Abstract
Human CYP27A1 is a mitochondrial cytochrome P450, which is principally found in the liver and plays important roles in the biological activation of vitamin D3 and in the biosynthesis of bile acids. We have applied a systematic analysis of hydrogen bonding patterns in 11 prokaryotic and mammalian CYP crystal structures to construct a homology-based model of CYP27A1. Docking of vitamin D3 structures into the active site of this model identified potential substrate contact residues in the F-helix, the β-3 sheet, and the β-5 sheet. Site-directed mutagenesis and expression in COS-1 cells confirmed that these positions affect enzymatic activity, in some cases shifting metabolism of 1α-hydroxyvitamin D3 to favor 25- or 27-hydroxylation. The results suggest that conserved hydrophobic residues in the β-5 hairpin help define the shape of the substrate binding cavity and that this structure interacts with Phe-248 in the F-helix. Mutations directed toward the β-3a strand suggested a possible heme-binding interaction centered on Asn-403 and a structural role for substrate contact residues Thr-402 and Ser-404.
INTRODUCTION
The use of vitamin D3 by the body is mediated through a series of mitochondrial cytochrome P450 enzyme reactions in which the parent vitamin is transformed sequentially by the hepatic 25-hydroxylase (CYP27A1) into 25-OH-D3 and the renal 1α-hydroxylase (CYP27B1) into the active hormone, 1α,25-(OH)2D3. Subsequent catabolism in vitamin D target tissues by the 1α,25-(OH)2D3-inducible 24-hydroxylase (CYP24A1) produces the biliary excretory product calcitroic acid (1). The active hormone and its precursors are transported between tissues complexed to vitamin D-binding protein. In the nucleus of cells in vitamin D target tissues, 1α,25-(OH)2D3 binds vitamin D receptor to transactivate, the expression of specific gene products affecting calcium homeostasis and the proliferation and differentiation of certain specialized cell types (2). Thus the structure of the vitamin D molecule helps define crucial interactions with at least five proteins, thereby influencing transport, activation, biological action, catabolism, pharmacokinetics, and the precise sequence of these events (3). However, research into the design of vitamin D prodrugs has shown that the activation sequence can be manipulated, and that a nonphysiological, 1α-hydroxylated vitamin D analog can be 25-hydroxylated and activated in a single cytochrome-mediated step (4).
The circulating level of 25-OH-D3 is a measure of vitamin D status (5), but the CYP enzymes regulating its formation are still not completely defined. The best studied 25-hydroxylase, CYP27A1, is well conserved across species and is a bifunctional enzyme that also expresses 27-hydroxylase activity toward cholesterol and related sterols in two separate bile acid biosynthetic pathways (6). Despite being the affected target in cerebrotendinous xanthomatosis (CTX) (7,8), a lipid storage disorder characterized by the accumulation of abnormal metabolites of bile acid precursors (9), the only indication of vitamin D deficiency associated with the loss of CYP27A1 is an increased risk of osteoporosis and bone fractures and in low 25-OH-D3 levels that fail to normalize with bile-acid replacement therapy (10). This suggests a redundant or compensatory 25-hydroxylase activity that is in agreement with one of the earliest studies of vitamin D 25-hydroxylation, which inferred the existence of a high specificity, low capacity (microsomal) enzyme and a low specificity, high capacity (mitochondrial) enzyme (11). Two of the candidates for the human microsomal 25-hydroxylase include CYP3A4, a hepatic drug metabolizing enzyme with broad substrate specificity (12,13), and CYP2R1, recently shown to have activity (14) and to be associated with a mild form of rickets (15). A similar microsomal 25-hydroxylase redundancy is seen in the rat enzymes CYP2J3 (16), CYP2R1, and CYP2C11 (17), the pig CYP2D25 (18), and the mouse CYP2R1 (19).
Since there are no crystal structures of mitochondrial cytochrome P450s, any model for CYP27A1 must be based upon one or more existing prokaryotic or microsomal CYP crystal structures. Although an important model of bovine mitochondrial CYP11A1 (side-chain cleavage enzyme, P450scc) (20) is available in the Protein Data Bank, there is disagreement concerning the existence of an ad hoc surface loop between the E- and F-helices hypothesized to mediate membrane binding in it since subsequent models of bovine CYP11A1 (21), human CYP11A1 (22), human CYP11B1 and CYP11B2 (23), and all cytochrome P450 crystal structures, including 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis (24,25) and the mammalian enzymes CYP2B4 (26), CYP2C5 (27,28), CYP2C8 (29), CYP2C9 (30), and CYP3A4 (31,32) show no evidence of this surface loop. Thus the quality of a previous model for CYP27A1 based on the original bovine model (7) remains in doubt.
It is generally recognized that the multisequence alignment is a basic prerequisite for homology modeling. In the case of the cytochrome P450 superfamily, obtaining a comprehensive alignment is difficult because sequence homologies between families are typically 10–20%. We have examined many P450 crystal structures, including a variety of mammalian cytochromes from related families, and used the observed secondary structure to refine the multisequence alignment process. This simplified the assignment of secondary structure in CYP27A1 to a degree that appears superior to theoretical prediction. On further examination of the crystal structures, we observed an extensive conservation of hydrogen bonding patterns and structural motifs acting to stabilize the folded structure. Consequently, this helped to align less conserved residues that merely contribute to local structure through backbone interactions. The cumulative effect of well-defined structural elements, extensive β-sheets and conserved helices enabled an unprecedented refinement of the multisequence alignment.
We have used this systematic analysis of P450 crystal structures to homology model CYP27A1 and to investigate substrate contact residues in the binding cavity. Using structure-guided, site-directed mutagenesis, we report the effects of mutations in the F-helix, β-3 sheet, and β-5 hairpin on the hydroxylation of the vitamin D prodrug 1α-OH-D3. We have also used the model to examine substrate recognition, the location of access channels in CYP27A1, and the structure-function relationships associated with the polymorphisms giving rise to CTX (CYP27A1) and VDDR-I (in CYP27B1) (1).
MATERIALS AND METHODS
Alignment and structural modeling
Protein sequences of selected cytochrome P450s and CYP crystallographic templates retrieved from GenBank were aligned using ClustalX (33) and the alignment edited manually using GeneDoc (34), according to observed secondary structure. Structural coordinates retrieved from the Brookhaven Protein Data Bank listed in Table 1 were aligned to the α-carbons of the heme-thiolate cysteine and I-helix of P450cam using the LSQKAB structural alignment utility (CCP4, Collaborative Computational Project, No. 4 Suite: Programs for Protein Crystallography, University of York, Heslington, UK). The aligned coordinates of P450bm3, P450cam, P450eryF, and P450terp were superimposed to give a cage-like consensus framework into which the human CYP27A1 sequence was threaded using Turbo Frodo (Architecture et Fonction des Macromolécules Biologiques-Centre National de la Recherche Scientifique, Marseille, France). This initial structure was annealed through a series of extensively constrained geometry minimizations using Gasteiger-Marsili atom charges in SYBYL v6.8 (Molecular Simulations, San Diego, CA). Constraints applied to maintain proper hydrogen-bond lengths in the α-helices, β-sheets, structural motifs, and hydrogen-bonding networks seen in 11 crystal structures were set to 2 Å and gradually relaxed during extensive geometry optimizations. Molecular dynamics simulations were performed using Insight II v2000 (Accelrys, San Diego, CA) and RMSD calculations were made using PyMOL (DeLano Scientific, San Carlos, CA). Ramachandran parameters were calculated with PROCHECK (European Bioinformatics Institute, Cambridge, UK). Crystal structures and models were visualized for detailed study using custom script files written for Chime (Molecular Simulations) and PyMOL.
TABLE 1.
CYP crystallographic templates
| Cytochrome P450 | PDB coordinates |
|---|---|
| P450bm3 | 2hpd.pdb, 1bu7.pdb, 1fah.pdb, 1jpz.pdb |
| P450cam | 1dz4.pdb, 1dz8.pdb, 1o76.pdb, 1pha.pdb |
| 1phb.pdb, 1phd.pdb, 4cpp.pdb, 6cpp.pdb | |
| 1noo.pdb, 1mpw.pdb, 1lwl.pdb,1k20.pdb | |
| lj51.pdb, 1iwi.pdb, 1gjm.pdb | |
| P450eryF | 1oxa.pdb, 1jio.pdb, 1jin.pdb, 1jip.pdb, |
| 1egy.pdb, 1eup.pdb | |
| P450terp | 1cpt.pdb |
| CYP119 | 1f4u.pdb, 1f4t.pdb |
| CYP51 | 1ea1.pdb, 1e9.pdb, 1x8v.pdb |
| CYP2B4 | 1po5.pdb (closed), 1su0.pdb |
| CYP2C5 | 1nr6.pdb, 1n6b.pdb |
| CYP2C8 | 1pq2.pdb |
| CYP2C9 | 1og2.pdb, 1og5.pdb |
| CYP3A4 | 1w0e.pdb, 1w0f.pdb, 1w0g.pdb, 1tqn.pdb |
Substrate structures including vitamin D3, 1α-hydroxyvitamin D3, cholesterol, and bile acid precursor 5β-cholestane-3α,7α,12α-triol were retrieved from the Cambridge Crystallographic Data Centre and modified and geometry optimized as required. The vitamin D structures were manually docked with c25 and c27 constrained at 4 Å above the heme A-ring nitrogen and cholesterol structures with c27 at 4 Å above the heme A-ring nitrogen according to the position of c5 in camphor bound to P450cam. For comparison, these structures were also docked in the binding cavity using AutoDock v3.05 (Scripps Research Institute, La Jolla, CA). Wild-type and mutant CYP27A1 structures used to assess the impact of each mutation on substrate binding were energy minimized under identical conditions (1000 iterations) from a single energy minimized structure.
Expression vectors and site-directed mutagenesis
Human CYP27A1 in pSG5 (4) was subcloned into the EcoRI-NotI site of the C-terminal tag vector pcDNA3.1 His-V5-“B” (Invitrogen, Carlsbad, CA) after introduction of these “in frame” restriction sites by amplification with Taq polymerase (Qiagen, Mississauga, Ontario, Canada) and oligonucleotide primers (Cortec DNA Service Laboratories, Kingston, Ontario, Canada) on a PerkinElmer (Wellesley, MA) Thermocycler. Restriction enzymes and other molecular biological reagents were obtained from Promega (Madison, WI) and Amersham-Pharmacia (Baie d'Urfe, Quebec, Canada). After confirmatory sequencing (Cortec DNA Service Laboratories), both vectors were used in site-directed mutagenesis reactions using a Chameleon Site-Directed Mutagenesis Kit (Stratagene, LaJolla, CA). The list of primers is as follows:
Phe-248-Met, 5′-CATAGAGTGAGTTCTGCATCATTAACCCGAT3′.
Thr-402-Phe, 5′-CCTTTTCTATGATCCGGGAGTTGAAGGGGACCACAGGGTAGAG-3′.
Asn-403-Thr, 5′-CTATGATCCGGGAGGTTGTGGGGACCA-3′.
Ser-404Ala, 5′-CCTTTTCTATGATCCGGGCGTTTGTGGGGACCACAGG-3′.
Ser-404-Thr, 5′-CCTTTTCTATGATCCGGGTGTTTGTGGGGACCACAGG-3′.
Ile-514-Phe, 5′-CTTATTGGGAACCAGGACAAAGCGGGCCA-CACTCTTCAACTC-3′.
Val515-Ile, 5′-CTTTCTTATTGGGAACCAGGATAATGCGGGCCACA-CTCTTCAA-3′.
Val-515-Leu, 5′-CTTTCTTATTGGGAACCAGGAGAATGCGGGCCACACT-CTTCAA-3′.
Leu-516-Val, 5′-CTTATTGGG-AACCACGACAATGCGGGCC-3′.
Site-directed mutations were verified by sequencing and the vectors purified using Wizard Maxi-prep resin (Promega). The Phe-248-Met, Thr-402-Phe, Asn-403-Thr, and Leu-516-Val mutations were transfected in the pSG5-CYP27A1 vector, whereas the Ser-404-Ala, Ser-404-Thr, Ile-514-Phe, Val-515-Ile, and Val-515-Leu mutations were transfected in the pcDNA3.1-CYP27A1 vector.
COS-1 transfection
African green monkey kidney COS-1 cells (ATCC CRL-1650) were grown on Nunc cell culture dishes using Dulbecco's modified Eagle's medium and 1× antibiotic-antimycotic (Gibco-BRL Canadian Life Technologies, Burlington, Ontario, Canada) supplemented with 10% CELLect Gold fetal bovine serum (ICN, Costa Mesa, CA) at 37°C. COS-1 cells were transiently cotransfected on 150 mm plates by the DEAE-dextran method (35) using a plasmid cocktail containing 10 μg each of CYP27A1 wild-type or mutant vector and vectors expressing bovine ferredoxin (pEAdx) (36), and ferredoxin reductase (pEAR) (37); 3 μg of a firefly luciferase expression plasmid (pRSVLuc) were included as a transfection control in each cocktail. Transfected cells were allowed to recover overnight in supplemented medium and subcultured onto triplicate 100 mm plates 1 day before CYP27A1 activity assay.
Vitamin D compounds
1α-OH-D3 and 1α,25-(OH)2D3 were gifts of Drs. Ernst Binderup and Anne-Marie Kissmeyer (Leo Pharmaceuticals, Ballerup, Denmark). Reference compound 1α,26-(OH)2D3 was synthesized and generously provided by Dr. Martin Calverley (Leo Pharmaceuticals).
CYP27A1 activity assay
Transfected COS-1 cells were cultured to late log phase, washed with 10 mL phosphate-buffered saline, and then incubated with 10 mL incubation medium/dish containing Dulbecco's modified Eagle's medium, 1% bovine serum albumin (Boehringer-Mannheim/Roche Diagnostics, Laval, Quebec, Canada), 1× antibiotics, 100 μM N,N-diphenylethylenediamine as an antioxidant (Sigma-Aldrich, Oakville, Ontario, Canada), 40 μg (10 μM) 1α-OH-D3, and 0.02% ethanol for 48 h. No-cell and dead-cell controls consisted of, respectively, an empty dish or dead cells (microwave treated) incubated as above. The incubation medium was extracted to isolate vitamin D compounds in the total lipid fraction using a modification of the method of Bligh and Dyer (38) in which chloroform was replaced by dichloromethane. The organic layer was evaporated to dryness under a stream of nitrogen, redissolved in hexane/isopropanol/methanol (91:7:2 v/v/v) and subjected to separation by HPLC (39).
High performance liquid chromatography
Straight-phase HPLC (40,41) was performed using Millennium32, an Alliance 2690 Separations Module, and a model 990 photodiode array detector (Waters Associates, Milford, MA). Vitamin D metabolites were separated on Zorbax-SIL (3 μ, 6.2 × 80 mm) columns using hexane/isopropanol/methanol 91/7/2 at a flow rate of 1 mL/min. and identified by the characteristic ultraviolet absorption spectrum of the triene chromophore (λmax = 265 nm). Retention times of metabolites varied on the different columns used. Metabolite peak areas were normalized to an HPLC internal standard (2 μg 25-OH-D3) and to the cell transfection efficiency, the latter determined from luciferase activity measurements acquired using cell culture lysis buffer, single luciferase assay reagents (Promega), and a Microlumat LB96V microplate luminometer (EG&G Berthold, Bad Wildbad, Germany).
RESULTS
Aligning the sequences
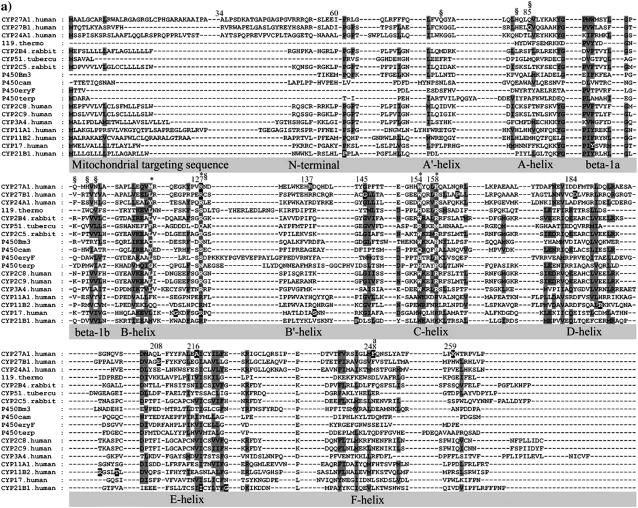
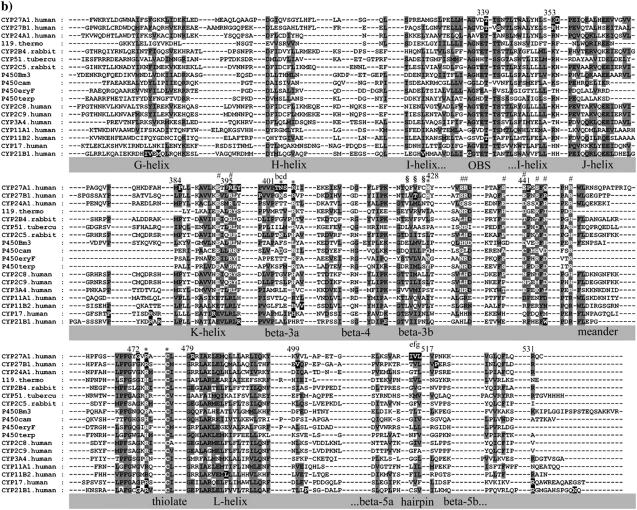
The observed α-helices, β-sheet, and loop secondary structure in eleven CYP crystallographic templates was used to manually parse the sequences in the alignment shown in Fig. 1. Gaps were introduced into the alignment to highlight these secondary structures. This revealed many functionally equivalent residues and conserved backbone interactions within secondary structures and tertiary structural motifs. Further refinements were made by systematically cataloging conserved hydrogen bonding patterns in the templates as shown in Figs. 2 and 3. As described below, there was extensive conservation of β-sheet structure and hydrogen bonding networks across all templates. The refined alignment was used to assign secondary structure in seven uncharacterized CYPs, including CYP27A1, using size of structural element, sequence homology, and side-chain polarity.
FIGURE 1.
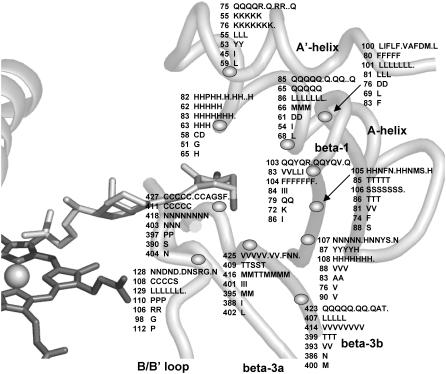
Multisequence alignment of CYP27A1, CYP27B1, CYP24A1, CYP11A1, CYP11B2, CYP17, and CYP21B1. Primary sequences were parsed and secondary structure predictions assigned using the observed secondary structure and conserved hydrogen bonding patterns seen in eleven CYP crystallographic templates. Shaded positions with dark text denote similar residues conserved in at least 9 of 18 aligned sequences. Shaded positions with white text and marked with “*” and “#” denote heme-binding and ERR-triad residues, respectively. Residues in the β-sheets structurally positioned toward substrate recognition in a pw2a access channel are denoted by “§”. Residues mutated in this study are indicated with white text on black and identified by lower case letters: a, Phe-248-Met; b, Thr-402-Phe; c, Asn-403-Thr; d, Ser-404-Ala, Ser-404-Thr; e, Ile-514-Phe; f, Val-515-Ile, Val-515-Leu; and g, Leu-516-Val. Sites of missense mutations causing cerebrotendinous xanthomatosis (CYP27A1), vitamin D-dependency rickets (CYP27B1), and autosomal deficiencies in CYP11B1, CYP17, and CYP21B1 are also indicated with white text on black shading. Gaps within secondary structures were added to emphasize structurally important landmarks.
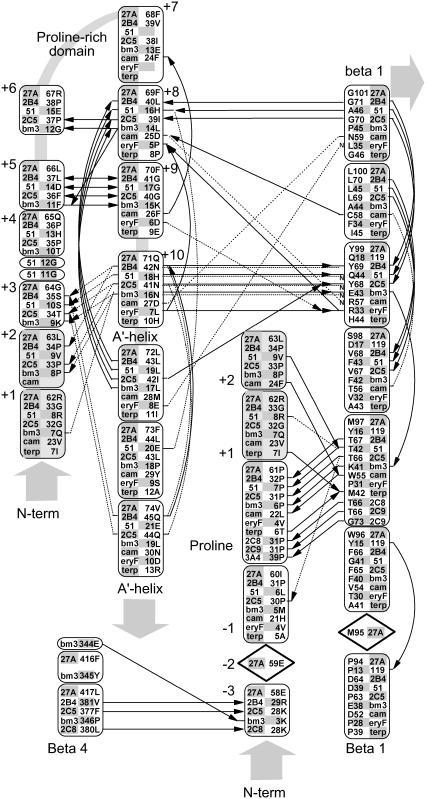
FIGURE 2.
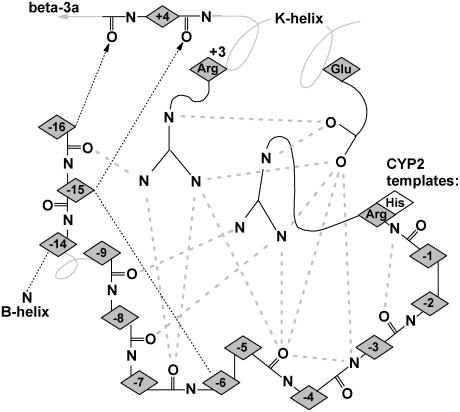
Hydrogen bonding patterns in the N-terminal domain of selected CYP crystallographic templates. Although the structure of the N-terminal domain is highly variable, a conserved hydrogen bond is seen in most templates between a conserved proline amide carbonyl and the amide nitrogen of an aligned β-1 residue. This proline is part of a PGP-motif or larger proline-rich domain found in many P450s but seen as Pro-61, Arg-62, and Leu-63 in Cyp27A1. This proline provides a useful landmark for alignment and modeling and helps position a highly structured hairpin near the β-1 sheet. Relative to this proline landmark, a hydrogen bond is seen from the −3proline amide carbonyl to the β-4 sheet in 4/11 templates. Other hydrogen bonds stabilize this hairpin through +5proline to +9proline double backbone interaction and a +8proline amide carbonyl to +11proline amide nitrogen. Additional hydrogen bonds connect the +10proline backbone and side chain (usually asparagine) to the +2proline and +3proline backbone and the amide nitrogen at a conserved position in the β-1 sheet position. The aligned +1proline and +2proline positions were repeated in the figure for clarity. Notes: i), hydrogen bonding patterns in CYP2C8, CYP2C9, and CYP3A4 are similar to CYP2B4 and CYP2C5; and ii), solid lines indicate backbone hydrogen bonds between amide nitrogen and amide carbonyl (indicated by an arrowhead). Double arrows indicate the presence of two backbone hydrogen bonds between the indicated residues. Dashed lines indicate hydrogen bonds involving side chains, and those interacting with amide nitrogen are marked with a small font “N”. Diamond placeholders indicate positions of aligned residues not involved in structural hydrogen bonding interactions.
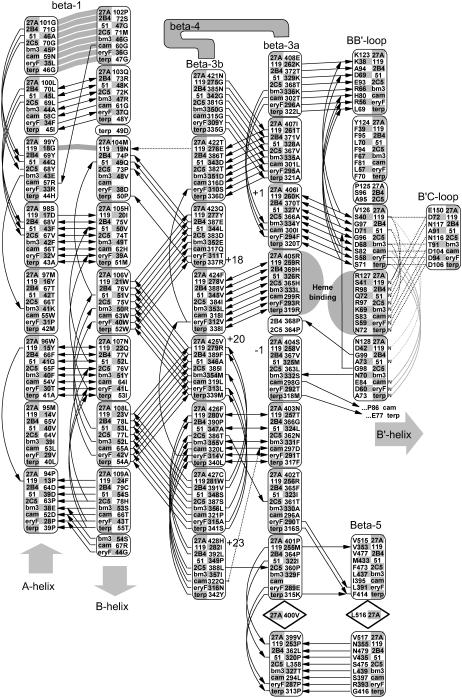
FIGURE 3.
Conserved backbone hydrogen bonding patterns in the extended β-sheet of eight selected crystal structures. As shown, there are 4–6 conserved hydrogen bonds between adjacent strands of the β-1 andβ-3 sheets. The shortened β-1 structure seen in the thermophillic archaeon CYP119 crystal structure retains the alignment of four out of five hydrogen bonds despite a four-residue truncation. Numbered positions denote distance from the heme-binding β-3 arginine or histidine-proline pair. The heme-binding residue in the β-3a strand hydrogen bonds to the heme A-ring propionate in all structures except P450Bm3, where the interaction is mediated through structural waters to the amide nitrogen and carbonyl of the adjacent residue, Ser-332. The symbols used are noted in Fig. 2.
Modeling the N-terminus region
CYP27A1 is expressed with a 33-residue amino terminal mitochondrial targeting sequence containing seven separated basic residues. Cleaved during mitochondrial import, the resulting mature protein begins at residue 34 with a hydrophilic, arginine-rich patch (Arg-51, Arg-52, Arg-53, Gln-54, Arg-55) preceding a conserved proline (Pro-61). Unfortunately, since none of the templates contain an analogous structure for this sequence, we extrapolated the N-terminal region back to Arg-53 to find that the arginine-rich patch could be within hydrogen bonding distance of polar residues between the B-B′ loop (e.g., Asn-128) and β-1 (e.g., Gln-103) on β-3b (e.g., Gln-423) and the F-G loop, suggesting possible influence on substrate access. The position of Pro-61 is predicted in 7 out of 11 (7/11) templates as the first proline in a PGP motif of a larger proline-rich sequence required for proper folding and heme incorporation in CYP27A1 and CYP11A1 (40) and in rat CYP2C11 where it may have been inadvertently disrupted experimentally due to a sequence alignment error (41). It is also the site of two P450 deficiency polymorphisms (P42S in CYP11B1 and P30L in CYP21B1). As shown in Fig. 2, the proline amide carbonyl forms a conserved hydrogen bond to the same β-1 sheet amide nitrogen position in 7/11 templates or an equivalent interaction to the same β-1 residue position in 2/4 of the remaining templates. This helps orient a hairpin structure between the PGP motif and the A′-helix region in 7/11 templates. Postulated to form a folding nucleus (42), the PGP motif and surrounding structure may also influence active site structure and a substrate access channel through interactions with the F-G loop and the A′-helix region.
Shielding and contacting part of the substrate binding site, the sequence along the 14–15 amino acid length of the A-helix is, for a given CYP, well conserved across species. A conserved glutamine in the middle of the CYP27A1 and CYP27B1 A-helix corresponds to a VDDR-I mutation (Q65H in CYP27B1). The orientation of the A-helix depends on CYP-specific polar interactions with the β-3 and β-5 sheets and the presence of a tyrosine-glycine pair at the A-helix terminus. The β-1 sheet begins at the end of the A-helix and is distinguished by a glycine at the β-1 hairpin and the pattern of hydrogen bond conservation seen in Fig. 3. Although conserved in the smaller β-1 sheet of the thermophile CYP119, what is especially remarkable about this pattern is how it integrates seamlessly through the β-3 sheet and B-B′ loop to two major heme-binding residues in all templates. In fact, this extended β-sheet encompasses conserved connections through the β-5 sheet to the D-E loop and across one side of the active site through the B-B′ loop to the B′-C loop.
Incorporating the heme into a structural network
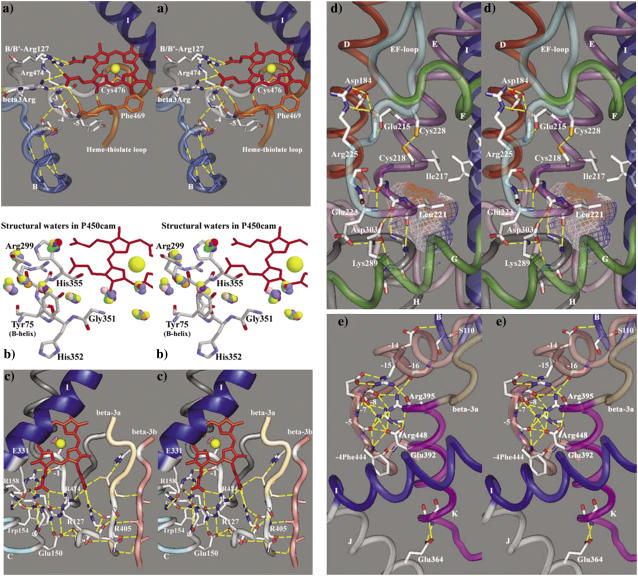
The heme-binding region is a rigid network of 20 or more hydrogen bonds and structural waters seen in Fig. 4, a–c, to connect the heme A- and D-ring propionates to the B-/B′ loop, B′-C loop, C-helix, β-3 sheet, and heme-thiolate loop. First described in P450Bm3, P450cam, and P450terp (43), the heme-binding region is remarkably conserved in all templates with functionally equivalent residues occupying an exclusive space and making an equivalent contribution. Careful maintenance of these heme-binding interactions during energy minimization stabilizes important substrate contact residues in adjacent structures.
FIGURE 4.
Stereo images of hydrogen bonding patterns in CYP27A1. Panel a shows heme-binding interactions with the heme propionates from residues in the heme-thiolate loop, β-3a strand, and B-B′ loop. The heme-thiolate loop is stabilized through a conserved hydrogen bond between the B-helix and the −3thiolate amide nitrogen. The hydrogen bond seen between the −6thiolate amide carbonyl and the A-ring propionate should be interpreted as an interaction between the −5thiolate amide nitrogen and the A-ring propionate, which is mediated by a structural water. The conservation of structural waters in the heme-binding region is illustrated in panel b, where selected crystallographic waters from 12 P450cam structures are colored differently and superimposed to reveal the conserved interactions with the −5thiolate amide nitrogen (His-352) and other stabilizing interactions with the heme-binding region. Panel c further illustrates how residues in the B-B′ loop, B′-C loop, C-and I-helices, β-3 sheet, and the heme-thiolate loop contribute to the hydrogen bonding in the heme-binding region in CYP27A1. Panel d shows the hydrogen bonding at the junction between the E- and H-helices that is conserved in 10/11 templates and mediated through the interaction of a basic G-helix residue with the conserved aspartate (Asp-303) at the beginning of the H-helix. The structural contribution of the Asp-84, Glu-215, and Arg-225 interaction to the stability of the E-F loop in CYP27A1 resembles similar interactions seen in the CYP2 templates. This panel also illustrates the position of a putative disulfide bond and a cavity analogous to a xenon-binding pocket seen in P450cam (49). Panel e shows the conserved hydrogen binding network surrounding the ERR-triad and a well conserved structural link to the start of the B-helix. Also shown is a well-conserved link between the J-helix Glu-354 and the K-helix.
Situated on the proximal side of the heme, the B-helix contains polar residues directed toward the protein surface and cofactor binding and hydrophobic residues directed inward toward the β-3 sheet. The B-helix flanks the heme-thiolate loop (containing the conserved cysteine thiol) and structurally supports it through a hydrogen bond between the penultimate B-helix residue carbonyl and the −3thiolate amide nitrogen (Fig. 4 a). In some of the templates, this hydrogen bonding appears to be mediated by structural waters, as shown in Fig. 4 b for 12 superimposed P450cam structures. Disruption of this interaction in CYP2R1 by an inactivating Leu-99-Pro polymorphism abolished all 25-hydroxylase activity toward vitamin D3 and left a small residual activity toward vitamin D2 (15). The mutation of the analogous residue in CYP51 (F105L) arose clinically in five Candida albicans isolates as fluconazole resistance (44). That structural integrity in this region is important is further illustrated by a CAH polymorphism (G424S in CYP21B1) at the −4thiolate position.
The heme iron is axially coordinated by the conserved cysteine thiol on the proximal side of the heme. This thiolate bond is stabilized by the surrounding residues, with the sulfur atom being nearly equidistant from the +1thiolate, +2thiolate, and +3thiolate amide nitrogens, and the tight turn at +2thiolate into the L-helix probably accounts for the near absolute conservation of glycine at this position. As shown in Fig. 4 a, the heme-thiolate loop is stabilized by four conserved hydrogen bonds: −1thiolate amide nitrogen to −4thiolate amide carbonyl; −6thiolate amide nitrogen to −9thiolate amide carbonyl; cysteine amide carbonyl to +3thiolate amide nitrogen; and cysteine amide nitrogen to −7thiolate amide carbonyl, the latter being a highly conserved phenylalanine influencing heme reduction potential (45). The heme-thiolate loop is further constrained by hydrogen bonds between a C-helix arginine (Arg-158 in CYP27A1) and the −1thiolate amide carbonyl in 9/11 templates (Fig. 4 c). This C-helix arginine also hydrogen bonds with the D-ring propionate, but the main C-helix to D-ring propionate link involves tryptophan one turn earlier (Trp-154 in CYP27A1) in the mitochondrial CYPs and 6/11 templates. In the remaining five templates, an equivalent polar side chain (arginine, glutamine, or histidine) is hydrogen bonded to the D-ring propionate, and in three of these it is also hydrogen bonded to −3thiolate amide carbonyl.
Another hydrogen bond to the D-ring propionate is made by the −2thiolate arginine (or histidine in CYP119, CYP51, P450cam, P450eryF, and P450terp) and mutation of this side-chain disrupted heme incorporation and enzyme activity in CTX (R474Q, R474W), VDDR-I (R453C), and P450 deficiency polymorphisms (R448H in CYP11B1, R440H in CYP17A1, and R426H in CYP21B1). The −2thiolate arginine also hydrogen bonds to amide carbonyls of key substrate contact residues in the B′-C loop and to the side chain at the start of the C-helix (Glu-150 in CYP27A1; Fig. 4 c). The latter is the site of a P450 deficiency polymorphism (N133H in CYP11B1) and the aspartate, glutamate, or asparagine side chain found there is bonded to the backbone of the B-B′ loop at precisely the location of a conserved arginine (asparagine, lysine, or serine in the prokaryotic templates). This B-B′ loop arginine hydrogen bonds to the A- and D-ring propionates, a fact that was not fully appreciated until CYP2C5 and other eukaryotic structures became available. B-B′ loop arginine is the site of CTX (R127Q and R127W), VDDR-I (R107H), and CAH (R96W in CYP17A1) polymorphisms. In all other templates, the hydrogen bonding of the aligned residue (asparagine, lysine, or serine) to the A-ring propionate is mediated through the nearby β-3 “heme-binding” arginine. The B-B′ loop is connected to the +1β3Arg by two β-sheet bonds in all templates. Polymorphisms affecting β-3 arginine cause CTX (R405Q, R405W), VDDR-I (R389C, R389G, R389H), and P450 deficiency (R384P in CYP11B2) because they disrupt a major structural link (usually a bidentate salt bridge) to the A-ring propionate. Conserved in the mitochondrial, bacterial (except P450bm3), and CYP3A4 enzymes, β-3 arginine is replaced by proline + histidine in the CYP2 templates. In many microsomal CYPs, this histidine binds heme by bridging the A-ring propionate to the B-B′ loop. In all enzymes with β-3 arginine though, the side chain is hydrogen bonded to both −1β3Arg amide carbonyl and via structural water to the penultimate B-helix carbonyl.
Other heme-binding interactions in the network are directed toward the A-ring propionate. Hydrogen bonds from the −1β3Arg amide nitrogen are seen in CYP51 (Met-325 mediated by Tyr-76), P450bm3 (Ser-332 via water), P450eryF (Thr-292 via water), and CYP3A4 (Glu-374 via water), through the −1β3Arg amide carbonyl in P450bm3 (Ser-332 via water) and CYP2 templates (proline amide carbonyl mediated through B-B′ loop arginine). In the other templates, the −2β3Arg side chain hydrogen bonds to the A-ring propionate, and this interaction is further stabilized by a side chain in the second β-3 strand. In CYP2C5, this corresponds to Asn-362 and +21β3Arg side chain (Thr-386), in P450cam as Asp-297 and +23β3Arg (Gln-322), and in P450eryF +23β3Arg (Asn-316). This −2β3Arg and +23β3Arg side-chain interaction with the A-ring propionate is especially relevant since the corresponding residues, Asn-403 and His-428 in CYP27A1, are completely conserved across species. In P450terp, the +23β3Arg side chain (Tyr-342) hydrogen bonds to the A-ring propionate directly. In all templates, another hydrogen bond to the A-ring propionate is made from the heme-thiolate loop by −5thiolate amide nitrogen. This is mediated by a structural water in all templates except P450terp (which bonds directly) and the CYP2 templates (mediated by the −6thiolate serine side chain).
The rigid structure of the heme-binding region is continuous with the B′-C loop in all templates (except CYP51, which is atypical), and this helps to position a critical substrate contact residue (Phe-147 in CYP27A1) above the D-ring propionate. Irrespective of B′-helix variability, the B′-C loop assumes a well-defined structure in the vicinity of a conserved glycine in 6/11 templates and at similarly aligned residues in other CYPs. The structural importance of the B′-C loop glycine in orienting the substrate contact residue is revealed in the existence of CTX (G145E) and VDDR-I (G125E) polymorphisms and in a study of inactivating site-directed mutants: G145A and G125A/V, respectively (46). The B′-C loop is internally stabilized through hydrogen bonding of the glycine amide carbonyl to +4B′Cglycine amide nitrogen in P450bm3, CYP2B4 and CYP3A4, to +4B′Cglycine side chain in the CYP2C templates, and to +3B′Cglycine amide nitrogens in 4/6 prokaryotic templates. In the same prokaryotic templates, stability is further enhanced by a +1B′Cglycine amide carbonyl to +4B′Cglycine amide nitrogen interaction. Spatially, the B′-C loop is oriented through conserved hydrogen bonds with adjacent structures. This includes B-B′ loop arginine (or equivalent) to +2B′Cglycine amide carbonyl (9/11 templates), −2thiolate arginine (or histidine) to +1B′Cglycine and +4B′Cglycine (10/11 templates), and an I-helix aspartate in CYP2C templates hydrogen bonded to +1B′Cglycine and +2B′Cglycine amide nitrogens. The latter I-helix interaction was deduced in CYP2D6 (47), and a functionally equivalent link between the I-helix and B′-C loop is seen in CYP3A4 and at least 3/6 prokaryotic templates. In fact, the alignment of a conserved glutamate at this position (Glu-331 in CYP27A1) in the mitochondrial vitamin D hydroxylases and aspartate or glutamate in many other mammalian CYPs (e.g., CYP2C11, CYP2J3, CYP2R1, CYP11, CYP17, and CYP21) further emphasizes the importance of a structural link between the I-helix and the B′-C loop.
Building the EH-gap motif, E-F loop, and a putative disulfide
Adjacent to the C-D loop is a highly structured region encompassing the end of the E-helix and the start of the H-helix. As shown in Fig. 4 d, two hydrogen bonds bridge the gap to join aligned residues and create the effect of a continuous helix in 10/11 templates. Centered on the interaction of a conserved aspartate at the start of the H-helix with a basic side chain four residues from the end of the G-helix, this EH-gap motif established these two residues as important landmarks in the alignment and positions the end of the G-helix. The EH-gap also contributed to the presence of a small cavity in the model similar to one recognized as a xenon binding site in P450cam (48,49).
In the CYP2 templates, where the E-F loop is long enough to parallel the D- and E-helices, hydrogen bonding residues near the middle of each helix interact with the second residue in the E-F loop (usually arginine) to further stabilize the region. Fig. 4 d shows this interaction in CYP27A1 with Asp-164 (D-helix), Arg-225 (E-F loop), and Glu-215 (E-helix). These residues are very well conserved in the vitamin D hydroxylases where the aspartate is the site of a VDDR-I mutation (D164N) and the glutamate is adjacent to a CTX mutation (A216P). Although the latter may be splicing mutant (50), it is also adjacent to Ile-217, which aligns with the most common mutation site in CYP21B1-related CAH deficiency (I172N) (51). The E-helix backbone near this Ile-217 is part of a small hydrogen bonded network in all templates that joins the I-helix oxygen binding site to a conserved L-helix glutamate (asparagine or glutamine) through a structurally conserved water molecule. Other structurally important landmarks nearby include the aspartate at the beginning of the E-helix (8/11 templates) and the conserved hydrogen bonds between the β-5 sheet and D-E loop.
The establishment of hydrogen bonding interactions to anchor the E-F loop in the model of CYP27A1 exposed a possible disulfide bond with the E-helix involving Cys-218 and Cys-228 (Fig. 4 d). These cysteines are conserved in CYP27A1 from human, dog, pig, rat, rabbit, baboon, and Xenopus, but the latter cysteine has been lost in bovine, chicken, mouse, and fugu. In addition to CYP27A1, there may be two disulfides in CYP27B1 (human) and three in CYP24A1 (human, pig, and rat). The possibility of native disulfide bonds in cytochrome CYPs has never been addressed before despite multiple possibilities associated with the D- and E-helices in CYP2C5, CYP2C8, and CYP2C9. Interestingly, one introduced into P450cam by site-directed mutagenesis appears to have connected the B′- and G-helices to affect substrate access and egress channels (49).
Positioning the F-helix
The alignment of the F-helix benefited significantly from detailed examination of the templates. Although the F-helix begins with an aspartate in 8/11 templates, neither it nor an apparently conserved phenylalanine in 9/11 templates was spatially conserved outside of P450bm3, CYP51, and the CYP2 templates. Thus the alignment of the F-helix for CYP3A4 and the prokaryotic templates in Fig. 1 is based on a spatially conserved α-carbon corresponding to CYP2 template phenylalanines. On the basis that the phenylalanine is conserved across species in the mitochondrial CYPs, the alignment of Phe-240 in CYP27A1 was used to model the height and spin orientation of the F-helix.
Linking the J-, K-, and L-helices
The alignment and modeling of the I- and J-helices was essentially unremarkable. The abrupt turn of the I-helix into the J-helix, however, is punctuated by sites of CTX (D354G) and VDDR-I (R335P) mutations. A second VDDR-I locus in the J-helix (L343F) is directed toward the K-helix. The structural disruption mediated by these mutations became apparent only after a conserved glutamate side chain (aspartate in P450eryF and P450terp) was found to be hydrogen bonded with conserved amide nitrogen(s) near the start of the K-helix in 9/11 templates. Disruption in CYP27A1 of this J- to K-helix link (involving the J-helix Glu-364 side chain and the K-helix Leu-385 and Leu-386 amide nitrogens), shown in Fig. 4 e, by an adjacent CTX mutation (P384L) probably destabilizes the J- and K-helices and J-K loop.
Two of four CAH mutations (P342T, R347H in CYP17A1 and R339H, R341W in CYP21B1) seen in the J-K loop correspond to a residue involved in a four-part hydrogen bonding network connecting the K- and L-helices in the CYP2 templates (three-part in CYP2C9). Three side chains of this four-part network in CYP2B4, CYP2C5, and CYP2C8 align with essential cationic charges in CYP17A1 necessary for lyase activity (52). Three side chains in this four-part network also align with residues that appear to provide a functionally equivalent four-part interaction in the vitamin D hydroxylases (Lys-387, Lys-391, Glu-483, and Gln-487). Overall, functionally equivalent KL-motifs are seen in 10/11 templates and appear to be very well conserved in the mitochondrial vitamin D hydroxylases and may stabilize a P450cam-type xenon binding pocket above it (49).
Building the ERR-triad
Integral to the interactions of the K-helix with surrounding secondary structures is its participation in the ERR-triad. Continuous with the β-3a strand, the K-helix also anchors the ERR-triad to the heme-region, the substrate-binding site, and the extended β-sheet. At the center of the ERR-triad, the conserved glutamate (Glu-392) and arginine (Arg-395) appearing as a ExxR motif in the K-helix (11/11 templates) are hydrogen bonded to an arginine (Arg-448) in the meander region of 7/11 templates or histidine in CYP2 templates. Originally described as “a structureless wandering of ∼20 amino acids” (43), the meander region contains some of the 16 residues preceding the arginine (Arg-448) that enveloped the ERR-triad in a highly coordinated hydrogen bonding network first described in P450Bm3, P450cam, and P450terp (43) and viewed in the CYP27A1 model shown in Fig. 4 e and depicted in Fig. 5. A pervasive structural element seen in all P450s, the ERR-triad may act as a folding motif, stabilize heme-binding, and play a role in redox-partner binding (43). It also anchored the start of the B-helix from a conserved aspartate numbered −14 from the meander arginine in all templates except in CYP51 where the aspartate is at position −11.
FIGURE 5.
Schematic view of the hydrogen bonding network in the ERR-triad. Shaded dashed lines indicate conserved hydrogen bonds between the triad residues and the surrounding backbone. Numbering in the K-helix is relative to the conserved glutamate, and numbering in the structure surrounding the triad is relative to the conserved arginine (histidine in CYP2 templates) near the start of the meander region. The thin dashed lines indicate additional side-chain interactions that stabilize the loop/helix structure at the end of the β-3b strand and the link to the B-helix.
Contribution of Phe-248 to the binding cavity
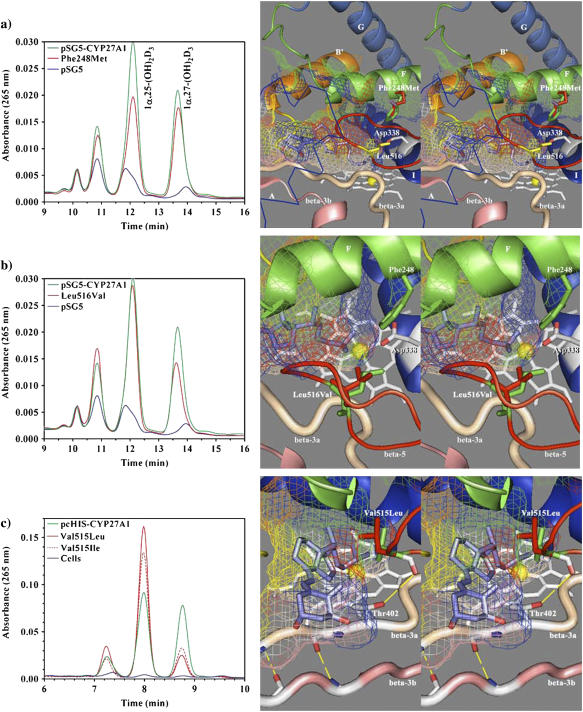
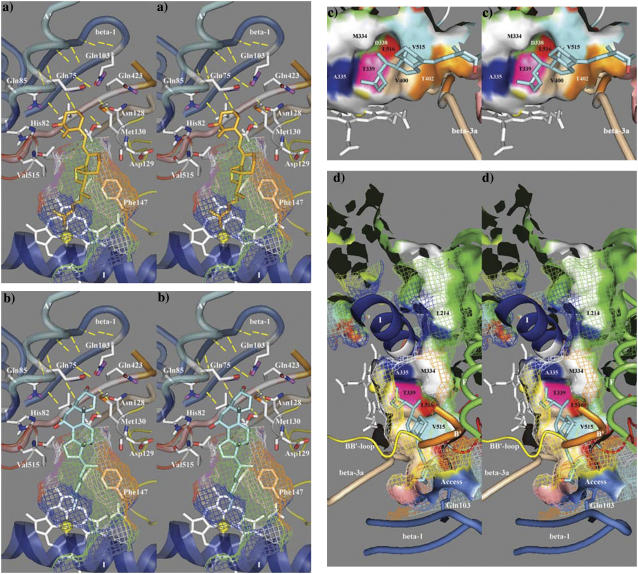
The F-helix forms the top of the binding cavity and contains a number of residues that are conserved in a subfamily-specific fashion. Phe-248 was selected for mutagenesis since it is conserved across species in all mitochondrial CYPs except CYP24A1 (conserved there as methionine) and was seen in the model to contribute a hydrophobic surface to the binding cavity. Although direct contact with substrate is possible, Phe-248 is positioned high above the heme at the back of the cavity where it could influence the shape of the β-5 hairpin or interact with residues near the conserved threonine in the I-helix. The mutation to methionine was chosen to mimic CYP24A1, and although Phe-248-Met did not confer any 24-hydroxylase-like activity, it did preferentially reduce 25-hydroxylase activity (Fig. 6 a). Table 2 shows how the 1α,25-(OH)2D3 to 1α,27-(OH)2D3 product ratio shifted from wild-type 49:36 to mutant 42:43. Interestingly, Pikuleva et al. also experimented with Phe-248 (their numbering; F215) and Ile-244 (I211) using lysine scanning and demonstrated that either mutant decreased turnover number and caused a minor accumulation of novel metabolites from the bile acid precursor substrate 5β-cholestane-3α,7α,12α-triol (53). Annalora et al. experimented with a nearby phenylalanine in rat CYP24A1 (Phe-249), which aligns with Ser-251 in CYP27A1, and concluded that mutation to alanine, threonine, or tyrosine disrupted the orientation and affinity of 24-oxidation intermediates in the multicatalytic oxidation sequence converting 1α,25-(OH)2D3 to calcitroic acid. It was postulated that phenylalanine contact with the vitamin D cis-triene was the determining factor in the accumulation of pathway intermediates (54). Overall, these studies suggest that the part of the F-helix around Phe-248 influences the orientation of bound substrate, possibly through interaction with Leu-516 as described below.
FIGURE 6.
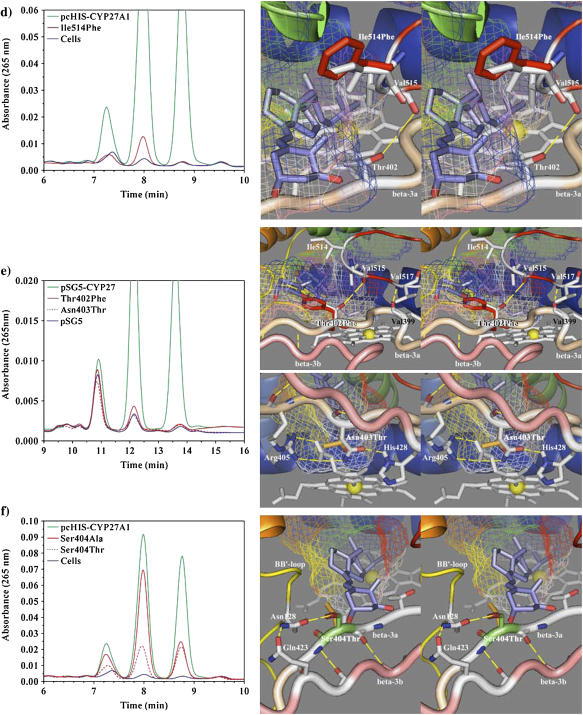
HPLC analysis of 1α-OH-D3 metabolites from COS-1 cells transiently transfected with wild-type and site-directed mutants of CYP27A1 and stereo views of the targeted mutations with docked substrate in the binding cavity. (a) Profile of metabolites from the Phe-248-Met mutant show a preferential loss in 25-hydroxylase activity with reduced 1α,25-(OH)2D3 formation relative to 1α,27-(OH)2D3. The stereo image illustrates the position of the wild-type phenylalanine (green) and mutant methionine (red) side chain in proximity to Leu-516 (yellow) in the β-5 sheet and Asp-338 (white) in the I-helix. Contact of Phe-248 with the binding cavity is minor, suggesting that the effect of the mutation may be mediated through contact with Leu-516 in the β-5 sheet or a gating effect at Asp-338. The 1α-OH-D3 substrate is shown inside the mesh framework of the binding cavity on the heme distal face. (b) The metabolism of 1α-OH-D3 by the Leu-516-Val mutant is opposite in effect to the Phe-248-Met mutation and shows a preferential reduction in 27-hydroxylase activity and a minor increase in the formation of a 24-hydroxylated product associated with reduced steric contact with substrate. (c) The enhanced 25-hydroxylase and reduced 27-hydroxylase activities associated with the Val-515-Leu (shown in the stereo view) and Val-515-Ile mutations in the β-5 hairpin appear to result from increased steric contact with the substrate D-ring. (d) The Ile-514-Phe mutation results in a considerable loss of enzyme activity consistent with an unfavorable steric conflict with the substrate A-ring, occlusion of the substrate access channel, or a distortion of the β-5 sheet through disruption of a structural link between Thr-402 hydroxyl and Val-515 amide carbonyl. (e) The loss of activity seen with the Thr-402-Phe mutation is consistent with steric conflict with the substrate A-ring, which would impede substrate access to the binding cavity and with disruption of the β-5 sheet through the loss of a structural hydrogen bond between Thr-402 amide nitrogen and/or hydroxyl with Val-515 amide carbonyl. The loss of activity associated with the Asn-403-Thr mutation is consistent with the loss of a heme-binding interaction to the A-ring propionate, which is stabilized by His-428 and conserved in CYP27A1 and CYP27B1 species homologs. (f) Metabolic profiles seen for the Ser-404-Ala and Ser-404-Thr mutations. Disruption of a hydrogen bond to Asn-128 in the Ser-404-Ala mutant is hypothesized to alter the orientation of the B-B′ loop and disrupt bound substrate orientations leading to better retention of 27-hydroxylation. The Ser-404-Thr mutation, shown in the stereo view, maintains the contact with Asn-128, but the threonine methyl group results in a loss of enzyme activity by sterically disrupting substrate access to the binding cavity.
TABLE 2.
Effect of mutations on the metabolism of 1α-OH-D3
| 1α,24-(OH)2D3 | 1α,25-(OH)2D3 | 1α,27-(OH)2D3 | Total product | Total activity | 24:25:27 hydroxylation ratio | |||
|---|---|---|---|---|---|---|---|---|
| (ng) | (ng) | (ng) | (ng) | % | % | % | % | |
| Vector | 36.9 ± 1.4 | 32.7 ± 4.2 | 13.1 ± 1.0 | 83 | ||||
| pSG27A1 | 110.9 ± 6.6 | 289.6 ± 25.3 | 202.3 ± 21.6 | 603 | 100 | 14 | 49 | 36 |
| Phe-248-Met | 99.5 ± 6.5 | 213.5 ± 20.5 | 199.2 ± 29.8 | 512 | 83 | 15 | 42 | 43 |
| Leu-516-Val | 136.1 ± 4.6 | 273.6 ± 36.9 | 137.9 ± 18.6 | 548 | 89 | 21 | 52 | 27 |
| Vector | 48.9 ± 1.6 | 14.1 ± 1.7 | 4.6 ± 0.2 | 68 | ||||
| pSG27A1 | 103.5 ± 2.9 | 298.6 ± 1.8 | 263.8 ± 2.1 | 666 | 100 | 9 | 48 | 43 |
| Thr-402-Phe | 50.4 ± 3.0 | 21.6 ± 2.1 | 7.4 ± 0.7 | 79 | 2 | 13 | 63 | 24 |
| Asn-403-Thr | 47.6 ± 2.6 | 13.2 ± 2.1 | 4.4 ± 1.8 | 65 | 0 | |||
| Vector | 17.5 ± 3.1 | 8.5 ± 0.8 | 4.9 ± 1.2 | 31 | ||||
| pcDNA-27 | 58.4 ± 12.7 | 264.7 ± 67.0 | 259.1 ± 65.0 | 582 | 100 | 7 | 46 | 46 |
| Ser-404-Ala | 57.9 ± 1.9 | 240.6 ± 10.2 | 81.8 ± 3.7 | 380 | 63 | 12 | 66 | 22 |
| Ser-404-Thr | 30.5 ± 0.3 | 74.6 ± 2.0 | 68.8 ± 2.0 | 174 | 26 | 9 | 46 | 45 |
| Ile-514-Phe | 17.2 ± 0.3 | 41.0 ± 1.4 | 4.5 ± 0.2 | 63 | 6 | 0 | 100 | 0 |
| Val-515-Ile | 86.1 ± 31.2 | 590.0 ± 89.3 | 123.4 ± 22.6 | 800 | 139 | 9 | 76 | 15 |
| Val-515-Leu | 118.4 ± 6.4 | 584.8 ± 27.2 | 87.0 ± 4.8 | 790 | 138 | 13 | 76 | 11 |
Substrate contacts in the β-5 hairpin
In the CYP27A1 model, the β-5 hairpin contributed a large and well-conserved hydrophobic surface to the binding cavity and contained three potential substrate contact residues of interest. The closest of these to the heme iron, Leu-516, was conserved across species in the CYP27 and CYP11 isoforms and in CYP24A1. A conservative mutation, Leu-516-Val, was selected to probe the effect of steric size on substrate contact, and HPLC analysis of the resulting 1α-OH-D3 metabolites shown in Fig. 6 b revealed that 27-hydroxylation was preferentially reduced. The hydroxylation product ratio shown in Table 2 was shifted from 14:49:36 (wild-type) to 21:52:27 (Leu-516-Val) with only an 11% loss of total activity. This was notably opposite to the effect of the F-helix Phe-248-Met mutation and suggests that these two residues act together to shape a substrate contact surface. The minor increase in 24-hydroxylation was also in agreement with an altered substrate orientation in the Leu-516-Val mutation. The residue preceding Leu-516 is less conserved and seen as valine in CYP27 isoforms and isoleucine in CYP24A1 and the CYP11 isoforms. Two conservative mutations at Val-515-Ile and Val-515-Leu were selected to probe the effect of a larger side chain at this position. These mutations dramatically shifted the products toward 25-hydroxlation at the expense of 27-hydroxylation (Fig. 6 c) and caused a 40% increase in total enzyme activity (Table 2). This indicated that the Val-515 side chain made a large contribution to the orientation of bound 1α-OH-D3, and the docked substrate shown in Fig. 6 c suggests that this occurs through contact near the substrate D-ring. The third residue in the β-5 hairpin is conserved as isoleucine in many CYP27A1 homologs, threonine in CYP27B1, glycine in CYP24A1, leucine in CYP11A1, and phenylalanine in CYP11B isoforms, and may represent a finely tuned determinant of substrate specificity. The mutation of this residue (Ile-514-Phe), completely shifted activity toward 25-hydroxylation (Fig. 6 d), but abolished 94% of the total activity. Western analysis of the Ile-514-Phe mutant revealed that it disrupts protein expression (data not shown), suggesting that it may destabilize correct folding of the β-5 hairpin and a functional enzyme. Coincidentally, a similar effect was seen in the Thr-402-Phe mutant described below.
Substrate contacts and heme-binding in the β-3 sheet
The β-3a strand connects the ERR-triad and β-5 hairpin to the heme-binding region. Along its length are structural, heme-binding, and potential substrate contact residues that are generally specific to a given CYP subfamily and conserved across species. In the CYP27 family, the β-3a strand starts with a PVVP motif in which the position of the first valine amide carbonyl hydrogen bonds to the amide nitrogen of an aligned β-5 position (Val-517) in all 11 templates. This backbone interaction helps position the side chain at the second valine position toward a substrate contact near the heme A-ring. In the CYP2 templates, the amide carbonyl at the second valine position also hydrogen bonds with the +3 amide nitrogen to structurally stabilize the heme-binding histidine at the +6 position. Following the PVVP motif in CYP27A1 are three residues, Thr-402, Asn-403, and Ser-404, which are as well conserved as Gly-Asn-Ser in CYP27B1 and Phe-Thr-Thr in CYP24A1. The first of these in CYP27A1, Thr-402, was seen in the model to make a potential hydrophobic contact through its methyl group with the vitamin D A-ring. Conserved in human, mouse, and rat, this interpretation is also consistent with valine in pig, bovine, and zebrafish. Additionally, computational docking of 1α-OH-D3 in the binding cavity suggested that the threonine hydroxyl interacts with the substrate 1α-hydroxyl and its methyl interacts with the substrate C- and D-rings. Interaction of a threonine hydroxyl in this vicinity with a substrate hydroxyl has also been implicated in substrate recognition in CYP27B1 (55). However, given the absolute conservation of phenylalanine in CYP24A1, the Thr-402-Phe mutation was chosen to see if a possible contact with the cis-triene or changes in the shape of the binding cavity would shift CYP27A1 activity toward 24-hydroxylation.
Similar to the effect of Ile-514-Phe, the Thr-402-Phe mutation abolished 98% of the activity toward 1α-OH-D3 (Table 2) and produced only a trace of 25-hydroxylated product (Fig. 6 e). As with the Ile-514-Phe mutant, Western analysis revealed that the Thr-402-Phe mutation also disrupts protein folding (data not shown). Given the proximity of Thr-402 and Ile-514 and their location in the model, the residual activity of these mutants may be attributed to effects on substrate orientation, forcing bound substrate deep into the cavity. The loss of detectable protein for these mutants suggests that either phenylalanine mutation may sterically disrupt a structural hydrogen bond between the β-5 and β3a sheets. A structural interaction here between the β-sheets is likely to be important in CYP27A1 since hydrogen bonding between these two loci is found in 8/11 templates. This is best seen from position 402 side chain (threonine) through a crystallographic water to position 515 amide carbonyl in CYP2C5, CYP2C9, and probably CYP2C8 and from position 402 side chain (serine) to position 514 amide carbonyl in P450terp. In other templates, this interaction involves the position 402 amide nitrogen to position 514 amide carbonyl in P450eryF, to position 515 amide carbonyl in P450cam and P450terp, and through a crystallographically conserved water to position 515 amide carbonyl in CYP51, CYP2C5, CYP2C9, and probably CYP2C8. In CYP119, an analogous hydrogen bond links position 401 amide nitrogen to position 515 amide carbonyl. Given that the β-5 hairpin is also secured by a hydrogen bond between the PVVP motif and the position 517 amide, the principal effect of hydrogen bonding at the Thr-402 position would be in the positioning of Ile-514 residue. However, the involvement of the side-chain hydroxyl (threonine) in this interaction is not crucial in P450eryF or possible in half of the CYP27A1 species homologs. Interestingly, the presence of threonine at position 514 in fish CYP27A1s and in the CYP27B1 homologs may provide the reciprocal hydrogen bond interaction to the β-3a strand.
All evidence points to the next β-3a residue, Asn-403, being an accessory heme-binding residue. Designated earlier as −2β3Arg, heme-binding with the A-ring propionate is clearly evident in CYP2C5 (Asn-362 stabilized by +21β3Arg Thr-386) and in P450cam (Asp-297 stabilized by +23β3Arg Gln-322). In CYP27A1, heme-binding by Asn-403 would be stabilized by +23β3Arg His-428, the latter being conserved across 15 species. Structurally, the −2β3Arg position is constrained in the β-sheet through two backbone hydrogen bonds (in 7/11 templates including CYP3A4) or a single backbone hydrogen bond within the β-3a strand in the CYP2 family (4/11 templates). In all templates except P450terp, this hydrogen bonding oriented the −2β3Arg side chain downward and negated any possible substrate contact. Although a serine or threonine at this position is only seen in 3/11 templates and only bound heme through a crystallographic water in P450eryF, the Asn-403-Thr mutation in CYP27A1 was selected to examine whether threonine (conserved in CYP24A1) could assume a heme-binding role. The resulting loss of activity, shown in Fig. 6 e, suggests that Asn-403-Thr cannot support a structurally viable heme-binding environment. Western analysis confirmed that Asn-403-Thr disrupts protein expression, and this is similar to the effect of a mutation at an adjacent heme-binding and CTX residue, Arg-405-Lys, which was shown spectrally to disrupt heme incorporation (46).
The third β-3a residue examined in this study was −1β3Arg Ser-404. From its proximity to the binding cavity and the heme A-ring, it is clear that Ser-404 is a potential substrate contact residue. Computational docking of 1α-OH-D3 in the binding cavity also suggested that the serine hydroxyl interacts with the substrate 3β-hydroxyl. Ser-404 also appears to be an integral structural component at the confluence of β-1, B-B′ loop, F-G loop, β-3a, and β-3b strands. This region is structurally constrained by extensive β-sheet structure and by heme-binding interactions through the β-3 arginine (Arg-405) and B-B′ loop arginine (Arg-127). Rooted in this well-defined structure, the B-B′ loop made additional contacts with the binding cavity and supported important determinants of substrate specificity and substrate access in the B′-helix, which are poorly represented in the crystallographic templates. Interactions of the B-B′ loop with the β-3a strand (in the vicinity of Ser-404) and β-3b strand occur in a number of templates. This is best seen in P450bm3 as a triad (−1β3Arg Ser-322, +1BB′Arg Asn-70, and +18β3Arg Glu-352), which is functionally conserved in CYP27A1 (across at least seven species) as Ser-404, Asn-128, and Gln-423. Interestingly, functionally similar interactions anchor the B-B′ loop in CYP3A4 (−1β3Arg Glu-374 to +1BB′Arg Arg-106), CYP2B4 (+2BB′Arg Lys-100 amide nitrogen to +18β3Arg Glu-387), and CYP2C5 (+2BB′Arg Ser-99 amide to +18β3Arg Asp-383). The hydrogen bonding pattern was more elaborate in P450cam (+2BB′Arg Cys-85, +4BB′Arg Phe-87 amide carbonyl, +18β3Arg Gln-317), and P450terp (+4BB′Arg Ser76 amide carbonyl, +1β3Arg Thr-320, +18β3Arg Arg-337).
Within this context, two site-directed mutations were engineered to explore the role of Ser-404 in CYP27A1. The first, Ser-404-Ala, was selected to structurally disrupt the hydrogen bond with the B-B′ loop without sterically interfering with substrate binding. As shown in Fig. 6 f, the Ser-404-Ala mutation preferentially suppressed the 27-hydroxylation of 1α-OH-D3 and reduced the total enzyme activity by 37%. This is consistent with steric effects of B-B′ loop on the bound substrate resulting from the loss a structural hydrogen bond at Ser-404. The second mutation, Ser-404-Thr, was selected to probe the steric effect of a methyl group with the substrate without disturbing the local structure of the cavity. Surprisingly, this reduced overall enzymatic activity by 64% (Fig. 6 f) without changing the product ratio (wild-type 7:46:46 versus mutant 9:46:45). This suggests that the unfavorable steric effect in Ser-404-Thr is exerted during substrate entry, probably with the substrate C- and D-rings, and not during the final orientation of bound substrate. These findings suggest that Ser-404-Thr retains the substrate recognition function and structurally stabilized the B-B′ loop and the bound substrate contact residues therein.
Substrate docking and molecular dynamics
The binding cavity was adequately proportioned to permit manual docking of vitamin D3, 1α-OH-D3 A-ring conformers, cholesterol, or the bile acid precursor 5β-cholestane-3α,7α,12α-triol with little energy minimization. This was confirmed by computational docking that placed 1α-OH-D3 in the binding cavity with c25 positioned 0.97 ± 0.37 Å from its average position at a distance of 3.79 ± 0.42 Å from the heme iron (9/10 structures). The substrate A-ring was especially invariant in 8/10 of the docked structures and not significantly different from the manually docked position, reflecting the fact that much of the substrate is a structurally rigid seco-steroid. Most interestingly, the computationally docked substrate sampled the conformational space above the heme by rotating the substrate side-chain bonds at c20-c22, c22-c23, c23-c24, and c24-c25. This allowed c24, c25, and c26/27 to sample metabolically relevant orientations near the hydroxylation site. The absence of the 1α-hydroxyl group in vitamin D substrates did not appear to affect binding orientation (data not shown), but a comparison of A-ring conformers suggested that the more planar β-chair conformer, displaying an axial 1α-hydroxyl, gave a better fit (data not shown). The computational docking of 1α-OH-D2 was more precise, reflecting the smaller conformational space afforded by the Δ22–23 in this substrate side chain. Nine docked structures positioned c24 in a physiologically relevant orientation 0.56 ± 0.31 Å from its average position at a distance of 4.37 ± 0.36 Å from the heme iron. This was especially exciting from a vitamin D standpoint since the 24-hydroxylation of vitamin D2 is a unique enzymatic property of the enzyme (4). Overall, we are confident that the manually docked substrate adequately exemplifies a metabolically relevant conformation in the active site, especially in the placement of the A-ring hydroxyls as molecular “handles” interacting with Thr-402, Ser-404, and other polar residues in the binding site.
We performed molecular dynamics simulations using Insight II on our docking model and observed structural convergence as early as 100 ps with an RMSD of 1.22 Å averaged over 479 alpha carbons. In light of the quick convergence, there is no need to continue the molecular dynamics calculations. We subsequently docked 1α-OH-D3 into this structure and subjected it to an additional molecular dynamics simulation; only after 50 ps, it converged completely and the RMSD decreased to 1.13 Å. Using PROCHECK, we found that 96.2% of residues are in allowed Ramachandran plots.
The inner part of the binding cavity lay close to the β-3a strand and was defined by residues in the oxygen binding site in the F- and I-helix, B-B′ and B′-C loops, and nonpolar residues in the β-5 hairpin. The outer part of the binding cavity and substrate access channel are defined by residues in the B-B′ loop, A′-, A-, B′- and F-helices, F-G loop, and the β-1 and β-5 hairpin. Proton acceptor residues there are presumed to help partition substrate into the active site and orient it during the reaction cycle through interactions with A-ring hydroxyl groups in vitamin D and 1α-OH-D3 and with the 3-, 7-, and 12-hydroxyl moieties of bile acid precursors. Additionally, a proton donor may be present to coordinate with the c3 carbonyl of cholestenone, a preferred substrate of CYP27A1 (6).
The outer part of the binding cavity near the β-sheets contains an abundance of asparagines, glutamines, and histidines, which suggests a structural network of hydrogen-bonded residues with a multifaceted capacity to recognize substrate. Quite intriguingly, the contact residues in the β-sheets and A-helix are located at discrete positions (shown in Fig. 1) and have identical side-chain orientations that are independently determined by cytochrome P450 structure. The cross species conservation of these discretely positioned residues in all known vitamin D hydroxylases is summarized in Fig. 7. What contributes to substrate specificity here is the identity of the residue at each position. What determines substrate contact is the size of the substrate, size of the residue, and the extent to which side chains are displaced by adjacent residues. What determine substrate recognition are proximity, side-chain polarity, and the presence of functional groups in the substrate. There are other substrate contact and substrate recognition residues in the B′- and F-helices and in the B-B′ and F-G loops and in the substrate access channel, but in CYP27A1, there are no proton acceptor residues in the F-G loop other than backbone carbonyls and a nonionized Lys-259 or Arg-262 side chain.
FIGURE 7.
Cross-species variation of selected residues predicted to orient toward substrate in CYP27A1 and vitamin D hydroxylases. Based on conserved structures and hydrogen bonding patterns in 11 crystallographic templates, discrete positions in the β-1 sheet, β-3b strand, and A′- and A-helices have side chains oriented into the substrate binding cavity near the mouth of the pw2a substrate access channel. In CYP27A1, a majority of these residues are polar, suggesting the presence of a structurally important hydrogen bonded network with a multifaceted capacity to recognize substrate. This may be especially true of His-82, which was predicted to simultaneously hydrogen bond with the backbone of β-5 and the 1α-hydroxyl moiety of 1α-OH-D3 and is conserved in CYPs 27A1, 27B1, 24A1, 2R1, and 2J3. Variation within species homologs suggests small structural differences in an otherwise functionally equivalent substrate contact region. The numbered residue pertains to the first species homolog in each line as CYP27A1 (human, pig, mouse, rabbit, rat, baboon, bovine, dog, chicken, Xenopus, fugu, zebrafish, rehesus monkey); CYP27B1 (human, pig, mouse, rat, fugu); CYP24A1 (human, pig, mouse, rat, guinea pig, chicken, fugu, dog); CYP2R1 (human, mouse, rat); CYP3A4 (human, pig); CYP2C11 (rat); and CYP2J3 (rat). Dotted gaps indicate incomplete sequence data for some species homologs.
Fig. 8 a shows the discretely conserved positions of selected proton acceptors within range of the A-ring hydroxyl groups on 1α-OH-D3 and Fig. 8 c shows the contribution of binding cavity residues near the substrate side chain. It is interesting to note that, for clarity, His-105, Asn-107, and Cys-427 are not shown, but the histidine and cysteine are also well placed to hydrogen bond with the 3β-hydroxyl and 1α-hydroxyl groups, respectively. One of the more interesting residues, Gln-85, is a VDDR-I locus (Q65H) and was conserved in the A-helix across species in CYP27A1 and CYP27B1. It has been implicated in substrate binding in CYP27B1 (55), but given the opposite orientations of substrate in these two enzymes, it seems unlikely that Gln-85 side-chain placement would be optimal for substrate recognition in both. Instead, Gln-85 may fulfill a structural function, such as participation in the structural network in the outer part of the binding cavity or blocking an access channel between the A′- and A-helices. Alternatively, the Q65H polymorphism in CYP27B1 may disrupt a structural hydrogen bond to another discretely positioned residue, Thr-409, which is also the site of a VDDR-I polymorphism (T409I). If Gln-85 does coordinate with the 1α-hydroxyl group on 1α-OH-D3, the model suggests that this function may be shared with Cys-427 and His-82. In fact, His-82 may be the more interesting residue because it is positioned to simultaneously form a structural hydrogen bond with β-5 Ile-514 amide carbonyl and contact the substrate 1α-hydroxyl group. This may explain why His-82 is conserved across species in CYP11 isoforms, rat CYP2J3, CYP2R1, and all mitochondrial vitamin D hydroxylases (except murine Cyp27a1 where it is a proline).
FIGURE 8.
Predicted substrate binding orientations of (a) 1α-OH-D3 and (b) 5β-cholestane-3α,7α,12α-triol in the binding cavity of CYP27A1. This figure shows potential substrate recognition residues (proton acceptors) capable of hydrogen bonding to the hydroxyl moieties of bound substrates in the vicinity of the substrate access channel. (c) Contribution of I-helix and β-sheet residues to the surface of the binding cavity with 1α-OH-D3 side chain docked over the heme A-ring. (d) The binding cavity of CYP27A1 showing a partially open pw2a-type substrate access channel and a putative pw3 egress channel between the F- and G-helices near Met-334.
The role of these discretely conserved proton acceptors in bile precursor recognition is illustrated in Fig. 8 b, where several were found to interact with the hydroxyl groups on 5β-cholestane-3α,7α,12α-triol. Given its docked orientation, residues such as Gln-423 or His-105 (not shown) are well positioned to interact with the c3-hydroxyl, His-82 with the c7-hydroxyl, and possibly a B-B′ loop backbone carbonyl (e.g., Asp-129) with the c12-hydroxyl. These same residues could also interact with the carbonyl of cholestenone.
Substrate access channel
Previously identified in P450cam (56), P450Bm3 (57), and P450terp (58) and modeled into aromatase (59), the substrate access channel connects the binding cavity to the protein surface. In all but a few crystal structures, this opening is hidden, even in the presence of bound substrate or inhibitor. Molecular dynamics simulations in P450Bm3, P450cam, and P450terp have examined the energetics of a number of molecular trajectories out of the binding cavity (60,61) to find concealed channels, and a recent review (49) discussed at least eight possible access routes or pathways: pw1, between the heme and I-helix (possible oxygen channel); pw2a, opening at F-G loop, B′-helix, β-1; pw2b, parallel to β-3a and opening between the B-B′ loop and β-1; pw2c, opening between the G- and I-helices and the B′-helix, B-C loop; pw2d opening between the A- and A′-helices; pw2e through the B-B′ and B-C′ loops; pw3, between the F- and G-helices or at the E-F loop; and a solvent channel between the E-, F-, and I-helices and β-5. Unfortunately, the unveiling of a substrate access channel along one of these pathways may be a transitory event coincident with substrate penetration or the conformational changes associated with reaction cycling and membrane association. Although the most likely access route in most CYPs is pw2a (49), it is not known whether different CYPs use different pathways, whether a single CYP uses different channels for different substrates, or whether separate pathways are used for substrate access and product egress.
The geometry of the pw2a substrate access channel found in the CYP27A1 docking model and shown in Fig. 8 d is not ideal and exhibited some pw2b-type character similar to that seen in CYP2C8 and P450Bm3. It was primarily occluded by A′-helix Gln-75, F-helix Phe-256, and B-B′ loop Met-130, possibly in part due to the uncertainties in modeling the F-G loop and the B′-helix. Although the structure of this channel does not affect how substrates are docked, it is desirable to objectively refine it to better understand substrate recognition during binding. Our model revealed a cluster of 10 aromatic residues above the heme that contributed a large hydrophobic surface to the binding cavity and substrate access channel. This cluster is surrounded by the F-G loop and included Trp-133 (B′-helix), Tyr-253, and Phe-256 (F-helix), Trp-260 and Trp-268 (F-G loop), Phe-267, Tyr-271, Trp-275 (G-helix), and possibly Phe-279 and His-136, and may interact with another cluster of aromatic residues in the A′-helix, the latter possibly analogous to hydrophobic surface patches in P450bm3 (57) and P450terp (58) postulated to be involved in substrate recognition. Curiously, the former cluster corresponded approximately to the location of a bound palmitic acid in CYP2C8 (29) and an allosterically bound progesterone near an analogous cluster of seven phenylalanine residues (Phe cluster) in CYP3A4 (31). Additional modeling may be necessary to fully understand the contribution of this aromatic cluster to pw2a geometry and CYP activity.
To further characterize the contribution of the F-G loop to the pw2a channel, we examined the model and site-directed mutagenesis and kinetic data of Murtazina et al., who showed that F-G loop residues in CYP27A1 influence cholesterol binding and 27-hydroxylation (62). Mutations in these sites were postulated to disturb membrane binding and proton availability during the reaction cycle and to restrict product egress. Our interpretation is that selected mutations might also disrupt the aromatic cluster above the heme and may variously affect the shape of the binding cavity and/or the pw2a access channel to affect the enzyme kinetics. Although the Murtazina et al. model shows some similarity to the model presented here, its major shortcoming is the lack of a binding cavity, which we attribute to an incorrect positioning of Gly-145 (a VDDR-I and CTX residue) in the B′-C loop, Phe-240 in the F-helix, and an improperly folded heme region.
The finding of an open pw3-type egress channel above the I-helix and centered on Met-334 was intriguing. That this channel is visible at all may result from uncertainty in the positions and separation of the F- and G-helices or the position of the E-F loop. If and how it should be closed in the model will permit refinement of local structure and has important implications for the movement of molecules through the enzyme. Synchronization of substrate binding and metabolite release through different channels is attractive because it would sequentially expose both ends of vitamin D to hydroxylation, in agreement with reports of CYP27A1 exhibiting both 25- and 1α-hydroxylase activities (63,64).
DISCUSSION
In this article, we present novel methodologies for modeling cytochrome P450s, which were applied to model CYP27A1, an important mitochondrial enzyme involved in bile acid metabolism and the 25-hydroxylation of vitamin D. We used this model to guide the selection of nine site-directed mutations directed toward seven putative substrate contact residues affecting the metabolism of the vitamin D prodrug, 1α-OH-D3. Changes in activity and the regioselectivity of side-chain hydroxylation were interpreted to result from conformational changes affecting cavity shape, steric conflicts with bound substrate, or disruption of a secondary heme-binding interaction, though we cannot rule out that small differences in total enzyme activity might be due to differences in enzyme expression. The model was also used to explore, in silico, broader aspects of substrate recognition in CYP27A1 and the location of potential channels governing substrate access and metabolite egress. In the remainder of the discussion, we have highlighted the important aspects revealed by our study, namely: a), the contribution of selected residues to the active site structure and catalytic activity of CYP27A1 toward 1α-OH-D3, and b), a modeling strategy based on the systematic analysis of conserved hydrogen bonding patterns and structural motifs in 11 CYP crystallographic templates.
The major objective of our metabolism studies was to structurally probe putative substrate contacts in the binding cavity of CYP27A1. In our opinion, one of the most interesting findings in this study was that subtle changes in bound orientation directly affect the ratio of 25- and 27-hydroxylated products formed. The hydroxylation at c25 implied that the substrate is bound deeper into the pocket. Mutations targeted to the β-3a strand, β-5 hairpin, and F-helix changed the structure predictably and altered CYP activity from positions that have not been previously observed in patients with CTX, VDDR-I, or any other P450 deficiency.
Mutations affecting the hydrophobic binding cavity surface at the β-5 hairpin
The effect of the Phe-248-Met mutation on CYP27A1 activity was unique and shifted the ratio of 1α-OH-D3 metabolism toward 27-hydroxylation. Phe-248 was predicted to be well within the F-helix and was clearly modeled into the top of the substrate binding site. Whereas Phe-248 may contact the substrate directly, its presence in CYP27B1 and CYP11 isoforms suggests that it plays a more generic function, such as shaping the hydrophobic surface of the binding cavity near Leu-516 in the β-5 hairpin. Phe-248 may deflect Leu-516 away from the substrate so that the mutation to methionine allows Leu-516 to have increased contact with c27 to prevent deep penetration of substrate. The small decreases in total enzyme activity associated with the Phe-248-Met and Leu-516-Val mutations may suggest that part of this hydrophobic surface is also critical to the gating function of Asp-338 in regulating proton access during the reaction cycle via the adjacent solvent access channel. Interestingly, the Phe-248-Lys mutation reported in a previous study altered the regioselectivity of bile acid precursor hydroxylation by inducing a novel demethylase activity and the formation of non-c27-oxidized and/or unsaturated products (53). Consumption of protons during the reaction cycle could make lysine a proton acceptor capable of directing a 27-hydroxylated product toward demethylation. This could also disrupt bound substrate orientations and/or Asp-338-regulated proton transport resulting in altered product formation and product release.
All of the β-5 hairpin mutations transformed CYP27A1 into a more efficient 25-hydroxylase, shifting the ratio of hydroxylation products toward 1α,25-(OH)2D3. In Leu-516-Val, the smaller side chain had reduced contact with the substrate, enlarged the binding cavity, allowed the deeper penetration of substrate leading to 25-hydroxylation, and may have decreased overall activity by disrupting Asp-338-regulated proton transport. In our model, the side chain of Val-515 made deep penetration of the substrate somewhat unfavorable through steric contact with the c18 D-ring methyl. A longer side chain could adopt rotomer conformations that reduce this contact, and this was seen in the increased activity and deeper penetration of substrate in Val-515-Ile and Val-515-Leu. The effect of the Ile-514-Phe mutation is more complicated because the large loss of activity can be interpreted as a large and unfavorable impact on the presumed substrate access channel. Resulting in a preferential loss of 27-hydroxylated product, the residual 25-hydroxylase activity of the Ile-514-Phe mutation implied that what little substrate was bound was deflected deep into the active site. Whether this was caused by conformational changes in the F-G loop, a productive interaction with the vitamin D cis-triene, or a deflection of the adjacent Val-515 is not known. Overall, the mutations in the β-5 hairpin study suggest that this structure forms a contoured hydrophobic surface that affects substrate orientation.
Mutations affecting structure and substrate contact in the β-3 sheet
In our model, it was found that the β-3a strand in CYP27A1 connected the β-5 sheet and B-B′ loop to an extended β-sheet structure, bound the heme A-ring propionate from two positions, and made significant contact with bound substrate. The Thr-402-Phe mutation severely impaired the activity of the enzyme. We hypothesize that this occurred through the loss of a structural link to the β-5 hairpin near Ile-514, which disrupted protein stability and the loss of a contact with the substrate 1α-hydroxyl. The Asn-403-Thr mutation appears to have ablated activity by disrupting a heme-binding interaction between the His-428-stabilized Asn-403 and the A-ring propionate that disrupts protein stability. The loss of a proton donor function associated with the Ser-404-Ala mutation disrupted the contact with the substrate 3β-hydroxyl and the B-B′ loop through the release of hydrogen bonding interactions, the latter centered on Asn-128 and Gln-423 side chains. The ensuing conformational change oriented bound substrate deeper into the active site, preferentially disrupting 27-hydroxylation. A second mutation at this position, Ser-404-Thr, did not influence bound substrate orientation toward 25- or 27-hydroxylation, but the methyl group sterically interfered with binding and caused a significant loss of activity.
Substrate recognition in the substrate access channel
A survey of proton donors capable of recognizing the hydroxyl groups on CYP27A1 substrates revealed a multifaceted hydrogen bonding network of conserved asparagines, glutamines, and histidines in the extended β-sheet and amino-terminal helices. It was found that these residues occupied discrete positions and have side-chain orientations that are independently determined by cytochrome P450 structure. Some of these residues may assist the partitioning of substrates down a substrate access channel into the active site through transient interactions with substrate hydroxyl groups and presumably provide a mechanism for substrate specificity by filtering or selecting against previously hydroxylated substrates.
Our model also revealed the existence of a previously unrecognized aromatic cluster over the binding cavity. A similar cluster in CYP3A4 formed a prominent hydrophobic core, which was highly ordered and composed of residues known to affect activity (31). In CYP27A1, this aromatic cluster appears to contribute a large surface at the junction of the binding cavity and a pw2a substrate access channel, and contains residues shown in previous studies to affect the kinetics of cholesterol hydroxylation (53,62).
Structural motif-based modeling methodology
In this study, we have used a new approach to homology modeling which was based upon a strategy which is broadly applicable to other cytochrome P450 proteins. We have used the observed secondary structure and a systematic analysis of hydrogen bonding patterns in 11 CYP crystal structures to refine the multisequence alignment and simplify the assignment of secondary structure to a new CYP sequence. This analysis revealed a number of residues and tertiary structural motifs that are functionally conserved in most of the available CYP templates and probably in many other CYPs as well. Although the function of some conserved elements has been correctly deduced in other studies, for example the PGP motif (42), our analysis revealed a high degree of commonality in known CYP structures and drew attention to functionally equivalent interactions that may be easily overlooked. These structural motifs were used to guide the application of distance constraints during energy minimization and molecular dynamics. This strategy maximizes the likelihood of successfully propagating known structure into regions that are difficult to model due to template heterogeneity. Finally, we have used this approach to model CYP27A1 and explore the enigmatic structure and location of access channels.
The finding of such a functionally conserved and highly structured heme-binding region was unexpected and emphasizes the need for objective input during modeling. The hydrogen bonding network around the propionates helped position the B- and C-helices, B-B′ and B′-C loops, and most of the β-sheet structure relative to the heme group. The extensive hydrogen bonding network surrounding the ERR-triad suggests an important structural core that stabilizes adjacent structures and may, for some, change how protein cofactor binding at the proximal heme face will be viewed. This is probably also true for the conserved links between the J- and K-helices and the K- and L-helices. The EH-gap motif was an interesting finding since it helps delineate and position the E-, G-, and H-helices and the E-F loop. The incorporation of this motif in our model of CYP27A1 revealed a putative disulfide bond and raises the question of the potential for formation of this structure in other CYPs. The spatially conserved phenylalanine at the start of the F-helix is an important alignment and modeling landmark, as is the well-conserved aspartate at the start of the E-helix. The unique aspects of cytochrome P450 structure revealed in this modeling approach can now be used to predict and study the binding pocket in other vitamin D-related hydroxylases.
Acknowledgments
The authors acknowledge the excellent technical assistance of Kerith R. Geh in the metabolism studies, the contributions of Dr. Donald F. Weaver, Kelly A. Dakin, and Oreola A. T. Donini in the preparation of a preliminary model of CYP27A1, the instruction of Dr. Michael J. Kuiper in the usage of the SYBYL software, and the assistance of Jimin Zheng and Robert Campbell. They also acknowledge data not shown, which was collected by Valarie Byford, Martin Kaufmann, Sonoko Masuda, and Shelly West.
This work was supported by grants MT-9475, MMA-38116, and MMA-69106 to G.J. from the Medical Research Council of Canada (now the Canadian Institutes for Health Research).
Abbreviations used: CYP27A1, cytochrome P450 25-hydroxylase; CTX, cerebrotendinous xanthomatosis; CYP, cytochrome P450; CYP24A1, 1α,25-dihydroxyvitamin D3-inducible cytochrome P450 24-hydroxylase; CYP27B1, cytochrome P450 1α-hydroxylase; HPLC, high performance liquid chromatography; VDDR-I, vitamin D-dependent rickets-type I; 1α-OH-D3, 1α-hydroxyvitamin D3; 1α,25-(OH)2D3, 1α,25-dihydroxyvitamin D3, 25-OH-D3, 25-hydroxyvitamin D3; RMSD, root mean-square deviation; congenital adrenal hyperplasia, CAH.
References
- 1.Prosser, D. E., and G. Jones. 2004. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem. Sci. 29:664–673. [DOI] [PubMed] [Google Scholar]
- 2.Whitfield, G. K., P. W. Jurutka, C. A. Haussler, and M. R. Haussler. 1999. Steroid hormone receptors: evolution, ligands, and molecular basis of biologic function. J. Cell. Biochem. 110–122. [DOI] [PubMed]
- 3.Jones, G., S. A. Strugnell, and H. F. DeLuca. 1998. Current understanding of the molecular actions of vitamin D. Physiol. Rev. 78:1193–1231. [DOI] [PubMed] [Google Scholar]
- 4.Guo, Y.-D., S. Strugnell, D. W. Back, and G. Jones. 1993. Transfected human liver cytochrome P-450 hydroxylates vitamin D analogs at different side-chain positions. Proc. Natl. Acad. Sci. USA. 90:8668–8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zittermann, A. 2003. Vitamin D in preventive medicine: are we ignoring the evidence? Br. J. Nutr. 89:552–572. [DOI] [PubMed] [Google Scholar]
- 6.Norlin, M., S. von Bahr, I. Bjorkhem, and K. Wikvall. 2003. On the substrate specificity of human CYP27A1: implications for bile acid and cholestanol formation. J. Lipid Res. 44:1515–1522. [DOI] [PubMed] [Google Scholar]
- 7.Lee, M.-H., S. Hazard, J. D. Carpten, S. Yi, J. Cohen, G. T.Gerhardt, G. Salen, and S. B. Patel. 2001. Fine-mapping, mutation analyses, and structural mapping of cerebrotendinous xanthomatosis in U.S. pedigrees. J. Lipid Res. 42:159–169. [PMC free article] [PubMed] [Google Scholar]
- 8.Moghadasian, M. H. 2004. Cerebrotendinous xanthomatosis: clinical course, genotypes and metabolic backgrounds. Clin. Invest. Med. 27:42–50. [PubMed] [Google Scholar]
- 9.Honda, A., G. Salen, Y. Matsuzaki, A. K. Batta, G. R. Xu, E. Leitersdorf, G. S. Tint, S. K. Erickson, N. Tanaka, and S. Shefer. 2001. Differences in hepatic levels of intermediates in bile acid biosynthesis between Cyp27−/− mice and CTX. J. Lipid Res. 42:291–300. [PubMed] [Google Scholar]
- 10.Berginer, V. M., S. Shany, D. Alkalay, J. Berginer, S. Dekel, G. Salen, G. S. Tint, and D. Gazit. 1993. Osteoporosis and increased bone fractures in cerebrotendinous xanthomatosis. Metab. Clin. Exp. 42:69–74. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharyya, M. H. and H. F. DeLuca. 1974. Subcellular localization of rat liver calciferol-25-hydroxylase. Arch. Biochem. Biophys. 160:58–62. [DOI] [PubMed] [Google Scholar]
- 12.Gupta, R. P., B. W. Hollis, S. B. Patel, K. S. Patrick, and N. H. Bell. 2004. Cyp3A4 is a human microsomal vitamin D 25-hydroxylase. J. Bone Miner. Res. 19:680–688. [DOI] [PubMed] [Google Scholar]
- 13.Gupta, R. P., Y. A. He, K. S. Patrick, J. R. Halpert, and N. H. Bell. 2005. CYP3A4 is a vitamin D 24- and 25-hydroxylase: analysis of structure function by site-directed mutagenesis. J. Clin. Endocrinol. Metab. 90:1210–1219. [DOI] [PubMed] [Google Scholar]
- 14.Shinkyo, R., T. Sakaki, M. Kamakura, M. Ohta, and K. Inouye. 2004. Metabolism of vitamin D by human microsomal CYP2R1. Biochem. Biophys. Res. Commun. 324:451–457. [DOI] [PubMed] [Google Scholar]
- 15.Cheng, J. B., M. A. Levine, N. H. Bell, D. J. Mangelsdorf, and D. W. Russell. 2004. Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc. Natl. Acad. Sci. USA. 101:7711–7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamasaki, T., S. Izumi, H. Ide, and Y. Ohyama. 2004. Identification of a novel rat microsomal vitamin D3 25-hydroxylase. J. Biol. Chem. 279:22848–22856. [DOI] [PubMed] [Google Scholar]
- 17.Rahmaniyan, M., K. S. Patrick, and N. H. Bell. 2005. Characterization of recombinant CYP2C11: a vitamin D 25-hydroxylase and 24-hydroxylase. Am. J. Physiol. Endocrinol. Metab. 288:E753–E760. [DOI] [PubMed] [Google Scholar]
- 18.Hosseinpour, F., I. Ibranovic, W. Tang, and K. Wikvall. 2003. 25-Hydroxylation of vitamin D3 in primary cultures of pig hepatocytes: evidence for a role of both CYP2D25 and CYP27A1. Biochem. Biophys. Res. Commun. 303:877–883. [DOI] [PubMed] [Google Scholar]
- 19.Cheng, J. B., D. L. Motola, D. J. Mangelsdorf, and D. W. Russell. 2003. De-orphanization of cytochrome P450 2R1: a microsomal vitamin D 25-hydroxylase. J. Biol. Chem. 278:38084–38093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vijayakumar, S. and J. C. Salerno. 1992. Molecular modeling of the 3-D structure of cytochrome P-450scc. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1160:281–286. [DOI] [PubMed] [Google Scholar]
- 21.Usanov, S. A., S. E. Graham, G. I. Lepesheva, T. N. Azeva, N. V. Strushkevich, A. A. Gilep, R. W. Estabrook, and J. A. Peterson. 2002. Probing the interaction of bovine cytochrome P450scc (CYP11A1) with adrenodoxin: evaluating site-directed mutations by molecular modeling. Biochemistry. 41:8310–8320. [DOI] [PubMed] [Google Scholar]
- 22.Lewis, D. F. V., and P. Lee-Robichaud. 1998. Molecular modelling of steroidogenic cytochromes P450 from families CYP11, CYP17, CYP19 and CYP21 based on the CYP102 crystal structure. J. Steroid Biochem. Mol. Biol. 66:217–233. [DOI] [PubMed] [Google Scholar]
- 23.Belkina, N. V., M. Lisurek, A. S. Ivanov, and R. Bernhardt. 2001. Modelling of three-dimensional structures of cytochromes P45011B1 and 11B2. J. Inorg. Biochem. 87:197–207. [DOI] [PubMed] [Google Scholar]
- 24.Podust, L. M., T. L. Poulos, and M. R. Waterman. 2001. Crystal structure of cytochrome P450 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc. Natl. Acad. Sci. USA. 98:3068–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Podust, L. M., J. Stojan, T. L. Poulos, and M. R. Waterman. 2001. Substrate recognition sites in 14α-sterol demethylase from comparative analysis of amino acid sequences and X-ray structure of Mycobacterium tuberculosis CYP51. J. Inorg. Biochem. 87:227–235. [DOI] [PubMed] [Google Scholar]
- 26.Scott, E. E., M. A. White, Y. A. He, E. F. Johnson, C. D. Stout, and J. R. Halpert. 2004. Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl) imidazole at 1.9-Å resolution: insight into the range of P450 conformations and the coordination of redox partner binding. J. Biol. Chem. 279:27294–27301. [DOI] [PubMed] [Google Scholar]
- 27.Wester, M. R., E. F. Johnson, C. Marques-Soares, S. Dijols, P. M. Dansette, D. Mansuy, and C. D. Stout. 2003. Structure of mammalian cytochrome P450 2C5 complexed with diclofenac at 2.1Å resolution: evidence for an induced fit model of substrate binding. Biochemistry. 42:9335–9345. [DOI] [PubMed] [Google Scholar]
- 28.Wester, M. R., E. F. Johnson, C. Marques-Soares, P. M. Dansette, D. Mansuy, and C. D. Stout. 2003. Structure of a substrate complex of mammalian cytochrome P450 2C5 at 2.3 Å resolution: evidence for multiple substrate binding modes. Biochemistry. 42:6370–6379. [DOI] [PubMed] [Google Scholar]
- 29.Schoch, G. A., J. K. Yano, M. R. Wester, K. J. Griffin, C. D. Stout, and E .F. Johnson. 2004. Structure of human microsomal cytochrome P450 2C8. evidence for a peripheral fatty acid binding site. J. Biol. Chem. 279:9497–9503. [DOI] [PubMed] [Google Scholar]
- 30.Williams, P. A., J. Cosme, A. Ward, H. C. Angove, D. M. Vinkovic, and H. Jhoti. 2003. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 424:464–468. [DOI] [PubMed] [Google Scholar]
- 31.Williams, P. A., J. Cosme, D. Matak Vinkovic, A. Ward, H. C. Angove, P. J. Day, C. Vonrhein, I. J. Tickle, and H. Jhoti. 2004. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 305:683–686. [DOI] [PubMed] [Google Scholar]
- 32.Yano, J. K., M. R. Wester, G. A. Schoch, K. J. Griffin, C. D. Stout, and E. F. Johnson. 2004. The structure of human microsomal cytochrome P450 3A4 determined by x-ray crystallography to 2.05-Å resolution. J. Biol. Chem. 279:38091–38094. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholas, K. B., H. B. Nicholas, and D. W. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News. 4:14. [Google Scholar]
- 35.Levesque, J.-P., P. Sinsilvestri, A. Hatzfeld, and J. Hatzfeld. 1991. DNA transfection in COS cells: a low-cost serum-free method compared to lipofection. BioTecniques. 11:313–318. [PubMed] [Google Scholar]
- 36.Brentano, S. T. and W. L. Miller. 1992. Regulation of human cytochrome P450scc and adrenodoxin messenger ribonucleic acids in JEG-3 cytotrophoblast cells. Endocrinology. 131:3010–3018. [DOI] [PubMed] [Google Scholar]
- 37.Brentano, S. T., S. M. Black, D. Lin, and W. L. Miller. 1992. cAMP post transcriptionally diminishes the abundance of adrenodoxin reductase mRNA. Proc. Natl. Acad. Sci. USA. 89:4099–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bligh, E. G. and W. J. Dyer. 1957. A rapid method of total lipid extraction and purification. Can. J. Biochem. 37:911–917. [DOI] [PubMed] [Google Scholar]
- 39.Makin, G., D. Lohnes, V. Byford, R. Ray, and G. Jones. 1989. Target cell metabolism of 1,25-dihydroxyvitamin D3 to calcitroic acid. Evidence for a pathway in kidney and bone involving 24-oxidation. Biochem. J. 262:173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kusano, K., N. Kagawa, M. Sakaguchi, T. Omura, and M. Waterman. 2001. Importance of a proline-rich sequence in the amino-terminal region for correct folding of mitochondrial and soluble microbial P450s. J. Biochem. (Tokyo). 129:271–277. [DOI] [PubMed] [Google Scholar]
- 41.Kusano, K., M. Sakaguchi, N. Kagawa, M. R. Waterman, and T. Omura. 2001. Microsomal P450s use specific proline-rich sequences for efficient folding, but not for maintenance of the folded structure. J. Biochem. (Tokyo). 129:259–269. [DOI] [PubMed] [Google Scholar]
- 42.Kemper, B. 2004. Structural basis for the role in protein folding of conserved proline-rich regions in cytochromes P450. Toxicol. Appl. Pharmacol. 199:305–315. [DOI] [PubMed] [Google Scholar]
- 43.Hasemann, C. A., R. G. Kurumbail, S. S. Boddupalli, J. A. Peterson, and J. Deisenhofer. 1995. Structure and function of cytochromes P450: A comparative analysis of three crystal structures. Structure. 3:41–62. [DOI] [PubMed] [Google Scholar]
- 44.Löffler, J., S. L. Kelly, H. Hebart, U. Schumacher, C. Lass-Flörl, and H. Einsele. 1997. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol. Lett. 151:263–268. [DOI] [PubMed] [Google Scholar]
- 45.Ost, T. W. B., C. S. Miles, A. W. Munro, J. Murdoch, G. A. Reid, and S. K. Chapman. 2001. Phenylalanine 393 exerts thermodynamic control over the heme of flavocytochrome P450BM3. Biochemistry. 40:13421–13429. [DOI] [PubMed] [Google Scholar]
- 46.Sawada, N., T. Sakaki, S. Kitanaka, S. Kato, and K. Inouye. 2001. Structure-function analysis of CYP27B1 and CYP27A1: studies on mutants from patients with vitamin D-dependent rickets type I (VDDR-I) and cerebrotendinous xanthomatosis (CTX). Eur. J. Biochem. 268:6607–6615. [DOI] [PubMed] [Google Scholar]
- 47.Hanna, I. H., M. S. Kim, and F. P. Guengerich. 2001. Heterologous expression of cytochrome P450 2D6 mutants, electron transfer, and catalysis of bufuralol hydroxylation: the role of aspartate 301 in structural integrity. Arch. Biochem. Biophys. 393:255–261. [DOI] [PubMed] [Google Scholar]
- 48.Pylypenko, O. and I. Schlichting. 2004. Structural aspects of ligand binding to and electron transfer in bacterial and fungal P450s. Annu. Rev. Biochem. 73:991–1018. [DOI] [PubMed] [Google Scholar]
- 49.Wade, R. C., P. J. Winn, I. Schlichting, and Sudarko. 2004. A survey of active site access channels in cytochromes P450. J. Inorg. Biochem. 98:1175–1182. [DOI] [PubMed] [Google Scholar]
- 50.Garuti, R., N. Lelli, M. Barozzini, R. Tiozzo, M. T. Dotti, A. Federico, A. M. Ottomano, A. Croce, S. Bertolini, and S. Calandra. 1996. Cerebrotendinous xanthomatosis caused by two new mutations of the sterol-27-hydroxylase gene that disrupt mRNA splicing. J. Lipid Res. 37:1459–1467. [PubMed] [Google Scholar]
- 51.Hsu, L. C., N. C. Hsu, J. A. Guzova, V. M. Guzov, S. F. Chang, and B. C. Chung. 1996. The common I172N mutation causes conformational change of cytochrome P450c21 revealed by systematic mutation, kinetic, and structural studies. J. Biol. Chem. 271:3306–3310. [DOI] [PubMed] [Google Scholar]
- 52.Lee-Robichaud, P., M. E. Akhtar, J. N. Wright, Q. I. Sheikh, and M. Akhtar. 2004. The cationic charges on Arg347, Arg358, and Arg449 of human cytochrome P450c17 (CYP17) are essential for the enzyme's cytochrome b5-dependent acyl-carbon cleavage activities. J. Steroid Biochem. Mol. Biol. 92:119–130. [DOI] [PubMed] [Google Scholar]
- 53.Pikuleva, I. A., A. Puchkaev, and I. Björkhem. 2001. Putative helix F contributes to regioselectivity of hydroxylation in mitochondrial cytochrome P450 27A1. Biochemistry. 40:7621–7629. [DOI] [PubMed] [Google Scholar]
- 54.Annalora, A., E. Bobrovnikova-Marjon, R. Serda, L. Lansing, M. L. Chiu, A. Pastuszyn, S. Iyer, C. B. Marcus, and J. L. Omdahl. 2004. Rat cytochrome P450C24 (CYP24A1) and the role of F249 in substrate binding and catalytic activity. Arch. Biochem. Biophys. 425:133–146. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto, K., E. Uchida, N. Urushino, T. Sakaki, N. Kagawa, N. Sawada, M. Kamakura, S. Kato, K. Inouye, and S. Yamada. 2005. Identification of the amino acid residue of CYP27B1 responsible for binding of 25-hydroxyvitamin D3 whose mutation causes vitamin D-dependent rickets type 1. J. Biol. Chem. 280:30511–30516. [DOI] [PubMed] [Google Scholar]
- 56.Poulos, T. L., B. C. Finzel, and A. J. Howard. 1986. Crystal Structure of substrate-free Pseudomonas putida Cytochrome P-450. Biochemistry. 25:5314–5322. [DOI] [PubMed] [Google Scholar]
- 57.Ravichandran, K. G., S. S. Boddupalli, C. A. Hasemann, J. A. Peterson, and J. Deisenhofer. 1993. Crystal structure of hemoprotein domain of P450bm-3, a prototype for microsomal P450s. Science. 261:731–736. [DOI] [PubMed] [Google Scholar]
- 58.Hasemann, C. A., K. G. Ravichandran, J. A. Peterson, and J. Deisenhofer. 1994. Crystal structure and refinement of cytochrome P450terp at 2.3 Å resolution. J. Mol. Biol. 236:1169–1185. [DOI] [PubMed] [Google Scholar]
- 59.Graham-Lorence, S., B. Amarneh, R. E. White, J. A. Peterson, and E. R. Simpson. 1995. A three-dimensional model of aromatase cytochrome P450. Protein Sci. 4:1065–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lüdemann, S. K., V. Lounnas, and R. C. Wade. 2000. How do substrates enter and products exit the buried active site of cytochrome P450cam? 1. Random expulsion molecular dynamics investigation of ligand access channels and mechanisms. J. Mol. Biol. 303:797–811. [DOI] [PubMed] [Google Scholar]
- 61.Winn, P. J., V. Lounnas, R. Gauges, V. Lounnas, and R. C. Wade. 2002. Comparison of the dynamics of substrate access channels in three cytochrome P450s reveals different opening mechanism and a novel functional role for a buried arginine. Proc. Natl. Acad. Sci. USA. 99:5361–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murtazina, D., A. V. Puchkaev, C. H. Schein, N. Oezguen, W. Braun, A. Nanavati, and I. A. Pikuleva. 2002. Membrane-protein interactions contribute to efficient 27-hydroxylation of cholesterol by mitochondrial cytochrome P450 27A1. J. Biol. Chem. 277:37582–37589. [DOI] [PubMed] [Google Scholar]
- 63.Araya, Z., M .Norlin, and H. Postlind. 1996. A possible role for CYP27 as a major renal mitochondrial 25- hydroxyvitamin D3 1α-hydroxylase. FEBS Lett. 390:10–14. [DOI] [PubMed] [Google Scholar]
- 64.Pikuleva, I. A., I. Björkhem, and M. R. Waterman. 1997. Expression, purification, and enzymatic properties of recombinant human cytochrome P450c27 (CYP27). Arch. Biochem. Biophys. 343:123–130. [DOI] [PubMed] [Google Scholar]