Abstract
The genome of Red clover necrotic mosaic virus (RCNMV) in the genus Dianthovirus is divided into two RNA molecules of RNA1 and RNA2, which have no cap structure at the 5′ end and no poly(A) tail at the 3′ end. The 3′ untranslated region (3′ UTR) of RCNMV RNA1 contains an essential RNA element (3′TE-DR1), which is required for cap-independent translation. In this study, we investigated a cap-independent translational mechanism of RNA2 using a firefly luciferase (Luc) gene expression assay system in cowpea protoplasts and a cell-free lysate (BYL) prepared from evacuolated tobacco BY2 protoplasts. We were unable to detect cis-acting RNA sequences in RNA2 that can replace the function of a cap structure, such as the 3′TE-DR1 of RNA1. However, the uncapped reporter RNA2, RNA2-Luc, in which the Luc open reading frame (ORF) was inserted between the 5′ UTR and the movement protein ORF, was effectively translated in the presence of p27 and p88 in protoplasts in which RNA2-Luc was replicated. Time course experiments in protoplasts showed that the translational activity of RNA2-Luc did not reflect the amount of RNA2. Mutations in cis-acting RNA replication elements of RNA2 abolished the cap-independent translational activity of RNA2-Luc, suggesting that the translational activity of RNA2-Luc is coupled to RNA replication. Our results show that the translational mechanism differs between two segmented genomic RNAs of RCNMV. We present a model in which only RNA2 that is generated de novo through the viral RNA replication machinery functions as mRNA for translation.
Many lines of evidence indicate that expression of the virus genome is temporally and spatially regulated, as this is necessary to ensure efficient virus proliferation. RNA viruses use various mechanisms to express viral proteins encoded in their genomes. The mechanisms include subgenomic RNA (sgRNA) production, ribosomal frameshifting, and read-through to translate viral proteins not encoded in the 5′ proximal open reading frames (ORFs) of multicistronic genomic RNA, polyprotein production followed by processing to generate functional viral proteins, and segmentation of the genomic RNA to locate an ORF to the 5′ proximal region of viral genomes.
Unlike other members of the family Tombusviridae, which have monopartite RNA genomes, the genome of Red clover necrotic mosaic virus (RCNMV) in the genus Dianthovirus is divided into two RNA molecules of RNA1 and RNA2 (Fig. 1A) (12, 17, 35). RNA1 and RNA2 have no cap structure at the 5′ end (33) and no poly(A) tail at the 3′ end (29, 54). RNA1 encodes the putative RNA replicase components, 27-kDa protein (p27) and 88-kDa protein (p88). p88, which has an RNA-dependent RNA polymerase motif (27), is produced by programmed −1 ribosomal frameshifting (24, 53). Both p27 and p88, together with replication-competent viral RNA, are required to suppress RNA silencing, probably through RNA replication (42). RNA1 also encodes a 37-kDa coat protein (CP), which is expressed from an sgRNA (56). Transcription of the CP sgRNA requires an intermolecular interaction between RNA1 and RNA2 and is probably generated by a premature termination mechanism (40, 43). The trans-activator (TA) of RNA2, which is located within the 35-kDa movement protein (MP)-coding region and required for an interaction between RNA1 and RNA2, also plays an essential role in the replication of RNA2, independent of the TA function (43). RNA2 is a monocistronic RNA that encodes a MP, which is required for viral cell-to-cell movement in plants (29, 52). cis-acting RNA elements necessary for the replication of RNA2 have been mapped to the 5′ and 3′ untranslated regions (UTRs) in addition to the TA in the protein-coding region (42, 43, 47).
FIG. 1.
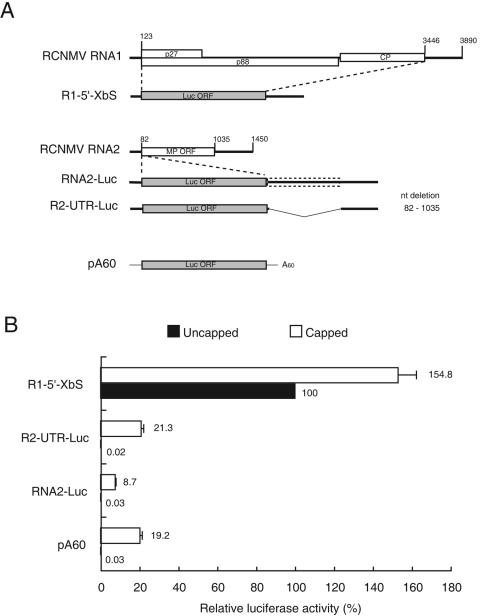
(A) Schematic diagram of RCNMV RNA1 and RNA2, reporter Luc mRNAs of RNA1 and RNA2 (R1-5′-XbS, RNA2-Luc, and R2-UTR-Luc), and Luc mRNA that contains vector-derived sequence (thin line) and poly(A) tail of 60 nt (pA60). The RCNMV genome is shown as a thick line with the protein-coding regions depicted as boxes. The dotted box indicates an untranslatable MP ORF. The bent line indicates the deleted region. Numbers indicate the positions of nucleotides. (B) Translational activities of capped and uncapped Luc RNA transcripts in cowpea protoplasts. Luc RNA was cotransfected with R-Luc mRNA that was used as an internal control. Transfected protoplasts were incubated at 17°C for 6 h. Luc activities are expressed as percentages of uncapped R1-5′-XbS activity. Error bars show the standard errors, and the mean values are given. Assays were performed at least three times.
In eukaryotic cells, the 5′ cap structure serves as the binding site for the eukaryotic initiation factor eIF4F, which is composed of eIF4G, eIF4E, and eIF4A, and assists the binding of 40S ribosomes to mRNA (14). The cap structure also interacts with poly(A)-binding protein through eIF4G, which stabilizes eIF4F binding to the 5′ cap (38). Consequently, the 5′ cap and 3′ poly(A) tail are critical for recruiting translation machinery and efficient translation of encoded proteins. However, neither of the segmented genomic RNAs of RCNMV have 5′ cap structures or 3′ poly(A) tails. Instead, RCNMV RNA1 contains an essential RNA element (3′TE-DR1) in the 3′ UTR, which is required for cap-independent translation (33). Such RNA elements have been detected in the 3′ UTRs of the genomic RNAs of viruses in the families Tombusviridae and Luteoviridae: Tobacco necrosis virus (genus Necrovirus) (39), Turnip crinkle virus (genus Carmovirus) (36), Hibiscus chlorotic ringspot virus (genus Carmovirus) (25), Tomato busy stunt virus (genus Tombusvirus) (6, 51), and Barley yellow dwarf virus (BYDV) (genus Luteovirus) (13, 48). It has been shown that a 5′-3′ RNA-RNA interaction is required for efficient translation of BYDV (13) and Tomato busy stunt virus (6) mRNAs. The 3′TE-DR1 of RCNMV RNA1 consists of five stem-loop (SL) structures, and SL-1 is conserved in the 3′ UTRs of genomic RNAs of BYDV and Tobacco necrosis virus as well as RNA1 of dianthoviruses (33, 48). In contrast, no nucleotide sequences or RNA secondary structures resembling 3′TE-DR1 are present in the 3′ UTR of RCNMV RNA2 (H. Mizumoto and T. Okuno, unpublished results).
In this study, we investigated the translational mechanism of RNA2 using a firefly luciferase (Luc) gene expression assay system in cowpea protoplasts (33) and a cell-free lysate (BYL) prepared from evacuolated tobacco BY2 protoplasts (26). Our results show that the translation mechanism of RNA2 differs from that of RNA1 and that the cap-independent translational activity of RNA2 is linked to RNA replication. We present a model in which only RNA2 that is generated de novo through the viral RNA replication machinery functions as mRNA for translation.
MATERIALS AND METHODS
Plasmid clones.
Full-length cDNA clones of RCNMV Australian strain RNA1 (pRC1|G) and RNA2 (pRC2|G) were kindly provided by S. A. Lommel (55). In the PCR primers given below, underlining indicates an introduced restriction enzyme site and boldface indicates a mutated nucleotide. All DNA fragments with incompatible ends used here were ligated after blunting with T4 DNA polymerase.
(i) pRNA2-Luc.
A cDNA fragment of RCNMV RNA2 from nucleotides (nt) 1 to 80 was amplified by PCR from pRC2|G using primers P/T7/AR2 (5′-AACTGCAGTTGTAATACGACTCACTATAGACAAACCTCGCTC-3′; the T7 promoter sequence is shown in italics) and Aus2-5′Nco (5′-AAAACCCATGGCCAAACCTCTTTGTATTG-3′). The amplified DNA fragment was cloned into pGEM-T Easy vector (Promega, Madison, WI), creating pGEMR2-5′. pGEMR2-5′ was digested with NdeI and NcoI and cloned into the corresponding enzyme sites of pSP-luc+ (Promega), creating pR2-5′. A cDNA fragment of RCNMV RNA2 from nt 1 to 96 was amplified by PCR from pRC2|G using primers P/T7/AR2 and RC2dMP− (5′-CCACATGAACAGCAATGCATCCAAACCTC-3′). A cDNA fragment of RCNMV RNA2 from nt 72 to 1450 was amplified by PCR from pRC2|G using primers RC2dMP+ (5′-GAGGTTTGGATGCATTGCTGTTCATGTGG-3′) and R-C/R2-3′ (5′-CGTCCCGGGGTGCCTAGCCGTTATACGAC-3′). Both DNA fragments were denatured and annealed, and the resulting mixture was used as a template for PCR (16). The newly synthesized DNA duplex was amplified by PCR using primers P/T7/AR2 and R-C/R2-3′. The amplified 1.5-kb DNA fragment was digested with PstI and SmaI and cloned into the corresponding enzyme sites of pUC118 (Takara Bio Inc., Otsu, Japan) to create pURC2fsMP. PstI/XbaI DNA fragment of pR2-5′ was cloned into PstI/EcoT22I sites of pURC2fsMP, creating pRNA2-Luc.
(ii) pRNA2-17/Luc.
A cDNA fragment of RCNMV RNA2 from nt 20 to 80 was amplified by PCR from pRC2|G using primers P/T7/AR2 (5′-AACTGCAGTTGTAATACGACTCACTATAGACAAAAGATTAATTGATTGGAAG-3′; the T7 promoter sequence is shown in italics) and Aus2-5′Nco (5′-AAAACCCATGGCCAAACCTCTTTGTATTG-3′). The amplified DNA fragment was digested with PstI and NcoI and cloned into the corresponding enzyme sites of pRNA2-Luc.
(iii) RNA2-MP3′D2/Luc.
NheI/XbaI DNA fragments of pfsMP3′-D2 (43) were cloned into the corresponding sites of pRNA2-Luc.
(iv) RNA2-3′SLm/Luc.
A cDNA fragment of RCNMV RNA2 from nt 675 to 1450 was amplified by PCR from pRC2|G using primers S/R2-int5′P (5′-TGACACAAGCAGGATGGAGA-3′) and 2-3′SLm− (5′-TACCCGGGGTGCCTAGCCGTATAACGACAT-3′). The amplified DNA fragment was digested with XbaI and SmaI and cloned into the corresponding enzyme sites of pRNA2-Luc.
(v) pR2-UTR-Luc.
XbaI/SmaI DNA fragments of pRC1|G were cloned into the corresponding sites of pR1-XbS (33), creating pLuc-A2. NdeI/NcoI DNA fragments of pGEMR2-5′ were cloned into the corresponding sites of pLuc-A2.
(vi) pUBRC1.
The small SacI-SmaI fragment of pUC118 (Takara) was replaced with the 3.9-kb SacI-SmaI fragment of pRC2|G to create pUCR1. The small EcoRI fragment of pBICBPBR2R (22) was treated with T4 DNA polymerase and then inserted into T4 DNA polymerase-treated KpnI site of pBICP35 (34) to create pBICP35R. The resulting plasmid encodes transcription cassettes containing the 35S promoter and terminator of Cauliflower mosaic virus (CaMV) and ribozyme sequence of a satellite RNA of Tobacco ringspot virus (sTRSV). A cDNA fragment of the 35S promoter sequence was amplified by PCR from pBICP35R using primers Sa35+ (5′-GCGAGCTCAACATGGTGGAGCACGACACGC-3′) and 35/RC1− (5′-CGTTTGTCCTCTCCAAATGAAATGAAC-3′). A cDNA fragment of RCNMV RNA1 from nt 1 to 1981 was amplified by PCR from pUCR1 using primers 35/RC1+ (5′-GGAGAGGACAAACGTTTTACCGGTTTG-3′) and S/R1-Int3′P (5′-TTGCGTGGCAATGCAAACCG-3′). The recombinant PCR product was digested with SacI and EcoRI and used to replace the corresponding region of pUCR1 to create pRCP35A1. A cDNA fragment of the sTRSV ribozyme and the 35S terminator sequence was amplified by PCR from pBICP35R using primers RC1/Rt+ (5′-GGTACCCCGTCACCGGATGTGTTTTCCG-3′) and RtSm+ (5′-TCCCCCGGGTATAGGGACTTTAGGTGATC-3′). A cDNA fragment of RCNMV RNA1 from nt 1852 to 3890 was amplified by PCR from pUCR1 using primers S/R1-Int5′P (5′-TGAGCAGATAAACCGCAATC-3′) and RC1/Rt− (5′-CCGGTGACGGGGTACCTAGCCGTTATAC-3′). The recombinant PCR product was digested with SacII and SmaI and used to replace the corresponding region of pRCP35A1 to create pUBRC1.
(vii) pUBRC1/3′D1.
A cDNA fragment of the sTRSV ribozyme and the 35S terminator sequence was amplified by PCR from pBICP35R using primers 35S/3D1+ (5′-GAATATTTCCCCGTCACCGGATGTGTTTTC-3′) and RtSm+ (5′-TCCCCCGGGTATAGGGACTTTAGGTGATC-3′). A cDNA fragment of RCNMV RNA1 from nt 1852 to 3862 was amplified by PCR from pUCR1 using primers S/R1-Int5′P (5′-TGAGCAGATAAACCGCAATC-3′) and 35S/3′D1− (5′-CCGGTGACGGGGTACCTAGCCGTTATAC-3′). The recombinant PCR product was digested with SacII and SmaI and used to replace the corresponding region of pUBRC1 to create pUBRC1/3′D1.
(viii) pUBRC1/p27fs.
pUBRC1 was digested with EcoRI, end filled with T4 DNA polymerase, and religated.
(ix) pUBRC1/CPfs.
The transcription vector of a CP frameshift mutant of RNA1 (pRNA1fsCP) (43) was digested with XhoI and SacII and used to replace the corresponding region of pUBRC1.
(x) pUBp27 and pUBp88.
pUBp27 and pUBp88 were constructed as previously described (42).
(xi) pUBGFPBam.
pUBGFPBam was constructed as previously described (41).
All constructs were verified by sequencing with an ABI 310 automated sequencer (Applied Biosystems, Foster City, CA).
RNA preparation.
RNA transcripts were synthesized in vitro from the linearized vector with T7 or SP6 RNA polymerase and purified with a Sephadex G-50 fine column (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). The RNA concentration was determined spectrophotometrically and its integrity verified by 1% agarose gel electrophoresis. RCNMV RNA1 was transcribed from SmaI-linearized pUCR1 with T7 RNA polymerase. Renilla luciferase (R-Luc) mRNA, Luc mRNA (pA60), R1-5′-XbS, and 3′TE-DR1/Lm1 were synthesized as described previously (33). RNA2-Luc, RNA2-17/Luc, RNA2-MP3′D2/Luc, RNA2-3′SLm/Luc, and R2-UTR-Luc (named for their parent plasmids minus the “p” prefix) were transcribed from SmaI-linearized plasmids with T7 RNA polymerase.
Protoplast transfection.
Cowpea plants (Vigna unguiculata cv. California Blackeye) were grown as previously described (7). Cowpea protoplasts were prepared from 12- to 16-day-old plants essentially as previously described (15), except that 0.5 M mannitol was used as an osmotic stabilizer in all solutions to which the protoplasts were exposed. Inoculation and incubation of protoplasts were carried out as previously described (28). Protoplasts (3.0 × 105) were inoculated with transcripts (2.0 pmol Luc mRNA and 0.7 pmol R-Luc mRNA) or plasmid DNA (10 μg) plus herring testes carrier DNA (20 μg) (BD Biosciences, Palo Alto, CA). Inoculated protoplasts were incubated at 17°C, an optimum temperature for RCNMV replication (32).
Dual-Luc assay.
Luciferase assays were performed using the dual-luciferase reporter assay system (Promega) as described previously (33). The luminescence of Luc was normalized to the luminescence of R-Luc. Each experiment was repeated at least three times with different batches of protoplasts.
Northern blot analysis.
Total RNAs extracted from protoplasts were subjected to Northern blot analysis as previously described (4). The digoxigenin (DIG)-labeled RNA probe specific to RCNMV 3′ UTR was previously described (32). The DIG-labeled RNA probe specific for the Luc gene was also previously described (33). The DIG-labeled RNA probe specific for negative-strand RNA2 was transcribed from SmaI-linearized pRC2|G with T7 RNA polymerase. The RNA signals were detected with a luminescent-image analyzer (LAS 1000 Plus; Fuji Photo Film, Japan), and the signal intensities were quantified with the Image Gauge program (Fuji Photo Film).
In vitro translation.
Preparation of cell extracts of evacuolated tobacco BY2 protoplasts (BYL) and in vitro translation reaction mixtures were performed by the method of Komada et al. (26). mRNAs carrying the luciferase gene (5 fmol) were translated in 25 μl of BYL translation reaction mixture. The reaction mixture was incubated at 17°C for 2.5 h. Aliquots (3 μl) of the reaction mixture were used to assay luciferase activity in the luciferase assay system (Promega). In other experiments, RNA1 mutants (190 fmol) were translated in the presence of RNA2-Luc (125 fmol) in 25 μl of BYL translation reaction mixture at 17°C for 2.5 h, and aliquots (10 μl) of the reaction mixture were used for immunoblot analysis. Aliquots (3 μl) of the reaction mixture were used to assay luciferase activity.
Immunoblot analysis.
Antibody production and immunoblot analysis were performed as described previously (42).
RESULTS
Nucleotide sequences of RNA2 alone are insufficient for cap-independent translation of RNA2.
Our preliminary experiments showed that reporter RNA2s, in which the firefly luciferase ORF replaced the MP ORF (R2-UTR-Luc) or was inserted between the 5′ UTR and the MP ORF (RNA2-Luc) (Fig. 1A), were not translated in the absence of the 5′ cap structure. These results suggest that RNA2 requires other factors in addition to RNA sequences for cap-independent translation and employs a translational strategy that differs from that of RNA1.
To investigate the translational mechanism of RNA2, we first compared the translational activities of capped and uncapped RNA2-Luc and R2-UTR-Luc with that of R1-5′-XbS, which is a reporter Luc mRNA with the 5′ and 3′ UTRs of RNA1 (33). In vitro-transcribed RNAs were transfected into cowpea protoplasts together with capped Renilla luciferase mRNA with a poly(A) tail (R-Luc), which was used as an internal control in the translation assays in protoplasts. Luciferase activities were measured at several intervals after transfection, and Luc activity was evaluated in reference to R-Luc activity. Results 6 h after transfection are shown in Fig. 1B. No Luc activity was detected after transfection with either RNA2-Luc or R2-UTR-Luc in the absence of the 5′ cap structure, but capped constructs of these RNAs were translated as efficiently as the capped reporter Luc mRNA that contains vector-derived sequence and a poly(A) tail of 60 nucleotides (pA60) (Fig. 1A). Similar results were obtained at all times for up to 16 h after transfection. These results confirmed our previous observations that nucleotide sequences of RNA2 alone are insufficient for cap-independent translation of RNA2 and that the cap-independent translation mechanism of RNA2 differs from that of RNA1.
Cap-independent translation of RNA2 requires RNA1.
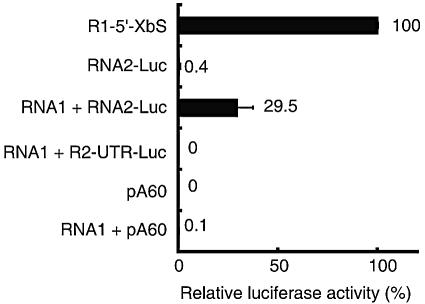
Because RNA2 has no 5′ cap structure (33), it must be translated in a cap-independent manner during viral infection. Therefore, we tested whether uncapped RNA2-Luc and R2-UTR-Luc are translated in the presence of RNA1. Uncapped transcripts of RNA2-Luc and R2-UTR-Luc were transfected into protoplasts together with RNA1 in an equal molar ratio (2 pmol each), and translational activity was measured 6 h after transfection. RNA1 did not enhance the translational activity of either uncapped R2-UTR-Luc or a capped reporter Luc mRNA (pA60) (Fig. 2). Interestingly, RNA2-Luc was translated in protoplasts (Fig. 2). In the protoplasts, RNA2-Luc was replicated together with RNA1 (data not shown) (see Fig. 4 and 5). R2-UTR-Luc was not amplified by RNA1 because of the absence of TA, which is an essential cis-acting replication element of RNA2 (43). These results suggest that replication of RNA2-Luc or RNA1, transcription of CP sgRNA, or CP expressed from the sgRNA is required for the cap-independent translation of RNA2-Luc. These results also indicate that translational enhancement by RNA1 is specific for the replication-competent RNA2-Luc that contains the entire RNA2 sequence.
FIG. 2.
Translational activity of uncapped Luc RNA transcripts in the presence or absence of RNA1. For other conditions, refer to the legend to Fig. 1B.
FIG. 4.
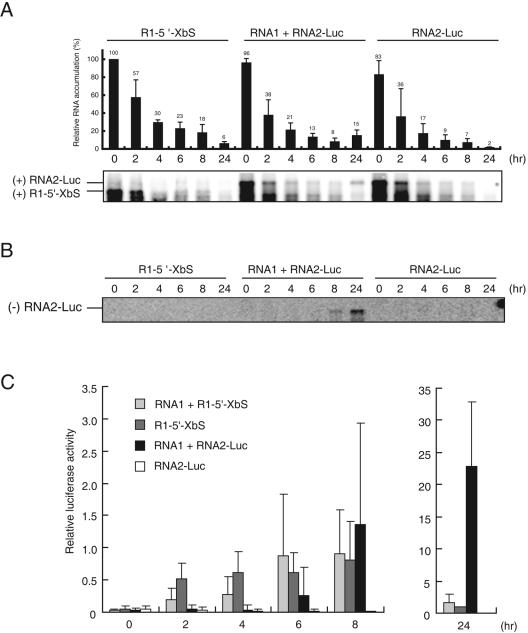
(A) Schematic diagrams of deletion or nucleotide substitution mutants of RNA2-Luc. The genome organization of RCNMV RNA2 is shown at the top. The bent line indicates the deleted region. The position of nucleotide substitution is indicated by the ▿ symbol. The dotted box indicates untranslatable MP ORF. (B) Translational activity of RNA2-Luc mutants in cowpea protoplasts. Transfected protoplasts were incubated at 17°C for 24 h. Luc activities for RNA2-Luc mutants are presented as Luc/R-Luc ratios. For other conditions, refer to the legend to Fig. 1B. (C) Accumulation of positive-strand RNA1 and sgRNA (top), positive-strand RNA2-Luc (middle), and negative-strand RNA2-Luc (bottom) in cowpea protoplasts. Total RNA was extracted from protoplasts 24 h after transfection. The membranes were then probed with DIG-labeled RNA specific for RCNMV RNA1 (top), Luc ORF (middle), and negative-strand RCNMV RNA2 (bottom). For other conditions, refer to the legend to Fig. 3C.
FIG. 5.
(A) Temporal changes in the accumulation pattern of Luc RNA. Total RNA was extracted from protoplasts immediately after transfection (0 h) and 2, 4, 6, 8, and 24 h after transfection, separated by gel electrophoresis, and blotted onto membranes. The membranes were then probed with DIG-labeled RNA specific for the Luc ORF. Relative values for the accumulation of Luc RNA (0 h in inoculation of R1-5′-XbS was defined as 100%) were calculated from four independent experiments, and a representative pattern on a Northern blot is shown below. Error bars show the standard errors, and mean values are given. (B) Temporal changes in the accumulation pattern of negative-strand RNA2-Luc. (C) Temporal changes in the translational activity of Luc RNA. Relative values for Luc activity (24 h in inoculation of R1-5′-XbS alone was defined as 1) were calculated from four independent experiments. Error bars show the standard errors.
p27 and p88 are sufficient to enhance the cap-independent translation of RNA2.
To determine the viral factors of RNA1 required for translational enhancement of RNA2-Luc, we used DNA vectors to consistently supply viral proteins or replication-incompetent RNA1. Protoplasts were transfected with RNA2-Luc together with pUBRC1, pUBRC1/3′D1, pUBRC1/CPfs, or pUBRC1/p27fs (Fig. 3A). pUBRC1/3′D1 produces RNA1 that lacks the 3′-terminal stem-loop structure and is expected not to replicate, but to function as mRNA for both p27 and p88 (33). pUBRC1/p27fs produces RNA1p27fs, which has a four-base insertion in the p27 ORF and produces a truncated p27. pUBRC1/CPfs produces a CP frameshift mutant of RNA1 (43), and pUBRC1 produces wild-type RNA1. Viral RNA accumulation and Luc activity were analyzed 24 h after transfection (Fig. 3B and C). RNA1 and CP sgRNA accumulated in protoplasts transfected with pUBRC1 or pUBRC1/CPfs, but not in those transfected with pUBRC1/3′D1 or pUBRC1/p27fs (Fig. 3C). RNA2-Luc accumulated in the protoplasts transfected with pUBRC1/3′D1, pUBRC1, or pUBRC1/CPfs, but accumulation in the protoplasts transfected with pUBRC1/3′D1 was lower than in those transfected with pUBRC1/CPfs or pUBRC1 (Fig. 3C). In these protoplasts, translational activity of RNA2-Luc was observed (Fig. 3B). No translational activity of RNA2-Luc was detected in protoplasts transfected with pUBRC1p27fs. These results indicate that RNA1 replication and CP are not essential for cap-independent translation of RNA2-Luc.
FIG. 3.
(A) Schematic diagrams of DNA-mediated expression plasmids. The pentagon labeled 35S represents the 35S promoter of CaMV. The boxes labeled Ribo and Ter represent the ribozyme of a sTRSV and the terminator of CaMV, respectively. The RCNMV genome is shown as a thick line with the protein-coding regions indicated by boxes. The four nucleotides used to generate the p27 and CP frameshift mutants are indicated by the ▿ symbol at the insertion sites. (B) Translational activity of RNA2-Luc cotransfected with DNA-mediated expression plasmids in cowpea protoplasts. Luc activities for RNA2-Luc are presented as Luc/R-Luc ratios. Transfected protoplasts were incubated at 17°C for 24 h. For other conditions, refer to the legend to Fig. 1B. (C) Accumulation of RNA1, sgRNA, and RNA2-Luc in cowpea protoplasts. Total RNA was extracted from protoplasts 24 h after transfection, separated by gel electrophoresis, and blotted onto membranes. The membranes were then probed with DIG-labeled RNA specific for RCNMV RNA1 (upper) and Luc ORF (lower).
To investigate whether p27 and p88 alone can enhance cap-independent translation of RNA2-Luc, we used pUBp27 and pUBp88 to completely uncouple the expression of p27 and p88 from RNA1 replication. pUBp27 and pUBp88 contain the p27 or p88 ORF between the CaMV 35S promoter and the terminator (Fig. 3A). Transfection of protoplasts with RNA2-Luc together with a mixture of pUBp27 and pUBp88 enhanced the translational activity of RNA2-Luc, whereas activity was not enhanced by transfection with either pUBp27 or pUBp88 (Fig. 3B). Northern blot analysis showed that RNA2-Luc accumulated in protoplasts transfected with a mixture of pUBp27 and pUBp88, but not in those transfected with pUBp27 or pUBp88 alone (Fig. 3C). These results indicate that p27 and p88 are both required and sufficient to enhance the cap-independent translational activity of RNA2-Luc.
RNA2 elements required for cap-independent translation.
The results presented in the previous section suggest a link between RNA2 replication and cap-independent translation. Replication of positive-strand RNA viruses consists of two main steps, negative-strand RNA synthesis and subsequent positive-strand RNA synthesis (2). cis-acting elements required for replication of RNA2 have been mapped to the 5′ and 3′ UTRs of RNA2 (47) and to the MP-coding region, including TA (43). The 3′ and 5′ UTRs of RNA2 are required for negative-strand RNA and positive-strand RNA synthesis, respectively (47). The TA may be required for negative-strand RNA synthesis, because the RNA2 mutant, which has a deletion in the MP-coding region that includes the TA, did not accumulate and failed to suppress RNA silencing in an Agrobacterium-mediated RNA silencing suppression assay (42) (M. Tsukuda and T. Okuno, unpublished results). To identify the most important step in RNA replication for cap-independent translation of RNA2-Luc, we used three RNA2-Luc mutants. RNA2-17/Luc and RNA2-MP3′D2/Luc have deletions in the 5′ UTR and the MP-coding region that includes the TA (43), respectively, and RNA2-3′SLm/Luc has a nucleotide substitution in the 3′ proximal stem-loop structure of RNA2 (Fig. 4A). These RNA2-Luc mutants were transfected together with RNA1 into protoplasts and were tested for translational activity and RNA accumulation. The translational activities of all three RNA2 mutants were 100 times less than that of RNA2-Luc (Fig. 4B). We were unable to detect positive- or negative-strand RNAs of any RNA2-Luc mutant 24 h after transfection in the Northern blot analysis (Fig. 4C). These results suggest that replication of RNA2-Luc is required for cap-independent translation of RNA2-Luc.
The amount of RNA2 does not reflect the translational activity of RNA2.
It should be noted that the results presented in the preceding section do not rule out the possibility that RNA replication merely increases the amount of RNA2-Luc but does not reflect cap-independent translational activity. To investigate this possibility, the relationship between accumulation level and translational activity of RNA2-Luc in the presence or absence of RNA1 was studied using time course analysis by the Northern blotting method or the Luc assay, respectively. R1-5′-XbS was used as a control. Protoplasts were transfected with RNA2-Luc or R1-5′-XbS alone or together with RNA1. Northern blot results showed that the quantity of RNA2-Luc and R1-5′-XbS decreased during incubation except for that of RNA2-Luc in the presence of RNA1 24 h after transfection (Fig. 5A). The rate of decrease in RNA for RNA2-Luc and R1-5′-XbS did not differ significantly in the absence of RNA1 (Fig. 5A). The presence of RNA1 did not significantly affect the stabilities of RNA2-Luc (Fig. 5A) and R1-5′-XbS (data not shown). Despite the decrease in RNA levels, the translational activity of R1-5′-XbS increased immediately after transfection (Fig. 5C). The presence of RNA1 did not significantly enhance the translational activity of R1-5′-XbS during incubation (Fig. 5C). In contrast to R1-5′-XbS, no increase in translational activity was detected in protoplasts transfected with RNA2-Luc alone at any time point during incubation (Fig. 5C). However, in the presence of RNA1, the translational activity of RNA2-Luc was detected 6, 8, and 24 h after transfection (Fig. 5C). At 6 and 8 h after transfection, the level of positive-strand RNA2-Luc was as low as that in the absence of RNA1 (Fig. 5A). It should be noted that negative-strand RNA2-Luc was detected 8 h after transfection (Fig. 5B). These results suggest that the RNA replication process, rather than the amount of RNA2-Luc, contributes to cap-independent translation. At 24 h after transfection, the translational activity of RNA2-Luc was 17 times that at 8 h after transfection, in contrast to 1.3-fold and 1.8-fold increases in the translational activity of R1-5′-XbS and R1-5′-XbS + RNA1, respectively. A drastic increase in the translational activity of RNA2-Luc observed 24 h after transfection probably reflects active RNA replication, because RNA2-Luc, but not R1-5′-XbS, is competent in replication. Taken together, the cap-independent translation of RNA2-Luc is likely dependent on the de novo production of RNA.
It should be noted that the Northern blot results shown in Fig. 5A might not accurately reflect RNA incorporated into protoplasts, because transfected protoplasts may also contain RNA inoculum remaining on the cell surface. Therefore, it is difficult to accurately compare translation efficiency between 3′TE-DR1-mediated translation for RNA1 and replication-linked translation for RNA2 in our experiments using protoplasts.
In vitro translation assay.
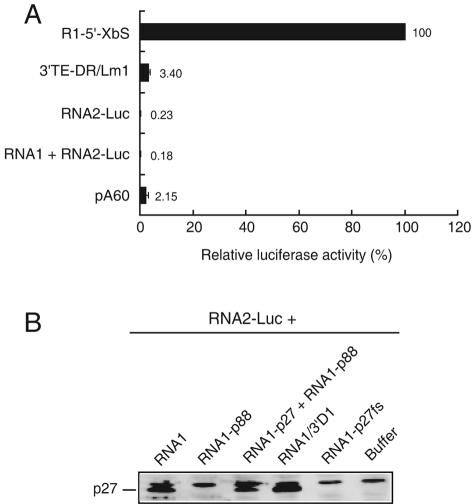
The in vitro translation assay is often used to investigate the translation mechanisms of eukaryotic mRNAs, including viral RNA (8-10, 49). To confirm the absence in RNA2 of cis-acting RNA elements equivalent to the 3′TE-DR1 present in RNA1, we analyzed the translational activity of RNA2-Luc and other Luc reporter mRNAs in vitro using BYL prepared from evacuolated tobacco BY2 protoplasts (26). BYL reflects the cap-independent translational activity of reporter Luc mRNAs with a series of mutations in the 3′ UTR of RNA1 (33) in cowpea protoplasts (data not shown), although the removal of the cap structure did not completely abolish the translational activities of pA60 and 3′TE-DR1/Lm1 (Fig. 6A). The construct of 3′TE-DR1/Lm1 is an R1-5′-XbS mutant with nucleotide substitutions in the 3′TE-DR1, and it does not exhibit translational activity in the absence of a cap structure in cowpea protoplasts (33). In BYL, the translational activity of uncapped RNA2-Luc was lower than that of uncapped pA60 and 3′TE-DR1/Lm1 (Fig. 6A). The addition of RNA1 did not enhance the translational activity of RNA2-Luc in BYL (Fig. 6A). The results from the in vitro translation assay support the results obtained in protoplasts, namely, that RNA2 has no cis-acting elements equivalent to the 3′TE-DR1 of RNA1 and that it requires additional factors for cap-independent translation. To confirm the expression of p27 from RNA1 in BYL, increasing amounts of RNA1 were translated in BYL together with RNA2-Luc. The immunoblot analysis using p27 antibody showed that RNA1 expressed p27 (Fig. 6B) and probably expressed p88 (53). Translational activity of RNA2-Luc was not enhanced by any RNA1 mutants (data not shown). A complete RCNMV replication system in BYL may help to elucidate the cap-independent translation mechanism of RNA2.
FIG. 6.
(A) Translational activity of uncapped Luc mRNAs in cell-free lysate (BYL) prepared from evacuolated tobacco BY2 protoplasts (26). Luc mRNAs (5 fmol) were translated at 17°C for 2.5 h. Luc activities were expressed as percentages of uncapped R1-5′-XbS activity; error bars show the standard errors from four independent experiments. (B) Western blot analysis of viral proteins in BYL using RCNMV p27 antibody. RNA2-Luc (125 fmol) was incubated together with RNA1 mutants (190 fmol) or a mixture of RNA1-p27 and RNA1-p88 (95 fmol each) at 17°C for 2.5 h.
DISCUSSION
These analyses of the translational activity of reporter Luc RNA2 mutants showed that the nucleotide sequence of RNA2 alone is insufficient for its cap-independent translation in vivo or in vitro. This suggests that the translation mechanism of RNA2 differs from that of RNA1. Unlike RNA1, RNA2 does not have an RNA element such as the 3′TE-DR1 in the 3′ UTR, which functions as an efficient translational enhancer in cap-independent translation of RNA1 in vivo and in vitro (Fig. 1B and 6A) (33). The 3′TE-DR1 of RNA1 appears to play a role similar to that of a cap structure, because mutations in the 3′TE-DR1 completely abolish the cap-independent translational activity of uncapped mutant mRNAs, but not that of capped mutant mRNAs (33). In contrast to RNA1, RCNMV RNA2 has no RNA elements like 3′TE-DR1 that can replace the function of a cap structure. However, the uncapped reporter RNA2, RNA2-Luc, was effectively translated in the presence of p27 and p88 in protoplasts in which RNA2-Luc was replicated (Fig. 3B). This suggests that the translational activity of RNA2-Luc is coupled to RNA replication.
In positive-strand RNA viruses, RNA replication and translation pathways conflict with each other, because genomic RNA can serve as a template for synthesis of negative-strand RNA by viral RNA replicase and as a template for translation of viral proteins by ribosomes. Therefore, positive-strand RNA viruses must control these two processes to allow efficient viral proliferation. Analysis of the interplay between translation and RNA replication in poliovirus shows that the binding of viral protein 3CD represses translation and facilitates negative-strand RNA synthesis (11). In pestivirus, efficient termination of translation is essential for efficient replication of viral genomic RNA (18). Surprisingly, our results indicate that the translational activity of uncapped RNA2-Luc is strongly coupled to replication of RNA2-Luc (Fig. 5). Stabilization of RNA2-Luc with p27 and p88 is not directly involved in the enhancement of its cap-independent translational activity, because the amount of RNA2-Luc was not significantly different in the presence or absence of RNA1 for at least 8 h after transfection in protoplasts (Fig. 5A). Moreover, there was no translational activity of RNA2-Luc or translational enhancement by RNA1 in the absence of RNA replication in the in vitro translation assay (Fig. 6A). A possible explanation for these observations is that only progeny RNA2-Luc generated de novo through the RNA replication pathway functions as mRNA. RNA2 replicated de novo and devoted to translation may have some modifications, including the addition of a cap structure. Indeed, capped RNA2-Luc was efficiently translated (Fig. 1B). However, if the modification is for a cap structure, the cap structure must be removed before encapsidation, because virion RNAs of RCNMV do not have a cap structure (33). A complete RCNMV RNA replication system in vitro may help to confirm this hypothesis.
Another possibility is that a viral RNA replication pathway and associated viral RNA replication complexes are required to recruit translation factors to RNA2, because RNA replication complexes of positive-strand RNA viruses often contain proteins that are involved as host factors in translation. For example, phage Qβ RNA polymerase complexes contain 30S ribosomal protein S1 and translation elongation factors EF-Tu (EF1A) and EF-Ts (EF1B) (1). In plant RNA viruses, the GCD 10-like subunit and the p41 subunit of eIF3 have been detected in purified RNA-dependent RNA polymerase complexes of Tobacco mosaic virus (TMV) (45) and Brome mosaic virus (37), respectively. eEF1A binds to the 3′ UTRs of TMV (57) and Turnip yellow mosaic virus (31). For RCNMV replication, we recently proposed a model, in which Dicer-like protein-1 (DCL1) or its homologue are possible candidates for the host factor that is directly or indirectly required to form viral replication complexes and to suppress RNA silencing (42). Proteins associated with DCL1 may be required for cap-independent translation of RNA2, because rabbit translation factor eIF2c, which is a member of the Argonaute (AGO) protein family, forms a complex with Dicer in human and mouse cells (5).
What does the link between the cap-independent translation of RNA2 and RNA replication mean for RCNMV infection? mRNA localization is a widespread and efficient means of targeting gene products to specific intracellular regions in a variety of organisms (19, 50). For example, in plants, prolamines, one of the major storage proteins of rice seed, are localized to a specific compartment of the endomembrane system, whereas the gluteins, another class of storage protein, are transported via the Golgi apparatus to a storage vacuole. These distinct protein localizations are controlled by specific targeting of corresponding mRNAs to different subdomains of the endoplasmic reticulum (3). Replication-dependent translation of RNA2 may assure that MP is synthesized near the RNA replication machinery, which localizes in the cortical and cytoplasmic endoplasmic reticulum in Nicotiana benthamiana cells (46). This may be important for intracellular viral movement for RCNMV, because TMV MP and the 126-kDa protein colocalize at the same subcellular sites and viral replication complexes, including MP, 126-kDa protein, and viral RNA, play an important role in intracellular and intercellular movement of viral RNA (23, 30).
The requirement of efficient RNA replication for cap-independent translation of RNA2 results in delayed expression of MP compared with that of p27 and p88, as was predicted by time course experiments using reporter mRNAs (Fig. 5). Suppression of MP production during the early phase of infection in a replication-dependent translation manner may be important for RCNMV infection. The replication-dependent translational control of MP expression may be unique to dianthoviruses whose genome is bipartite, unlike other viruses in the families Tombusviridae and Ludeoviridae, whose genomes are monopartite. These viruses with a monopartite genome produce sgRNAs, which are coterminally transcribed from the genomic RNAs. Therefore, these sgRNAs have a 3′ UTR identical to that of the genomic RNA (36, 49). These 3′ UTRs contain RNA elements required for cap-independent translation. These viruses may regulate viral gene expression at transcriptional steps, because accumulation patterns of the genomic RNA and two sgRNAs of a member of the tombusvirus group differ at different time points in infection (21, 44, 58).
Genomic RNA is divided into several segments in many RNA viruses. Segmentation of the genome may have several selective advantages. Translational control of viral gene expression in viruses with divided genomes has been reported for rotaviruses. Eleven proteins encoded in the rotavirus genome are not synthesized at equivalent levels in infected cells, nor do the levels of individual proteins correspond with the levels of cognate mRNAs (20). As described and discussed in this report, the translational mechanism differs between two segmented genomic RNAs of RCNMV. RCNMV also controls CP expression at the transcriptional stage through intermolecular interaction between RNA1 and RNA2 (40, 43). Thus, genome segmentation contributes to separate early and late gene expression through translational and transcriptional steps in RCNMV infection.
Acknowledgments
We are grateful to S. A. Lommel for pRC1|G and pRC2|G. We are also grateful to K. Komoda and M. Ishikawa for advice on preparation of cell extracts of evacuolated tobacco BY2 protoplasts.
This work was supported in part by a Grant-in-Aid for Scientific Research (A) (13306005) from the Japan Society for the Promotion of Science and in part by a Grant-in-Aid for Scientific Research on priority area (A) (12052201) “Molecular Mechanisms of Plant-Microbe Interaction toward Production of Disease Resistant Plants” from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
REFERENCES
- 1.Blumenthal, T., and G. G. Carmichael. 1979. RNA replication: function and structure of Qβ-replicase. Annu. Rev. Biochem. 48:525-548. [DOI] [PubMed] [Google Scholar]
- 2.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi, S. B., C. Wang, D. G. Muench, K. Ozawa, V. R. Franceschi, Y. Wu, and T. W. Okita. 2000. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407:765-767. [DOI] [PubMed] [Google Scholar]
- 4.Damayanti, T. A., H. Nagano, K. Mise, I. Furusawa, and T. Okuno. 2002. Positional effect of deletions on viability, especially on encapsidation, of brome mosaic virus D-RNA in barley protoplasts. Virology 293:314-319. [DOI] [PubMed] [Google Scholar]
- 5.Doi, N., S. Zenno, R. Ueda, H. Ohki-Hamazaki, K. Ui-Tei, and K. Saigo. 2003. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 13:41-46. [DOI] [PubMed] [Google Scholar]
- 6.Fabian, M. R., and K. A. White. 2004. 5′-3′ RNA-RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mRNA: a potential common mechanism for Tombusviridae. J. Biol. Chem. 279:28862-28872. [DOI] [PubMed] [Google Scholar]
- 7.Fujita, Y., K. Mise, T. Okuno, P. Ahlquist, and I. Furusawa. 1996. A single codon change in a conserved motif of a bromovirus movement protein gene confers compatibility with a new host. Virology 223:283-291. [DOI] [PubMed] [Google Scholar]
- 8.Gallie, D. R. 2002. The 5′-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Res. 30:3401-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallie, D. R., and K. S. Browning. 2001. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J. Biol. Chem. 276:36951-36960. [DOI] [PubMed] [Google Scholar]
- 10.Gallie, D. R., D. E. Sleat, J. W. Watts, P. C. Turner, and T. M. Wilson. 1987. The 5′-leader sequence of tobacco mosaic virus RNA enhances the expression of foreign gene transcripts in vitro and in vivo. Nucleic Acids Res. 15:3257-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gould, A. R., R. I. B. Francki, T. Hatta, and M. Hollings. 1981. The bipartite genome of red clover necrotic mosaic virus. Virology 108:499-506. [DOI] [PubMed] [Google Scholar]
- 13.Guo, L., E. M. Allen, and W. A. Miller. 2001. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell 7:1103-1109. [DOI] [PubMed] [Google Scholar]
- 14.Hershey, J. W. B., and W. C. Merrick. 2000. The pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Hibi, T., G. Rezelman, and A. Van Kammen. 1975. Infection of cowpea mesophyll protoplasts with cowpea mosaic virus. Virology 64:308-318. [DOI] [PubMed] [Google Scholar]
- 16.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiruki, C. 1987. The dianthoviruses: a distinct group of isometric plant viruses with bipartite genome. Adv. Virus Res. 33:257-300. [DOI] [PubMed] [Google Scholar]
- 18.Isken, O., C. W. Grassmann, R. T. Sarisky, M. Kann, S. Zhang, F. Grosse, P. N. Kao, and S. E. Behrens. 2003. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 22:5655-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansen, R. P. 2001. mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol. 2:247-256. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, M. A., and M. A. McCrae. 1989. Molecular biology of rotaviruses. VIII. Quantitative analysis of regulation of gene expression during virus replication. J. Virol. 63:2048-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston, J. C., and D. M. Rochon. 1995. Deletion analysis of the promoter for the cucumber necrosis virus 0.9-kb subgenomic RNA. Virology 214:100-109. [DOI] [PubMed] [Google Scholar]
- 22.Kaido, M., M. Mori, K. Mise, T. Okuno, and I. Furusawa. 1997. Auto-cleavable ribozyme sequences attached to brome mosaic virus cDNAs enhances accumulation of viral RNAs transcribed in vivo from the cDNAs. Ann. Phytopathol. Soc. Jpn. 63:95-98. [Google Scholar]
- 23.Kawakami, S., Y. Watanabe, and R. N. Beachy. 2004. Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. USA 101:6291-6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, K. H., and S. A. Lommel. 1994. Identification and analysis of the site of −1 ribosomal frameshifting in red clover necrotic mosaic virus. Virology 200:574-582. [DOI] [PubMed] [Google Scholar]
- 25.Koh, D. C., D. X. Liu, and S. M. Wong. 2002. A six-nucleotide segment within the 3′ untranslated region of hibiscus chlorotic ringspot virus plays an essential role in translational enhancement. J. Virol. 76:1144-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komoda, K., S. Naito, and M. Ishikawa. 2004. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl. Acad. Sci. USA 101:1863-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 28.Kroner, P., D. Richards, P. Traynor, and P. Ahlquist. 1989. Defined mutations in a small region of the brome mosaic virus 2a gene cause diverse temperature-sensitive RNA replication phenotypes. J. Virol. 63:5302-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lommel, S. A., M. Weston-Fina, Z. Xiong, and G. P. Lomonossoff. 1988. The nucleotide sequence and gene organization of red clover necrotic mosaic virus RNA-2. Nucleic Acids Res. 16:8587-8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mas, P., and R. N. Beachy. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 147:945-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda, D., S. Yoshinari, and T. W. Dreher. 2004. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology 321:47-56. [DOI] [PubMed] [Google Scholar]
- 32.Mizumoto, H., Y. Hikichi, and T. Okuno. 2002. The 3′-untranslated region of RNA1 as a primary determinant of temperature sensitivity of red clover necrotic mosaic virus Canadian strain. Virology 293:320-327. [DOI] [PubMed] [Google Scholar]
- 33.Mizumoto, H., M. Tatsuta, M. Kaido, K. Mise, and T. Okuno. 2003. Cap-independent translational enhancement by the 3′ untranslated region of red clover necrotic mosaic virus RNA1. J. Virol. 77:12113-12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mori, M., G. H. Zhang, M. Kaido, T. Okuno, and I. Furusawa. 1993. Efficient production of human gamma interferon in tobacco protoplasts by genetically engineered brome mosaic virus RNAs. J. Gen. Virol. 74:1255-1260. [DOI] [PubMed] [Google Scholar]
- 35.Okuno, T., C. Hiruki, D. B. Rao, and G. C. Figueired. 1983. Genetic determinants distributed in two genomic RNAs of sweet clover necrotic mosaic, red clover necrotic mosaic and clover primary leaf necrosis viruses. J. Gen. Virol. 64:1907-1914. [Google Scholar]
- 36.Qu, F., and T. J. Morris. 2000. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J. Virol. 74:1085-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quadt, R., C. C. Kao, K. S. Browning, R. P. Hershberger, and P. Ahlquist. 1993. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 90:1498-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sachs, A. 2000. Physical and functional interactions between the mRNA cap structure and poly(A) tail, p. 447-465. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Shen, R., and W. A. Miller. 2004. The 3′ untranslated region of tobacco necrosis virus RNA contains a barley yellow dwarf virus-like cap-independent translation element. J. Virol. 78:4655-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281:829-832. [DOI] [PubMed] [Google Scholar]
- 41.Takeda, A., K. Sugiyama, H. Nagano, M. Mori, M. Kaido, K. Mise, S. Tsuda, and T. Okuno. 2002. Identification of a novel RNA silencing suppressor, NSs protein of tomato spotted wilt virus. FEBS Lett. 532:75-79. [DOI] [PubMed] [Google Scholar]
- 42.Takeda, A., M. Tsukuda, H. Mizumoto, K. Okamoto, M. Kaido, K. Mise, and T. Okuno. 2005. A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J. 24:3147-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatsuta, M., H. Mizumoto, M. Kaido, K. Mise, and T. Okuno. 2005. The Red clover necrotic mosaic virus RNA2 trans-activator is also a cis-acting RNA2 replication element. J. Virol. 79:978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tavazza, M., A. Lucioli, A. Calogero, A. Pay, and R. Tavazza. 1994. Nucleotide sequence, genomic organization and synthesis of infectious transcripts from a full-length clone of artichoke mottle crinkle virus. J. Gen. Virol. 75:1515-1524. [DOI] [PubMed] [Google Scholar]
- 45.Taylor, D. N., and J. P. Carr. 2000. The GCD10 subunit of yeast eIF-3 binds the methyltransferase-like domain of the 126 and 183 kDa replicase proteins of tobacco mosaic virus in the yeast two-hybrid system. J. Gen. Virol. 81:1587-1591. [DOI] [PubMed] [Google Scholar]
- 46.Turner, K. A., T. L. Sit, A. S. Callaway, N. S. Allen, and S. A. Lommel. 2004. Red clover necrotic mosaic virus replication proteins accumulate at the endoplasmic reticulum. Virology 320:276-290. [DOI] [PubMed] [Google Scholar]
- 47.Turner, R. L., and K. W. Buck. 1999. Mutational analysis of cis-acting sequences in the 3′- and 5′-untranslated regions of RNA2 of red clover necrotic mosaic virus. Virology 253:115-124. [DOI] [PubMed] [Google Scholar]
- 48.Wang, S., K. S. Browning, and W. A. Miller. 1997. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 16:4107-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, S., L. Guo, E. Allen, and W. A. Miller. 1999. A potential mechanism for selective control of cap-independent translation by a viral RNA sequence in cis and in trans. RNA 5:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wickens, M., E. B. Goodwin, J. Kimble, S. Strickland, and M. Hentze. 2000. Translational control of developmental decisions, p. 295-370. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Wu, B., and K. A. White. 1999. A primary determinant of cap-independent translation is located in the 3′-proximal region of the tomato bushy stunt virus genome. J. Virol. 73:8982-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong, Z., K. H. Kim, D. Giesman-Cookmeyer, and S. A. Lommel. 1993. The roles of the red clover necrotic mosaic virus capsid and cell-to-cell movement proteins in systemic infection. Virology 192:27-32. [DOI] [PubMed] [Google Scholar]
- 53.Xiong, Z., K. H. Kim, T. L. Kendall, and S. A. Lommel. 1993. Synthesis of the putative red clover necrotic mosaic virus RNA polymerase by ribosomal frameshifting in vitro. Virology 193:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiong, Z., and S. A. Lommel. 1989. The complete nucleotide sequence and genome organization of red clover necrotic mosaic virus RNA-1. Virology 171:543-554. [DOI] [PubMed] [Google Scholar]
- 55.Xiong, Z. G., and S. A. Lommel. 1991. Red clover necrotic mosaic virus infectious transcripts synthesized in vitro. Virology 182:388-392. [DOI] [PubMed] [Google Scholar]
- 56.Zavriev, S. K., C. M. Hickey, and S. A. Lommel. 1996. Mapping of the red clover necrotic mosaic virus subgenomic RNA. Virology 216:407-410. [DOI] [PubMed] [Google Scholar]
- 57.Zeenko, V. V., L. A. Ryabova, A. S. Spirin, H. M. Rothnie, D. Hess, K. S. Browning, and T. Hohn. 2002. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J. Virol. 76:5678-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, G., V. Slowinski, and K. A. White. 1999. Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. RNA. 5:550-561. [DOI] [PMC free article] [PubMed] [Google Scholar]