Abstract
The linear, positive-stranded RNA genome of flaviviruses is thought to adopt a circularized conformation via interactions of short complementary sequence elements located within its terminal regions. This process of RNA cyclization is a crucial precondition for RNA replication. In the case of mosquito-borne flaviviruses, highly conserved cyclization sequences (CS) have been identified, and their functionality has been experimentally confirmed. Here, we provide an experimental identification of CS elements of tick-borne encephalitis virus (TBEV). These elements, termed 5′-CS-A and 3′-CS-A, are conserved among various tick-borne flaviviruses, but they are unrelated to the mosquito-borne CS elements and are located at different genomic positions. The 5′-CS-A element is situated upstream rather than downstream of the AUG start codon and, in contrast to mosquito-borne flaviviruses, it was found that the entire protein C coding region is not essential for TBEV replication. The complementary 3′-CS-A element is located within the bottom stem rather than upstream of the characteristic 3′-terminal stem-loop structure, implying that this part of the proposed structure cannot be formed when the genome is in its circularized conformation. Finally, we demonstrate that the CS-A elements can also mediate their function when the 5′-CS-A element is moved from its natural position to one corresponding to the mosquito-borne CS. The recognition of essential RNA elements and their differences between mosquito-borne and tick-borne flaviviruses has practical implications for the design of replicons in vaccine and vector development.
Flaviviruses, i.e., members of the genus Flavivirus, family Flaviviridae, have a genome that is a single-stranded, ∼11-kb-long RNA molecule with positive-strand polarity (43). It carries a 5′-terminal Cap structure but no 3′-terminal poly(A) tail (61, 62) and serves as the only viral mRNA, encoding all of the viral proteins in a single long open reading frame (ORF). After uptake of the viral particle into the cytoplasm of the host cell and release of the genome from the nucleocapsid, the RNA is translated into a single polyprotein. This precursor protein is processed by a viral protease, as well as host cell proteases, to yield three structural proteins (C, prM, and E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5), each of which is an essential component of the viral replication complex (37, 39, 63). After the formation of the replication complex, negative-sense genome-length RNA is synthesized, which serves as a template for the synthesis of new positive-strand genomic molecules. RNA replication is an asymmetric process in which the plus-sense RNA is synthesized in 10- to 100-fold excess over minus-sense RNA (12, 47, 54). The newly synthesized plus-sense RNA is subsequently used for translation of further viral proteins or synthesis of additional minus-sense RNA, or it becomes incorporated into new viral particles. Thus, the genome RNA has at least three different functions (translation, replication, and association with nascent viral particles), which need to be regulated and coordinated during the viral replication cycle.
The terminal regions of the flavivirus genome, i.e., the approximately 100- to 130-nucleotide (nt) 5′-noncoding region (NCR) plus the adjacent N terminus of the protein C coding region, as well as the approximately 400- to 800-nt 3′-NCR, contain RNA sequence motifs and secondary structure elements that are involved in these regulatory processes (43). In most cases, a functional importance was originally proposed based on theoretical studies in which conserved sequence or folding patterns were detected. More recently, some of these predictions have been confirmed by in vitro and in vivo studies that directly addressed the functionalities of such elements by mutational analysis (for a review, see reference 43).
It is known for a number of cellular and viral RNA molecules that they can adopt a circular conformation in which their termini are brought into close proximity, and this cyclization has functional importance (5, 19, 20, 25, 60). In the case of flaviviruses, such a process is thought to be mediated by specific inverse repeated sequence motifs located in the terminal regions of the RNA genome. Specific base pairing between the 5′-element and the complementary 3′-element of this cyclization sequence (CS) is thought to induce cyclization of the genome into a “panhandle” structure (18, 43).
The majority of flaviviruses are transmitted by mosquitoes unto their vertebrate hosts. This group of viruses includes important human pathogens such as yellow fever virus (YFV), West Nile virus (WNV), Japanese encephalitis virus (JEV), Saint Louis encephalitis virus (SLEV), and the four serotypes of dengue virus (DV) (37). The presence of complementary sequence elements in the genomes of mosquito-borne flaviviruses was first noted by Hahn et al. (18), who identified an 8-nt core sequence, which is conserved among this entire group of viruses, and who proposed that these sequences induce genome cyclization. The 5′-element of this CS is located within the 5′-terminal region of the viral ORF coding for the amino-terminal part of protein C (approximately 40 nt downstream of the start codon). The complementary 3′-CS is present in the 3′-NCR, just upstream of a 3′-terminal stem-loop structure (3′-SL), which shows a high degree of conservation among all flaviviruses (43). In silico studies based on thermodynamic folding of the terminal regions or, more recently, entire flavivirus genomes, have subsequently corroborated the notion that specific base pairing between the 5′- and the 3′-CS elements can put the genomic termini into close proximity (3, 28, 57, 64). Some studies have also identified additional sequences that may take part in such long-range interactions (3, 57). Several mutational studies (in vitro and in vivo) performed with Kunjin virus (a subtype of WNV), YFV, WNV, and DV have provided direct experimental evidence that complementarity of the 5′- and 3′-CS is an essential prerequisite for viral replication (1, 2, 28, 38, 46). Mutations of either CS element that destroyed base pairings were lethal, whereas mutations in both elements that reconstituted base pairing were tolerated.
A smaller group of flaviviruses includes viruses that are transmitted to their vertebrate hosts by ticks rather than mosquito vectors. The most important human pathogen within this group is tick-borne encephalitis virus (TBEV), which is endemic in parts of Europe and Asia and causes thousands of cases of severe neurological illness every year (7). Although tick-borne flaviviruses have the same genome organization as the mosquito-borne relatives, they lack homologous CS elements. Instead, sequence comparisons among tick-borne flaviviruses revealed alternative conserved motifs that occur as complementary inverted repeats in the terminal regions of the genome (28, 41, 57). These putative tick-borne CS elements, termed 5′-CS-A and 3′-CS-A, are located within the 5′-NCR and the stem of the 3′-SL structure, respectively. Thus, the putative tick-borne 5′- and 3′-CS-A elements are located closer to the genomic termini than the mosquito-borne CS elements. Thermodynamic folding of the terminal regions confirmed the probability for long-range interactions of the tick-borne 5′- and 3′-CS-A but also predicted additional interactions of another pair of sequences (termed 5′- and 3′-CS-B) whose components are located at genomic positions more similar to those of the mosquito-borne CS, namely, within the amino-terminal coding region of protein C and upstream of the 3′-SL, respectively (28). Thus, tick-borne flaviviruses have two different pairs of putative complementary CS elements, CS-A and CS-B, but experimental evidence concerning their functional importance has not yet been obtained for either of them.
This study now provides a functional analysis of the tick-borne flavivirus CS elements. Studying the replication properties of various TBEV-derived replicons, we provide clear evidence that complementarity of 5′- and 3′-CS-A is essential, whereas no crucial function seems to be connected with the complementarity of the CS-B elements. In fact, the data indicate that the entire protein C-coding-region of TBEV, including its amino-terminal part containing the 5′-CS-B element, can be removed without loss of RNA replication competence. This result is in pointed contrast to observations with various mosquito-borne flaviviruses, all of which demonstrated a stringent requirement for this domain. Finally, we demonstrate that the functionality of 5′-CS-A is not strictly dependent on its location outside the ORF but can also provide for functional replication when it is moved from its natural position into the location of 5′-CS-B.
MATERIALS AND METHODS
Analysis of base-pairing probabilities.
Predictions of conserved secondary structure were performed by using the computer program alidot-p, an ANSI C implementation of the “Vienna RNA Package” available at http://www.tbi.univie.ac.at/RNA/ (22). This algorithm is designed to reach consensus structures by combining thermodynamic information (in the form of independent structure predictions for individual sequences) and phylogenetic comparison (as multiple sequence alignment) as discussed in previous publications (21, 23). Secondary structure predictions for individual sequences were calculated by using RNAfold-p, a further program of the Vienna RNA Package. The algorithm reaches plausible base-pair probabilities by using McCaskill's partition function algorithm (45) based on previously published energy parameters (44). Sequence alignments were obtained by CLUSTAL W (56). The following complete genomic sequences of tick-borne flaviviruses were included (GenBank accession numbers are in parentheses): TBEV strains 263 (TEU27491), Neudoerfl (TEU27495), Hypr (TEU39292), Oshima 5-10 (AB062063), Sofjin (AB062064), and Vasilchenko (NC_001809); Louping ill virus (NC_001809); Langat virus (NC_003690); and Powassan virus (L06436).
Plasmids and cells.
All new plasmid constructs described in the present study are derivatives of the infectious cDNA clone of TBEV Western subtype prototypic strain Neudoerfl (40). Two mutants of strain Neudoerfl, used as controls in the present study, were derived from previously described plasmids. One is mutant prM(ΔR88), which has a full-length genome with a single amino acid deletion mutation in the furin cleavage site of protein prM. This mutant is fully competent for viral RNA replication and translation, but, due to its defect in prM cleavage, is released from cells in the form of a noninfectious immature viral particle (15). The other control is mutant ΔNS5, which is deficient for RNA replication, due to a large deletion in which about one-fifth of protein NS5 (the viral replicase) and part of the 3′-NCR have been removed (29, 31-33). All of the previously described and new plasmids are derivatives of pBR322.
Cell culture experiments were performed with BHK-21 cells that were grown and maintained under standard conditions as described previously (40).
Mutant construction.
Replicon C17 was generated by the replacement of most of the structural protein coding region by an in-frame insertion of a small artificial sequence element (multiple cloning site [MCS]). All of the other new mutants were derivatives of this initial construct. For the exact deletion boundaries, inserted nucleotide sequence, and additional mutations, please refer to the corresponding text and figures in the Results section. Detailed cloning protocols and primer sequences are available from the authors by request. Briefly, C17 was generated by first generating a partial cDNA clone containing the region extending from a unique SalI restriction site (upstream of the genomic 5′ terminus and the preceding T7 promoter) to the unique ClaI site at position 3155 within the NS1 coding region and containing the desired deletion and artificial sequence insertion. This clone was obtained from two PCR fragments generated with appropriate mutagenic primers and with wild-type TBEV cDNA as a template. Subsequently, the SalI-ClaI fragment of the wild-type full-length cDNA clone pTNd/c (40) was replaced by the modified fragment to generate replicon C17. Further manipulations within the 5′-terminal region were performed in this partial clone, either by PCR mutagenesis with appropriate mutagenic primers or by use of the commercially available GeneTailor site-directed mutagenesis system (Invitrogen). The mutations were transferred into C17 by taking advantage of a unique NotI site (within the MCS) and the 5′-terminal SalI site. Similarly, manipulations of the 3′-terminal regions were first performed on a partial cDNA clone containing sequences from genome position 8949 to the 3′ terminus (position 11141) and then transferred into the C17 backbone by fragment swapping, using unique restriction sites for AgeI at position 10796 and for AatII adjacent to the 3′ terminus.
All plasmids were amplified in Escherichia coli strain HB101, and small- and large-scale plasmid preparations were made by using QIAGEN purification systems. New constructs were checked by sequence analysis with an automated DNA sequencing system (PE Applied Biosystems, GA310 or GA3100). For all clones, the sequences of the 5′-terminal region up to at least the position of the unique ClaI (3155) site, as well as the entire 3′NCR, were checked in both strands prior to functional analysis.
RNA transcription, transfection, and sequence confirmation.
In vitro RNA transcription and transfection of BHK-21 cells by electroporation was performed as in previous studies (31-33, 40). Capped RNA was synthesized with m7GpppG Cap analogue and reagents from the T7 Megascript kit (Ambion) according to the manufacturer's protocol. After the transcription reaction, template DNA was digested by DNase I incubation and the quality of the RNA was checked by formalin-denaturing 1% agarose gel electrophoresis. For real-time PCR analysis, RNA was purified and separated from unincorporated nucleotides by using a RNeasy Mini kit (QIAGEN) and then quantified by spectrophotometric measurement. Equal amounts of RNA were used for all transfections. Electroporation was performed by using a GenePulser apparatus (Bio-Rad) with previously described settings that typically yield a very high (up to almost 100%) transfection efficiency (40).
To confirm the presence of the originally engineered sequences in some replication competent mutants at the end of the replication experiment, RNA was isolated from cells 96 h posttransfection and subjected to sequence analysis. Reverse transcription-PCR (RT-PCR) was performed with a commercial cDNA synthesis system (Roche Applied Science) and appropriate primers according to standard protocols. For the analysis of the genomic 3′ terminus, the replicon RNA was polyadenylated with E. coli poly(A) polymerase (Ambion), purified with the RNeasy Mini kit, and then amplified by RT-PCR as described by others (59). PCR products were sequenced on both strands with an automated DNA sequencing system (PE Applied Biosystems, GA310 or GA3100).
Immunofluorescence staining.
Expression of viral proteins was determined by immunofluorescence staining of nonstructural protein NS1. After transfection with RNA, BHK-21 cells were seeded into 24-well tissue culture plates and supplied with growth medium containing 5% fetal calf serum (FCS). The medium was replaced at 20 h posttransfection with maintenance medium containing only 1% FCS to slow cell growth. Immunofluorescence analysis was performed 3, 7, and 14 days posttransfection. Cells were permeabilized by acetone-methanol (1:1) fixation, and the presence of protein NS1 was visualized by successive incubations with a mouse anti-protein NS1 monoclonal antibody (26) and fluorescein isothiocyanate-conjugated anti-mouse antibody. Immunofluorescence staining was evaluated by visual inspection with a Nikon Microphot miscroscope.
RNA quantification by real-time PCR analysis.
BHK-21 cells were transfected with equimolar amounts of RNA (corresponding to approximately 2 × 1012 copies of RNA) by electroporation. To remove noninternalized RNA, cells were washed twice by taking them up in growth medium containing 5% FCS and collecting them again by low-speed centrifugation. Then cells were transferred into growth medium, and aliquots of approximately 106 cells were seeded into flasks (25 cm2). At 20 h posttransfection, the FCS concentration was reduced to 1%. Cells were detached by trypsin incubation and collected from individual flasks at various time points ranging from 3 to 96 h posttransfection as indicated in the Results section. They were washed twice with phosphate-buffered saline (PBS; pH 7.5) containing 1% bovine serum albumin and counted with a Casy 1 TT cell counter (Schärfe Systems). Aliquots of 105 cells were then suspended in 175 μl of lysis buffer (50 mM Tris-Cl [pH 8.0], 140 mM NaCl, 1.5 mM MgCl2, 1 mM dithiothreitol) and incubated for 5 min on ice. Lysates were cleared by low-speed centrifugation and stored for further analysis at −80°C. RNA was purified from one-fifth (corresponding to 2 × 104 cells) of each lysate by using RNeasy mini kits and, again, one-fifth of each RNA preparation (corresponding to 4 × 103 cells) was subjected to RT using the reagents and protocol supplied with the iScriptcDNA Synthesis kit (Bio-Rad). One-half of each cDNA preparation (corresponding to 2 × 103 cells) was used for quantification by real-time PCR. Amplification and detection were carried out on an ABI 7300 Real Time PCR System (PE Applied Biosystems) with TaqMan Universal PCR Mastermix (PE Applied Biosystems), and primers specific for part of the TBEV NS5 coding region: 25 pmol of the forward (NS5forward, 5′-GCG GAG GCT GAA CAA CT-3′) and the reverse primers (NS5reverse, 5′-TCA CGT TCC GTC TCC AG-3′) and 10 pmol of TaqMan probe (5′-FAM-TGT GTA CAG GCG CAC CGG CA-TAMRA-3′). The temperature profile of the reaction was: 3 min at 50°C, 10 min at 95°C, followed by 45 cycles of 15 s at 95°C, 30 s at 55°C, and 31 s at 72°C. RNA was quantified by comparison of the results to a standard curve prepared from a log10 dilution series of spectrophotometrically quantified, in vitro-transcribed RNA. The lower cutoff value of the reaction was set at 101 copies of RNA because values below this limit were observed to have poor reproducibility. Logarithmic means of the obtained RNA copy numbers were calculated and plotted with error bars indicating maximum and minimum values with the program GraphPad Prism (version 3.0; GraphPad Software, Inc.). The kinetics of RNA increase or decrease was evaluated by using the formula Nt = N0.eλt, where N0 is the number of molecules at the beginning and Nt is the number of molecules at the end of time period t. Doubling times were calculated as td = ln2/λ.
RESULTS
In silico analysis of 5′- and 3′-terminal regions of TBEV.
Previous studies have provided predictions on various secondary structure elements in both terminal regions of the TBEV genome (17, 41, 51, 53) and putative CS elements that may be involved in long-range interactions between the 5′- and 3′-terminal regions (28, 41, 57). As a starting point for our mutation analysis, we reevaluated those predictions by using an algorithm that allows folding of entire RNA molecules corresponding in length to the flavivirus genome and calculates base-pairing probabilities on the basis of both thermodynamic and phylogenetic parameters (see Materials and Methods). Figure 1 shows a compilation of the current knowledge on the sequence of TBEV prototypic strain Neudoerfl. Secondary structure is shown as predicted when neglecting the possibility of long-range interactions between nucleotides of the 5′- and 3′-terminal regions.
FIG. 1.
Secondary structures in the 5′- and 3′-terminal regions of TBEV and putative long-range interactions between the two regions of the strain Neudoerfl sequence (numbering corresponding to the wild-type sequence, GenBank accession no. U27495). The stem-loop structures (5′-SL1, -2, -3, and -4 and 3′-SL1, -2, -3, -4, and -5) are shown as predicted for the linear genome, i.e., without consideration of potential long-range interactions between 5′- and 3′-proximal sequence motifs. Predicted ΔG values of individual SL structures are given for the tick-borne consensus sequence (no ΔG value is shown for 5′-SL1 due to considerable variation among different tick-borne flavivirus sequences). Base-pair assignment corroborated by compensatory mutations in other tick-borne flavivirus genomes are indicated by circles. Base pairs that are absent in one or two of the other tick-borne sequences are marked by asterisks. The AUG start codon in 5′-SL3 is marked with “Start.” Potential long-range interactions of putative cyclization elements (CS) are highlighted in color and are indicated by connecting lines. Solid lines depict long-range interactions predicted by the algorithm used in the present study (see Materials and Methods), whereas dashed lines refer to previously predicted interactions not confirmed in the present analysis.
Potential long-range interactions between various putative CS elements (as also indicated in the figure) would clearly interfere with the formation of some of the shown stem-loop structures. One pair of previously predicted complementary sequences (termed CS-A) (28, 41, 57) has its 5′-element (5′-CS-A) in the 5′-SL2 structure and its 3′-element (3′-CS-A) in the base of structure 3′-SL1. Our reevaluation of base-pairing probabilities confirmed a high likelihood for long-range interactions between the two complementary CS-A elements, albeit with sequence boundaries slightly different from previous predictions. Another previously described pair of complementary sequences (28) (here termed CS-B) which has its 5′-element coinciding with 5′-SL4, and its 3′-complement (3′-CS-B) overlapping with the structures 3′-SL4 and 3′-SL5, was not confirmed by the present in silico analysis. Furthermore, there are three additional sequence elements in the vicinity of or overlapping with CS-A or CS-B which according to previous or the present computer predictions may participate in long-range interactions (termed here CS-a, CS-b1, and CS-b2). Figure 1 and Table 1 summarize the sequences, exact position numbers, and calculated ΔG values of all of these potential CS elements in the TBEV genome.
TABLE 1.
Putative TBEV cyclization sequences
| Designation | Genome positiona
|
ΔG (kcal)d | Reference | |
|---|---|---|---|---|
| 5′ element | 3′ element | |||
| CS-ab | 109-111 | 11139-11141 | −0.3 | 57 |
| CS-Ac | 115-129 | 11057-11071 | −24.4 | 28, 41, 57 |
| CS-b1 | 157-166 | 10772-10781 | −20.8 | |
| CS-b2b | 166-168 | 10956-10958 | 57 | |
| CS-B | 164-174 | 10949-10958 | 28 | |
Nucleotide numbers refer to positions in the wild-type genome of TBEV strain Neudoerfl.
Designated as P1′ and P2 in reference (57).
Previous studies predicted slightly different lengths and ΔG values.
As determined in the present study. No values are listed for CS elements not confirmed in the present analysis.
Construction and characterization of TBEV replicon C17.
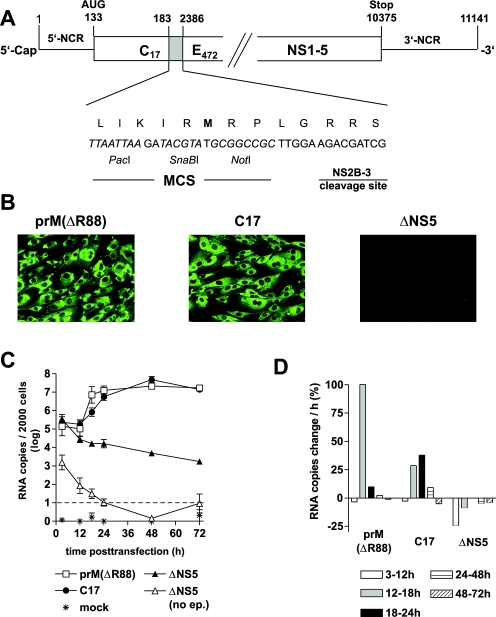
To study the role of the putative CS elements of TBEV independently of the processes of particle formation, egress, and secondary rounds of infection, we constructed replicon C17, which is a derivative of the previously described full-length infectious cDNA clone of TBEV strain Neudoerfl. As shown in Fig. 2A, we removed almost the entire structural protein coding region, except for the parts coding for the first 17 amino acid residues of protein C (and thus retaining all of 5′-SL4 and 5′-CS-B) and the internal signal sequence at the carboxy terminus of protein E (to ensure proper processing of protein NS1). In its place, a short artificial sequence, containing an MCS and a NS2B/3 cleavage site was inserted. The original reading frame was maintained in the C17 sequence and thermodynamic analysis of the replicon sequence predicted essentially the same local and long-range secondary structures as they are depicted for the full-length genome in Fig. 1.
FIG. 2.
Organization and characterization of replicon C17. (A) Schematic drawing of C17 (not to scale). The nucleotide sequence between positions 183 and 2386 (corresponding to amino acid residue 18 of protein C and 471 of protein E) of the wild-type TBEV genome was replaced by an artificial sequence that includes an MCS with recognition sequences for the restriction enzymes PacI, SnaBI, and NotI and a sequence coding for a potential cleavage site of the viral protease NS2B/3. (B) Immunofluorescence analysis of cells transfected with RNA of replicon C17, the prM-cleavage-deficient TBEV mutant prM(ΔR88) (positive control), and the replication-deficient mutant ΔNS5 (negative control). Immunofluorescence staining was performed 3 days posttransfection with a monoclonal antibody recognizing protein NS1. (C) Replication kinetics of C17 and control RNAs. RNA isolated from 2,000 cells was quantified at various time points after transfection by real-time PCR. Logarithmic means from two experiments are shown (the error bars indicate the maximum and minimum values). ΔNS5 (no ep.) refers to control experiments in which cells were incubated with RNA but not electroporated to quantify the number of RNA molecules that remain attached to the cell surface. This value dropped below the cutoff limit of 101 (indicated by a dashed line) at 24 h posttransfection. (D) Average increase or decrease in RNA copies in percent per hour between the individual measurements of the experiments.
To characterize the ability of replicon C17 to replicate and be translated, protein expression was examined by immunofluorescence, using a previously described full-length noninfectious TBEV mutant, prM(ΔR88), for comparison. prM(ΔR88) differs from the wild-type TBEV sequence only by the deletion of a single amino acid, which abolishes furin-mediated cleavage of protein prM (15). This mutant therefore only forms immature, noninfectious particles. We chose this mutant as a positive control for the characterization of replicon C17 because it should exhibit wild-type functionality except for its inability to cause a secondary round of infection. As a negative control, the replication-deficient mutant ΔNS5 was used. Equal amounts of in vitro-synthesized RNAs of C17, prM(ΔR88), and ΔNS5 were introduced into BHK-21 cells by electroporation, and expression of protein NS1 was visualized by immunofluorescence staining 3 days posttransfection. As shown in Fig. 2B, both C17 and prM(ΔR88) yielded bright immunofluorescence staining, whereas ΔNS5 was negative, indicating that, in the absence of RNA replication, translation from incoming molecules is insufficient to yield a positive stain in the immunofluorescence analysis. On the other hand, since no RNA replication can occur in the absence of translation, the positive result obtained for C17 means that this replicon was competent for both translation and replication.
For a quantitative comparison of RNA replication, a real-time PCR assay was established. RNA was isolated from cell lysates at various time points posttransfection and quantified by PCR. The results are shown in Fig. 2C and D. The levels of both C17 and prM(ΔR88) RNA remained almost constant during the first 12 h. After this lag phase, the amount of both RNAs increased about 100-fold to reach approximately equal final levels. In contrast, the replication-deficient control RNA ΔNS5 exhibited a distinct decline starting between 3 and 12 h and continuing over the entire observation period. As a further control, ΔNS5 RNA was added to cells, but no electric pulse was applied so that the RNA would not be internalized by the cells. As can be seen in Fig. 2C, the values for this control remained at least 2 orders of magnitude below those of the other samples and decreased below the detection limit within the first 24 h, indicating that noninternalized input RNA did not significantly contribute to the quantitative RNA values determined in our assay.
A close inspection of the data indicates that the increase of C17 RNA was somewhat delayed compared to prM(ΔR88). prM(ΔR88) replicated most efficiently in the time interval between 12 and 18 h posttransfection with an average increase of more than a 100% per hour (i.e., its average doubling time during that period was less than an hour) (Fig. 2D). During the later time intervals, the amount of prM(ΔR88) remained almost constant. In contrast, C17 showed a steady increase between 12 and 24 h, with a maximum average increase of 38% between 18 and 24 h (corresponding to a doubling time of approximately 2 h) (Fig. 2D). Thus, C17 replicated its RNA to the same level as prM(ΔR88), but with a somewhat lower replication rate. This suggests that C17 contains all of the essential regulatory elements for protein translation and RNA replication, but the deletion of the structural protein coding region caused a moderate impairment of its replicative ability compared to the full-length control. This may be explained by the loss of some auxiliary function exerted by one of the structural proteins or RNA elements of the deleted region.
The N-terminal protein C-coding region of TBEV is nonessential.
Because it contains the 5′-element of the CS, the region coding for the N-terminal part of protein C is essential for replication of mosquito-borne flaviviruses (1, 2, 10, 27, 28, 38, 46). In the TBEV genome, this region contains the putative 5′-CS-B element (as well as the alternative, overlapping elements 5′-CS-b1 and 5′-CS-b2, see Fig. 1 and Table 1) and also two predicted stem-loop structures (5′-SL3 and 5′-SL4) of potential functional importance. To investigate whether any of these elements were necessary for translation or replication of TBEV, gradually increasing deletions were introduced into replicon C17 (Fig. 3C). In C17 the codons for the first 17 amino acid residues of protein C were maintained. Now this region was further truncated to retain only the first 12, 10, 7, or 3 codons (the mutants were accordingly named C12, C10, C7, and C3). In another construct, the deletion was extended to retain only the AUG start codon (C1), and in the final one, the entire 5′-SL3 was deleted (Δ5′-SL3). Equal amounts of in vitro-transcribed RNAs of all of these mutants, as well as C17 and ΔNS5 RNAs (positive and negative controls) were introduced into BHK-21 cells, and protein NS1 expression was visualized by immunofluorescence staining 3 days after transfection. Surprisingly, all of the mutants (except for the negative control ΔNS5) yielded a positive staining pattern (Fig. 3B and C). There was only a minor difference in immunofluorescence staining intensity between C17 and the replicons with truncations of 12, 10, or 7 codons of protein C. Thus, the removal of part or all of the 5′-CS-B (and 5′-CS-b1 and 5′-CS-b2 as well) and 5′-SL4 was apparently tolerated, indicating that neither these sequence elements nor the 5′-SL4 secondary structure are essential for replication or translation of the replicon RNA. However, further truncations affecting the start-codon-containing 5′-SL3 (C3, C1 and Δ5′-SL3) reproducibly caused a marked reduction in the brightness and overall number of immunofluorescence-positive cells, which also exhibited a more focal and granular staining pattern. Nevertheless, even removal of the entire 5′-SL3, including the authentic start codon, yielded a positive expression pattern. In fact, in several independent experiments, this mutant yielded more positive cells and somewhat more intense staining than mutant C1. Apparently, an alternative start codon, possibly the in-frame AUG codon that was part of the artificially introduced stretch of restriction enzyme recognition sequences (MCS) that had been inserted into C17 (Fig. 2A), could be utilized by this mutant. In conclusion, the data indicate that for TBEV, translation and replication can take place in the absence of the region encoding the N terminus of protein C.
FIG. 3.
Analysis of protein expression from deletion mutants derived from replicon C17. (A) Schematic drawing of secondary structure and potential CS elements. For details, refer to the legend to Fig. 1. (B to D) Schematic representation of the 5′- and 3′-terminal regions of various deletion mutants, indicating potential CS elements in color (yellow, CS-a, CS-b1 and CS-b2; red, CS-A; blue, CS-B) and the AUG-containing 5′-SL3 in gray. Deletions are represented by dashed lines. Mutant designations are given on the left, together with the exact boundaries of the deletions, as listed in parentheses (numbers correspond to nucleotide positions on the wild-type genome). On the right, protein expression is shown as determined by anti-NS1 immunofluorescence staining at 3 days posttransfection. (B) Positive and negative controls. The position of the AUG start codon is depicted. (C) Mutants with sequential truncations of the protein C coding region and 5′-SL3. (D) Mutants with deletions of 5′ or 3′ copies of putative CS elements.
Deletion of CS-A elements, but not CS-B elements, is deleterious.
The previous results implied that 5′-CS-B was not an essential sequence element for TBEV. To confirm this finding, two additional mutants that had an in-frame deletion of the 5′-CS-B (Δ5′-CS-B) or of the 3′-CS-B (Δ3′-CS-B) were constructed and analyzed (Fig. 3D). Δ5′-CS-B yielded a staining pattern indistinguishable from the positive control, C17. Mutant Δ3′-CS-B also yielded clearly positive cells, albeit in much lower numbers and with not as bright immunofluorescence staining as C17. This result was confirmed by immunofluorescence staining 7 and 14 days after transfection (data not shown). Various deletions in this region of the 3′-NCR were previously shown to significantly impair viability of TBEV (42, 50), and it has been proposed that these effects were primarily due to alterations of its secondary structure rather than the loss of particular sequence motifs (52).
Next, we investigated, whether CS-A or CS-a were essential sequence elements (Fig. 3D). Deletions were introduced into the 5′-NCR, removing 5′-CS-A (Δ5′-CS-A), 5′-CS-a (Δ5′-CS-a), or both of these elements (Δ5′-CS-a+A). In another mutant, the 3′-element of CS-A was deleted (Δ3′-CS-A). Protein expression was monitored by immunofluorescence staining as before. Cells transfected with mutants Δ5′-CS-A, Δ5′-CS-a+A, and Δ3′-CS-A were completely negative by 3 days (Fig. 3D), as well as at 7 and 14 days (not shown) after transfection. In contrast, cells transfected with Δ5′-CS-a were clearly positive for replication and translation.
In summary, neither the small 5′-CS-a nor the 5′-and 3′-CS-B elements are essential for TBEV replication, but 5′- and 3′-CS-A appear to have an important function.
Sequence complementarity of CS-A elements is crucial.
Deletion of 5′-CS-A has the additional effect of destroying 5′-SL2 (compare Fig. 1). To distinguish between mutations affecting circularization and 5′-SL2 formation, mutants carrying point mutations that would impede potential long-range base pairing by eliminating complementarity with 3′-CS-A while maintaining internal complementarity within 5′-SL2 were constructed and analyzed. Two mutants, carrying four or seven individual nucleotide changes in 5′-CS-A were designed (Fig. 4). These mutations interfered with approximately one-fourth or one-half of the predicted long-range base pairings, respectively. Thermodynamic analysis was in agreement with the assumption that both sets of mutations abolished the ability of the 5′- and 3′-CS-A elements to interact with each other but conserved the propensity for 5′-SL2 formation. No protein expression was observed with these mutants, termed 5′-4mut and 5′-7mut (Fig. 5c and f), indicating that preservation of the 5′-SL2 structure was not itself sufficient to maintain functionality. In the converse experiment, mutants with four and seven nucleotide changes in the 3′-CS-A (Fig. 4) were constructed and tested. Also these mutants, 3′-4mut and 3′-7mut, did not express protein (Fig. 5d and g).
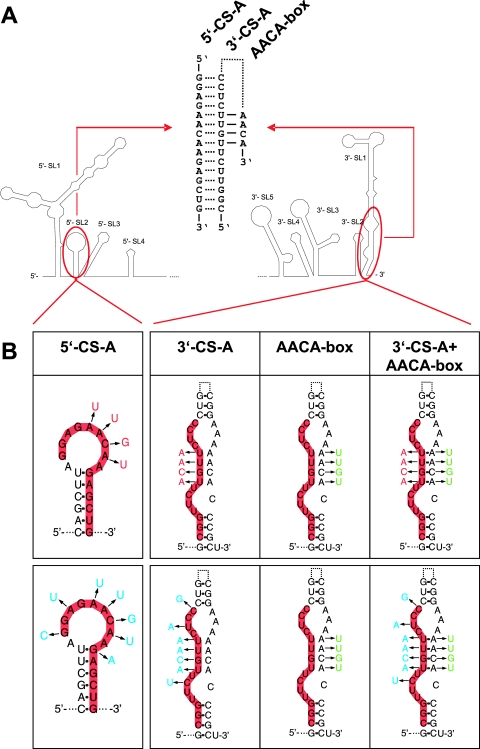
FIG. 4.
Synopsis of nucleotide changes introduced to analyze the functionality of CS-A. (A) Schematic drawing of the 5′- and 3′-terminal regions. In a cyclic conformation of the genome, the 5′- and 3′-elements of CS-A are predicted to form 15 bp (as indicated by dotted lines in the central part of the panel). In a noncircularized conformation, 3′-CS-A is assumed to form base-pairs as part of the 3′-SL1 stem structure (indicated by a dotted line). A stretch of four predicted base pairs with the sequence motif 5′-AACA-3′ is indicated by solid lines. (B) Four (red) or seven (blue) nucleotide changes were engineered into the 5′-element and/or the 3′-element of CS-A. In addition, mutants with an altered sequence of the AACA-box (green) were engineered together with a wild-type 3′-CS-A or a mutated 3′-CS-A.
FIG. 5.
Protein expression analysis of various CS-A mutants by immunofluorescence staining 3 days posttransfection. The CS-A sequences, the AACA-box and various mutations therein are depicted as in Fig. 4A. Potential base pairings between 5′-CS-A and 3′-CS-A (dotted lines) and/or between 3′-CS-A and the AACA box (solid lines) are indicated. (A) Positive (a) and negative controls (b); four or seven nucleotide changes were introduced either into 5′-CS-A only (c and f), 3′-CS-A only (d and g), or both CS-A elements to restore complementarity (e and h). (B) The AACA-box was mutated in the context of wild-type CS-A sequences (i), a mutated 5′-CS-A (j and m), a mutated 3′-CS-A (k and n), or both elements of CS-A with compensatory mutations (l and o).
Next, four or seven complementary changes were introduced in both the 5′- and 3′-elements of CS-A (the corresponding mutants were named 5′+3′-4mut and 5′+3′-7mut). According to thermodynamic analysis these mutants can establish base pairings between the two elements of CS-A with probability values at least as high as calculated for the wild-type sequence. Transfection with these mutant RNAs yielded a clearly positive (although somewhat more focal) expression pattern than the wild-type replicon (Fig. 5e and h). This result demonstrated that complementarity between the 5′- and 3′-CS-A sequences rather than the primary sequence or local secondary structure of these elements itself was essential for RNA replication.
To obtain a more detailed picture of the properties of these mutants, the RNA replication of mutants 5′-4mut, 3′-4mut, and 5′+3′-4mut, together with C17 and ΔNS5 (as positive and negative controls), was analyzed by quantitative PCR (Fig. 6A and B). 5′-4mut and 3′-4mut, in which four potential base pairs between the two elements of CS-A were destroyed, exhibited the same gradual decrease in RNA copy number as seen with the negative control ΔNS5, indicating that these mutants were indeed entirely incompetent for RNA replication. In contrast, mutant 5′+3′-4mut, which had these four base pairs restored by compensatory mutations, replicated to approximately the same level as the parental replicon C17. However, there was a clear delay in the onset of replication, suggesting that the primary sequence of the 5′- and 3′-CS-A may also contribute toward some functionality important for RNA replication. Whereas the rate of replication for C17 reached its maximum during the 18 to 24 h observation period (with an increase of ∼27% per hour, corresponding to a doubling time of ∼3 h), mutant 5′+3′-4mut showed almost no increase at early times (3 to 18 h) and then amplified its RNA until 48 h posttransfection with a rate of ca. 15 to 17% per hour (doubling time of 4.5 h). RT-PCR sequence analysis of RNA isolated 96 h posttransfection confirmed the nucleotide sequences of C17 and mutant 5′+3′-4mut throughout both CS-A elements and adjacent regions, excluding the possibility that unintended mutations arose and were selected during the course of the replication experiment (data not shown).
FIG. 6.
Kinetics of RNA replication of various CS-A mutants. C17 and ΔNS5 were used as positive and negative controls, respectively. (A and C) RNA copies isolated from 2,000 cells were quantified at various time points posttransfection by real-time PCR. Logarithmic means from two experiments are shown (the error bars indicate maximum and minimum values). (B and D) Average increase or decrease of RNA copies in percent per hour between the individual measurement points of the experiment. Panels A and B show results obtained with mutants having four nucleotide changes in the 5′-CS-A (5′-4mut), the 3-CS-A (3′-4mut), or both elements of CS-A (5′+3′-4mut) to restore complementarity. Panels C and D show results from mutants with a mutated AACA-box in the context of wild-type CS-A (AACAmut), mutated 5′-CS-A (5′-4mut+AACAmut), mutated 3′-CS-A (3′-4mut+AACAmut), or both elements of CS-A (5′+3′-4mut+AACAmut).
In conclusion, the results indicate that the most important feature of the 5′- and 3′-CSA-elements lies in their sequence complementarity, corroborating the notion that they are the main cyclization sequences of the TBEV genome.
Nucleotide sequence in the stem of 3′-SL1, rather than base pairing, seems to be important.
The nucleotide substitutions introduced into 3′-CS-A disrupted four predicted base pairs in the bottom part of the stem of the 3′-terminal 3′-SL1 structure (Fig. 4). The replication competence of the mutants 5′+3′-4mut and 5′+3′-7mut indicated that these predicted base pairs within 3′-SL1 are not essential. Indeed, when 3′-CS-A is engaged in long-range interactions with its complement, the 5′-CS-A element, this part of the 3′-SL1 stem clearly does not exist in the predicted form. Nevertheless, we hypothesized that functionality may also be associated with the noncyclic form of the genome and that, in this conformation, the base pairing of the 3′-SL1 stem may be relevant. To investigate whether restoration of these potential base pairs would improve the functionality of 5′+3′-4mut and 5′+3′-7mut, we constructed two mutants in which, in addition to four or seven nucleotide changes in both elements of CS-A, the 3′-proximal sequence 5′-AACA-3′ was modified to allow for base pairing in 3′-SL1 (Fig. 4). Unexpectedly, analysis of these mutants (5′+3′-4mut+AACAmut and 5′+3′-7mut+AACAmut) indicated that these additional modifications had a negative, rather than an enhancing, effect on the observed protein expression (Fig. 5l and o). Mutant 5′+3′-4mut+AACAmut exhibited fewer positive cells with very little diffuse cytoplasmic staining and more focal accumulations (Fig. 5l) than mutant 5′+3′-4mut without the AACA modification (Fig. 5e). This was observed in several independent experiments (data not shown). Even more strikingly, mutant 5′+3′-7mut+AACAmut was completely negative (Fig. 5o). Sequence analysis of the entire genome of this mutant excluded the possibility of a sequence mistake, and the negative result was confirmed by immunofluorescence staining 7 and 14 days posttransfection (data not shown). The AACA-box was also mutated in the context of wild-type CS-A elements. This mutant, termed AACAmut, exhibited a positive, but weaker, staining than C17 (Fig. 5i). Furthermore, the AACA box was mutated in the contexts of four or seven nucleotide changes in 5′-CS-A or 3′-CS-A (mutant designations: 5′-4mut+AACAmut, 3′-4mut+AACAmut, 5′-7mut+AACAmut and 3′-7mut+AACAmut). As can be seen in the figure, all of these mutants were negative in the immunofluorescence staining (Fig. 5Bj, k, m, and n).
The immunofluorescence results were further supported by quantitative PCR (Fig. 6C and D). Mutations interfering with cyclization remained lethal, whether or not base pairing in 3′-SL1 was restored by mutation of the AACA-box. The mutant with compensatory changes in both elements of CS-A plus mutation of the AACA-box (5′+3′-4mut+AACAmut) showed a distinct decrease at early times but eventually reached wild-type RNA levels. The mutant with wild-type 5′- and 3′-CS-A sequences plus the AACA-box mutations (AACAmut) exhibited a significant delay in the onset of replication. At later times, RNA was replicated efficiently, but the amount of RNA remained below the level achieved with replicon C17 at all times. Sequence analysis of the RT-PCR fragments obtained from RNA isolated 96 h posttransfection confirmed the presence of all originally engineered sequences and revealed no additional mutations in the vicinity of the manipulated sequence elements (data not shown).
Taken together, the data supported the view that the complementarity of the 3′-element of CS-A with its 5′-counterpart is crucial for functionality, whereas the potential to base pair at the stem of 3′-SL1 is not essential.
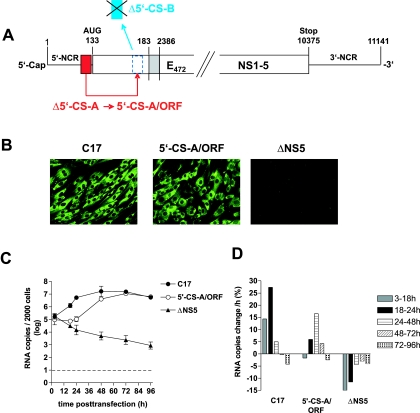
The 5′-element of CS-A is also functional when placed within the ORF.
Finally, we investigated whether the functionality of CS-A is dependent on its specific position. To address this question, we constructed a mutant in which 5′-CS-A was moved from its natural position in the 5′-NCR to the amino-terminal part of the protein C coding region, where it replaced the 5′-CS-B sequence (Fig. 7A). Indeed, this mutant (termed 5′-CS-A/ORF) was found by immunofluorescence staining to be competent for protein expression (Fig. 7B). Kinetic analysis (Fig. 7C and D) demonstrated replication of the mutant RNA. Sequence analysis indicated no changes in the sequence of mutant 5′-CS-A/ORF at 96 h posttransfection (data not shown). Compared to the parental replicon C17, the onset of replication was delayed, with a maximum rate of only 16.5% RNA increase per hour (doubling time of ∼4.5 h) during the 24- to 48-h time window. Eventually, however, both replicons reached the same intracellular RNA level. These observations indicate that the 5′-element of CS-A is still able to function when removed from its normal position, albeit with a reduced efficiency.
FIG. 7.
Construction and characterization of replicon 5′-CS-A/ORF. (A) Schematic drawing of the construction of 5′-CS-A/ORF from replicon C17 (not to scale). 5′-CS-A was deleted from its original position and used to replace the 5′-CS-B element within the ORF, which was not frame shifted by this manipulation. (B) Immunofluorescence analysis of cells transfected with RNA of replicon C17 (positive control), 5′-CS-A/ORF, or ΔNS5 (negative control); Immunofluorescence staining was performed 3 days posttransfection with a monoclonal antibody recognizing protein NS1. (C) Replication kinetics of 5′-CS-A/ORF and control RNAs. RNA isolated from 2,000 cells was quantified at various time points posttransfection by real-time PCR. The logarithmic means from two experiments are shown (error bars indicate maximum-minimum values). (D) Average increase or decrease of RNA copies in percent per hour between the individual measurement points of the experiment.
DISCUSSION
Different sets of potential cyclization sequences had been predicted for TBEV (28, 41, 57). In the present study, we obtained evidence that complementarity of the so-called CS-A elements, located, respectively, in the 5′-NCR and the bottom part of the 3′-terminal 3′-SL1 structure, is required for TBEV replication. The 5′- and 3′-CS-B elements, located at positions more similar to the CS of mosquito-borne flaviviruses, were found not to be essential. The mosquito-borne CS includes a core sequence that is highly conserved among members of that group (18, 28, 43), but no homologue of this sequence is present in tick-borne flavivirus genomes. Conversely, the CS-A elements show high conservation among tick-borne flaviviruses (41) but lack a homologous counterpart in members of the mosquito-borne group. Mosquito-borne and tick-borne flavivirus genomes share a great deal of organizational similarity, but these viruses differ with respect to their host range in arthropod cells (34). Their distinct CS sequences are possibly involved in this host range difference. Although our study did not reveal an involvement of the 5′- and 3′-CS-B elements or any of the other sequence elements that were identified in silico as possibly participating in RNA long-range interactions (CS-a, CS-b1, and CS-b2), an auxiliary function of these elements in these interactions can currently not be excluded. However, it is already clear that, in contrast to mosquito-borne flaviviruses, none of the regions coding for the amino terminus of protein C is absolutely required for TBEV RNA replication. Surprisingly, we observed that 5′-CS-A moved into the position of 5′-CS-B, i.e., within the protein C coding region, was still able to support RNA replication. Whether or not such a major genomic rearrangement would also be compatible with viral viability in the context of a full-length RNA genome is questionable and requires further investigation.
Sequences beyond the originally defined 8-nt core sequence that are important for cyclization of mosquito-borne flaviviruses have been identified (3, 57). Importantly, a recent study with DV2 demonstrated that the established CS elements are necessary but not sufficient for genome cyclization and revealed additional sequence elements (termed UAR for upstream AUG region) that are required for this process (3). According to that study, the 5′-UAR is located immediately upstream of the start codon of the ORF and the 3′-UAR is located within the bottom part of the 3′-SL, which is reminiscent of the locations of the tick-borne CS-A elements. These authors also succeeded for the first time in visualizing individual RNA molecules in their circularized or noncircularized conformations and provided evidence that cyclization of the DV2 genome can occur in the absence of viral or cellular proteins, at least under the artificial in vitro conditions used by their method. Long-range interactions of the UAR had already previously been proposed by a theoretical study (57), and deletion analysis of the DV genome had also indicated an essential role for this region (8). Furthermore, in vitro studies implied an involvement of the DV 5′-NCR in interactions with the 3′-terminal region (64). Thus, it appears that, as with TBEV, at least some mosquito-borne flaviviruses also use 5′-NCR sequences for genome cyclization, but there seems to be no sequence homology between the mosquito-borne UAR sequences and the TBEV CS-A elements.
Genome circularization has been observed for a wide range of cellular and viral RNA molecules (5, 19, 20, 25, 60). In principle, circularization could be involved in any of the main functionalities of the RNA, of which there are at least three in the case of the flavivirus genome: translation, replication, and packaging. RNA replication, as measured in our study by quantitative PCR, is dependent on prior translation to generate the components of the viral replication complex. Thus, it was difficult to distinguish to what degree each mutation affected translation versus replication. However, there is a considerable body of evidence from studies with mosquito-borne flaviviruses indicating that sequence complementarity, i.e., genome circularization, is relevant for RNA replication (1-3, 6, 28, 38, 46, 48, 64, 65). In contrast, translation does not require complementarity of 5′ and 3′ sequences (2, 9, 38) but is modulated by a number of primary sequence motifs and secondary structure within both the 5′- and the 3′-terminal regions (9, 13, 24, 35). We observed that replicons of TBEV with compensatory mutations in 5′- and 3′-CS-A elements were competent for RNA replication but with an initial delay of the onset of replication. Similar observations were reported for analogous studies in which compensatory mutations were introduced in the CS of mosquito-borne flaviviruses (2, 28). One possible explanation for this observation is that the primary sequence changes, either by themselves or by inducing perturbations of local secondary structure, may have impaired translation thus causing a delay in the formation of functional replication complexes. On the other hand, the deletions introduced into the amino-terminal part of the protein C coding region, in particular the deletions affecting the AUG-containing 5′-SL3 structure, very likely affected primarily protein translation. For a more sophisticated analysis of this region, it will be necessary to develop systems for TBEV that allow translation and replication to be distinguished, as has been achieved for other flaviviruses (2, 9, 11, 38). However, we have already established that in the absence of 5′-SL3 and the authentic start codon, translation can occur from an alternative start codon and is sufficiently effective to produce a detectable signal in the immunofluorescence assay. Several mutants with deletions affecting the N-terminal region of the capsid protein and mutants with compensatory mutations in the 5′- and 3′-CS-A and/or in the AACA-box, exhibited immunofluorescence staining patterns that appeared to be different from the parental C17 replicon, suggesting an altered cellular distribution of viral nonstructural proteins. The molecular basis of this observation, however, remains to be elucidated.
The 3′-terminal SL structure (3′-SL1) is a universal characteristic of flavivirus genomes, and functional studies have demonstrated its involvement in RNA translation and replication (6, 9, 14, 24, 43, 58, 66). In addition to the highly conserved CACAG motif, which is exposed on a side loop, certain sequence motifs and defined bulges in its long stem structure have been proposed to provide specific protein binding sites (4, 16, 30, 36, 43, 49, 55). We observed that mutations disrupting four predicted base pairs in the bottom part of the TBEV 3′-SL1-stem were tolerated. If complementarity within this stem was restored by changing the sequence of a 3′-proximal AACA-box, the functionality was impaired rather than improved, suggesting that probably the primary sequence, rather than secondary structure, was relevant at this particular location. This notion is in agreement with mutational studies on mosquito-borne flaviviruses, which have indicated that in the bottom part of the stem, the primary sequence is more important than the secondary structure, whereas in the top part of the stem, the opposite was found to be the case (66, 67). In fact, involvement of the tick-borne CS-A elements or the mosquito-borne UAR in long-range interactions implies that the bottom part of the 3′-SL1 stem cannot form when the genome adopts a cyclic conformation. Thus, in contrast to the widely accepted view of the flavivirus 3′-SL1, the most 3′-terminal nucleotides would not be involved in 3′-SL1 base pairing in the circularized conformation. One may imagine instead that this region represents an easily accessible binding site for protein factors. For DV 2, the bottom part of the 3′-SL1 was observed to support replication in a host-cell-specific manner (67). One may speculate that the tick-borne flavivirus CS-A elements and the mosquito-borne flavivirus UAR, both of which are located in this bottom part but have different nucleotide sequences, are involved in the host-cell-specific functionality of this region.
The identification of functional RNA elements is also of considerable importance for flavivirus vaccine development. Using TBEV, we have recently introduced a replicon vaccine candidate that is based on the introduction of a large deletion into the protein C coding region (31). Defining the role of RNA elements that are part of this region is instrumental for the further development of this vaccine approach and its intended application to different flaviviruses. For this purpose it will be necessary to take into account the different sequence requirements for genome cyclization of tick-borne versus mosquito-borne flaviviruses.
Acknowledgments
We thank Ines Pree, Jasmin Speil, Christian Riepl, Sabrina Schrauf, David Weissmann, and Harald Rouha for their participation in mutant construction, which was performed as part of a laboratory course. Paul Breit is gratefully acknowledged for photographic artwork. Finally, we want to thank Steven L. Allison for helpful discussions and critical reading of the manuscript.
The project was funded by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (grant P17584-B14). Cardine Thurner was supported by the Austrian GEN-AU bioinformatics integration network sponsored by bm:bwk (grant no. 200.067/3-VI/1/2002).
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, D. E., A. L. Lella Ezcurra, S. Fucito, and A. V. Gamarnik. 2005. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 339:200-212. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez, D. E., M. F. Lodeiro, S. J. Luduena, L. I. Pietrasanta, and A. V. Gamarnik. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 79:6631-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell, J. L., and M. A. Brinton. 1995. BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J. Virol. 69:5650-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenthal, T., and G. G. Carmichael. 1979. RNA replication: function and structure of Qbeta-replicase. Annu. Rev. Biochem. 48:525-548. [DOI] [PubMed] [Google Scholar]
- 6.Bredenbeek, P. J., E. A. Kooi, B. Lindenbach, N. Huijkman, C. M. Rice, and W. J. Spaan. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 84:1261-1268. [DOI] [PubMed] [Google Scholar]
- 7.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1125. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 8.Cahour, A., A. Pletnev, M. Vazielle-Falcoz, L. Rosen, and C. J. Lai. 1995. Growth-restricted dengue virus mutants containing deletions in the 5′ noncoding region of the RNA genome. Virology 207:68-76. [DOI] [PubMed] [Google Scholar]
- 9.Chiu, W. W., R. M. Kinney, and T. W. Dreher. 2005. Control of translation by the 5′- and 3′-terminal regions of the dengue virus genome. J. Virol. 79:8303-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corver, J., E. Lenches, K. Smith, R. A. Robison, T. Sando, E. G. Strauss, and J. H. Strauss. 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 77:2265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deas, T. S., I. Binduga-Gajewska, M. Tilgner, P. Ren, D. A. Stein, H. M. Moulton, P. L. Iversen, E. B. Kauffman, L. D. Kramer, and P. Y. Shi. 2005. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J. Virol. 79:4599-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamond, M. S., D. Edgil, T. G. Roberts, B. Lu, and E. Harris. 2000. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J. Virol. 74:7814-7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgil, D., M. S. Diamond, K. L. Holden, S. M. Paranjape, and E. Harris. 2003. Translation efficiency determines differences in cellular infection among dengue virus type 2 strains. Virology 317:275-290. [DOI] [PubMed] [Google Scholar]
- 14.Elghonemy, S., W. G. Davis, and M. A. Brinton. 2005. The majority of the nucleotides in the top loop of the genomic 3′ terminal stem-loop structure are cis-acting in a West Nile virus infectious clone. Virology 331:238-246. [DOI] [PubMed] [Google Scholar]
- 15.Elshuber, S., S. L. Allison, F. X. Heinz, and C. W. Mandl. 2003. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 84:183-191. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Montalvo, B. M., F. Medina, and R. M. del Angel. 2004. La protein binds to NS5 and NS3 and to the 5′ and 3′ ends of Dengue 4 virus RNA. Virus Res. 102:141-150. [DOI] [PubMed] [Google Scholar]
- 17.Gritsun, T. S., K. Venugopal, P. M. Zanotto, M. V. Mikhailov, A. A. Sall, E. C. Holmes, I. Polkinghorne, T. V. Frolova, V. V. Pogodina, V. A. Lashkevich, and E. A. Gould. 1997. Complete sequence of two tick-borne flaviviruses isolated from Siberia and the UK: analysis and significance of the 5′ and 3′-UTRs. Virus Res. 49:27-39. [DOI] [PubMed] [Google Scholar]
- 18.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33-41. [DOI] [PubMed] [Google Scholar]
- 19.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewlett, M. J., R. F. Pettersson, and D. Baltimore. 1977. Circular forms of Uukuniemi virion RNA: an electron microscopic study. J. Virol. 21:1085-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofacker, I. L., M. Fekete, C. Flamm, M. A. Huynen, S. Rauscher, P. E. Stolorz, and P. F. Stadler. 1998. Automatic detection of conserved RNA structure elements in complete RNA virus genomes. Nucleic Acids Res. 26:3825-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofacker, I. L., W. Fontana, P. F. Stadler, S. Bonhoeffer, M. Tacker, and P. Schuster. 1994. Fast folding and comparison of RNA secondary structure. Monatsh. Chem. 125:167-188. [Google Scholar]
- 23.Hofacker, I. L., and P. F. Stadler. 1999. Automatic detection of conserved base pairing patterns in RNA virus genomes. Comput. Chem. 23:401-414. [DOI] [PubMed] [Google Scholar]
- 24.Holden, K. L., and E. Harris. 2004. Enhancement of dengue virus translation: role of the 3′ untranslated region and the terminal 3′ stem-loop domain. Virology 329:119-133. [DOI] [PubMed] [Google Scholar]
- 25.Hsu, M. T., J. D. Parvin, S. Gupta, M. Krystal, and P. Palese. 1987. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl. Acad. Sci. USA 84:8140-8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iacono-Connors, L. C., J. F. Smith, T. G. Ksiazek, C. L. Kelley, and C. S. Schmaljohn. 1996. Characterization of Langat virus antigenic determinants defined by monoclonal antibodies to E, NS1 and preM and identification of a protective, non-neutralizing preM-specific monoclonal antibody. Virus Res. 43:125-136. [DOI] [PubMed] [Google Scholar]
- 27.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiermayr, S., R. M. Kofler, C. W. Mandl, P. Messner, and F. X. Heinz. 2004. Isolation of capsid protein dimers from the tick-borne encephalitis flavivirus and in vitro assembly of capsid-like particles. J. Virol. 78:8078-8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, S. M., and Y. S. Jeong. 2006. Polypyrimidine tract-binding protein interacts with the 3′ stem-loop region of Japanese encephalitis virus negative-strand RNA. Virus Res. 115:131-140. [DOI] [PubMed]
- 31.Kofler, R. M., J. H. Aberle, S. W. Aberle, S. L. Allison, F. X. Heinz, and C. W. Mandl. 2004. Mimicking live flavivirus immunization with a noninfectious RNA vaccine. Proc. Natl. Acad. Sci. USA 101:1951-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kofler, R. M., F. X. Heinz, and C. W. Mandl. 2002. Capsid protein C of tick-borne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. J. Virol. 76:3534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kofler, R. M., A. Leitner, G. O'Riordain, F. X. Heinz, and C. W. Mandl. 2003. Spontaneous mutations restore the viability of tick-borne encephalitis virus mutants with large deletions in protein C. J. Virol. 77:443-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrie, C. H., N. Y. Uzcategui, M. Armesto, L. Bell-Sakyi, and E. A. Gould. 2004. Susceptibility of mosquito and tick cell lines to infection with various flaviviruses. Med. Vet. Entomol. 18:268-274. [DOI] [PubMed] [Google Scholar]
- 35.Li, W., and M. A. Brinton. 2001. The 3′ stem-loop of the West Nile virus genomic RNA can suppress translation of chimeric mRNAs. Virology 287:49-61. [DOI] [PubMed] [Google Scholar]
- 36.Li, W., Y. Li, N. Kedersha, P. Anderson, M. Emara, K. M. Swiderek, G. T. Moreno, and M. A. Brinton. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76:11989-12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 38.Lo, M. K., M. Tilgner, K. A. Bernard, and P. Y. Shi. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 77:10004-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackenzie, J. M., A. A. Khromykh, M. K. Jones, and E. G. Westaway. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203-215. [DOI] [PubMed] [Google Scholar]
- 40.Mandl, C. W., M. Ecker, H. Holzmann, C. Kunz, and F. X. Heinz. 1997. Infectious cDNA clones of tick-borne encephalitis virus European subtype prototypic strain Neudoerfl and high virulence strain Hypr. J. Gen. Virol. 78:1049-1057. [DOI] [PubMed] [Google Scholar]
- 41.Mandl, C. W., H. Holzmann, C. Kunz, and F. X. Heinz. 1993. Complete genomic sequence of Powassan virus: evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology 194:173-184. [DOI] [PubMed] [Google Scholar]
- 42.Mandl, C. W., H. Holzmann, T. Meixner, S. Rauscher, P. F. Stadler, S. L. Allison, and F. X. Heinz. 1998. Spontaneous and engineered deletions in the 3′ noncoding region of tick-borne encephalitis virus: construction of highly attenuated mutants of a flavivirus. J. Virol. 72:2132-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Markoff, L. 2003. 5′- and 3′-noncoding regions in flavivirus RNA. Adv. Virus Res. 59:177-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 45.McCaskill, J. S. 1990. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 29:1105-1119. [DOI] [PubMed] [Google Scholar]
- 46.Molenkamp, R., E. A. Kooi, M. A. Lucassen, S. Greve, J. C. Thijssen, W. J. Spaan, and P. J. Bredenbeek. 2003. Yellow fever virus replicons as an expression system for hepatitis C virus structural proteins. J. Virol. 77:1644-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muylaert, I. R., T. J. Chambers, R. Galler, and C. M. Rice. 1996. Mutagenesis of the N-linked glycosylation sites of the yellow fever virus NS1 protein: effects on virus replication and mouse neurovirulence. Virology 222:159-168. [DOI] [PubMed] [Google Scholar]
- 48.Nomaguchi, M., T. Teramoto, L. Yu, L. Markoff, and R. Padmanabhan. 2004. Requirements for West Nile virus minus-and plus-strand subgenomic RNA synthesis In vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli. J. Biol. Chem. 279:12141-12151. [DOI] [PubMed] [Google Scholar]
- 49.Nova-Ocampo, M., N. Villegas-Sepulveda, and R. M. del Angel. 2002. Translation elongation factor-1α, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology 295:337-347. [DOI] [PubMed] [Google Scholar]
- 50.Pletnev, A. G. 2001. Infectious cDNA clone of attenuated Langat tick-borne flavivirus (strain E5) and a 3′ deletion mutant constructed from it exhibit decreased neuroinvasiveness in immunodeficient mice. Virology 282:288-300. [DOI] [PubMed] [Google Scholar]
- 51.Proutski, V., E. A. Gould, and E. C. Holmes. 1997. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 25:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Proutski, V., T. S. Gritsun, E. A. Gould, and E. C. Holmes. 1999. Biological consequences of deletions within the 3′-untranslated region of flaviviruses may be due to rearrangements of RNA secondary structure. Virus Res. 64:107-123. [DOI] [PubMed] [Google Scholar]
- 53.Rauscher, S., C. Flamm, C. W. Mandl, F. X. Heinz, and P. F. Stadler. 1997. Secondary structure of the 3′-noncoding region of flavivirus genomes: comparative analysis of base pairing probabilities. RNA 3:779-791. [PMC free article] [PubMed] [Google Scholar]
- 54.Shi, P. Y., M. Tilgner, and M. K. Lo. 2002. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology 296:219-233. [DOI] [PubMed] [Google Scholar]
- 55.Ta, M., and S. Vrati. 2000. Mov34 protein from mouse brain interacts with the 3′ noncoding region of Japanese encephalitis virus. J. Virol. 74:5108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thurner, C., C. Witwer, I. L. Hofacker, and P. F. Stadler. 2004. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 85:1113-1124. [DOI] [PubMed] [Google Scholar]
- 58.Tilgner, M., T. S. Deas, and P. Y. Shi. 2005. The flavivirus-conserved penta-nucleotide in the 3′ stem-loop of the West Nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology 331:375-386. [DOI] [PubMed] [Google Scholar]
- 59.Tilgner, M., and P. Y. Shi. 2004. Structure and function of the 3′ terminal six nucleotides of the West Nile virus genome in viral replication. J. Virol. 78:8159-8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wells, S. E., P. E. Hillner, R. D. Vale, and A. B. Sachs. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2:135-140. [DOI] [PubMed] [Google Scholar]
- 61.Wengler, G., and G. Wengler. 1981. Terminal sequences of the genome and replicative-from RNA of the flavivirus West Nile virus: absence of poly(A) and possible role in RNA replication. Virology 113:544-555. [DOI] [PubMed] [Google Scholar]
- 62.Wengler, G., G. Wengler, and H. J. Gross. 1978. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology 89:423-437. [DOI] [PubMed] [Google Scholar]
- 63.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276:15581-15591. [DOI] [PubMed] [Google Scholar]
- 65.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for Dengue virus: initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274:33714-33722. [DOI] [PubMed] [Google Scholar]
- 66.Yu, L., and L. Markoff. 2005. The topology of bulges in the long stem of the flavivirus 3′ stem-loop is a major determinant of RNA replication competence. J. Virol. 79:2309-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng, L., B. Falgout, and L. Markoff. 1998. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J. Virol. 72:7510-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]