Abstract
The Mll gene is a member of the mammalian trithorax group, involved with the antagonistic Polycomb group in epigenetic regulation of homeotic genes. MLL contains a highly conserved SET domain also found in various chromatin proteins. In this study, we report that mice in which this domain was deleted by homologous recombination in ES cells (ΔSET) exhibit skeletal defects and altered transcription of particular Hox genes during development. Chromatin immunoprecipitation and bisulfite sequencing analysis on developing embryo tissues demonstrate that this change in gene expression is associated with a dramatic reduction in histone H3 Lysine 4 monomethylation and DNA methylation defects at the same Hox loci. These results establish in vivo that the major function of Mll is to act at the chromatin level to sustain the expression of selected target Hox genes during embryonic development. These observations provide previously undescribed evidence for the in vivo relationship and SET domain dependence between histone methylation and DNA methylation on MLL target genes during embryonic development.
Keywords: histone methyltransferase, MLL-SET domain, homeotic transformations
The control of cell identity during development is specified, in large part, by the unique expression patterns of multiple homeobox-containing (Hox) genes in specific segments of the embryo (1). The trithorax and polycomb groups (trx-G and PcG) were identified for their role in faithfully maintaining the transcriptional states of these key developmental regulators, providing an epigenetic mechanism of cellular memory (2–4).
The gene expression maintenance function of the trxG and PcG proteins is highly conserved. Mixed lineage leukemia (Mll), a human homolog of Drosophilatrithorax and a member of the trxG family, was identified first for its involvement in chromosomal translocations associated with lymphoid and myeloid acute leukemia in infants and adults (5, 6). Mll encodes a 3,969-aa nuclear protein with multiple domains, including three AT-hook motifs, a DNA methyltransferase homology domain (DNMT) in the amino-terminal half of the protein, a central zinc finger (PHD) region, and a highly conserved 130-aa carboxyl-terminal SET domain. The MLL protein was shown to be proteolytically processed into two portions (MLLN and MLLC) with antagonistic transcriptional effector properties, that reassociate and stabilize each other (7–9). The MLL protein is critical for proper regulation of the Hox genes during embryonic development (10). In Mll null mutant mice (Mll−/−), Hox gene expression is correctly initiated but is not sustained as the function of Mll becomes necessary (11), leading to embryonic lethality.
It is strongly believed that maintenance of the transcriptional status of target genes by PcG and trxG proteins is achieved through chromatin modifications (12). The structure similarity between some trxG/PcG and suppressors or enhancers of position effect variegation (PEV) further substantiates this point. One of the most remarkable shared domains within these chromatin proteins is the SET domain. Present at the C terminus of the Trithorax and MLL proteins, this motif also is found in a large number of other proteins, including the PEV modifier SU(VAR)3–9 and the PcG protein E(z) (13, 14). Many SET domain-containing proteins have been demonstrated to mediate lysine-directed histone methylation (15–19). These proteins decorate the histones at specific position, providing a recognition site for activating or repressing proteins. In particular, the methylation on lysine-9 of histone H3 is associated with transcriptional repression (20, 21). Di- and trimethylation on lysine-4 of histone H3 (H3K4me2 and H3K4me3), on the other hand, are associated with a permissive and transcriptionally active state of the chromatin, respectively (22, 23). MLL was shown in vitro to bind to the promoter regions of some active Hox genes, where it recruits a very large multiprotein complex carrying several chromatin modifying and remodeling activities. They include a histone H3 lysine-4 methyltransferase activity conferred by the SET domain of MLL (24, 25) and histone acetyltransferase activities through the recruitment of MOF, a member of the MYST family of histone acetyltransferases (HATs), which acetylates histone H4 at Lys-16 (26, 27).
To gain more insight into the in vivo function of the MLL-SET domain during development, we have generated mice in which the SET domain of Mll was deleted by homologous recombination (ΔSET). In these conditions a SET domain-truncated allele of Mll is normally expressed. ΔSET mutant mice are viable and fertile. We show that they exhibit developmental skeletal defects and an alteration in the maintenance of the proper transcription levels of several target Hox loci during development. Importantly, these changes in gene expression levels are associated with a reduction of histone H3K4 monomethylation (H3K4me1) and altered DNA methylation patterns at the same Hox loci. These results demonstrate in vivo an essential role for the MLL-SET domain on chromatin structure and Hox gene regulation. They provide evidence for epigenetic relationship in the maintenance of Hox genes activation during embryonic development.
Results
Targeted Disruption of the Mll-SET Domain.
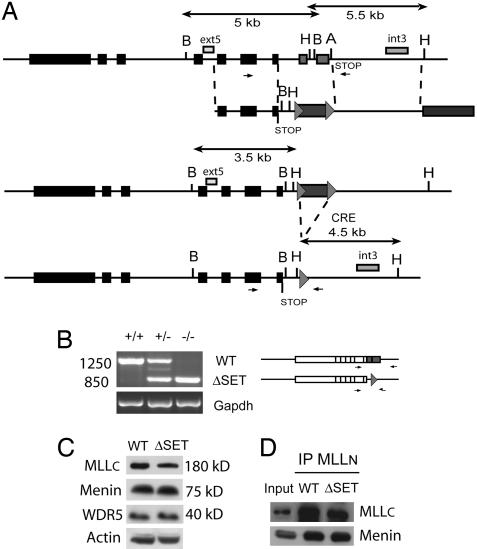
To investigate the function of the SET domain of the Mll gene during development, we have deleted this motif by homologous recombination in ES cells by replacing the SET-encoding region by a floxed neomycin-resistance (neo) gene. A “stop” codon was introduced in frame to allow appropriate termination of protein synthesis in the absence of the SET domain (Fig. 1A). Homologous recombinant clones were obtained and transfected in vitro with a CRE expressing vector to excise the neo cassette. Two independent ΔSET+/− ES cell clones were at the origin of several chimeras, which transmitted the truncated Mll allele to their offsprings. Southern analysis on BamHI- and HindIII-digested tail DNA from selected mice, using the appropriate DNA probes (Fig. 1A), confirmed that the SET domain was deleted in mutant mice (Fig. 6A, which is published as supporting information on the PNAS web site). RT-PCR by using RNA extracted from WT (+/+), ΔSET+/−, and −/− mouse embryonic fibroblasts [MEFs; derived from 12.5 days postcoitum (dpc) embryos] demonstrated that the ΔSET allele was transcribed at similar level than the WT allele (Fig. 1B). Immunofluorescence studies on WT and ΔSET −/− MEFs confirmed that the deletion of the SET domain did not affect the level of expression or distribution of the truncated MLL protein in the nucleus. Consistent with previous studies (24), a punctuate pattern was observed (Fig. 6B). The biochemical purification of MLL complex identified multiple components, including the tumor suppressor Menin, an essential protein within the MLL complex, required for appropriate Hox gene expression. This complex is highly similar to that of the yeast and human SET1 complex (COMPASS) (9). Upon deletion of the SET domain of MLL, the global levels of MLLC and MLL-associated proteins WDR5 and Menin were not significantly affected (Fig. 1C). Total protein extract were prepared from WT and ΔSET MEFs for Western blot analysis. MLLC, WDR5, and Menin levels were assessed while Actin estimated the equivalence of protein loading. Immunoprecipitation-Western blot experiments by using an antibody directed against MLLN further demonstrated that the SET domain truncated MLLC is still associated in a complex with MLLN and Menin (Fig. 1D). Altogether these analyses show that the loss of the SET domain of MLL does not significantly alter the stability of the MLL complex.
Fig. 1.
Generation of the MllΔSET allele. (A) Mll-SET domain targeting vector with homology arms, LoxP sites (triangles) flanking the neomycin resistance cassette. Partial restriction map of the Mll locus (A, ApaI; B, BamHI; H, HindIII) is shown before and after targeted replacement of the SET domain encoding region by the neomycin resistance cassette. Southern strategy to detect homologous recombination (BamHI digestion and “ext5” probe) and CRE deletion of the neomycin resistance cassette (HindIII digestion and “int3” probe) is indicated. RT-PCR primers encompassing the SET domain region are indicated by arrows. (B) The expression of the WT and MLL SET truncated alleles was evaluated by RT-PCR analysis in embryonic fibroblasts generated from 12.5 dpc embryos. Primers overlap the SET domain region, distinguishing the WT and truncated alleles. Equivalent loading of RNA was controlled by using GAPDH primers. (C) Western blot analysis of selected MLL-associated factors: MLLc, Menin, and WDR5 in WT and MLLΔSET MEFs. Molecular sizes of marker proteins are shown on the right. (D) Coimmunoprecipitation of MLL with associated factors. Nuclear extracts of WT and mutant MEFs (lane 1 input) were subjected to immunoprecipitation (IP) by using an antibody specific for MLLN. The immunoprecipitates were fractionated in SDS/PAGE and immunoblotted with the antibodies indicated to the left of the panels (anti-MLLC and anti-Menin)
Homeotic Transformations in ΔSET Mice.
Three Mll-disruption alleles have been previously generated (10, 28, 29); all result in embryonic lethality in homozygotes. In sharp contrast, in the ΔSET mutant mice, the mating of heterozygous mice yielded WT, heterozygous, and homozygous offspring at the expected Mendelian ratios, indicating no significant embryonic lethality. At birth, homozygotes were indistinguishable from their WT and heterozygous littermates and did not present significant weight or growth differences [12.1 ± 1.5 g (n = 28) and 12.8 ± 0.5 g (n = 35), for 3-week-old WT and ΔSET mice, respectively].
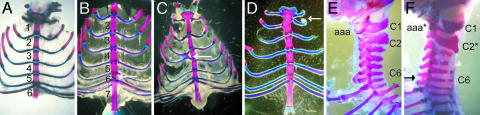
Mice heterozygous for the Mll-lacZ allele displayed bidirectional homeotic transformations of the axial skeleton and sternal malformations. These homeotic transformations were associated with a posterior shift of the anterior boundaries of expression of several Hox genes in heterozygous mutant animals (10, 11). Examination of 3-week-old homozygous ΔSET mutant skeletons showed defects in the formation of the vertebral column (Fig. 2). Mutant mice displayed an additional ossification center between sternebra 5 and the xiphoid process (Fig. 2B, arrow). In consequence, they exhibited seven sternebras instead of six in WT animals (Fig. 2A). Additional typical sternal malformations included uni- or bilateral fusions between ribs (Fig. 2 C and D, arrows). The cervical region of the ΔSET mutant mice was also affected; the atlas (C1) and the axis (C2) presented abnormal shape, the axis looking more like the atlas, whereas the anterior arch of the atlas (aaa) was often missing or reduced in size (Fig. 2F, compare with WT, Fig. 2E). Moreover mutant mice displayed a typical posterior transformation of C6 and C7 revealed by the presence of rudimentary ribs. These observations show that the SET domain can account, at least in part, for the role of Mll in defining the proper identity of cells in the segments along the anteroposterior axis. With the exception of the sternal malformations, the defects are specific for the ΔSET allele, suggesting that several domains of MLL are involved in establishing a proper segment identity in mice.
Fig. 2.
Skeletal abnormalities in ΔSET mice. Views are shown of the thoracic (A–D) and cervical (E and F) regions of cleared skeletons of WT (A and E) and ΔSET mutant mice (B–D and F). Typical sternal abnormalities were detected in mutant mice. The additional ossification center is indicated by an arrow in B. Two different fusions between ribs are shown (C and D, indicated by arrows). Lateral view of the cervical region of a WT (E) and ΔSET mouse (F) is shown. ΔSET mice present bone abnormalities in the cervical region. The C2 vertebra is broadened (C2∗). The anterior arch of the atlas is severely reduced (aaa∗). C6 (black arrow) show posterior transformation to C7.
Hox Gene Expression Defects in ΔSET Mutant Embryos.
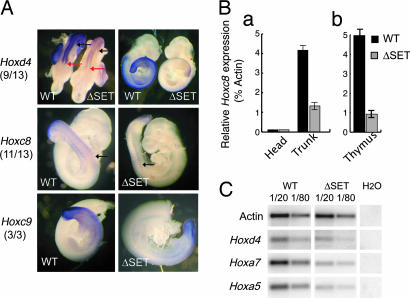
Regulation of both Hox gene expression boundaries and dosage are important for correct specification of segment identity (30). Knockout mice have revealed that Mll is required for maintenance rather than establishment of Hox gene expression early in embryogenesis. Downstream targets of Mll are activated appropriately in the absence of Mll but require Mll for sustaining their expression between 8.5 and 9 dpc (10, 11). Therefore, we have examined the expression patterns of some representative Hox genes, known to be regulated by trxG and PcG genes, in 9.5 dpc ΔSET embryos (24, 31). RNA in situ hybridization detected Hoxd4, Hoxc8, and Hoxc9 transcripts at their normal anterior boundaries both in mesodermal and neuroectodermal tissues (Fig. 3A). This observation suggests that the SET domain function is not required for the spatial regulation of Hox genes expression along the anteroposterior axis. However, the general level of expression of those genes was consistently reduced in mutants as compared with WT mice (Fig. 3A). To confirm the reduction of Hox genes expression, real-time quantitative RT-PCR was used on RNA extracted from 9.5 dpc embryos and from thymocytes (number of thymocytes were equivalent in WT and mutant mice). In these experiments, 9.5 dpc WT and ΔSET embryos were divided at the level of the otic vesicle (indicated by asterisks in Fig. 3A Top). The head (Hoxd4 and c8 negative) and trunk (Hoxd4 and c8 positive) were used for RNA extraction. As shown on Fig. 3B, MllΔSET trunk presented a 4-fold decrease of Hoxc8 mRNA level as compared with WT embryos and a 6-fold reduction in thymus. Similarly, expression levels of Hoxd4, Hoxa7, and Hoxa5 were significantly reduced in thymocytes from 3-week-old mutant mice (Fig. 3C). However, some Hox genes, including Hoxb9 and Hoxd11, did not show any significant reduction in their level of transcription (data not shown), suggesting a specific rather than global alteration of Hox gene expression in ΔSET mice. Altogether, these results indicate that the SET domain may be involved in maintaining proper Hox gene expression levels during development.
Fig. 3.
Altered expression of Hox genes in ΔSET mice. (A) Hoxd4, Hoxc8, and Hoxc9 whole-mount in situ hybridization on 9.5 dpc, somite-matched, ΔSET and WT embryos. Red arrows mark the anterior limit of neurectoderm expression, and black arrows mark the anterior limit of mesodermal expression. The numbers of ΔSET −/− embryos presenting decreased levels of Hox gene expression are indicated. (B) Quantitative PCR analysis of Hoxc8 expression in 9.5 dpc embryos (a) and thymocytes (b). (C) Semiquantitative RT-PCR analysis of Hox gene expression in thymocytes (Hoxd4, Hoxa7, and Hoxa5). Actin was used as a control to standardize RNA input and serial dilutions (1/20 and 1/80) of RNA used to compare expression levels.
Reduced Histone H3K4 Monomethylation at Target Hox Loci in ΔSET Mutant Embryos.
Recently it was shown that the MLL-SET domain possesses an intrinsic H3K4 methyltransferase activity (24, 25). Using cell transfection and biochemical assays, Milne et al. (24) demonstrated that this methyltransferase activity is associated with Hox gene activation and is stimulated by acetylation of Lys-9 or -14 of H3 peptides.
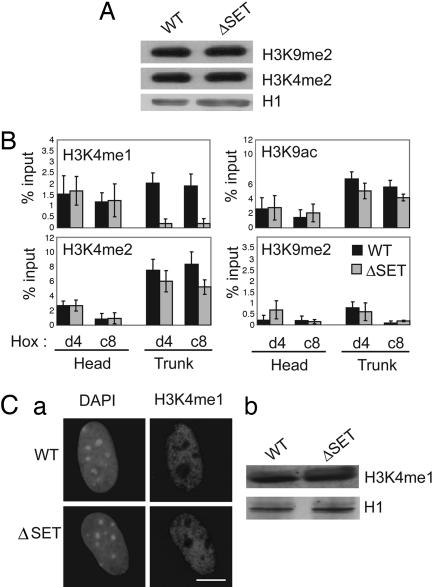
We first examined by immunofluorescence on WT and ΔSET embryonic fibroblasts the abundance and distribution of specific hallmarks of euchromatin. In these experiments, the relative abundance of histone modifications typical of transcriptionally active (H3K4me2 and H3K9ac) and inactive (H3K9me2) chromatin was compared by using a TCS Leica laser-scanning confocal microscope. The microscope settings and laser power were kept constant so that the relative abundance of each modification within 100 WT and ΔSET MEF nuclei could be directly compared and quantified by using metamorph 4.0. No overall change in histone methylation levels or distribution was detected (Fig. 7, which is published as supporting information on the PNAS web site), and Western blot analysis confirmed this observation (Fig. 4A). For this analysis, total protein extracts were prepared from WT and ΔSET MEFs. H3K4me1 (Fig. 4Cb), H3K4me2, and H3K9me2 levels were assessed, and Histone H1 estimated the equivalence of protein loading. This result is consistent with a locus specific effect of the ΔSET mutation at selective target genes and suggests that most features of euchromatin are probably preserved in ΔSET MEFs.
Fig. 4.
H3 lysine-4 methylation pattern alteration at Hox loci in ΔSET mice. (A) Western blots confirming the steady levels of H3K4me2 and H3K9me2 in the ΔSET mutant MEFs. (B) Chromatin immunoprecipitation analysis in which the abundance of histone modifications (H3K4me1, H3K4me2, H3K9ac, and H3K9me2) were compared at Hoxd4 P1 and Hoxc8 promoters in the head and trunk of WT (black histograms) and ΔSET (gray histograms) 9.5 dpc embryos. Values shown are the mean of five independent experiments with average deviation. (C) Immunofluorescence (a) and Western blots (b) confirming the steady levels of H3K4me1 in the ΔSET mutant MEFs. Whole-cell extracts were prepared from MEFs cultures, and histone H1 was used as a control to standardize protein loading.
Chromatin immunoprecipitation analysis was performed to compare the chromatin structure at the promoter of Hox genes whose expression was altered in ΔSET thymocytes and embryos. To evaluate chromatin changes more accurately during embryonic development, chromatin immunoprecipitation was performed on developing embryo tissue. In these experiments, 9.5 dpc WT and ΔSET embryos were divided at the level of the otic vesicle (indicated by asterisks in Fig. 3A Top). The head (Hoxd4 and c8 negative) and trunk (Hoxd4 and c8 positive) were used for the extraction of chromatin samples, and the relative abundance of H3K4me1, H3K4me2, H3K4me3, H3K9ac, and H3K9me2 was compared at the promoter regions of Hoxd4 (P1) and Hoxc8. As shown in Fig. 4B, chromatin immunoprecipitation analysis revealed a very significant decrease in H3K4me1 in ΔSET mutant trunks at both Hoxd4 and Hoxc8 promoters (≈10-fold). H3K4me2 and H3K9ac, two other marks associated with transcriptional activation, were mildly affected at both Hox genes (20–40% reduction), whereas H3K4me3 levels were not affected at these Hox genes in the trunk (data not shown). H3K9me2 levels, associated with gene silencing, were unaffected by the deletion of the SET domain of Mll (Fig. 4B), and no significant changes in histone modification was detected in the head in the ΔSET mutant embryos. Interestingly, in WT animals, both H3K4me2 and H3K9ac levels were significantly higher in the trunk, where Hoxd4 and c8 genes are expressed. H3K4me1 levels were reduced but comparable, and H3K9me2 levels were low both in the head and trunk. Consistent with transcriptional activation in the trunk, RNA Pol II was associated with Hoxd4 and Hoxc8 promoters in the trunk but was absent from the head region (data not shown).
Our results establish in vivo the specific histone H3 lysine-4 methylation activity of the MLL-SET domain at target loci. In addition, we show that H3K4 mono-, di- and trimethylation are differentially affected, suggesting a role for the SET domain of MLL in monomethylation at lysine-4 of histone H3 in vivo at the Hoxd4 and Hoxc8 loci during embryonic development. The chromatin state in the head and trunk is consistent with gene activity in 9.5 dpc embryos.
DNA Methylation Defects in ΔSET Mutant Mice.
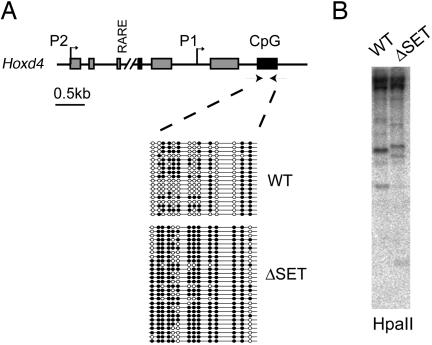
DNA methylation at CpG dinucleotides in regulatory regions is associated with silencing of gene expression (32–34). To verify the status of DNA methylation in ΔSET mutants, we performed PCR amplification of sodium bisulfite-treated DNA isolated from embryonic fibroblasts. The CpG-rich region of Hoxd4, in the vicinity of exon 2 (Fig. 5A) was abnormally methylated, with 10 of 14 CpGs found to be hyper- or hypomethylated in ΔSET MEFs (compare with WT MEFs, Fig. 5A). Changes in DNA methylation pattern were confirmed by Southern blot analysis with the methylation-sensitive enzyme HpaII (Fig. 5B). Similar results were consistently obtained on a different portion of Hoxd4, on promoter P2 (Fig. 8A, which is published as supporting information on the PNAS web site). These modifications are gene-specific rather than global because the analysis of a lymphocyte-specific gene, CD3ε in MEFs, did not show any change in the DNA methylation pattern (Fig. 8B). In addition, immunofluorescence stainings by using an antibody directed against 5-methyl cytosine did not reveal any change in global levels of DNA methylation (Fig. 8C). These results demonstrate that the loss of the SET domain has a significant effect on DNA methylation at the Hoxd4 locus. However, the finding that both DNA methylation and demethylation alterations coexist suggests that the SET domain is not directly responsible for these modifications. Altogether, these epigenetic changes at Hox genes could account for decreased level of expression, and they demonstrate that the SET domain, presumably through histone H3K4 methylation, influences DNA methylation.
Fig. 5.
DNA methylation changes at Hox loci in ΔSET mice. (A) A partial map of the mouse Hoxd4 gene is shown. Methylation of cytosines was assessed by bisulfite sequencing upstream of exon 6, a CpG rich region of the Hoxd4 gene. A summary of methylation data from MEFs is shown where each line represents a separate clone. Methylated CpGs are represented by filled beads, and unmethylated CpGs are represented by open beads. (B) Southern blot analysis of DNA methylation changes at Hoxd4 in ΔSET embryonic cells (trunk from embryonic day 9.5 embryos) by using the methylation-sensitive enzyme HpaII.
Discussion
We report here the generation and characterization of a previously undescribed mutant allele of Mll, the mammalian homolog of the Drosophila trithorax gene. The ΔSET mutation deleted the two last exons of the Mll gene, which encodes the highly conserved SET domain. In contrast to Mll-null mutant mice, ΔSET−/− animals survive embryogenesis. Three previously reported Mll alleles have suggested that the N terminus of MLL protein is functionally important, the phenotype being less severe as a larger segment of the N terminus of the MLL protein is expressed (10, 28). Recently, several studies showed that MLL can be proteolytically processed into two fragments and that postcleavage association of the N-terminal (p320) and C-terminal (p180) fragments confers its stability and subnuclear localization to MLL (8, 25). This process, unaffected by the deletion of the SET domain, may confer normal stability and subnuclear localization to the truncated MLL protein, allowing it to achieve important SET-independent functions necessary for survival.
We show that ΔSET mutant mice present homeotic skeletal defects and that the level of expression of some Hox genes is reduced during development. On the other hand, the anterior limit of Hox gene expression along the anteroposterior axis is not altered in mutant embryos. We propose that the SET domain function of Mll is required for the maintenance of optimal levels of Mll target gene expression levels (24). The spatial regulation of Hox genes expression may be achieved through an Mll-SET-independent mechanism, probably mediated by the N terminus of the protein. Proper levels of Hox genes expression are important for specific developmental processes (35) and could account for the homeotic transformations detected in ΔSET mutant skeleton.
It was shown previously that the MLL protein is responsible for the maintenance rather than activation of Hox genes expression (11, 36). In Mll−/− embryos, the pattern of Hox genes expression is established normally and lost between embryonic days 8.5 and 9 when the MLL protein is required to maintain this pattern (11). This development stage may be indicative of a period of epigenetic transition at Mll target genes, leading to either silencing or maintained expression of target loci. The SET domain of MLL possesses a H3K4 histone methyltransferase activity (24). H3K4 mono/dimethylation occurs at both inactive and active euchromatic genes, whereas trimethylation is preferentially and strongly associated with transcriptionally active genes (37, 38). It is therefore the presence of a trimethylated K4 that defines an active state of gene expression, whereas mono/dimethylated K4 may distinguish transcriptionally permissive chromatin (23). The SET domain of MLL is important for H3K4 HMT activity and transcriptional activation at MLL target loci. Recombinant MLL SET domain was shown to have methyltransferase activity toward unmodified H3 peptides but appeared to lack the ability to catalyze the conversion from di- to trimethylation (24). Recently, a stable complex containing both MLL and the histone acetyltransferase MOF was purified from HeLa cells (26). This complex was shown to effect H3K4 methylation (mono-, di-, and tri-) and H4K16 acetylation, resulting in transcriptional activation of MLL target Hox genes. In these experiments, dimethylated H3K4 peptide is a better substrate than unmodified and monomethylated peptides. Our results suggest that the deletion of MLL-SET affects primarily H3K4 monomethylation at Hox genes. A reduction in H3K9 acetylation levels mirrors the reduction in H3K4 dimethylation and no increase in H3K9 methylation levels could be detected. This altered histone modification pattern at both Hoxd4 and Hoxc8 loci may be responsible for the sustained but reduced levels of transcriptional gene expression in the ΔSET embryos. Our results favor a model in which the SET domain has mono-H3K4 histone methyltransferase activity. A SET domain-dependent mechanism/interaction may actively dimethylate H3K4, whereas a SET domain-independent mechanism would mediate the conversion from di- to trimethylation via the recruitment of specific enzymatic activity in the same way that the MLL–WDR5 complex recruits histone acetyltransferase activity via a SET domain-independent interaction with MOF (26). However, we cannot formally exclude that another trithorax-group protein with a redundant function is compensating for the loss of the Mll-SET domain and is able to di- and trimethylate H3K4.
Interestingly, appropriate chromatin modifications distinguish the transcriptional status of Hox genes in regions where they are active (trunk) and silent (head). Hoxd4 and Hoxc8 loci in the trunk bear modifications associated with transcriptional activation (di- and trimethylation of H3K4 and acetylation of H3K9), and these features are reduced to absent in the head at the same loci. These results are consistent with previously reported study of chromatin states in anterior and posterior compartments during embryonic development (39). The difference in monomethylation of lysine-4 of H3 between the head and trunk are not significant, suggesting that H3K4me1 is not directly involved in transcriptional activation. In addition, transcriptional expression of Hox genes is maintained in the trunk of ΔSET embryos despite reduced H3K4me1 levels. We propose that H3K4me1 could be a mark for poised chromatin, which is established at Hoxd4 and Hoxc8 loci through a MLL-dependent manner in the trunk and a MLL-independent manner in the nonexpressing, anterior region of the embryo. In this context, the SET domain of MLL would be involved in the maintenance of a permissive state of the chromatin after 8.5 dpc (putatively through establishment/maintenance of H3K4me1 marks), whereas other factors recruited within the MLL complex are required to generate further chromatin modifications and proper transcriptional activation of target loci (9, 26).
An increasing number of studies suggest that DNA methylation could be regulated, at least partly, by H3K9 modifications, possibly by creating a mark that can be targeted by proteins involved in DNA methylation (40–43). Milne et al. (24) have shown that the Hoxc-8 locus is extensively DNA methylated in Mll−/− cells, and the bisulfite sequencing data we present suggest that in the absence of Mll-SET domain, reduced levels of H3K4me1 are associated with abnormal DNA methylation patterns. The loss of histone H3K4 monomethylation in MllΔSET could affect the recruitment or exclusion of proteins involved in the regulation of DNA methylation. It was demonstrated that the PcG SET domain containing protein EZH2 interacts with DNA methyltransferases and that EZH2 is required for DNA methylation of EZH2-target promoters (44). In the same way, MLL could recruit DNA methyltransferases at target genes; Alternatively, the N terminus of MLL contains a region with sequence similarity to the DNA methyltransferase DNMT1, which binds in vitro to unmethylated CpG-rich DNA (45–47), suggesting a more direct role of MLL in the regulation of DNA methylation at target genes. The biochemical mechanism underlying the process of aberrant methylation in MllΔSET mutant mice remains an open question.
Taken together, our data show that the loss of the MLL SET domain is sufficient to induce histone and DNA methylation defects at the promoter of several Hox genes in mutant mice and that expression of these same Hox genes is down-regulated. Viability despite the altered genomic methylation and reduction of methylated H3K4 histone marks at target loci makes the MllΔSET mutant an ideal model system for the study of epigenetic regulation during developmental processes and tumorigenesis.
Materials and Methods
Generation of ΔSET Mutant Mice.
Targeting vector DNA (Fig. 1A; see also Supporting Materials and Methods, which is published as supporting information on the PNAS web site, for a detailed description of the Mll-SET targeting vector generation) was electroporated in 3 × 107 129/ola ES cells cultured in DMEM (Invitrogen) supplemented with 15% FCS/1,000 units/ml of LIF and antibiotics. The transfected cells were selected with 0.3 mg/ml G418, and resistant colonies were screened for homologous recombination by Southern blot analysis. Mll-SET targeted ES clones (MllΔSET+/−neo) were transiently transfected with a vector encoding the CRE recombinase (pMC-CRE) and expanded in the absence of G418 selection for 7 days. Individual clones were screened by PCR and Southern blot for the deletion of the neomycin resistance gene (MllΔSET+/− ES cells). Three independent MllΔSET+/− ES clones were injected into embryonic day 3.5 blastocysts and transferred into the uteri of pseudopregnant mice. Resulting chimeric mice were mated with BALB/c mice, and germ-line transmission was confirmed by PCR and Southern blot analysis of tail DNA.
Establishment of MEFs.
MEFs were obtained from WT, MllΔSET+/− and −/− mutant embryos at developmental stage of 12.5 dpc. Cells were cultured in DMEM supplemented with 10% FCS and antibiotics. Cells were frozen as stocks at the second passage and used for the subsequent studies at the third passage.
RT-PCR Analysis.
Total RNA was extracted with RNAzol B, and cDNA synthesis was performed by using oligo (dT)12–18 (Invitrogen). PCR amplification was performed by using Standard TaqDNA polymerase (Qiagen, Valencia, CA) and the following cycles: (94°C for 30 s, Tm for 30 s, and 72°C for 60 s). For Analysis of Hox gene expression, OneStep RT-PCR kit (Qiagen) was used according to manufacturer's instructions. The primers used for PCR amplification are available in Table 1, which is published as supporting information on the PNAS web site.
Coimmunoprecipitation and Western Blot Analysis.
Indirect immunofluorescence labeling was performed by using the following primary antibodies: rabbit anti-MLL (kind gift from A. G. Fisher, Medical Research Council, London), rabbit anti-H3K9ac, anti-H3K4me2, and anti-H3K9me2 (Upstate Biotechnology). Secondary antibodies were purchased from Molecular Probes. For Western blot and coimmunoprecipitation analysis, the primary antibodies used were as follows: anti-MLLN [Abcam, Inc. (Cambridge, U.K.) ab17959], anti-MLLC and anti-Menin (Bethyl Laboratories, A300–086A; A300–374A; A300–105A), anti-H3K4me2, anti-H3K9me2, anti-H1 (Upstate Biotechnology), and WDR5 (kind gift of W. Herr, Cold Spring Harbor Laboratory, Plainview, NY). Whole-cell extracts were prepared by direct lysis of cells in SDS-loading buffer. After electrophoresis, blots were probed with primary antibodies and visualized by using an Amersham Pharmacia ECL detection kit. Coimmunoprecipitations were performed by using Nuclear Complex Co-IP Kit (active motif).
Immunostaining and Microscopy.
Immunostaining samples were fixed for 20 min in 2% paraformaldehyde, permeabilized in 0.4% Triton, and incubated 30 min in a blocking solution (2.5% BSA/0.05% Tween 20 in PBS), incubated in primary antibody (1.5 h at room temperature), washed, and incubated in fluorochrome-labeled secondary antibody (45 min at room temperature). Nuclei were counterstained by using DAPI (1 μg/ml), and slides were mounted in Vectashield before analysis by using a TCS Leica laser-scanning confocal microscope and quantification by using metamorph 4.0.
Whole-Mount RNA in Situ Hybridization.
Whole-mount in situ hybridization was performed as described in refs. 48 and 49. See Supporting Materials and Methods for a detailed protocol.
Skeletal Analysis.
Dissected animals were fixed 12 h in 100% ethanol. Cartilaginous tissues were stained in an alcian blue solution (0.15 mg/ml in acetic acid/ethanol solution) for 24 h. Skeleton were rinsed for 1 h in 95% ethanol and incubated for 24 h in a 2% KOH solution. Bones were stained with a 1% KOH/75 mg/ml alizarin red solution for 12–24 h. Stained skeletons were stored in a 20% glycerol/1% KOH solution and preserved in a glycerol/ethanol (50/50) solution.
Chromatin Immunoprecipitation on Embryos and Real-Time PCR.
The 9.5 dpc embryos were carefully dissected into anterior (head) and posterior (trunk) regions. A section was made by using fine forceps at the base of the otic vesicle, at the boundary of r6/7 for the Hoxd-4 analysis and at the level of the forelimb bud for Hoxc-8 (7th somite). Chromatin immunoprecipitation experiments were performed according to a protocol provided by Upstate Biotechnology. The cells (106) were lysed for 15 min on ice and incubated with 200 μl of SDS lysis buffer for 20 min on ice to release the chromatin. After sonication, the chromatin was precleared with salmon sperm DNA-protein A-agarose beads for 1 h and incubated overnight with specific antibodies (5 μg per experiment). Chromatin was eluted from the beads, and cross-links were reversed at 65°C for 6 h. Phenol/chloroform-extracted DNA was ethanol precipitated and used as a template for real-time PCR (in triplicate) by using the SYBR Green Taq ReadyMix kit for quantitative PCR (Sigma) and the 5700 detection system from Applied Biosystems. Data are expressed as a percentage of the input.
Bisulphite Sequencing.
Genomic DNAs from embryonic WT and ΔSET embryonic fibroblasts were modified by sodium bisulfite treatment as described in ref. 50. The primers used for PCR amplification of treated DNAs are available in Table 1. PCR cycling conditions consisted of 4 min at 95°C followed by 30 cycles of 40 sec denaturation at 94°C, 45 sec annealing at 54°C, 45 sec of extension at 72°C, with final extension at 72°C for 10 min.
Supplementary Material
Acknowledgments
We thank J. Hess and P. Ernst for critical reading of the manuscript and for helpful discussions. This work was supported by grants from Centre National de la Recherche Scientifique, Association pour la Recherche sur le Cancer, Institut National du Cancer, and Fondation de France.
Abbreviations
- dpc
days postcoitum
- MEF
mouse embryonic fibroblast.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ingham P. W. Int. J. Dev. Biol. 1998;42:423–429. [PubMed] [Google Scholar]
- 2.Moehrle A., Paro R. Dev. Genet. 1994;15:478–484. doi: 10.1002/dvg.1020150606. [DOI] [PubMed] [Google Scholar]
- 3.Paro R. Trends Genet. 1995;11:295–297. doi: 10.1016/s0168-9525(00)89081-2. [DOI] [PubMed] [Google Scholar]
- 4.Paro R., Strutt H., Cavalli G. Novartis Found. Symp. 1998;214:51–61. doi: 10.1002/9780470515501.ch4. discussion 61–66, 104–113. [DOI] [PubMed]
- 5.Djabali M., Selleri L., Parry P., Bower M., Young B. D., Evans G. A. Nat. Genet. 1992;2:113–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 6.Gu Y., Nakamura T., Alder H., Prasad R., Canaani O., Cimino G., Croce C. M., Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh J. J., Cheng E. H., Korsmeyer S. J. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh J. J., Ernst P., Erdjument-Bromage H., Tempst P., Korsmeyer S. J. Mol. Cell. Biol. 2003;23:186–194. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokoyama A., Wang Z., Wysocka J., Sanyal M., Aufiero D. J., Kitabayashi I., Herr W., Cleary M. L. Mol. Cell. Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu B. D., Hess J. L., Horning S. E., Brown G. A., Korsmeyer S. J. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 11.Yu B. D., Hanson R. D., Hess J. L., Horning S. E., Korsmeyer S. J. Proc. Natl. Acad. Sci. USA. 1998;95:10632–10636. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon J. A., Tamkun J. W. Curr. Opin. Genet. Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- 13.Breiling A., Orlando V. Nat. Struct. Biol. 2002;9:894–896. doi: 10.1038/nsb1202-894. [DOI] [PubMed] [Google Scholar]
- 14.Alvarez-Venegas R., Avramova Z. Gene. 2002;285:25–37. doi: 10.1016/s0378-1119(02)00401-8. [DOI] [PubMed] [Google Scholar]
- 15.Rea S., Eisenhaber F., O'Carroll D., Strahl B. D., Sun Z. W., Schmid M., Opravil S., Mechtler K., Ponting C. P., Allis C. D., Jenuwein T. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 16.Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., Kouzarides T. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 17.Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 18.Tachibana M., Sugimoto K., Fukushima T., Shinkai Y. J. Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 19.Jenuwein T., Allis C. D. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 20.Heard E., Rougeulle C., Arnaud D., Avner P., Allis C. D., Spector D. L. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama J., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 22.Bernstein B. E., Humphrey E. L., Erlich R. L., Schneider R., Bouman P., Liu J. S., Kouzarides T., Schreiber S. L. Proc. Natl. Acad. Sci. USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santos-Rosa H., Schneider R., Bernstein B. E., Karabetsou N., Morillon A., Weise C., Schreiber S. L., Mellor J., Kouzarides T. Mol. Cell. 2003;12:1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- 24.Milne T. A., Briggs S. D., Brock H. W., Martin M. E., Gibbs D., Allis C. D., Hess J. L. Mol. Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura T., Mori T., Tada S., Krajewski W., Rozovskaia T., Wassell R., Dubois G., Mazo A., Croce C. M., Canaani E. Mol. Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 26.Dou Y., Milne T. A., Tackett A. J., Smith E. R., Fukuda A., Wysocka J., Allis C. D., Chait B. T., Hess J. L., Roeder R. G. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 27.Wysocka J., Swigut T., Milne T. A., Dou Y., Zhang X., Burlingame A. L., Roeder R. G., Brivanlou A. H., Allis C. D. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 28.Yagi H., Deguchi K., Aono A., Tani Y., Kishimoto T., Komori T. Blood. 1998;92:108–117. [PubMed] [Google Scholar]
- 29.Ayton P., Sneddon S. F., Palmer D. B., Rosewell I. R., Owen M. J., Young B., Presley R., Subramanian V. Genesis. 2001;30:201–212. doi: 10.1002/gene.1066. [DOI] [PubMed] [Google Scholar]
- 30.Deschamps J., van Nes J. Development (Cambridge, U.K.) 2005;132:2931–2942. doi: 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- 31.Core N., Bel S., Gaunt S. J., Aurrand-Lions M., Pearce J., Fisher A., Djabali M. Development (Cambridge, U.K.) 1997;124:721–729. doi: 10.1242/dev.124.3.721. [DOI] [PubMed] [Google Scholar]
- 32.Yeivin A., Razin A. EXS. 1993;64:523–568. doi: 10.1007/978-3-0348-9118-9_24. [DOI] [PubMed] [Google Scholar]
- 33.Razin A., Cedar H. Microbiol. Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bird A. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 35.Ohnemus S., Bobola N., Kanzler B., Mallo M. Mech. Dev. 2001;108:135–147. doi: 10.1016/s0925-4773(01)00502-0. [DOI] [PubMed] [Google Scholar]
- 36.Ernst P., Mabon M., Davidson A. J., Zon L. I., Korsmeyer S. J. Curr. Biol. 2004;14:2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 37.Zhu B., Mandal S. S., Pham A. D., Zheng Y., Erdjument-Bromage H., Batra S. K., Tempst P., Reinberg D. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krogan N. J., Dover J., Khorrami S., Greenblatt J. F., Schneider J., Johnston M., Shilatifard A. J. Biol. Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 39.Rastegar M., Kobrossy L., Kovacs E. N., Rambaldi I., Featherstone M. Mol. Cell. Biol. 2004;24:8090–8103. doi: 10.1128/MCB.24.18.8090-8103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamaru H., Selker E. U. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 41.Tamaru H., Zhang X., McMillen D., Singh P. B., Nakayama J., Grewal S. I., Allis C. D., Cheng X., Selker E. U. Nat. Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- 42.Malagnac F., Bartee L., Bender J. EMBO J. 2002;21:6842–6852. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuks F., Hurd P. J., Deplus R., Kouzarides T. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vire E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J. M., et al. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 45.Birke M., Schreiner S., Garcia-Cuellar M. P., Mahr K., Titgemeyer F., Slany R. K. Nucleic Acids Res. 2002;30:958–965. doi: 10.1093/nar/30.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuks F., Burgers W. A., Brehm A., Hughes-Davies L., Kouzarides T. Nat. Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 47.Bestor T. H. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 48.Conlon R. A., Rossant J. Development (Cambridge, U.K.) 1992;116:357–368. doi: 10.1242/dev.116.2.357. [DOI] [PubMed] [Google Scholar]
- 49.Bel-Vialar S., Core N., Terranova R., Goudot V., Boned A., Djabali M. Dev. Biol. 2000;224:238–249. doi: 10.1006/dbio.2000.9791. [DOI] [PubMed] [Google Scholar]
- 50.Clark S. J., Harrison J., Paul C. L., Frommer M. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.