1) (see Table S1 in the supplemental material). This includes all ORFs beginning with an ATG and encoding a protein of at least 100 aa. When an ORF completely overlaps a larger ORF (49 ORFs), it is included only if it is encoded on the opposite cDNA strand (31 ORFs) or in a different reading frame on the same strand (18 ORFs). Ten ORFs of less than 100 aa are also included since there is significant homology (>20% at the aa level) to a previously annotated CMV ORF. Finally, 11 ORFs have homology with CMV genes that have been previously reported to be spliced.
FIG. 1.
Genome organization of rhCMV 180.92. The scale in kilobase pairs is shown at the top of each line. Protein-coding regions are indicated by leftward or rightward arrows according to the DNA strand on which they are encoded. Introns are shown as narrow white bars. Names of leftward ORFs are shown above the genome, and names of rightward ORFs are shown below the genome. rhCMV 180.92 ORFs with sequence homologues in hCMV (>20% aa similarity) are shown in pink and labeled with numbers in gray boxes, minus the rhUL prefix for ORFs rhUL6 to rhUL148 located between 15 and 166 kbp and the rhUS prefix for ORFs rhUS1 to rh US32 located between 178 and 212 kbp. rhORF6 is a homologue to the previously reported hCMV ORF6 (14). rhCMV 180.92 ORFs with no homologues in hCMV are shown in blue and labeled with plain numbers, minus the rh prefix. rhCMV 180.92 ORFs not found in rhCMV68.1 are shown in yellow, with the other half of the box coded pink or blue for the presence or absence of sequence homology to hCMV ORFs. The arrowhead at nt 166333 indicates the comparable position where 10 ORFs found in 68.1 but not in 180.92 are located. A fusion between rhUL148 and rh167 in rhCMV 180.92 is indicated by the interruption in the ORF.
rhCMV isolates 180.92 and 68.1 have an overall 97% nucleotide homology, but the rhCMV 180.92 genome is 5,781 nucleotides shorter than the rhCMV 68.1 genome (221,459 nucleotides [nt]) (9). Comparing the ORFs in 68.1 and 180.92 is complicated by the use of different criteria for identifying ORFs and different nomenclature for naming the ORFs. When we revisited the 68.1 sequence with the same criteria described above, we found a total of 260 instead of 230 ORFs in 68.1; (i) 34 additional 68.1 ORFs were identified (see Table S2 in the supplemental material) and (ii) we noted a net change of −4 to the total number of 68.1 ORFs originally reported, due to minor changes of the original 68.1 genome annotation (see Comment S1 in the supplemental material).
rhCMV isolate 68.1 ORFs were named numerically in order from left to right in the genome, beginning with an Rh (initial letter capitalized) if the ORF had a sequence homologue in hCMV (9). This convention failed to identify the hCMV homologues and made it difficult to incorporate newly identified rhCMV ORFs. In an effort to maximize information and maintain consistency, all rhCMV 180.92 ORFs are identified with the prefix rh. rhCMV 180.92 ORFs with significant homology to hCMV ORFs are identified using the rh prefix followed by the hCMV name (Fig. 1, ORFs shown in pink), e.g., the homologue of hCMV UL11 is identified as rhUL11. rhCMV 180.92 ORFs with significant homology to rhCMV 68.1 ORFs but with no homology to hCMV ORFs are identified using the 68.1 numbering (Fig. 1, blue ORFs), e.g., the homologue of rhCMV 68.1 rh2 is identified as rh2 in the rhCMV180.92 genome. When a new rhCMV ORF is identified and has no homology to hCMV, the rhCMV ORF is identified using the number of the immediately upstream rhCMV 68.1 ORF, followed by a decimal point and additional digit, e.g., a new rhCMV 180.92 ORF located immediately downstream to rhCMV 68.1 ORF rh13 is identified as rh13.1. Exons are identified using the name of the ORF they are part of followed by “ex1,” “ex2,” etc.; e.g., the two exons encoding the homologue of UL36 are identified as rhUL36ex1 and rhUL36ex2.
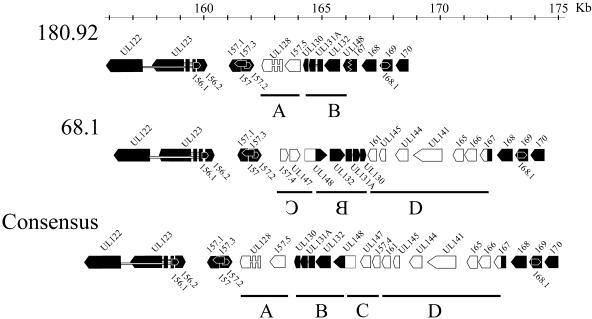
The overall amino acid homology between isolate 180.92 and 68.1 ORFs is 98%. The most significant differences between 180.92 and 68.1 are (i) 10 ORFs encoded in 68.1 but not 180.92 (Fig. 1, the arrowhead at nt 166333 indicates the comparable location of the 10 ORFs in 68.1) and (ii) 8 ORFs encoded in 180.92 but not found in 68.1 (Fig. 1, yellow ORFs). Most of these differences are encompassed within a divergent region between approximately nt 160000 and 175000 (Fig. 2). The region of the 180.92 genome, including ORFs from rhUL122 to rh170, is approximately 5 kb smaller than the comparable region in 68.1. Starting from the left end of this region, rhUL122 through rh157.2 are present in both genomes. However, in 180.92, there are two ORFs, rhUL128 and rh157.5, immediately adjacent to rh157.2 that are not present in 68.1 (region A in Fig. 2). UL128 is a protein with sequence characteristics of a CC-chemokine that confers endothelial cell tropism (1, 8). The gene is frequently disrupted in CMV isolates passaged through fibroblasts in vitro; for example, chCMV has a frame shift in the UL128 homologue, and in Toledo UL128, exon 3 is missing due to an inversion of the region between UL128 and UL133 (1, 6, 17).
FIG. 2.
Major region of divergence between rhCMV 68.1 and 180.92 genomes. ORFs present in both rhCMV 68.1 and 180.92 are represented by black arrows, whereas ORFs found in only one rhCMV genome are represented by white arrows. A consensus wild-type rhCMV genome organization for this region is shown at the bottom. Regions A, B, C, and D described in the text are designated by the lines. Inversion of regions B and C is demonstrated by the reversed letters.
Moving to the right of rh157.5 in isolate 180.92 (Fig. 2, region B), there are three leftward ORFs homologous to UL130, UL131A, and UL132. A striking difference is that this group of ORFs is rightward in 68.1, a dilemma similar to the situation in AD169 and Toledo strains where this group of genes is present in opposite orientations in the two hCMV strains. In AD169, they are located in the same orientation relative to, for example, the immediate-early (IE) genes (UL122/123), and in Toledo, they are oriented in the opposite direction of the IE genes (1, 6, 17). This issue was resolved by studying low-passage hCMV strains and sequencing chCMV, which revealed leftward UL130, UL131A, and UL132 ORFs oriented in the same direction as the IE genes, suggesting that this region is inverted in the Toledo strain (1, 6). Thus, by homology, we interpret that rhUL130, rhUL131A, and rhUL132 are normally positioned in the same orientation as the IE genes and have probably been inverted in 68.1.
Interestingly, the regions immediately to the left and right of region B in isolate 68.1, regions C and D in Fig. 2, have both been deleted from 180.92. Furthermore, 180.92 has a fusion between the 5′ end of rh167 and the 3′ end of rhUL148. In 68.1, rhUL147 and rhUL148 from region C are rightward, whereas they are leftward genes in Towne hCMV and chCMV (6, 17). This combination of events can be most directly explained if both region B and region C have been inverted in 68.1. In this case, the defect in 180.92 could be explained by a single deletion of region C and D from the putative wild-type consensus sequence where the breakpoints were located in rhUL148 and rh167, resulting in a fusion of the two ORFs (Fig. 2).
This region accounts for all 10 ORFs found in isolate 68.1 but not found in 180.92, and 2 of the 8 ORFs found in 180.92 but not in 68.1. The other six ORFs unique to 180.92 have significant nucleotide homology to comparable regions in 68.1, but complete ORFs are not present in 68.1 due to minor nucleotide sequence differences (see Table S3 in the supplemental material). It is not clear whether these represent natural strain variants, alterations during in vitro passage, or technical issues related to cloning or sequencing.
These studies suggest that a consensus wild-type rhCMV genome may be best constructed by comparing multiple genome sequences, similar to the use of chCMV and multiple isolates of hCMV for the description of the hCMV gene repertoire (6, 7, 14, 15). In this case, a consensus wild-type rhCMV would encode 268 ORFs, with 112 of these ORFs having recognized homology to hCMV genes. By contrast, 163 of 168 chCMV genes have homologues in hCMV (6, 14). This analysis is complicated by the use of different criteria for identifying and annotating CMV ORFs, but the results still reflect significant divergence of rhCMV from hCMV. The reason for the dramatic difference between rhCMV and hCMV is not clear. Perhaps some of the “unique” rhCMV ORFs have functional, but not sequence, homology with known or yet to be identified hCMV genes (16).
These studies demonstrate that the rhCMV genome is more complex than previously appreciated. Comparisons with a second rhCMV isolate highlight genomic instability in a common region for both isolates that shares a striking similarity to the genetic changes that arise in passaged hCMV isolates. We cannot exclude the possibility that some portions of the rhCMV genome may have been deleted from both isolates. Recognition that some of the rhCMV genome may be lost or rearranged during in vitro passage suggests that primate CMVs are not free of the genomic instability that plagues hCMV. An approach similar to the use of chCMV and multiple hCMV sequences to develop a consensus hCMV ORF repertoire may be necessary to define the complete ORF repertoire for rhCMV.
Nucleotide sequence accession number.
The complete annotation of the rhCMV 180.92 genome sequence was submitted to GenBank under the accession number DQ120516.
Supplementary Material
Acknowledgments
This research was supported by grants from the Public Health Service (CA68051 and AI43890), the New England Primate Research Center base grant (USPHS P51RR00168), and a developmental core award from the Partners/Fenway/Shattuck Center for AIDS Research (CFAR), an NIH-funded program (P30 AI42851).
We thank M. D. Daniel for the original isolation of the 180.92 rhCMV.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Akter, P., C. Cunningham, B. P. McSharry, A. Dolan, C. Addison, D. J. Dargan, A. F. Hassan-Walker, V. C. Emery, P. D. Griffiths, G. W. Wilkinson, and A. J. Davison. 2003. Two novel spliced genes in human cytomegalovirus. J. Gen. Virol. 84:1117-1122. [DOI] [PubMed] [Google Scholar]
- 2.Baskin, G. B. 1987. Disseminated cytomegalovirus infection in immunodeficient rhesus monkeys. Am. J. Pathol. 129:345-352. [PMC free article] [PubMed] [Google Scholar]
- 3.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 5.Dargan, D. J., F. E. Jamieson, J. MacLean, A. Dolan, C. Addison, and D. J. McGeoch. 1997. The published DNA sequence of human cytomegalovirus strain AD169 lacks 929 base pairs affecting genes UL42 and UL43. J. Virol. 71:9833-9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 7.Dolan, A., C. Cunningham, R. D. Hector, A. F. Hassan-Walker, L. Lee, C. Addison, D. J. Dargan, D. J. McGeoch, D. Gatherer, V. C. Emery, P. D. Griffiths, C. Sinzger, B. P. McSharry, G. W. Wilkinson, and A. J. Davison. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301-1312. [DOI] [PubMed] [Google Scholar]
- 8.Hahn, G., M. G. Revello, M. Patrone, E. Percivalle, G. Campanini, A. Sarasini, M. Wagner, A. Gallina, G. Milanesi, U. Koszinowski, F. Baldanti, and G. Gerna. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 78:10023-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen, S. G., L. I. Strelow, D. C. Franchi, D. G. Anders, and S. W. Wong. 2003. Complete sequence and genomic analysis of rhesus cytomegalovirus. J. Virol. 77:6620-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaur, A., M. D. Daniel, D. Hempel, D. Lee-Parritz, M. S. Hirsch, and R. P. Johnson. 1996. Cytotoxic T-lymphocyte responses to cytomegalovirus in normal and simian immunodeficiency virus-infected rhesus macaques. J. Virol. 70:7725-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaur, A., C. L. Hale, B. Noren, N. Kassis, M. A. Simon, and R. P. Johnson. 2002. Decreased frequency of cytomegalovirus (CMV)-specific CD4+ T lymphocytes in simian immunodeficiency virus-infected rhesus macaques: inverse relationship with CMV viremia. J. Virol. 76:3646-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaur, A., N. Kassis, C. L. Hale, M. Simon, M. Elliott, A. Gomez-Yafal, J. D. Lifson, R. C. Desrosiers, F. Wang, P. Barry, M. Mach, and R. P. Johnson. 2003. Direct relationship between suppression of virus-specific immunity and emergence of cytomegalovirus disease in simian AIDS. J. Virol. 77:5749-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lockridge, K. M., G. Sequar, S. S. Zhou, Y. Yue, C. P. Mandell, and P. A. Barry. 1999. Pathogenesis of experimental rhesus cytomegalovirus infection. J. Virol. 73:9576-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy, E., I. Rigoutsos, T. Shibuya, and T. E. Shenk. 30 October 2003. Reevaluation of human cytomegalovirus coding potential. Proc. Natl. Acad. Sci. USA 100:13585-13590. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 1 December 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 100:14976-14981. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pande, N. T., C. Powers, K. Ahn, and K. Fruh. 2005. Rhesus cytomegalovirus contains functional homologues of US2, US3, US6, and US11. J. Virol. 79:5786-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prichard, M. N., M. E. Penfold, G. M. Duke, R. R. Spaete, and G. W. Kemble. 2001. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev. Med. Virol. 11:191-200. [DOI] [PubMed] [Google Scholar]
- 18.Rivailler, P., Y. G. Cho, and F. Wang. 2002. Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a new world primate: a defining point in the evolution of oncogenic lymphocryptoviruses. J. Virol. 76:12055-12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sequar, G., W. J. Britt, F. D. Lakeman, K. M. Lockridge, R. P. Tarara, D. R. Canfield, S. S. Zhou, M. B. Gardner, and P. A. Barry. 2002. Experimental coinfection of rhesus macaques with rhesus cytomegalovirus and simian immunodeficiency virus: pathogenesis. J. Virol. 76:7661-7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.