Abstract
The orientation of a β-sheet membrane peptide in lipid bilayers is determined, for the first time, using two-dimensional (2D) 15N solid-state NMR. Retrocyclin-2 is a disulfide-stabilized cyclic β-hairpin peptide with antibacterial and antiviral activities. We used 2D separated local field spectroscopy correlating 15N-1H dipolar coupling with 15N chemical shift to determine the orientation of multiply 15N-labeled retrocyclin-2 in uniaxially aligned phosphocholine bilayers. Calculated 2D spectra exhibit characteristic resonance patterns that are sensitive to both the tilt of the β-strand axis and the rotation of the β-sheet plane from the bilayer normal and that yield resonance assignment without the need for singly labeled samples. Retrocyclin-2 adopts a transmembrane orientation in dilauroylphosphatidylcholine bilayers, with the strand axis tilted at 20° ± 10° from the bilayer normal, but changes to a more in-plane orientation in thicker 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidyl-choline (POPC) bilayers with a tilt angle of 65° ± 15°. These indicate that hydrophobic mismatch regulates the peptide orientation. The 2D spectra are sensitive not only to the peptide orientation but also to its backbone (φ, ψ) angles. Neither a bent hairpin conformation, which is populated in solution, nor an ideal β-hairpin with uniform (φ, ψ) angles and coplanar strands, agrees with the experimental spectrum. Thus, membrane binding orders the retrocyclin conformation by reducing the β-sheet curvature but does not make it ideal. 31P NMR spectra of lipid bilayers with different compositions indicate that retrocyclin-2 selectively disrupts the orientational order of anionic membranes while leaving zwitteronic membranes intact. These structural results provide insights into the mechanism of action of this β-hairpin antimicrobial peptide.
INTRODUCTION
The orientation of membrane peptides in lipid bilayers is an important aspect of the three-dimensional structure of these molecules. Solid-state NMR spectroscopy is a well-established tool for determining the orientation of α-helical membrane peptides. The most common approach is to measure the 15N chemical shift and 15N-1H dipolar couplings of macroscopically oriented peptides bound to lipid membranes (1). These two 15N interaction tensors are approximately parallel to the helical axis, thus their frequencies reflect the orientation of the helical axis relative to the magnetic field. When the alignment axis is parallel to the magnetic field, this is also the helix orientation relative to the bilayer normal. To determine the peptide orientation with high angular resolution, two-dimensional (2D) separated-local field (SLF) spectroscopy correlating the 15N chemical shift with 15N-1H dipolar coupling is particularly powerful. Due to the small misalignment between the N-H bonds and the helical axis, the 2D SLF spectra of multiply 15N-labeled helical peptides give characteristic wheel-like patterns whose positions and sizes are exquisitely sensitive to the tilt angle of the helix from the membrane normal (2,3). The peaks on these wheel patterns follow the helical wheel projection in a well-defined fashion, so that they can be assigned readily as long as one of the peaks is identified using a site-specifically labeled sample.
Compared to advances in the orientation determination of α-helical membrane peptides, knowledge about the orientation and insertion of β-sheet peptides in lipid bilayers is scarce. Although theoretical analyses of the 2D spectra of β-sheet peptides were given recently (4,5), no experimental study of β-sheet peptide orientation using this 2D approach has been reported. The fact that the N-H bonds in β-sheet peptides are perpendicular rather than parallel to the strand axis further makes it unclear whether 15N NMR is adequate for determining β-sheet peptide orientations.
Disulfide-stabilized β-hairpin antimicrobial peptides (6) are promising systems both for understanding β-sheet peptide binding to lipid membranes and for testing the applicability of the 15N solid-state NMR method. These peptides are potent microbicidal molecules present in many animals and plants as part of their innate immune system (7–9). The most common mechanism of action of these small cationic peptides is the disruption of the microbial cell membrane. D-enantiomers of these peptides show similar activities as their L-counterparts, indicating that the targets of these peptides are the achiral lipids of the membrane rather than protein receptors in the membrane or inside the cell (10,11).
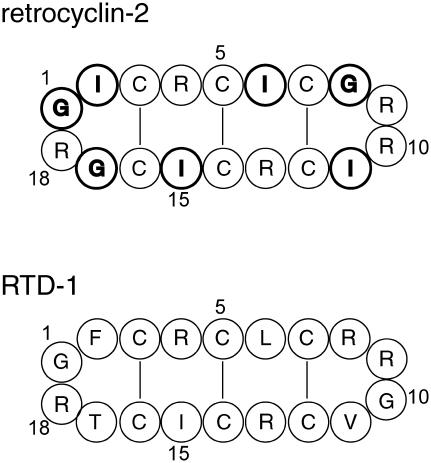
Retrocyclin-2 is a circular 18-residue antimicrobial peptide encoded in the human genome by a θ-defensin pseudogene (12). It exhibits antibacterial activities and inhibits human immunodeficiency virus entry into human cells (13–15). Similar to the monkey homolog, rhesus θ-defensin (RTD) (16), retrocyclin-2 has a β-hairpin structure stabilized by three cross-strand disulfide bonds. Its five Arg residues and the hydrophobic residues are distributed at nearly identical positions as in RTD-1 (Fig. 1). Thus, the solution NMR structure of RTD-1 to a good approximation is applicable to retrocyclin-2. Retrocyclin-2 has a high affinity to carbohydrate-containing cell surface molecules (17) and is localized on the cell membrane based on confocal microscopy (13). However, the high-resolution structure of the peptide bound to the lipid bilayer is not yet known.
FIGURE 1.
Amino acid sequences of retrocyclin-2 and RTD-1. The 15N-labeled residues in retrocyclin-2 are shown in bold.
In this work, we report the orientation determination of multiply 15N-labeled retrocyclin-2 using 2D 15N dipolar-shift correlation NMR combined with macroscopic alignment of lipid membranes. We show that the 2D spectra of β-hairpin peptides are extremely sensitive to the orientations of the β-strand axis and β-sheet plane relative to the bilayer normal. Moreover, the spectral patterns yield nondegenerate values of the orientation angles, so that no site-specific labeling-based assignment is necessary to resolve angular ambiguity. Experimental spectra indicate that retrocyclin-2 is transmembrane in dilauroylphosphatidylcholine (DLPC) bilayers but changes to a more in-plane orientation in POPC bilayers. Further, the 2D spectra are sensitive to the backbone (φ, ψ) angles. Neither a bent conformation present in solution nor an ideal cyclic conformation with coplanar strands agrees with the experimental data. These provide the first insights into the conformation and orientation of this class of cyclic β-hairpin peptides in the membrane.
MATERIALS AND METHODS
Materials
All lipids, including DLPC, POPC, 1-palmitoyl-2-oleoyl-phosphatidylethanolamine (POPE), and 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)]) (POPG), were purchased from Avanti Polar Lipids (Alabaster, AL) and used without further purification. Retrocyclin-2 (GICRCICGRG ICRCICGR) was synthesized on a 0.25-mmol scale with an ABI 431A peptide synthesizer using FastMoc chemistry (18). All residues were double coupled to insure optimal yield. Gly-1, Ile-2, Ile-6, Gly-8, Ile-11, Ile-15, and Gly-17 were 15N labeled (Fig. 1). 15N-labeled amino acids were purchased from Cambridge Isotope Laboratory (Andover, MA) and converted to the Fmoc derivatives by AnaSpec (San Jose, CA). The crude reduced peptide was purified by reverse-phase high-performance liquid chromatography, oxidized with dimethylsulfoxide buffer, purified again to remove unreacted peptide, then cyclized (13). Glass cover slides with a thickness of 0.08 mm were obtained from Marienfeld Laboratory (Lauda-Koenigshofen, Germany) and cut to 6 × 12 mm2 rectangles.
Membrane sample preparation
Glass-plate-oriented membrane mixtures were prepared using a naphthalene-incorporated procedure described recently (19). The peptide was dissolved in trifluoro-ethanol and mixed with a chloroform solution of the lipids with the desired molar ratio. The mixture was dried under a stream of N2 gas and the dried film was redissolved in a 1:1 mixture of chloroform/TFE containing a twofold excess of naphthalene with respect to the lipids. The solution was deposited on 10–30 glass plates with an area concentration of 0.01–0.02 mg/mm2, air dried for 2 h, and then vacuum dried for 5 h to remove all solvents and naphthalene. About 1 μL of water was added directly to each glass plate, then the sample was hydrated indirectly at a relative humidity of 98% over a saturated solution of K2SO4 for 1–2 weeks. The glass plates were stacked, wrapped in parafilm, and sealed in a polyethylene bag to prevent dehydration during the NMR experiments.
Solid-state NMR
NMR experiments were carried out on a Bruker DSX-400 spectrometer (Karlsruhe, Germany) operating at a resonance frequency of 162.12 MHz for 31P and 40.58 MHz for 15N. A static double-resonance probe with a home-built rectangular coil with the dimension of 6 × 12 × 5 mm3 was used for the oriented membrane samples. The 15N chemical shift was referenced to the isotropic signal of N-acetyl-valine at 122 ppm. The 31P chemical shift was referenced to 85% phosphoric acid at 0 ppm. The 2D SLF experiments used the MREV-8 sequence (20,21) to decouple the 1H-1H dipolar interaction during t1, although other homonuclear decoupling schemes are also applicable. The MREV-8 90° pulse length was 3.8 μs. A 1H decoupling field strength of 50 kHz was used during 15N detection. The 1H-15N CP contact time was 1 ms. The 2D spectra were acquired using 22–24 t1 slices with a dwell time of 45.6 μs, resulting in a maximum evolution time of slightly over 1 ms. A total of 1280 and 3072 scans were averaged for each t1 time point for the DLPC- and POPC-bound peptide samples, respectively.
Orientation simulations
2D 15N-1H/15N correlation spectra were calculated using two FORTRAN programs. The first program defines a molecule-fixed coordinate system that reflects the β-strand axis and β-sheet plane geometry and calculates the anisotropic frequencies based on the orientation of the magnetic field (B0) in this coordinate system. The z axis of this reference system, the β-strand axis, is defined and calculated as the average orientation of an even number of consecutive C′i−1–Ni bonds (Fig. 2 a). We used the six peptide bonds of residues 2–7 for this purpose. The y-z plane, the local β-sheet plane, is defined as the common plane containing the z axis and a specific C=O vector. The C=O bond of residue 2 was used. The tilt angle τ is between the β-strand (z) axis and the B0 field, whereas the rotation angle ρ is defined as between the y axis and the projection of B0 onto the x-y plane. ρ = 0° indicates that B0 is parallel to the β-sheet (y-z) plane. The molecular bonds necessary for defining the orientations of the 15N chemical shift and N-H dipolar tensors, including the N-HN, C′-N, and N-Cα bonds, were extracted from the Protein Data Bank (PDB) coordinates of RTD-1 (1HVZ). The chemical shift and dipolar coupling frequencies were calculated from the scalar products between B0 and the respective tensors as B0 is rotated through all combinations of (τ, ρ) angles. The unique angular range of τ is 0°–90°, whereas ρ is sampled over the entire 360° range. We refer to this β-sheet-based program as the relative orientation program.
FIGURE 2.
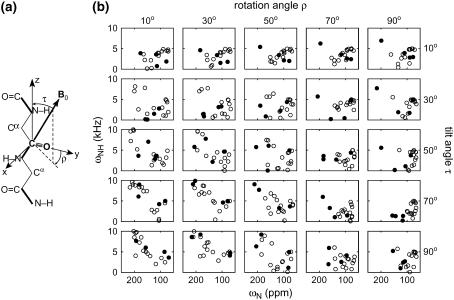
(a) Definition of the tilt angle τ and rotation angle ρ in a β-strand peptide. (b) Calculated 2D 15N-1H/15N correlation spectra as a function of τ and ρ for an 18-residue β-hairpin molecule, using structure 15 of RTD-1. The resonances of all 18 residues are shown. Filled and open circles represent the resonances of β-turn and β-strand residues, respectively.
To accurately visualize the results of the orientation calculation and to determine the orientation of nonideal β-hairpins, whose β-sheet axis and β-sheet plane are ill-defined, a second FORTRAN program without an internal β-sheet reference system was used. The program defines the B0 orientation by a polar angle β and an azimuthal angle α in the default PDB coordinate system (22). This program is referred to as the absolute orientation program. The best-fit (α, β) angles were converted into the Cartesian coordinates (sinßcosα, sinßsinα, cosβ) of a vector from the origin and added to the PDB file. This vector, the bilayer normal, was rotated together with the molecule until it was vertical on the screen, thus giving the exact orientation of the β-sheet peptide (22).
The two FORTRAN programs were checked for consistency by calculating the spectra for a transmembrane (τ = 0°, ρ = 0°) and an in-plane (τ = 90°, ρ = 90°) extended strand (φ = ψ = 180°) using the relative orientation program, then fitting these spectra using the absolute orientation program. The best-fit (α, β) angles were then visualized in Insight II to confirm that the molecules have the desired orientations.
Input 15N chemical shift and N-H dipolar tensors for the simulations were as follows. The z axis of the 15N chemical shift tensor is 17° from the N-H bond (23), whereas the x axis is 25° from the peptide plane (24). The rigid-limit N-H dipolar coupling was 10 kHz, corresponding to a bond length of 1.07 Å. Literature 15N chemical shift principal values of (64, 77, 217) ppm (23) were used to simulate the general orientation-dependent spectra of Figs. 2 and 3. To fit the experimental 2D spectra of retrocyclin-2, the chemical shift difference between Ile and Gly was taken into account. The principal values were estimated from the 15N magic-angle spinning (MAS) sideband intensities of unoriented retrocyclin-2 in DLPC bilayers. For Ile, the 15N principal values were (75, 76, 221) ppm, whereas for Gly, the principal values were (42, 86, 202) ppm. These were used for most of the simulations in Figs. 4, 6, 7, and 8. Calculations show that the use of literature 15N chemical shift tensor values did not change the best-fit (τ, ρ) angles, but the fit to the experimental spectra is actually somewhat better using the standard 15N tensor values.
FIGURE 3.
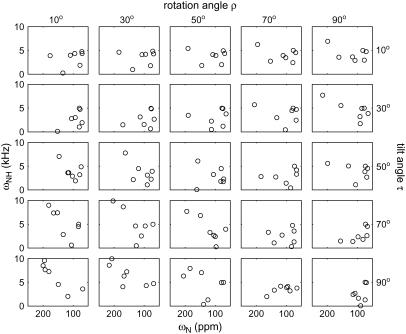
Calculated 2D 15N-1H/15N correlation spectra for the seven 15N-labeled residues in retrocyclin-2. The spectra are subsets of those in Fig. 2. Note the clear difference between the transmembrane (τ = 10°, ρ = 10°) and in-plane (τ = 90°, ρ = 90°) orientations.
FIGURE 4.
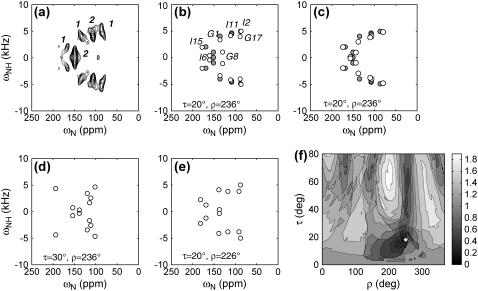
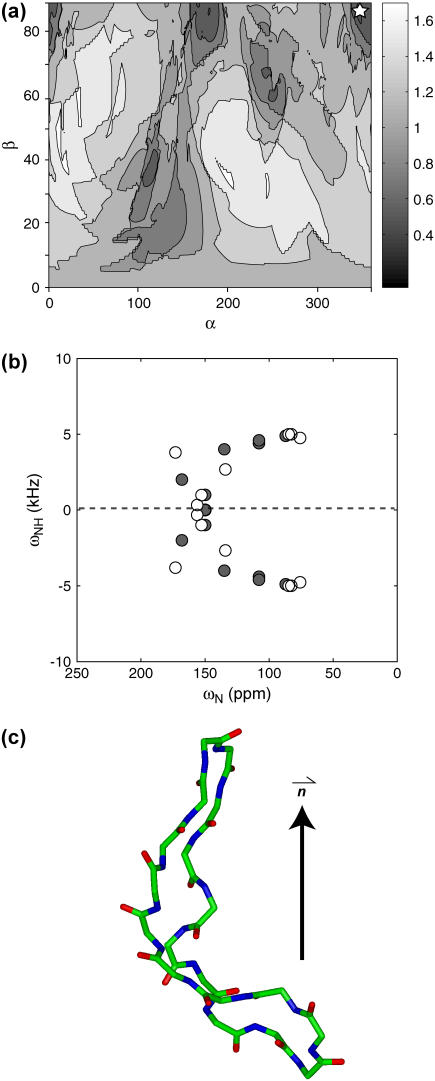
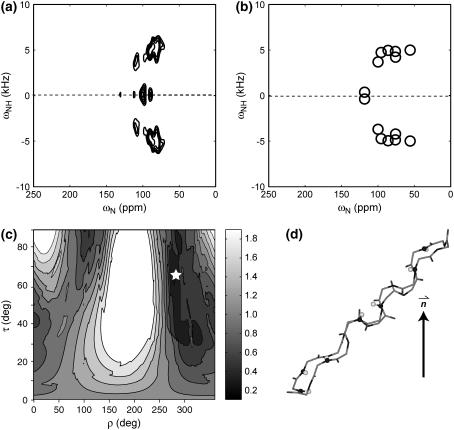
(a) Experimental 2D 15N-1H/15N correlation spectrum of retrocyclin-2 in DLPC bilayers (P/L = 1:25). The relative volumes of the resolved peaks are indicated. (b) Best-fit spectrum (open circles) using the measured 15N chemical shift principal values of the peptide, which are (42, 86, 202) ppm for Gly and (75, 76, 221) ppm for Ile. Best-fit angles: τ = 20°, ρ = 236°. Resonance assignment is indicated. For comparison, the idealized experimental spectrum (solid circles) is superimposed. (c) Best-fit spectrum using standard 15N chemical shift tensor values of (64, 77, 217) ppm (23) for all sites. The same best-fit angles as (b) are obtained, but the agreement with the experimental spectrum is better than (c), especially for the Gly-8 position. (d) Simulated 2D spectrum with τ = 30°, ρ = 236°. (e) Simulated 2D spectrum for τ = 20°, ρ = 226°. (f) RMSDs between the experiment and simulations as a function of (τ, ρ) angles. The minimum RMSD occurs at τ = 20°, ρ = 236° (star).
FIGURE 6.
(a) RMSD between the experiment and simulations using a bent hairpin structure of RTD-1. The minimum RMSD of 0.47, which is significantly higher than the experimental RMS noise of 0.20, occurs at β = 88°, α = 347° (star). (b) Best-fit simulation (open circles) superimposed with the experimental spectrum (solid circles). The two differ significantly. (c) RTD-1 structure 13 used for the simulations, showing significant curvature in the β-hairpin. The peptide is shown in its best-fit orientation, which happens to be transmembrane.
FIGURE 7.
(a) Simulated 2D 15N-1H/15N correlation spectra (open circles) of an ideal β-hairpin as a function of (τ, ρ) angles. Only the frequencies of the seven labeled residues are shown. Gray lines illustrate the orientation-dependent elliptical patterns on which the strand resonances fall. The best-fit spectrum, near (τ = 70°, ρ = 220°), does not fit the experimental spectrum (solid circles) well. (b) The ideal hairpin conformation.
FIGURE 8.
(a) Experimental 2D 15N-1H/15N correlation spectrum of retrocyclin-2 in POPC bilayers (P/L = 1:25). The peak shift to lower chemical shifts compared to the DLPC spectrum (Fig. 4 a) and the strong overlap both indicate a more in-plane orientation of the peptide. (b) Best-fit spectrum with τ = 65° and ρ = 278°. (c) RMSD between the experiment and simulations as a function of (τ, ρ). The minimum RMSD position is indicated by a star. (d) Orientation of retrocyclin-2 in POPC bilayers.
The solution NMR structure of RTD-1 (PDB code: 1HVZ) was used to represent the retrocyclin-2 structure. The RTD-1 structure has a significant backbone RMSD of 1.55 Å due to dynamic disorder in the middle of the strands (25). As a result, some of the minimum-energy structures show a substantial curvature of the β-sheet plane. We chose structure 15 to represent the straight β-hairpin population and structure 13 to represent the bent hairpin. The average (φ, ψ) angles for the strand residues (2–8 and 11–17) in structure 15 are (−103°, 99°), whereas the average (φ, ψ) angles for the strand residues in structure 13 are (−66°, 63°), which deviate significantly from the ideal β-strand geometry.
An “ideal” 18-residue β-hairpin was constructed in Insight II to further assess the sensitivity of the 2D spectra to backbone conformation. This ideal β-hairpin has uniform torsion angles of (φ = −137°, ψ = 135°) for the strand residues and (φi = −45°, ψi = 85°) and (φi+1 = 155°, ψi+1 = −20°) for the turn residues. The turn-residue torsion angles were optimized to make the two strands coplanar.
Best fits to the experimental spectrum were determined by finding the minimum root-mean squared deviation (RMSD) between the experiment and the simulations:
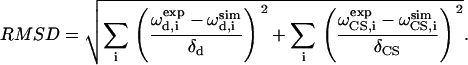 |
(1) |
Here 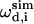 and
and 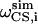 are the calculated dipolar coupling and chemical shift of residue i, respectively, whereas
are the calculated dipolar coupling and chemical shift of residue i, respectively, whereas 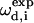 and
and 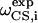 represent the frequencies of an experimental peak closest to the calculated frequencies of residue i. The frequency differences were normalized by the rigid-limit anisotropy, δd and δCS, of the interactions. The goodness of fit was assessed by comparing the RMSD with the experimental root-mean-square (RMS) noise, which was obtained by replacing
represent the frequencies of an experimental peak closest to the calculated frequencies of residue i. The frequency differences were normalized by the rigid-limit anisotropy, δd and δCS, of the interactions. The goodness of fit was assessed by comparing the RMSD with the experimental root-mean-square (RMS) noise, which was obtained by replacing 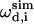 and
and 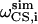 in Eq. 1 with frequencies that reflect the readout uncertainty of each peak. Resonance assignment was obtained from the best-fit spectrum: an experiment peak whose frequencies are closest to the calculated frequencies of residue i was assigned to residue i
in Eq. 1 with frequencies that reflect the readout uncertainty of each peak. Resonance assignment was obtained from the best-fit spectrum: an experiment peak whose frequencies are closest to the calculated frequencies of residue i was assigned to residue i
RESULTS
Fig. 2 b shows the calculated spectra for a range of τ and ρ-angles based on the RTD-1 structure 15. All 18 resonances are shown. The strand and turn residues are shown as unfilled and filled circles, respectively. As expected, the turn residues often give rise to outlier peaks in the 2D spectra due to their distinct N-H bond orientations from the strand residues, thus they serve as useful identifiers of the peptide orientation. The unique range of the ρ-angle is 360°; but for clarity only a 90° range is shown. Significant peak dispersion is observed in these spectra, both when the strand axis is nearly parallel (e.g., τ = 10°, ρ = 10°) and when it is perpendicular (e.g., τ = 90°, ρ = 10°) to B0. This differs from α-helical peptides, which have no dispersion when the helical axis is parallel to the bilayer normal (τ = 0°) and very limited spectral dispersion when τ = 90°. The frequency dispersion in Fig. 2 results from both the inherent twist of the β-strand and the presence of the turn residues. The largest spectral dispersion is obtained when (τ, ρ) approach (90°, 0°), which corresponds to the case where the strand axis is perpendicular to the bilayer normal whereas the β-sheet plane is parallel to the membrane normal. This orientation is unlikely for a small monomeric β-strand peptide but possible as part of a β-barrel protein. Fig. 2 shows that the transmembrane (τ→0°) and in-plane orientations (τ = 90°, ρ = 90°) have sufficient frequency differences to be distinguishable, even though in theory both their N-H bonds are perpendicular to B0. The in-plane orientation has more limited spectral dispersion, with the peaks clustered at the 90° edge of both dimensions. Fig. 3 shows the calculated spectra for the seven 15N-labeled residues in retrocyclin-2. For this subset of signals, the transmembrane (e.g., τ = 10°, ρ = 10°) and in-plane (τ = 90°, ρ = 90°) spectra are even more distinguishable.
Fig. 2 indicates that the 2D spectra of β-hairpin peptides depend sensitively on both the τ and ρ-angles. The resonance patterns are much less symmetric than the PISA wheels of α-helices, whose shape and position depend primarily on τ but not on ρ. The ρ-angle of a β-sheet peptide can be uniquely determined without resonance assignment, as long as the number of residues is smaller than the periodicity dictated by the β-sheet twist. Such periodicity can range from 25 to 108 residues depending on the β-sheet (φ, ψ) angles (4). Since typical β-strands have <15 residues, this condition is usually satisfied. Therefore, from the 2D 15N-1H/15N correlation spectra of β-sheet peptides, nondegenerate values of (τ, ρ) angles can be obtained without the need for singly labeled samples for resonance assignment, in contrast to α-helices. For β-hairpin peptides, the presence of the turn residues further facilitates orientation determination and resonance assignment.
The experimental 2D spectrum of retrocyclin-2 bound to DLPC bilayers (1:25 molar ratio) is shown in Fig. 4 a. The spectrum shows five resolved peaks, two of which contain overlapping resonances based on the peak volumes. The best-fit spectrum was obtained at (τ = 20°, ρ = 236°) and shown in Fig. 4 b along with the assignment. The simulated spectrum, calculated using 15N chemical shift principal values estimated from the MAS sideband spectrum of the peptide, agrees well with the experimental pattern for all peaks except for Gly-8, whose chemical shift differs by ∼10 ppm between the two. However, it is well known that the Gly 15N chemical shift tensor values are less well defined than the other amino acids. Indeed, when standard literature 15N chemical shift tensor values of (64, 77, 217) ppm (23) were used in the simulation, much better agreement in the Gly-8 position was obtained (Fig. 4 c), whereas the best-fit angles remain unchanged at (τ = 20°, ρ = 236°).
To assess the angular uncertainty of the measured orientation angles, we show two simulated spectra near the best fit, with τ and ρ each differing by 10° (Fig. 4, d and e). Both give spectral patterns distinctly different from the experimental spectrum. Fig. 4 f shows the 2D RMSD map as a function of (τ, ρ) at 1° increments. The global minimum at (τ = 20°, ρ = 236°) has an RMSD value of 0.26, comparable to the experimental RMS noise (0.20), whereas the two alternative simulations in Fig. 4, d and e, have much higher RMSD values of 0.63 and 0.40, confirming that the (τ, ρ) uncertainties are within ±10°. Moreover, the 2D RMSD contour plot shows a single global minimum, indicating the uniqueness of the angles determined due to the multiple frequency constraints available in the spectrum.
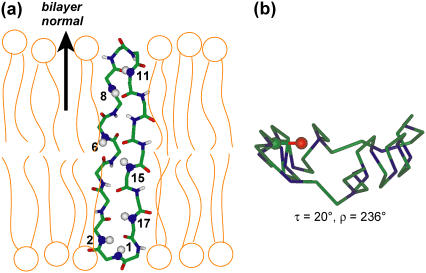
Fig. 5 shows the orientation of retrocyclin-2 in DLPC bilayers. Since the peptide is almost completely transmembrane, the plane of the β-sheet is roughly parallel to the bilayer normal despite the large ρ-angle of 236°. In other words, the peptide inserts into the membrane in a way that encounters low resistance.
FIGURE 5.
Retrocyclin-2 orientation in DLPC bilayers. (a) Viewed from the side of the DLPC bilayer. The end-to-end backbone length of the β-hairpin is 27 Å, comparable to the P-P distance of 31 Å for liquid-crystalline DLPC bilayers. (b) Viewed from the top of the lipid bilayer. The C=O bond of residue 2 used for defining the ρ-angle is highlighted. The β-sheet plane is relatively straight.
Since the 20 minimum-energy structures of RTD-1 show considerable variations in the curvature of the β-sheet, to assess whether the orientation solution depends on the peptide backbone conformation, we carried out further simulations using a bent hairpin structure (Fig. 6 c). Since it was not possible to define a meaningful β-strand axis and β-sheet plane for this structure, the simulation was carried out using the absolute orientation program. The resulting RMSD map between the simulated and the experimental spectra is shown in Fig. 6 a. The best-fit orientation occurs at (β = 88°, α = 347°) and has an RMSD value (0.47) that is more than twice the experimental uncertainty (0.20), indicating a poor fit. This can be seen clearly in the superposition of the experimental and best-fit simulated spectra in Fig. 6 b. The lack of a good fit rules out the bent hairpin conformation of retrocyclin-2 in the membrane.
The possibility of fast exchange between the straight and the bent β-hairpin conformations can be reasonably ruled out, because the 15N chemical shift anisotropies of the peptide in DLPC bilayers are close to the rigid limit values, and moreover five out of seven 15N-labeled residues are located at the rigid β-turns of the peptide.
The solution NMR structure (Fig. 5) used to fit the experimental spectrum has strand (φ, ψ) angles that span a large range: the φ angles range from −155° to −67°, whereas ψ angles range from 59° to 158°. The turn residues in this structure have torsion angles close to those of a type-II β-turn: (φ18 = −60°, ψ18 = 94°), (φ1 = 131°, ψ1 = −25°); and (φ9 = −67°, ψ9 = 80°), (φ10 = 128°, ψ10 = −18°). To test if the peptide exists as a more ideal β-hairpin in the membrane, we simulated the 2D spectra for a model β-hairpin structure (Fig. 7 a) with uniform (φ, ψ) angles. The strand torsion angles correspond to those of the classical antiparallel β-sheet. The turn residues' torsion angles were modified from those of the RTD-1 structure so that the two strands are coplanar. As expected, the calculated spectra for this ideal hairpin show much less frequency dispersion than the spectra of the actual RTD-1 structure in Fig. 3, and the resonances fall on predictable elliptical patterns indicated as gray lines in Fig. 7 (4). The global best fit using this ideal β-hairpin, near (τ = 70°, ρ = 220°), disagrees noticeably with the experimental data (solid circles). The minimum RMSD is 0.43, again significantly higher than the experimental RMS noise. Thus, the 2D data rule out this ideal β-hairpin structure for the membrane-bound retrocyclin-2.
To investigate the orientation dependence of retrocyclin-2 on membrane thickness, we performed the 2D 15N-1H/15N correlation experiment on retrocyclin-2 oriented in POPC bilayers, which have a P-P distance of ∼45 Å. The experimental spectrum (Fig. 8 a) shows smaller 15N chemical shifts and lower spectral resolution than those of the DLPC/retrocyclin-2 membrane, suggesting that the peptide has changed to a more in-plane orientation. Indeed, simulations yield best-fit angles of (τ = 65 ± 15°, ρ = 278 ± 20°) (Fig. 8, b and c), which differ substantially from the peptide orientation in DLPC bilayers. Thus, the β-hairpin orientation becomes more parallel to the membrane plane in POPC bilayers (Fig. 8 d) due to the increase of the membrane thickness.
To investigate the selective disruption of microbial membranes by retrocyclin-2, we measured the 31P spectra of oriented lipid bilayers of different compositions in the presence of 4% peptide. The zwitterionic POPC membrane retained good orientational order at this peptide concentration, whereas the mixed anionic membrane, POPC/POPG and POPE/POPG (3:1 molar ratio), showed significant powder intensities, indicating that retrocyclin-2 preferentially destroys the orientational order of anionic membranes. This resembles the behavior of RTD-1 (26). Since bacterial membranes are rich in phosphatidylglycerol lipids whereas eukaryotic membranes are not, this indicates that an important mechanism of retrocyclin action is electrostatic in origin.
DISCUSSION
The above data show that 15N 2D dipolar-shift correlation NMR is a sensitive technique for determining the orientation and restraining the secondary structure of β-sheet and β-hairpin peptides in lipid bilayers. Several aspects of this orientation determination differ from the case of α-helical peptides. First, because β-strands lack cylindrical symmetry around the main molecular axis, their 2D spectra depend characteristically on both the tilt angle of the strand axis and the rotation angle of the β-sheet plane. In contrast, the PISA wheel patterns of α-helical peptides are distinguished mostly by the tilt angle of the helical axis and not by the helix rotation angle (2). The unique dependence of the β-sheet spectra on both τ and ρ, or the lack of angular degeneracy, means that resonance assignment is not necessary for orientation determination but can be obtained, if desired, from spectral fitting directly without additional singly labeled samples. The second aspect unique to β-hairpin peptides is the presence of outlier resonances of the turn residues because of their different N-H bond orientations from the strand residues. These peaks further enhance the spectral differences between different (τ, ρ) angles and facilitate orientation determination. Third, although both transmembrane and in-plane β-hairpin peptides have N-H bonds approximately perpendicular to the bilayer normal, in practice they give distinguishable spectra. This is a result of the inherent twist of the β-sheet, the nonideality of the β-hairpin, and the unique orientations of the turn residues. For α-helical peptides, the situation is quite different: the transmembrane and in-plane orientations resonate at completely different frequencies, but both show much less spectral dispersion than β-hairpin peptides.
The 2D 15N-1H/15N correlation technique employed here can be conducted in a number of ways: for example, the 1H-15N polarization transfer can be achieved using the PISEMA sequence (27), and 1H homonuclear decoupling can be carried out using alternative sequences such as FSLG (28). These should improve the resolution in the dipolar dimension but do not change the orientation-dependent spectral patterns. The fact that both the DLPC- and POPC-bound peptide spectra have a single global best fit (Figs. 4 f and 8 c) indicates the uniqueness of the orientation determination due to the multiple frequency constraints brought about by the multiple 15N labels. Thus, the combination of extensive 15N labeling, uniaxial membrane alignment, and 2D SLF NMR, is a powerful approach for determining β-sheet peptide orientations with high precision.
The transmembrane orientation of retrocyclin-2 in DLPC bilayers determined from this study has several implications. First, how does the peptide satisfy hydrogen bonding to reduce the number of polar backbone groups exposed to the hydrophobic membrane? About half the residues in retrocyclin-2 form cross-strand intramolecular hydrogen bonds, leaving 10 residues with remaining polar N-H and C=O groups exposed to the lipid bilayer. One possibility is that retrocyclin-2 may be oligomerized so that the number of polar groups per molecule is reduced. This could be tested by a recently developed spin diffusion experiment that determines the aggregation number of peptides in lipid membranes (29). Second, the transmembrane orientation of retrocyclin-2 in DLPC bilayers is stabilized by the hydrophobic matching between the peptide backbone length and the membrane thickness. The end-to-end length of the straight hairpin is ∼27 Å, whereas the P-P distance of DLPC bilayers is 31 Å (30,31). The similar hydrophobic length supports the transmembrane orientation. Moreover, the transmembrane orientation allows the three cationic Arg sidechains at the two β-turns (residues 9, 10, 18) to interact favorably with the anionic phosphate headgroups. These favorable hydrophobic and electrostatic interactions may help to overcome the energetic cost of inserting the remaining nonhydrogen-bonded polar groups into the membrane.
When the thicker POPC bilayer is used, retrocyclin-2 changes its tilt angle to ∼65°, much closer to the in-plane orientation. Similar dependences of peptide orientations on the membrane thickness have been reported in the literature (30,32–34). For peptides much shorter than the membrane thickness, an in-plane orientation is expected. For example, the 10-residue cyclic β-hairpin peptide gramicidin S was found to be oriented parallel to the plane of dimyristoylphosphatidylcholine bilayers (35). The change of retrocyclin-2 orientation from transmembrane in 12:0 DLPC bilayers to nearly surface bound in 16:0–18:1 POPC bilayers indicates that retrocyclin-2 structure depends on the lipid environment. In principle, the headgroup structure and charge can also affect the peptide orientation. Since retrocyclin-2 disrupts the orientational order of bacteria-mimicking anionic membranes (Fig. 9), it is difficult to determine its orientation in these lipid bilayers using the aligned-membrane approach. It is also possible that retrocyclin-2, like the lipids in these anionic membranes, may adopt a distribution of orientations in these environments.
FIGURE 9.
31P spectra of retrocyclin-2 bound to various oriented lipid bilayers at a peptide concentration of 4%. (a) POPC. (b) POPC/POPG. (c) POPE/POPG.
Acknowledgments
This work is supported by the National Institutes of Health grant GM-066976 to M. Hong and grants AI-22839 and AI-37945 to A. J. Waring and R. I. Lehrer.
References
- 1.Opella, S. J., A. Nevzorov, M. F. Mesleb, and F. M. Marassi. 2002. Structure determination of membrane proteins by NMR spectroscopy. Biochem. Cell Biol. 80:597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marassi, F. M., and S. J. Opella. 2000. A solid-state NMR index of helical membrane protein structure and topology. J. Magn. Reson. 144:150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang, J., J. Denny, C. Tian, S. Kim, Y. Mo, F. Kovacs, Z. Song, K. Nishimura, Z. Gan, R. Fu, J. R. Quine, and T. A. Cross. 2000. Imaging membrane protein helical wheels. J. Magn. Reson. 144:162–167. [DOI] [PubMed] [Google Scholar]
- 4.Marassi, F. M. 2001. A simple approach to membrane protein secondary structure and topology based on NMR spectroscopy. Biophys. J. 80:994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vosegaard, T., and N. C. Nielsen. 2002. Towards high-resolution solid-state NMR on large uniformly 15N- and [13C,15N]-labeled membrane proteins in oriented lipid bilayers. J. Biomol. NMR. 22:225–247. [DOI] [PubMed] [Google Scholar]
- 6.Lehrer, R. I., and T. Ganz. 2002. Defensins of vertebrate animals. Curr. Opin. Immunol. 14:96–102. [DOI] [PubMed] [Google Scholar]
- 7.Epand, R. M., and H. J. Vogel. 1999. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta. 1462:11–28. [DOI] [PubMed] [Google Scholar]
- 8.Hancock, R. E., and M. G. Scott. 2000. The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. USA. 97:8856–8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bechinger, B. 1999. The structure, dynamics, and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta. 1462:157–183. [DOI] [PubMed] [Google Scholar]
- 10.Wade, D., A. Boman, B. Wahlin, C. M. Drain, D. Andreu, H. G. Boman, and R. B. Merrifield. 1990. All D-amino acid containing channel forming antibiotic peptides. Proc. Natl. Acad. Sci. USA. 87:4761–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Owen, S. M., D. Rudolph, W. Wang, A. M. Cole, M. A. Sherman, A. J. Waring, R. I. Lehrer, and R. B. Lal. 2004. A theta-defensin composed exclusively of D-amino acids is active against HIV-1. J. Pept. Res. 63:469–476. [DOI] [PubMed] [Google Scholar]
- 12.Cole, A. M., W. Wang, A. J. Waring, and R. I. Lehrer. 2004. Retrocyclins: using past as prologue. Curr. Protein Pept. Sci. 5:373–381. [DOI] [PubMed] [Google Scholar]
- 13.Cole, A. M., T. Hong, K. M. Boo, T. Nguyen, C. Zhao, G. Bristol, J. A. Zack, A. J. Waring, O. O. Yang, and R. I. Lehrer. 2002. Retrocyclin: a novel primate peptide that protects cells from infection by T- and M-tropic strains of HIV-1. Proc. Natl. Acad. Sci. USA. 99:1813–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munk, C., G. Wei, O. O. Yang, A. J. Waring, W. Wang, T. Hong, R. I. Lehrer, N. R. Landau, and A. M. Cole. 2003. The theta-defensin, retrocyclin, inhibits HIV-1 entry. AIDS Res. Hum. Retroviruses. 19:875–881. [DOI] [PubMed] [Google Scholar]
- 15.Yasin, B., W. Wang, M. Pang, N. Cheshenko, T. Hong, A. J. Waring, B. C. Herold, E. A. Wagar, and R. I. Lehrer. 2004. Theta defensins protect cells from infection by herpes simplex virus by inhibiting viral adhesion and entry. J. Virol. 78:5147–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang, Y. Q., J. Yuan, G. Osapay, K. Osapay, D. Tran, C. J. Miller, A. J. Ouellette, and M. E. Selsted. 1999. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science. 286:498–502. [DOI] [PubMed] [Google Scholar]
- 17.Wang, W., A. M. Cole, T. Hong, A. J. Waring, and R. I. Lehrer. 2003. Retrocyclin, an antiretroviral theta-defensin, is a lectin. J. Immunol. 170:4708–4716. [DOI] [PubMed] [Google Scholar]
- 18.Fields, C. G., D. H. Lloyd, R. L. Macdonald, K. M. Ottenson, and R. L. Nobel. 1991. HBTU activation for automated Fmoc solid-phase peptide synthesis. Pept. Res. 4:95–101. [PubMed] [Google Scholar]
- 19.Hallock, K. J., K. Henzler Wildman, D. K. Lee, and A. Ramamoorthy. 2002. An innovative procedure using a sublimable solid to align lipid bilayers for solid-state NMR studies. Biophys. J. 82:2499–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhim, W.-K., D. D. Elleman, and R. W. Vaughan. 1973. Analysis of multiple-pulse NMR in solids. J. Chem. Phys. 59:3740–3749. [Google Scholar]
- 21.Mansfield, P. 1971. Symmetrized pulse sequences in high resolution NMR in solids. J. Phys. C Solid State Phys. 4:1444–1452. [Google Scholar]
- 22.Yamaguchi, S., T. Hong, A. Waring, R. I. Lehrer, and M. Hong. 2002. Solid-state NMR investigations of peptide-lipid interaction and orientation of a beta-sheet antimicrobial peptide, protegrin. Biochemistry. 41:9852–9862. [DOI] [PubMed] [Google Scholar]
- 23.Wu, C. H., A. Ramamoorthy, L. M. Gierasch, and S. J. Opella. 1995. Simultaneous characterization of the amide 1H chemical shift, 1H–15N dipolar, and 15N chemical shift interaction tensors in a peptide bond by three-dimensional solid-state NMR. J. Am. Chem. Soc. 117:6148–6149. [Google Scholar]
- 24.Hong, M., J. D. Gross, W. Hu, and R. G. Griffin. 1998. Determination of the peptide torsion angle phi by 15N chemical shift and 13Ca-1Ha dipolar tensor correlation in solid-state MAS NMR. J. Magn. Reson. 135:169–177. [DOI] [PubMed] [Google Scholar]
- 25.Trabi, M., H. J. Schirra, and D. J. Craik. 2001. Three-dimensional structure of RTD-1, a cyclic antimicrobial defensin from rhesus macaque leukocytes. Biochemistry. 40:4211–4221. [DOI] [PubMed] [Google Scholar]
- 26.Buffy, J. J., M. J. McCormick, S. Wi, A. Waring, R. I. Lehrer, and M. Hong. 2004. Solid-state NMR investigation of the selective perturbation of lipid bilayers by the cyclic antimicrobial peptide RTD-1. Biochemistry. 43:9800–9812. [DOI] [PubMed] [Google Scholar]
- 27.Wu, C. H., A. Ramamoorthy, and S. J. Opella. 1994. High-resolution heteronuclear dipolar solid-state NMR spectroscopy. J. Magn. Reson. A. 109:270–272. [Google Scholar]
- 28.Bielecki, A., A. C. Kolbert, H. J. M. de Groot, R. G. Griffin, and M. H. Levitt. 1990. Frequency-switched Lee-Goldburg sequences in solids. Adv. Magn. Reson. 14:111–124. [Google Scholar]
- 29.Buffy, J. J., A. J. Waring, and M. Hong. 2005. Determination of peptide oligomerization in lipid membranes with magic-angle spinning spin diffusion NMR. J. Am. Chem. Soc. 127:4477–4483. [DOI] [PubMed] [Google Scholar]
- 30.Harroun, T. A., W. T. Heller, T. M. Weiss, L. Yang, and H. W. Huang. 1999. Experimental evidence for hydrophobic matching and membrane-mediated interactions in lipid bilayers containing gramicidin. Biophys. J. 76:937–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kucerka, N., Y. Liu, N. Chu, H. I. Petrache, S. A. Tristram-Nagle, and J. F. Nagle. 2005. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using x-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys. J. 88:2626-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harzer, U., and B. Bechinger. 2000. Alignment of lysine-anchored membrane peptides under conditions of hydrophobic mismatch: a CD, 15N and 31P solid-state NMR spectroscopy investigation. Biochemistry. 39:13106–13114. [DOI] [PubMed] [Google Scholar]
- 33.de Planque, M. R., and J. A. Killian. 2003. Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring. Mol. Membr. Biol. 20:271–284. [DOI] [PubMed] [Google Scholar]
- 34.Park, S. H., and S. J. Opella. 2005. Tilt angle of a trans-membrane helix is determined by hydrophobic mismatch. J. Mol. Biol. 350:310–318. [DOI] [PubMed] [Google Scholar]
- 35.Salgado, J., S. L. Grage, L. H. Kondejewski, R. S. Hodges, R. N. McElhaney, and A. S. Ulrich. 2001. Membrane-bound structure and alignment of the antimicrobial β-sheet peptide gramicidin S derived from angular and distance constraints by solid state 19F-NMR. J. Biomol. NMR. 21:191–208. [DOI] [PubMed] [Google Scholar]