Abstract
We have applied tethered particle microscopy (TPM) as a single molecule analysis tool to studies of the conformational dynamics of poly-uridine(U) messenger (m)RNA and 16S ribosomal (r)RNA molecules. Using stroboscopic total internal reflection illumination and rigorous selection criteria to distinguish from nonspecific tethering, we have tracked the nanometer-scale Brownian motion of RNA-tethered fluorescent microspheres in all three dimensions at pH 7.5, 22°C, in 10 mM or 100 mM NaCl in the absence or presence of 10 mM MgCl2. The addition of Mg2+ to low-ionic strength buffer results in significant compaction and stiffening of poly(U) mRNA, but not of 16S rRNA. Furthermore, the motion of poly(U)-tethered microspheres is more heterogeneous than that of 16S rRNA-tethered microspheres. Analysis of in-plane bead motion suggests that poly(U) RNA, but less so 16S rRNA, can be modeled both in the presence and absence of Mg2+ by a statistical Gaussian polymer model. We attribute these differences to the Mg2+-induced compaction of the relatively weakly structured and structurally disperse poly(U) mRNA, in contrast to Mg2+-induced reinforcement of existing secondary and tertiary structure contacts in the highly structured 16S rRNA. Both effects are nonspecific, however, as they are dampened in the presence of higher concentrations of monovalent cations.
INTRODUCTION
RNA is a negatively charged biopolymer that fulfills a central functional role in all aspects of cellular gene expression (for some recent reviews, please see Noller (1), Zamore and Haley (2), and Moore (3)). Among its many capabilities, it is a pliable informational biomolecule that carries the genetic code en route from the genome to the protein-making machinery in the form of messenger (m)RNA, and as ribosomal (r)RNA, it catalyzes the peptidyl transferase reaction necessary for peptide synthesis within the ribosome (1,4,5). An impressive variety of secondary and tertiary structures are adopted by RNA to carry out such diverse functions, considering its limited repertoire of four nucleobase side chains (reviewed in Leontis and Westhof (6)). The versatility of RNA lies in its ability to rapidly transform from a disordered extended chain into a compact, correctly folded structure. Solving the conundrum of how it overcomes the associated RNA folding problem requires understanding how the strong repulsion of the negatively charged backbone phosphates is overcome (7).
RNA binds monovalent and divalent cations that neutralize the backbone phosphate charges and are required for proper folding (reviewed in (8–11)). In general, RNA folding is aided by association with monovalent cations such as Na+ or K+, which screen backbone charges and promote basepairing. In addition, binding of divalent cations, usually Mg2+, is thought to be required for the formation of specific tertiary contacts. Kinetic ensemble folding experiments have demonstrated that large structured RNAs often undergo a fast compaction step after a secondary structure is adopted but before a stable tertiary structure is formed (12–15). Such electrostatic relaxation requires high concentrations of monovalent cations often in excess of 100 mM (12) to effect charge neutralization or ∼10- to 100-fold lower concentrations of Mg2+ (13).
A large structured RNA that has been of particular interest in recent years is 16S rRNA, a highly conserved 1542-nucleotide RNA molecule that comprises the bulk of the small (30S) ribosomal subunit in bacteria (Fig. 2). With multiple reports of ribosomal crystal structures in recent years, it has become important to understand the assembly process of the ribosomal subunits that ultimately form the complex protein biosynthesis machine. The order of assembly of ribosomal 30S subunit proteins on “naked” 16S rRNA in vitro has been known for decades (16,17). Conformational changes in portions of 16S rRNA have been studied both in isolation and in the presence of protein ligands (18–20), yet little is known about the global structural properties of the naked 16S rRNA polymer, which represents the binding scaffold for the first layer of ribosomal proteins.
FIGURE 2.
Secondary structure of 16S rRNA from E. coli as determined by comparative sequence analysis (68).
In this study, we have applied tethered particle microscopy (TPM), a single molecule technique which is emerging as a valuable tool, to the study of two conformationally distinct RNA molecules, poly(U) mRNA and 16S rRNA. More specifically, we have observed the Brownian motion of a microsphere attached to the end of a single immobilized RNA molecule at varying ionic strength (Fig. 1) and have analyzed the bead motion to reveal information about the folded state of the RNA tether. TPM previously has been used to measure the rate of repressor-induced loop formation and breakdown in DNA (21) to characterize the relationship between the kinetics of transcription to rates of single polymerase molecules (22) and to study the strength of DNA-protein interactions (23). Recently, TPM has been extended to RNA; protein synthesis by single ribosomes has been observed via microspheres attached to poly(U) mRNAs (24). However, to our knowledge, the utility of this method for observation of metal ion induced RNA compaction and folding has not before been tested. A single molecule approach such as TPM has the advantage over bulk solution methods in that it allows for the observation of individual molecules instead of an averaged ensemble, so that it can resolve potential heterogeneity in the folding pathway among a population of molecules. Recently, single molecule fluorescence resonance energy transfer has proven to be a powerful technique in the study of RNA tertiary structure formation (reviewed in (25–27)). TPM complements FRET in the characterization of the folding of large RNAs since FRET methods are generally limited to probing distances of <10 nm (25–31).
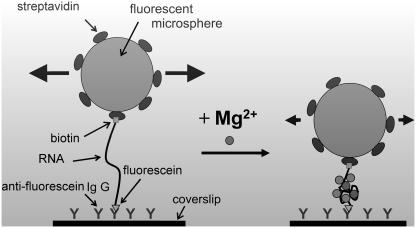
FIGURE 1.
Schematic of RNA-tethered fluorescent microspheres in a shallow-penetrating evanescent field (shaded) generated by total-internal reflection fluorescence (TIRF) illumination. The fluorescent microspheres are streptavidin coated and bind tightly to the 3′-biotinylated RNA tether. The 5′ terminus of the RNA is fluorescein labeled and adheres to the glass substrate via goat anti-fluorescein IgG. Upon addition of magnesium ions and subsequent compaction of the RNA tether, the Brownian motion of the microsphere becomes attenuated.
Employing TPM, we observe a slight attenuation of motion upon addition of Mg2+ to beads tethered by Escherichia coli 16S rRNA under low-ionic strength conditions (10 mM Na+). We observe a much more pronounced Mg2+-induced effect for beads tethered by less structured poly(U) mRNA, implying that this RNA undergoes a larger degree of compaction upon addition of divalent cations. These data confirm that Mg2+ efficiently promotes collapse of our representative RNA molecules into a more rigid and compact form, irrespective of a defined tertiary structure, and corroborate evidence from bulk solution biophysical methods, suggesting that large RNAs experience global collapse before specific tertiary structure is formed (15). In addition, we observe that the degree of motion for poly(U) tethered microspheres is much less homogeneous than that of their 16S rRNA-tethered counterparts, both in the absence and presence of Mg2+. This suggests that the defined secondary (and tertiary) structure of 16S rRNA, as compared with poly(U) mRNA, generally dictates a more homogeneous global fold, giving rise to more uniform tethered bead behavior. These observations are consistent with a high number of degrees of freedom afforded by poly(U), which results in a large conformational space of transient hydrogen bonds and base stacking interactions between sequentially remote regions of the mRNA. These interactions, which are likely to be different from molecule to molecule, are stabilized by the presence of Mg2+ and thus give rise to the heterogeneous behavior of poly(U)-tethered beads. Spring constants derived from the energy profiles of bead displacement show that, in general, the poly(U), but not the 16S rRNA tethers become significantly stiffer upon Mg2+-induced compaction. Finally, we find that poly(U) mRNA, but less so 16S rRNA, can realistically be modeled both in the presence and absence of Mg2+ by a simple Gaussian polymer model (32,33).
MATERIALS AND METHODS
RNA purification and labeling
16S rRNA was purified from whole E. coli ribosomal RNA (Boehringer, Mannheim, Germany) by electrophoresis using a denaturing 4%, 8 M urea, polyacrylamide gel. Poly(U) mRNA (Sigma, St. Louis, MO and Midland, TX) ranging in size from ∼600 nt to 6000 nt was also separated by molecular weight on such a gel. An RNA ladder (Promega, Madison, WI) was loaded on the gel adjacent to the poly(U) mRNA to allow for size discrimination. All RNA bands were visualized with ethidium bromide staining, and the proper bands were excised and crushed. Five narrow size ranges of poly(U) mRNA were obtained from gel fractionation: 1.0–1.2 kilobases (kb), 1.4–1.9 kb, 1.9–2.6 kb, 3.6–5.0 kb, and >5.0 kb (the upper limit of this RNA sample varied from lot to lot but was unlikely to contain RNA longer than ∼10,000 nt). The RNA was eluted from the gel fragments in buffer (0.5 M ammonium acetate, 0.1% sodium dodecyl sulfate (SDS), 1 mM EDTA) overnight at 4°C while tumbling. The eluted RNA was chloroform extracted to remove SDS and was subsequently ethanol precipitated.
The recovered RNA fractions were 5′ end labeled with fluorescein via a two-step reaction as described (34). More specifically, the 5′ phosphate was reacted with EDC (1-ethyl 3-(3-diethylaminopropyl) carbodiimide) and carbohydrazide (to create a strong nucleophile for the coupling with 5-fluorescein isothiocyanate (5-FITC)) under the following reaction conditions: 10 mM MgCl2, 50 mM NaCl, 18 μM carbohydrazide, 5.2 μM EDC, and 72 nM 16S rRNA. The reaction was incubated at 10°C in the dark and ethanol precipitated. In the second step, the RNA was resuspended in 100 μL of 70 mM HEPES-KOH, pH 7.0, 10 mM MgCl2, 30 mM NaCl; 5-FITC was added to a final concentration of 10 mM. The reaction was incubated at 22°C for 3 h and subsequently phenol extracted. The RNA was again ethanol precipitated.
Next, the RNA was 3′ end labeled with biotin in another two-step reaction (34). First, RNA was dried and resuspended in 100 μL of oxidation buffer consisting of 10 mM HEPES-KOH, pH 7.5, 1 mM MgCl2. After the addition of potassium periodate (KIO4) to a final concentration of 6.14 mM, the reaction was incubated at 0°C for 20 min. This reaction converts the vicinal 2′ and 3′ hydroxyls of the 3′ terminal nucleotide into an electrophilic aldehyde functionality. The RNA was again ethanol precipitated and resuspended in 100 μL 50 mM sodium acetate, pH 5.0, in preparation for the second reaction. A total of 1 μL of a saturated biotin hydrazide solution in water was added and the reaction incubated at 0°C overnight. The RNA was then ethanol precipitated three times to remove excess biotin.
Preparation of flow chambers and TPM samples
Two 1.1-mm holes were drilled into a glass microscope slide and the slide was subsequently rinsed with methanol. Tygon tubing was threaded through each hole, secured with epoxy resin, and trimmed to form the inlet and outlet ports of the flow cell. Double-sided sticky tape was mounted onto the surface of the slide, and a narrow channel was cut through the center to connect the two ports. Glass coverslips, previously washed with a 1:2 mixture of 30% (v/v) hydrogen peroxide and concentrated sulfuric acid, were rinsed and shaken for 1.5 h in a 0.00025% polyethylene glycol (10,000 Da) solution at room temperature. After drying, the coverslips were adhered to the surface of the tape, creating a fully enclosed channel subsequently sealed with epoxy resin on all sides.
Antifluorescein IgG was immobilized on the surface of the coverslip by pushing dilute (0.05 μg/μL) antibody stock solution (∼30 μL) through the inlet tube and incubating at room temperature for >3 h, after which the flow channels were flushed with phosphate-buffered saline, pH 7.4, and a 1:20 diluted blocking solution (Block Aid, Molecular Probes, Eugene, OR). A total of 200 μg of unlabeled poly(U) mRNA (Sigma) was added to the blocking solution to suppress a small residual RNase activity found in Block Aid (D. Möll and P. Guo, Purdue University, 2003, personal communication). The blocking mixture was allowed to incubate in the flow cell for >1 h. In parallel to this antibody immobilization, microspheres were tethered to RNA in the following way. A total of 0.56 μM green-yellow fluorescent streptavidin-coated polystyrene beads (Bangs Laboratories, Fishers, IN) were washed in TE-NaCl buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 10 mM NaCl or 100 mM NaCl (depending on desired experimental conditions)) and sonicated to prevent aggregation; labeled biotinylated RNA was added to the bead suspension to a final concentration of >1 nM and tumbled at room temperature for ∼3 h. The bead solution was then injected into the flow chamber and tumbled overnight at room temperature in the dark.
TPM data collection
Objective-type total internal reflection fluorescence (TIRF) illumination with stroboscopic excitation was used to generate an evanescent field on the bottom surface of the flow cell, as described (35). Briefly, backward-propagating fluorescence emission was focused onto a Photometrics (Tuscon, AZ) Cascade 650 charge-coupled device camera (CCD) (each pixel corresponds to 91 nm × 91 nm) and images were captured using WinSpec32 software. Data were recorded at 22°C with a frame rate of 10–100 Hz, depending on the size of the region of interest. Before data collection, TE-NaCl buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, and either 10 mM NaCl or 100 mM NaCl, depending on desired background monovalent cation conditions) was pushed through each cell to flush out free-floating microspheres. After proper focus was attained, ∼40 s of video were collected for a chosen field of view with several tethered beads. Then, buffer was exchanged and data collected on the same field of view so that the same tether could be observed under different Mg2+ concentrations. For all data presented in this study, the high Mg2+ concentration buffer consisted of sterile-filtered Tris-HCl buffer, pH 7.5, 10 mM MgCl2, and either 10 mM or 100 mM NaCl, depending on the desired ionic strength for an individual experiment.
Data analysis
A suite of processing scripts was written in-house for use with MATLAB 6.0 software (MathWorks, Natick, MA) to analyze the acquired video files. For each selected microsphere, in-plane and out-of-plane positions were extracted. The penetration depth of the evanescent field was ∼200 nm as calibrated using an optically trapped microsphere (35); z-position values, including standard deviation, are therefore expressed in nanometers. A moving-average was subtracted from all position measurements to correct for slow-scale drift and photobleaching. The filtered position data were used to calculate the standard deviation of the bead motion, the minimum and maximum excursion of the bead in the x- and y-dimensions, and the average in-plane distance of the bead from the anchor point. In addition, symmetry values symp and symi were calculated that represent ratios of the length of the major and minor axes of an ellipse described by the x-,y-position scatter plot and the bead image, respectively, as described (35). A symmetry value of 1, corresponding to a perfect circle, is ideal. Time constants were computed from a single-exponential decay fit to the autocorrelation of the position data.
Nonspecifically and multiply tethered beads have been previously observed in TPM studies (36). Microspheres that were likely to be tethered specifically by a single RNA molecule were therefore identified using the following selection criteria, as described (35): 1), minimum root mean square (rms) motion of 50 nm; this eliminates stuck beads and beads whose motion is severely restricted by multiple tethers; 2), averaged image symmetry value, symi, <1.5; this eliminates bead aggregates from the data set; this statistic required a relatively high threshold for selection because the microspheres themselves were often not perfectly spherical; and 3), number of dropped frames <10% of total frames; this criterion eliminates beads from the data set that may not have good contrast with the background. These criteria were found to be useful in distinguishing true tethers from those interacting nonspecifically with the glass surface. As additional control, the position symmetry value, symp, was computed; 81% of all data sets were found to have symp values of <1.5, indicating free Brownian motion of the tethered beads, and providing evidence for a single tether anchoring each of the microspheres (on average, symp increased with tether length). The overall percentages of data sets identified as acceptable for each RNA tether length using the imposed selection criteria are reported in Table 1. The same selection criteria were used for data collection at both magnesium chloride concentrations. For a given Mg2+ concentration, all microspheres that passed the selection criteria were included in the data analysis, although not all of the microspheres were observable at both concentrations.
TABLE 1.
Total number of analyzed data sets and number of acceptable data sets that passed the selection criteria for each RNA tether length
| 0 mM Mg2+
|
10 mM Mg2+
|
|||
|---|---|---|---|---|
| Total No. | No. of acceptable data sets | Total No. | No. of acceptable data sets | |
| Poly(U) mRNA, 1.0–1.2 kb | 111 | 41 (27*) (36.9%) | 90 | 41 (45.5%) |
| Poly(U) mRNA, 1.4–1.9 kb | 185 | 21 (4*) (11.4%) | 187 | 25 (13.4%) |
| Poly(U) mRNA, 1.9–2.6 kb | 93 | 36 (11*) (38.7%) | 80 | 18 (22.5%) |
| Poly(U) mRNA, 3.6–5.0 kb | 116 | 12 (0*) (10.3%) | 122 | 3 (2.5%) |
| Poly(U) mRNA, 5.0–7.0 kb | 277 | 21 (7*) (7.6%) | 324 | 20 (6.2%) |
| Poly(U) mRNA, 4.5–5.5 kb (in 100 mM NaCl) | 37 | 18 (16*) (48.6%) | 35 | 18 (51.4%) |
| 16S rRNA, 1.542 kb | 200 | 35 (10*) (17.5%) | 199 | 25 (12.5%) |
An experiment performed at higher than standard monovalent cation concentration is indicated. Numbers in parentheses with an asterisk (*) denote the numbers of beads for which data were obtained at both Mg2+ concentrations.
RESULTS
Poly(U) mRNA undergoes largely nonspecific compaction and stiffening upon binding Mg2+
In aqueous solution, uridine is the nucleoside that is least likely to self-associate and stack (37). Therefore, poly(U) is more likely than any other RNA sequence to adopt random coil configurations in solution rather than more defined single-stranded helices. The inherent bias of poly(U) toward a random coil makes it an excellent molecule to contrast with the highly structured 16S rRNA. To examine the physical properties of five different size ranges of poly(U) mRNA, 1.0–1.2 kb, 1.4–1.9 kb, 1.9–2.6 kb, 3.6–5.0 kb, and 5.0–7.0 kb (see Materials and Methods), the Brownian motion of fluorescent microspheres tethered to single RNA molecules was observed via nanometer scale position fluctuations of the attached beads under different ionic conditions. Based on an average rise of 4 Å per nucleotide in single-stranded helical RNA (38), we can expect the maximum length in solution of a poly(U) molecule from each of our size ranges to be ∼480 nm, ∼760 nm, ∼1,040 nm, ∼2,000 nm, and ∼3,000 nm, respectively. Indeed, we find our RNA tethers to be considerably shorter; the raw x-position data as a function of time for a typical bead tethered by a 5.0–7.0 kb poly(U) mRNA molecule is shown in Fig. 3 a. A histogram of these data is shown in Fig. 3 b with a Gaussian function fit to the data distribution, consistent with a stochastic nature of the bead fluctuations (39). A scatter plot of y- as a function of x-position (Fig. 3 c) finds a symmetric distribution of the position about the center of the plot. The value for the position symmetry statistics, symp (see Materials and Methods), approaches 1.0, the ideal value for a bead anchored to the substrate at a single point, and demonstrates that the microsphere is likely tethered by a single RNA molecule (40). In addition, energy profiles (in units of kBT, where kB is the Boltzmann constant and T is the temperature), as derived from Boltzmann statistics
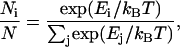 |
(1) |
are shown for the representative particle in Fig. 3 d. These symmetric energy profiles demonstrate that the bead motion is bound by a harmonic potential, which again is consistent with a single anchoring point for the tether. Spring constants, k, in pN/μM, were extracted from quadratic fits of the energy potentials (35) and are discussed below.
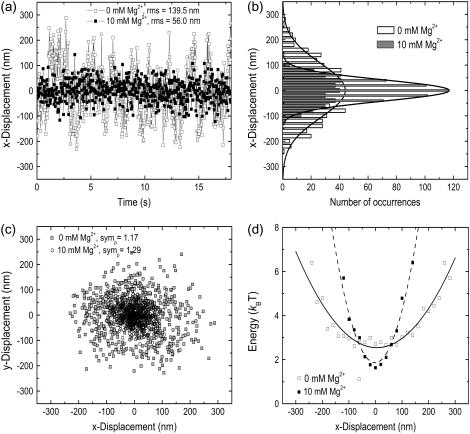
FIGURE 3.
Analysis of microsphere motion for a 5.0–7.0 kb poly(U) mRNA tether at 0 mM and 10 mM Mg2+ (in 10 mM Tris-HCl, pH 7.5, 10 mM NaCl, at 22°C). (a) Plot of raw x-position time course for 0 mM Mg2+ (open squares) and 10 mM Mg2+ (solid squares). rms x-,y-motion is labeled for both magnesium ion concentrations. (b) Histograms of position data shown in (a), fit with Gaussian distributions (curves). The open bars and fitted solid curve represent 0 mM Mg2+ data; the gray shaded bars and associated curve represent 10 mM Mg2+ data. (c) Scatter plot of x-position versus y-position for the same microsphere. Position symmetry statistics (symp) for each magnesium ion concentration are shown. Modest dampening of bead motion, a sign of RNA tether compaction, is observed upon addition of Mg2+. (d) Energy profiles for the same particle, calculated for both 0 mM and 10 mM Mg2+ data sets and fit with quadratic functions to yield spring constants reported in the text (solid curve, 0 mM Mg2+; dashed curve, 10 mM Mg2+).
To measure the Mg2+-induced degree of compaction of the poly(U) mRNA, flow chambers that allow for exchange of the buffer environment were used so that the same tethered bead could be observed under different magnesium ion concentrations. The observation of a single tether under different solution conditions ensured that any Mg2+-induced variations were attributable to changes in the structure or dynamic properties of the RNA tether and not induced by bead-to-bead or sample-to-sample variations. In general, the addition of 10 mM MgCl2 resulted in a decrease in the rms translational motion of the bead (labeled “rms,” Fig. 3 a). This can be observed as a narrowing of the Gaussian profile of the position data (Fig. 3 b) and a comparatively smaller diameter for the circle described by the x-,y-scatter plot (Fig. 3 c). Microspheres were also observed after the addition of 100 mM Mg2+, but higher levels of magnesium did not induce further compaction (data not shown). For simplicity, single molecule TPM data are grouped into two categories for each poly(U) size range, 0 mM and 10 mM Mg2+.
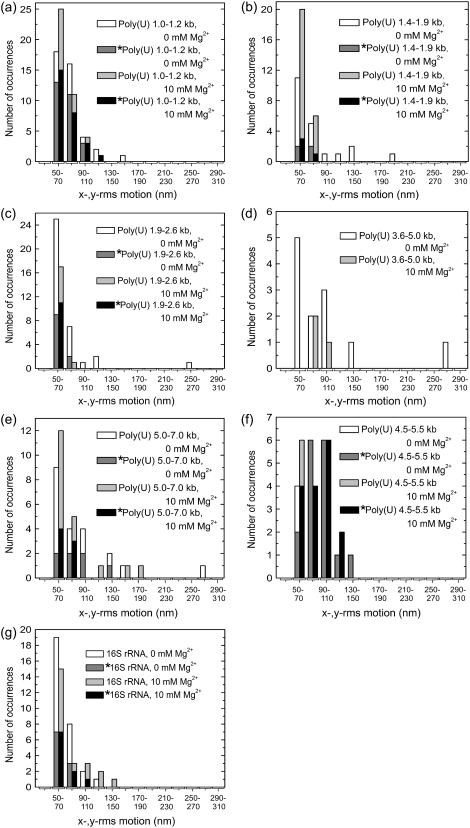
General trends in the motion observed for the beads tethered by poly(U) mRNA are summarized in Table 2 and as histograms of average rms motion for each poly(U) group in Fig. 4. We observed a relatively broad range of x-,y-rms motion values for each poly(U) size group at 0 mM Mg2+, indicating that the behavior of the RNA tethers is heterogeneous over the population. The range of rms values observed at 0 mM Mg2+, but not the mean, was found to be correlated with the tether length; this may be related to the increasingly heterogeneous behavior among longer poly(U) tethers. Furthermore, for each size group, the range of rms values as well as their mean generally decreases with the addition of 10 mM Mg2+.
TABLE 2.
Mean values and ranges of x-,y-rms motion values observed for poly(U) mRNA and 16S rRNA tethers of different lengths
| Mg2+ concentration (mM) | Mean x-,y- rms motion ± SD (nm) | Range or difference between smallest and largest rms values in the group (nm) | |
|---|---|---|---|
| Poly(U) mRNA, 1.0–1.2 kb | 0 | 75 ± 21 | 103 |
| 10 | 70 ± 14 | 60 | |
| Poly(U) mRNA, 1.4–1.9 kb | 0 | 81 ± 37 | 142 |
| 10 | 64 ± 9 | 31 | |
| Poly(U) mRNA, 1.9–2.6 kb | 0 | 73 ± 36 | 201 |
| 10 | 58 ± 8 | 35 | |
| Poly(U) mRNA, 3.6–5.0 kb | 0 | 99 ± 64 | 226 |
| 10 | 87 ± 8 | 14 | |
| Poly(U) mRNA, 5.0–7.0 kb | 0 | 94 ± 52 | 230 |
| 10 | 77 ± 33 | 119 | |
| 16S rRNA, 1.542 kb | 0 | 69 ± 17 | 69 |
| 10 | 76 ± 23 | 80 | |
| Poly(U) mRNA, 4.5–5.5 kb (in 100 mM NaCl)
|
0 | 89 ± 21 | 85 |
| 10 | 84 ± 22 | 73 |
An experiment performed at higher than standard monovalent cation concentration is indicated.
FIGURE 4.
Histograms of x-,y-rms motion for poly(U) mRNA 1.0–1.2 kb (a), 1.4–1.9 kb (b), 1.9–2.6 kb (c), 3.6–5.0 kb (d), and 5.0–7.0 kb in length (e), and for 16S rRNA (g) tethers in 10 mM NaCl, and 0 mM or 10 mM Mg2+, as indicated. Analogous plots for 4.5–5.5 kb poly(U) mRNA in 100 mM NaCl are shown in panel f. Data are grouped in two 20-nm bins, where the left and right bins correspond to 0 mM and 10 mM Mg2+ conditions, respectively. The darker shaded portion of each bar (denoted by an asterisk (*)) represents the subset of total beads in that particular bin for which data were obtained at both Mg2+ concentrations. These histograms demonstrate that the range of motion for poly(U) mRNA-tethered beads decreases with the addition of magnesium ions, whereas the range of motion for 16S rRNA-tethered beads largely remains unchanged.
Mg2+-induced compaction of the poly(U) mRNA was also observed through analysis of relative motion of attached microspheres in the z-dimension, as possible through TIRF illumination and evaluation of fluorescence fluctuations due to varying excitation depths within the evanescent field, which rapidly decays with distance from the substrate surface (35). Plots of the standard deviation in z-position as a function of rms motion in the x-,y-dimensions for representative poly(U) length ranges at 0 mM Mg2+ and 10 mM Mg2+ demonstrate that, in general, the x-,y-rms motion decreases more with the addition of metal ions than the corresponding z-standard deviation does (data not shown). This indicates that in these experiments, the x-,y-position information is more sensitive to fluctuations in bead motion and is likely a more reliable indicator of tether end-to-end distance and thus, RNA conformation. Previous studies of DNA tethers have also shown increased sensitivity of in-plane bead motion as compared with out-of-plane position fluctuations (35). These observations were attributed in part to the inability of the microsphere to penetrate the anchoring surface, which imposes an effective force on the tether (35).
Whereas changes in x-,y-rms motion of an attached microsphere report on changes in length and overall volume of the tether, time constants (τ) obtained from autocorrelation analysis of the bead's translational motion reveal information concerning the dynamic properties of the RNA. More specifically, time constants describe the rate of motion of the harmonically bound particle in its viscous environment. Theoretical models predict that, for a bead-and-spring system such as ours, dynamic behavior of the tether may either be dominated by the bead, the polymer (RNA, in our case), or both (39). The time constant can be expressed as a function of polymer length (N) and relative size of the tethered particle (σ), where
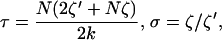 |
(2) |
and k is the spring constant of the polymer. Since the volume of the bead in our system is very large relative to that of the RNA, the frictional coefficient for the bead motion through the solvent (ζ′) will contribute significantly to the overall friction coefficient of the tether construct and thus significantly affect the relaxation time constant (τ). However, according to the theoretical model described by Qian and Elson (39), which takes into account the relative size of the bead, the RNA tethers used in our study consist of a large enough number (N) of nucleotides (or segments, each with frictional coefficient ζ) to affect τ-values significantly as well. Using Stokes law,
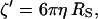 |
(3) |
where RS is the Stokes radius of the particle (0.23 μm) and η is the viscosity of the buffer (∼0.001 Ns/m2 for an aqueous solution at 22°C), we may calculate the frictional coefficient of the bead in our system to be ∼4.7 × 10−9. Using this number, and τ, k, and N-values for the data set shown in Fig. 3 with 0 mM Mg2+, we solve for the frictional coefficient of the RNA nucleotide segment and find that ζ ≈ 1.7 × 10−12. Considering that the RNA is composed of ∼5000–7000 such segments, we find that the frictional forces of the bead and the RNA polymer have comparable effects on relaxation of the coupled system. Our τ-values, therefore, fall within the regime where contributions from both the bead drag and the polymer are significant and relaxation times may be used to infer dynamic properties of the RNA. Fig. 5 a shows a typical autocorrelation plot for raw x-position as a function of time for a representative poly(U) 1.9–2.6 kb bead at 0 mM and 10 mM Mg2+. The autocorrelation function falls much more rapidly to zero for the 10 mM Mg2+ than the 0 mM Mg2+ data set. Such plots for each particle were fit with single exponential decay functions; the time constants extracted from the fits describe the relaxation time of the bead. Fig. 5 c shows plots of these time constants (τadj, x-) derived for the moving-averaged x-position as a function of x-,y-rms motion for a representative poly(U) length range and two different Mg2+ concentrations. At 0 mM Mg2+, the time constants are approximately linearly proportionate to the x-,y-rms motion values; that is, we find time constants extracted from the autocorrelation plots at low ionic strength to increase with the rms values, demonstrating that relaxation properties are at least partially defined by RNA tether length. By contrast, this proportionality between time constants and x-,y-rms motion values is lost at 10 mM Mg2+. Fig. 5 c shows that the relaxation time constants are considerably shorter in the presence of divalent cations, indicating that addition of Mg2+ accelerates tether relaxation.
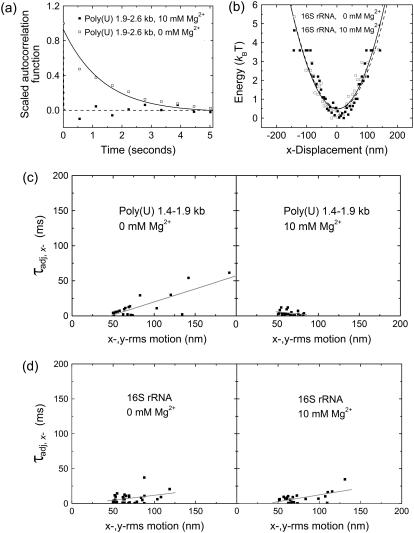
FIGURE 5.
Stiffness of poly(U) mRNA and 16S rRNA tethers. (a) Representative autocorrelation plots of the x-position shown with single exponential fits to extract time constants from a microsphere tethered by a single 1.9–2.6 kb poly(U) mRNA, at 0 mM and 10 mM Mg2+, as indicated. (b) Energy profiles for a representative 16S rRNA-tethered bead in 0 mM and 10 mM Mg2+, as indicated. (c,d) Adjusted time constants obtained from the autocorrelation function of the moving-averaged x-position and plotted as a function of x-,y-rms motion for 1.4–1.9 kb poly(U) mRNA (c) and 16S rRNA (d). Data in panels c and d are reasonably well represented by linear regression fits, as shown.
A complementary approach is to approximate the poly(U) mRNA tether as a linear spring characterized by a spring constant k, which is defined by the entropic elasticity of the RNA tether; the elasticity in turn depends on the flexibility and length of the tether (39). Spring constants extracted from the fits of the energy profiles for individual molecules (as represented in Fig. 3 b) indicate that the elasticity of poly(U) mRNA changes upon compaction. For example, the 5.0–7.0 kb poly(U) tether shown in Fig. 3 yields spring constants (extracted from the quadratic fit of the energy profile) of 0.025 ± 0.004 pN/μm and 0.125 ± 0.010 pN/μm at 0 mM and 10 mM Mg2+, respectively. Such stiffening must contribute to the observed differences in the in-plane bead relaxation time constants at 0 mM and 10 mM Mg2+. Although this stiffening upon Mg2+ addition is observed for all poly(U) size ranges (see, for example, the 1.4–1.9 kb poly(U) data in Fig. 5 c), additionally the time constants increase with the extent of x-,y-rms motion (Fig. 5c), suggesting that they are also at least partially influenced by the length of the RNA tethers.
To test whether the compaction observed in the poly(U) tethers upon addition of Mg2+ is due primarily to an increase in ionic strength, or due instead to a divalent specific effect, we carried out additional TPM measurements in a high-ionic strength background of 100 mM instead of 10 mM NaCl, using long poly(U) tethers of 4.5–5.5 kb. Ionic strength (I) of a solution is given by the following relation:
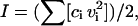 |
(4) |
where ci is the ion concentration and vi is the ion valency.
The ionic strength of the low-salt and high-salt buffers, in the absence of Mg2+, corresponds to I = 5 mM and I = 50 mM, respectively. Upon addition of 10 mM Mg2+, these values increase to I = 25 mM and I = 70 mM, respectively. The general trends in the motion observed for the 4.5–5.5 kb RNA tethers in high ionic strength are summarized in Table 2 and plotted as histograms of average rms motion in Fig. 4 g. Both the range and mean of x-,y-rms values for these 4.5–5.5 kb poly(U) tethers in 100 mM NaCl (in the absence of Mg2+) are lower than the values found for their similarly sized (3.6–5.0 and 5.0–7.0 kb) counterparts in l0 mM NaCl and 0 mM Mg2+ (Table 2), demonstrating that poly(U) mRNA undergoes similar compaction in the presence of higher concentrations of monovalent cations as it does in the presence of Mg2+. This finding suggests that Na+ can replace Mg2+ to a large extent and that cation binding is rather nonspecific. Consistent with this notion, tethers in 100 mM NaCl background undergo a relatively small compaction (∼6% of total length) upon addition of 10 mM Mg2+ (Table 2, Fig. 4 f), which corresponds to a 40% increase in ionic strength. This length change is significantly less pronounced than the analogous change in x-,y-rms motion (∼15% of total length) observed for tethers in 10 mM NaCl background upon addition of 10 mM Mg2+, a 500% increase in ionic strength (Table 2, Fig. 4 e). These data are consistent with previous observations that Mg2+, by virtue of its divalent character, screens the negatively charged RNA backbone more efficiently than do monovalent cations (7,10,11,13), thus allowing anionic RNA to compact further.
16S ribosomal RNA is compact and stiff at low ionic strength in the absence of Mg2+
Data for 16S rRNA tethers were collected in an identical manner to those for poly(U) mRNA. Histograms summarizing average x-,y-rms motion values for 16S rRNA-tethered beads in 0 mM and 10 mM Mg2+ are shown in Fig. 4 g. We observe no significant narrowing of the distribution of rms motion values upon addition of Mg2+. This is also reflected in Table 2, where the vast majority of 16S rRNA-tethered microspheres demonstrate similarly small x-,y-, and z-motions before and after magnesium ion binding. This insignificant attenuation of the Brownian motion of the tethered beads demonstrates that there is very little change in the overall end-to-end distance of single 16S rRNA molecules when 10 mM Mg2+ is added. Notably, the natively folded bacterial 30S ribosomal subunit is highly structured such that the 5′ and 3′ ends of their 16S rRNA component are only <10 nm apart (41), suggesting that under our experimental conditions, in both the absence and presence of Mg2+, naked 16S rRNA may already have a similarly compact structure.
Dynamic properties of 16S rRNA as observed from calculated time constants of bead motion reveal that the 16S rRNA tethers behave more homogeneously at 0 mM Mg2+ than their similarly sized poly(U)-tethered counterparts (Fig. 5). Additionally, plots of τadj, x- as a function of average x-,y-rms motion are virtually identical in 0 mM and 10 mM MgCl2 (Fig. 5 d), suggesting that 16S rRNA does not noticeably stiffen with the binding of divalent cations, in contrast to poly(U) mRNA. To further verify this conclusion, we examined energy profiles for particles tethered by 16S rRNA at 0 mM and 10 mM Mg2+ and found that, in general, the extracted spring constants are similar under both conditions. For example, spring constants obtained from the quadratic fit of the energy profiles for a representative 16S rRNA tether shown in Fig. 5 b are within error of each other at 0 mM and 10 mM Mg2+, with 0.12 ± 0.02 pN/μm and 0.11 ± 0.02 pN/μm, respectively. Taken together, we infer from these data that the addition of divalent metal ions does not result in a significant decrease in the elasticity of the highly structured 16S rRNA; it is already quite stiff at low ionic strength in the absence of Mg2+, most likely because of the presence of substantial secondary structure.
A simple polymer model describes structural properties before and after Mg2+ binding of poly(U) mRNA but not of 16S rRNA
Various studies of conformational changes in DNA have utilized the Gaussian polymer model to describe the dynamic behavior of this polyanion (see, e.g., Rivetti et al. (42) and Blumberg et al. (43)). Since the RNA molecules in our study are not stretched by constant force, it is not necessary to take into account the bending stiffness of the RNA on a short length scale, for which a worm-like chain model would be the better model. In fact, recent studies of the elastic properties of poly(U) mRNA in monovalent cations demonstrate that homopolymeric single-stranded RNA molecules are well described by a simple Gaussian model that includes polymer elasticity (44). However, the two molecules at the focus of this study, 16S rRNA and poly(U) mRNA, exemplify distinct structural properties and differ greatly in their response to magnesium ions. We therefore have investigated the applicability of a statistical polymer model to both RNAs in the presence and absence of Mg2+.
For an ideal polymer in a Gaussian-chain conformation where the contour length is significantly longer than the persistence length (42,45), the rms end-to-end distance R scales as
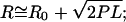 |
(5) |
where P is the persistence length, L the contour length of the RNA, and R0 is the offset added by the attached bead. Whereas the persistence length for double-stranded DNA is 45–50 nm (38), it is ∼50% larger for double-stranded RNA (∼72 nm) (46), most likely because the RNA's 2′ hydroxyl groups sterically hinder the flexibility of the helix backbone. By contrast, in the case of single-stranded (ss) nucleic acids ssRNA is more flexible, with a persistence length of ∼0.8–0.9 nm (44,47–49), in comparison to ssDNA with a persistence length of ∼1.5–3 nm (50,51).
In our studies, the end-to-end distance R for the RNA tethers is proportional to the rms value for motion in the x-,y-dimension, or r in polar coordinates. We use rms motion in the transverse plane for our analysis, since Mg2+-induced differences in motion were more pronounced in-plane than out-of-plane. Each value of r, therefore, is defined as the projection of the end-to-end distance R for the RNA tether in the transverse plane:
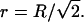 |
(6) |
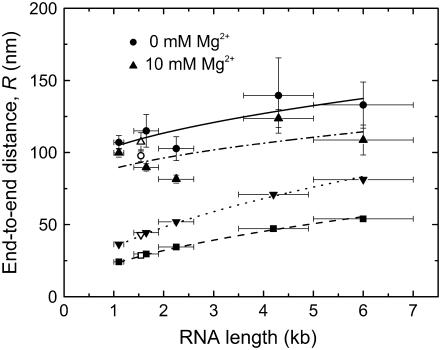
Fig. 6 shows a plot of R, calculated from the mean rms motion for each size range of poly(U) mRNA as well as 16S rRNA, as a function of average RNA length for both magnesium ion concentrations. The values of R for poly(U) mRNA are generally ∼30 nm greater in the absence than in the presence of Mg2+. Fitting these data with square root functions (Eq. 5, with an estimated offset R0 of 75 nm) yields apparent persistence lengths of 0.35 ± 0.06 nm and 0.12 ± 0.05 nm in the absence and presence of Mg2+, respectively, which is consistent with RNA compaction upon Mg2+ binding. For comparison, R-values predicted from the Gaussian chain model for our average RNA lengths N are also shown in Fig. 6, calculated using P = 0.855 nm (44) and L = N × 0.70 nm and L = N × 0.31 nm for extended (random coil) and more compact (helical) ssRNA conformers, respectively (38,52); these values are thus predicted by the square root function in Eq. 5 for P = 0.855 nm. Interestingly, although the R-values predicted by the simple polymer model are shorter than the measured mean particle motion, the difference between the two models (random coil versus helix) closely approximates the difference in R-values observed for poly(U) tethers in the presence and absence of Mg2+, respectively. This is consistent with the idea that poly(U) mRNA is well modeled as a random coil in the absence and as a more compact, stiffer single strand in the presence of Mg2+. The 280-nm radius of the attached bead adds to the R-values observed in our TPM measurements in the form of the offset R0; R0 is smaller than the bead radius due to the effective force imposed in the z-direction by the inability of the microsphere to penetrate the anchoring surface. We therefore conclude that Gaussian chain behavior is consistent with the dynamic properties of single-stranded poly(U) mRNA, both in the absence and presence of magnesium ions. In contrast, experimentally derived R-values for 16S rRNA in the presence and absence of magnesium ions (open shapes, Fig. 6) do not seem to follow the trend we expect for a Gaussian polymer chain. In fact, the experimental R-values found for 16S rRNA at 10 mM Mg2+ are slightly larger, not smaller, than the end-to-end distances in the absence of Mg2+. This suggests that 16S rRNA is not particularly well described by simple polymer models, in both the absence and presence of divalent cations, most likely due to the complexity and stability of its secondary (and perhaps tertiary) structure even under our low ionic strength conditions (10 mM NaCl).
FIGURE 6.
RNA 5′-to-3′ end distance R as measured by TPM, plotted as a function of average RNA length in kilobases. Poly(U) size ranges are represented as closed circles (0 mM Mg2+) and closed triangles (10 mM Mg2+) and are fit with solid and dashed-dotted curves, respectively. The fitted curves represent the following function: 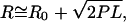 where R0 is the offset in the end-to-end distance of the polymer (R) due to the attached bead. R0 offsets for the curves described by the closed circles (0 mM Mg2+) and closed triangles (10 mM Mg2+) were found to be ∼75 nm. R-values calculated from persistence length (P) for a single-stranded RNA polymer and end-to-end distance for either an extended RNA chain (closed upside-down triangles) or a single-stranded helix (closed squares) are also plotted as a function of polymer length and fit with dotted and dashed curves, respectively, using the equation described above (except that R0 was set to zero). Open shapes represent experimental and predicted R-values for 16S rRNA.
where R0 is the offset in the end-to-end distance of the polymer (R) due to the attached bead. R0 offsets for the curves described by the closed circles (0 mM Mg2+) and closed triangles (10 mM Mg2+) were found to be ∼75 nm. R-values calculated from persistence length (P) for a single-stranded RNA polymer and end-to-end distance for either an extended RNA chain (closed upside-down triangles) or a single-stranded helix (closed squares) are also plotted as a function of polymer length and fit with dotted and dashed curves, respectively, using the equation described above (except that R0 was set to zero). Open shapes represent experimental and predicted R-values for 16S rRNA.
DISCUSSION
Magnesium ions are indispensable to many cellular processes, due in part to the central roles they play in RNA structure and function. Magnesium exists predominantly as hexa-hydrated 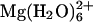 ions in solution, which localize nonspecifically to negatively charged pockets within an RNA tertiary structure and exchange rapidly with bulk solvent ions. In this work, we have introduced TPM as a method to study metal ion-induced compaction in single RNA molecules. TPM has an advantage over bulk solution methods such as small-angle x-ray scattering because molecules are observed individually via attached microspheres so that the behavior of RNA tethers may be assessed on an individual basis instead of as an ensemble average. TPM as a single molecule method has allowed us to assess the heterogeneity in the structural and dynamic behavior among a population of poly(U) messenger RNA molecules, as compared with their structurally better-defined and more homogeneous 16S ribosomal RNA counterparts. Two additional advantages of TPM over commonly used biophysical techniques are that only subnanomole amounts of RNA are required and that conformational changes can be detected in real-time on a length scale much larger than that accessible by single molecule FRET (<10 nm) (reviewed in (26–29)).
ions in solution, which localize nonspecifically to negatively charged pockets within an RNA tertiary structure and exchange rapidly with bulk solvent ions. In this work, we have introduced TPM as a method to study metal ion-induced compaction in single RNA molecules. TPM has an advantage over bulk solution methods such as small-angle x-ray scattering because molecules are observed individually via attached microspheres so that the behavior of RNA tethers may be assessed on an individual basis instead of as an ensemble average. TPM as a single molecule method has allowed us to assess the heterogeneity in the structural and dynamic behavior among a population of poly(U) messenger RNA molecules, as compared with their structurally better-defined and more homogeneous 16S ribosomal RNA counterparts. Two additional advantages of TPM over commonly used biophysical techniques are that only subnanomole amounts of RNA are required and that conformational changes can be detected in real-time on a length scale much larger than that accessible by single molecule FRET (<10 nm) (reviewed in (26–29)).
We note that two characteristic observations are associated with compaction of a TPM tether: shortening of the end-to-end distance of the RNA, and stiffening of the RNA. We have assessed these properties by comparison of x-deviation as a function of time, mean x-,y-rms motion values, range of rms motion values, relaxation time constants, and elastic spring constants. Heterogeneity of the mean x-,y-rms motion values over the different poly(U) mRNA size groups resulted in large standard deviations (Table 2); therefore, we found the range of values more useful in comparing the overall end-to-end distances for the different RNA size ranges. Individual mean x-,y-rms motion values did, however, prove a valuable parameter when comparing the degree of overall compaction between single tethers. Stiffness of the RNA molecules was best assessed by computing the spring constant of the tether. Since the viscous drag on the attached bead contributed to the extracted spring constants, the stiffness values obtained from this method were only used to compare the relative RNA stiffness. Spring constants extracted from quadratic fits of the energy profiles of the tethers produced low errors (between ∼8% and ∼18% of the value) and were thus effective in distinguishing subtle differences between tethers with similar mean x-,y-rms motion. Relaxation time constants were employed for assessing the relative “compactness” of a tether and were found to be affected by both end-to-end distance and stiffness of the RNA. Consequently, it was difficult to dissect the relative contributions of the two parameters to the measured time constants. In addition, the autocorrelation curves for the more compact tethers contain few data points before the function fully decays (Fig. 5), making an accurate exponential fit difficult and perpetuating relatively large errors. In summary, we conclude that the end-to-end distance of the tethers was most accurately assessed by the mean x-,y-rms motion values, and the stiffness of the RNA was best described by the spring constants extracted from the energy profiles of the beads.
Different roles for magnesium ions in folding of poly(U) mRNA and 16S rRNA: quantifying the impact on structure and dynamics
16S ribosomal RNA, which comprises the bulk of the small ribosomal subunit in bacteria, adopts a highly ordered compact scaffold stabilized by magnesium ions and the small subunit ribosomal proteins. Crystal structures of both 30S subunits (41,53) and intact ribosomes (54–56) depict many specifically bound Mg2+ ions within the complex 16S rRNA fold. In general, large RNAs require magnesium ion binding to assume a correctly folded structure (8–11). The resulting aggregate magnesium ion dissociation constant in the Tetrahymena group I ribozyme, for example, is ∼500 μM (57). This concentration appears to be a general threshold for RNA tertiary structure formation, as it has been identified as the critical concentration of Mg2+ required for proper folding of other RNAs as well (e.g., (58,59)).
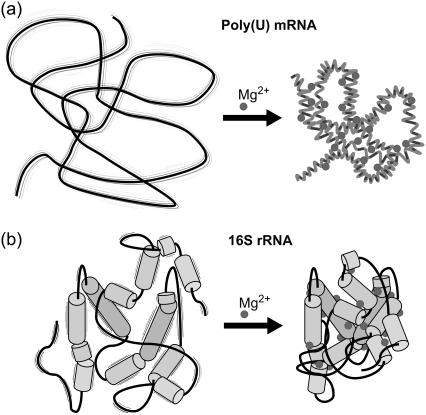
Distinct folding intermediates have been identified in large structured RNAs, such as the Tetrahymena group I and RNase P RNA ribozymes, in the presence of various magnesium ion concentrations, by both bulk solution and single molecule methods (12,57,60–62). At the resolution of our TPM studies of 16S rRNA, however, no discrete steps were distinguished in x-, y-, or z-dimension bead motion as Mg2+ concentrations were sequentially increased by several orders of magnitude, starting at 1 μM (data not shown). We therefore conclude that the secondary structure of 16S rRNA is, at least at this resolution, largely formed in the presence of 10 mM monovalent cations in our buffer. This may explain why 16S rRNA behavior is much more homogeneous than that of our poly(U) mRNA tethers; most 16S rRNA molecules are likely folded into similarly compact conformations. The small differences in bead motion upon Mg2+ addition may then be due to slight rearrangements of helices and stabilization of specific tertiary contacts that occurs upon association with both specifically and nonspecifically bound Mg2+ ions (Fig. 7).
FIGURE 7.
Summary of proposed magnesium-induced structural changes in poly(U) mRNA (a) and 16S rRNA (b).
Homopolymeric RNA molecules such as polyuridylic acid, or poly(U), are thought to form partially ordered single-stranded helices (63) because the resulting base stacking is enthalpically favorable. However, single-stranded stacks are only moderately stable in solution, and for any given single-stranded RNA segment, an equilibrium exists between stacked helical and unstacked random coil conformations (64). Solution conditions such as temperature, ionic strength, and pH can shift this equilibrium. In comparison to 16S rRNA, poly(U) is therefore generally quite unstructured in solution under low ionic strength conditions. Although hydrogen bonds and stacking interactions may form between distal regions of the polymer chain, these interactions are likely transient due to the absence of a defined surrounding network of hydrogen bonds to stabilize them or provide the basis for a cooperative folding pathway. Locally, however, stable U-U pairs may indeed form (for an example from a highly structured RNA see Theimer et al. (65)), although a homopolymeric RNA molecule can of course form a tremendous number of such intrastrand interactions. Such a combination of ill-defined long- and short-range interactions is expected to give rise to an extremely diverse set of structures, providing a possible explanation for the heterogeneous behavior we observe in the degree of motion for microspheres tethered by single poly(U) mRNA molecules. Upon addition of 10 mM Mg2+, we observe an overall compaction of the tether. Complementary studies performed at higher concentrations of monovalent cations (100 mM) resulted in a similar compaction of the poly(U) tethers as the addition of 10 mM Mg2+, suggesting that the role of magnesium ions in compaction can be fulfilled by monovalent cations and that cation binding is thus nonspecific. Still, our studies indicate that Mg2+-induced compaction may occur at a lower ionic strength (I = 25 mM) than does compaction induced by monovalent cations (I = 50 mM). It is known that, on average, ∼0.49 Mg2+ ions are retained per phosphate in poly(U)-poly(A) A-form helices (10,66). We propose that this efficient charge neutralization by Mg2+ (or monovalents) stabilizes any transient intrastrand interactions so that a low-energy single-stranded A-form-like helical structure is assumed (38) that traps randomly formed regions of helical stacks into more compact conformations. This is summarized in Fig. 7; poly(U) is unstructured and flexible in the presence of low concentrations of monovalents and compacts substantially upon addition of 10 mM Mg2+ (or higher concentrations of monovalents).
Recent biomolecular packing calculations by Voss and Gerstein (67) have led to the realization that RNA structures are generally more densely packed than protein structures. However, these calculations did not include structurally ill-defined RNA homopolymers such as poly(U). Our TPM studies demonstrate that Mg2+ induces a similar degree of compaction in 1.4–1.9 kb poly(U) mRNA as observed for the 1,542-nt 16S rRNA (Table 2). We therefore propose that randomly structured RNAs can be tightly arranged in space in the presence of sufficient concentrations of divalent (or monovalent) cations, similar to “globular” structured RNAs. Spring constants extracted from energy profiles of such Mg2+-compacted poly(U) tethers indicate that their rigidity is about equal to that measured for 16S rRNA in the presence of Mg2+. From these data we infer a correlation between degree of RNA compactness and elasticity, where the end-to-end distance of the RNA is inversely proportional to the spring constant.
CONCLUSIONS
We have established here TPM as a powerful method for the study of the structural and mechanical properties of large RNA molecules. Microspheres tethered by single 16S rRNA molecules, a highly structured RNA, showed relatively homogeneous behavior; the addition of Mg2+ induced only slight changes in overall rms motion and very little change in the relaxation time constants and spring constants. These findings are consistent with negligible changes in the 5′-to-3′ end distance and the flexibility of the 16S rRNA, implying that little structural change occurs upon addition of Mg2+. We propose that the global secondary structure of 16S rRNA is already assumed in low concentrations of monovalents (10 mM); additional secondary and tertiary contacts may form upon binding of Mg2+, but with little effect on the end-to-end distance of the RNA. In contrast, we observe considerable heterogeneity over a population of poly(U) mRNA molecules, coupled with significant attenuation of tethered bead motion (a decrease of ∼7.5–30% in mean rms motion over the entire size range of poly(U) mRNA) and a significant increase in spring constant upon addition of Mg2+. This indicates that divalent cations induce a significant degree of compaction in poly(U) mRNA. Relaxation time and spring constants in the presence of divalents reveal that poly(U) mRNA also indicates that the RNA becomes stiffer after addition of 10 mM Mg2+. We find that higher concentrations (100 mM) of monovalent cations induce similar compaction in poly(U) mRNA, indicating that cation binding is relatively nonspecific. Finally, we find that randomly structured RNA such as poly(U) mRNA, but not the highly structured 16S rRNA, may be accurately described in both its Mg2+ free and bound forms by a Gaussian polymer model derived from statistical physics. These findings exemplify the practicality and usefulness of TPM as a biophysical technique for the study of RNA conformation and dynamics.
Acknowledgments
The authors thank Saskia Thomas and Katarzyna Chmielinska for technical support, and Phillip T. Sekella and Rajalakshmi Nambiar for pioneering contributions to this project. We are also grateful to all members of the Meiners and Walter Labs for helpful input, to Dieter Möll for his useful insights, to Troy Lionberger for critical reading of the manuscript, and to anonymous reviewers for their helpful suggestions for revisions.
This work was funded by grants from the National Aeronautics and Space Administration (No. NNA04CD01G and No. NNC04AA21A) and the National Institutes of Health (NIH) (RO1 GM065934 to J.C.M. and RO1 GM062357 to N.G.W.). M.N.L. is the recipient of a Ruth Kirschstein National Research Service Award postdoctoral fellowship from the NIH.
References
- 1.Noller, H. F. 2005. RNA structure: reading the ribosome. Science. 309:1508–1514. [DOI] [PubMed] [Google Scholar]
- 2.Zamore, P. D., and B. Haley. 2005. Ribo-gnome: the big world of small RNAs. Science. 309:1519–1524. [DOI] [PubMed] [Google Scholar]
- 3.Moore, M. J. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science. 309:1514–1518. [DOI] [PubMed] [Google Scholar]
- 4.Sievers, A., M. Beringer, M. V. Rodnina, and R. Wolfenden. 2004. The ribosome as an entropy trap. Proc. Natl. Acad. Sci. USA. 101:7897–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinger, J. S., K. M. Parnell, S. Dorner, R. Green, and S. A. Strobel. 2004. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat. Struct. Mol. Biol. 11:1101–1106. [DOI] [PubMed] [Google Scholar]
- 6.Leontis, N. B., and E. Westhof. 2003. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 13:300–308. [DOI] [PubMed] [Google Scholar]
- 7.Misra, V. K., R. Shiman, and D. E. Draper. 2003. A thermodynamic framework for the magnesium-dependent folding of RNA. Biopolymers. 69:118–136. [DOI] [PubMed] [Google Scholar]
- 8.Pyle, A. M. 2002. Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 7:679–690. [DOI] [PubMed] [Google Scholar]
- 9.DeRose, V. J. 2003. Metal ion binding to catalytic RNA molecules. Curr. Opin. Struct. Biol. 13:317–324. [DOI] [PubMed] [Google Scholar]
- 10.Draper, D. E. 2004. A guide to ions and RNA structure. RNA. 10:335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodson, S. A. 2005. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 9:104–109. [DOI] [PubMed] [Google Scholar]
- 12.Takamoto, K., R. Das, Q. He, S. Doniach, M. Brenowitz, D. Herschlag, and M. R. Chance. 2004. Principles of RNA compaction: insights from the equilibrium folding pathway of the P4–P6 RNA domain in monovalent cations. J. Mol. Biol. 343:1195–1206. [DOI] [PubMed] [Google Scholar]
- 13.Das, R., L. W. Kwok, I. S. Millett, Y. Bai, T. T. Mills, J. Jacob, G. S. Maskel, S. Seifert, S. G. J. Mochrie, P. Thiyagarajan, S. Doniach, L. Pollack, and D. Herschlag. 2003. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the tetrahymena ribozyme. J. Mol. Biol. 332:311–319. [DOI] [PubMed] [Google Scholar]
- 14.Russell, R., I. S. Millett, S. Doniach, and D. Herschlag. 2000. Small angle x-ray scattering reveals a compact intermediate in RNA folding. Nat. Struct. Biol. 7:367–370. [DOI] [PubMed] [Google Scholar]
- 15.Russell, R., X. W. Zhuang, H. P. Babcock, I. S. Millett, S. Doniach, S. Chu, and D. Herschlag. 2002. Exploring the folding landscape of a structured RNA. Proc. Natl. Acad. Sci. USA. 99:155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima, S., and M. Nomura. 1970. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 226:1214–1218. [DOI] [PubMed] [Google Scholar]
- 17.Grondek, J. F., and G. M. Culver. 2004. Assembly of the 30S ribosomal subunit: positioning ribosomal protein S13 in the S7 assembly branch. RNA. 10:1861–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Recht, M. I., and J. R. Williamson. 2001. Central domain assembly: thermodynamics and kinetics of S6 and S18 binding to an S15-RNA complex. J. Mol. Biol. 313:35–48. [DOI] [PubMed] [Google Scholar]
- 19.Recht, M. I., and J. R. Williamson. 2004. RNA tertiary structure and cooperative assembly of a large ribonucleoprotein complex. J. Mol. Biol. 344:395–407. [DOI] [PubMed] [Google Scholar]
- 20.Adilakshmi, T., P. Ramaswamy, and S. A. Woodson. 2005. Protein-independent folding pathway of the 16S rRNA 5′ domain. J. Mol. Biol. 351:508–519. [DOI] [PubMed] [Google Scholar]
- 21.Finzi, L., and J. Gelles. 1995. Measurement of lactose repressor-mediated loop formation and breakdown in single DNA-molecules. Science. 267:378–380. [DOI] [PubMed] [Google Scholar]
- 22.Schafer, D. A., J. Gelles, M. P. Sheetz, and R. Landick. 1991. Transcription by single molecules of RNA-polymerase observed by light-microscopy. Nature. 352:444–448. [DOI] [PubMed] [Google Scholar]
- 23.Koch, S. J., A. Shundrovsky, B. C. Jantzen, and M. D. Wang. 2002. Probing protein-DNA interactions by unzipping a single DNA double helix. Biophys. J. 83:1098–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanzi, F., S. Vladimirov, C. R. Knudsen, Y. E. Goldman, and B. S. Cooperman. 2003. Protein synthesis by single ribosomes. RNA. 9:1174–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuang, X. W., H. Kim, M. J. B. Pereira, H. P. Babcock, N. G. Walter, and S. Chu. 2002. Correlating structural dynamics and function in single ribozyme molecules. Science. 296:1473–1476. [DOI] [PubMed] [Google Scholar]
- 26.Zhuang, X. W., and M. Rief. 2003. Single-molecule folding. Curr. Opin. Struct. Biol. 13:88–97. [DOI] [PubMed] [Google Scholar]
- 27.Zhuang, X. 2005. Single-molecule RNA science. Annu. Rev. Biophys. Biomol. Struct. 34:399–414. [DOI] [PubMed] [Google Scholar]
- 28.Walter, N. G., and J. M. Burke. 2000. Fluorescence assays to study structure, dynamics, and function of RNA and RNA-ligand complexes. Methods Enzymol. 317:409–440. [DOI] [PubMed] [Google Scholar]
- 29.Walter, N. G. 2001. Structural dynamics of catalytic RNA highlighted by fluorescence resonance energy transfer. Methods. 25:19–30. [DOI] [PubMed] [Google Scholar]
- 30.Bokinsky, G., D. Rueda, V. K. Misra, M. M. Rhodes, A. Gordus, H. P. Babcock, N. G. Walter, and X. Zhuang. 2003. Single-molecule transition-state analysis of RNA folding. Proc. Natl. Acad. Sci. USA. 100:9302–9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rueda, D., G. Bokinsky, M. M. Rhodes, M. J. Rust, X. Zhuang, and N. G. Walter. 2004. Single-molecule enzymology of RNA: essential functional groups impact catalysis from a distance. Proc. Natl. Acad. Sci. USA. 101:10066–10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kratsky, O., and G. Porod. 1949. Röntgenuntersuchung aufgelöster Fadenmoleküle. Recueil. 68:1016–1022. [Google Scholar]
- 33.Flory, P. J. 1969. Statistical Mechanics of Chain Molecules. Interscience, New York.
- 34.Bakin, A. V., O. F. Borisova, I. N. Shatsky, and A. A. Bogdanov. 1991. Spatial-organization of template polynucleotides on the ribosome determined by fluorescence methods. J. Mol. Biol. 221:441–453. [DOI] [PubMed] [Google Scholar]
- 35.Blumberg, S., A. Gajraj, M. W. Pennington, and J. C. Meiners. 2005. Three-dimensional characterization of tethered microspheres by total internal reflection fluorescence microscopy. Biophys. J. 89:1272–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh-Zocchi, M., S. Dixit, V. Ivanov, and G. Zocchi. 2003. Single-molecule detection of DNA hybridization. Proc. Natl. Acad. Sci. USA. 100:7605–7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ts‘o, P. O. P. 1974. Bases, nucleosides, and nucleotides. In Basic Principles in Nucleic Acid Chemistry. P. O. P. Ts’o, editor. Academic Press, New York. 453–584.
- 38.Saenger, W. 1983. Principles of Nucleic Acid Structure. Springer-Verlag, New York.
- 39.Qian, H., and E. L. Elson. 1999. Quantitative study of polymer conformation and dynamics by single-particle tracking. Biophys. J. 76:1598–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pouget, N., C. Dennis, C. Turlan, M. Grigoriev, M. Chandler, and L. Salome. 2004. Single-particle tracking for DNA tether length monitoring. Nucleic Acids Res. 32:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wimberly, B. T., D. E. Brodersen, W. M. Clemons, R. J. Morgan-Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature. 407:327–339. [DOI] [PubMed] [Google Scholar]
- 42.Rivetti, C., C. Walker, and C. Bustamante. 1998. Polymer chain statistics and conformational analysis of DNA molecules with bends or sections of different flexibility. J. Mol. Biol. 280:41–59. [DOI] [PubMed] [Google Scholar]
- 43.Blumberg, S. A., V. Tkachemko, and J. C. Meiners. 2005. Effect of tension on DNA looping. Biophys. J. 88:1692–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seol, Y., G. M. Skinner, and K. Visscher. 2004. Elastic properties of a single-stranded charged homopolymeric ribonucleotide. Phys. Rev. Lett. 93:118102. [DOI] [PubMed] [Google Scholar]
- 45.Landau, L. D., and E. M. Lifshitz. 1980. Statistical Physics, 3rd ed. Pergamon Press, New York.
- 46.Kebbekus, P., D. E. Draper, and P. Hagerman. 1995. Persistence length of RNA. Biochemistry. 34:4354–4357. [DOI] [PubMed] [Google Scholar]
- 47.Vanzi, F., Y. Takagi, H. Shuman, B. S. Cooperman, and Y. E. Goldman. 2005. Mechanical studies of single ribosome/mRNA complexes. Biophys. J. 89:1909–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caliskan, G., C. Hyeon, U. Perez-Salas, R. M. Briber, S. A. Woodson, and D. Thirumalai. 2005. Persistence length changes dramatically as RNA folds. Phys. Rev. Lett. 95:268303. [DOI] [PubMed] [Google Scholar]
- 49.Abels, J. A., F. Moreno-Herrero, T. van der Heijden, C. Dekker, and N. H. Dekker. 2005. Single-molecule measurements of the persistence length of double- stranded RNA. Biophys. J. 88:2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy, M. C., I. Rasnik, W. Cheng, T. M. Lohman, and T. J. Ha. 2004. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys. J. 86:2530–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thirumalai, D., and B. Y. Ha. 1988. Statistical mechanics of semi-flexible chains. In Theoretical and Mathematical Models in Polymer Research. A. Grosberg, editor. Academic Press, San Diego, CA. 1–35.
- 52.Harlepp, S., T. Marchal, J. Robert, J. F. Leger, A. Xayaphoummine, H. Isambert, and D. Chatenay. 2003. Probing complex RNA structures by mechanical force. Eur. Phys. J. E. 12:605–615. [DOI] [PubMed] [Google Scholar]
- 53.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 407:340–348. [DOI] [PubMed] [Google Scholar]
- 54.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 angstrom resolution. Science. 289:905–920. [DOI] [PubMed] [Google Scholar]
- 55.Schluenzen, F., A. Tocilj, R. Zarivach, J. Harms, M. Gluehmann, D. Janell, A. Bashan, H. Bartels, I. Agmon, F. Franceschi, and A. Yonath. 2000. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 102:615–623. [DOI] [PubMed] [Google Scholar]
- 56.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. D. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 angstrom resolution. Science. 292:883–896. [DOI] [PubMed] [Google Scholar]
- 57.Sclavi, B., S. Woodson, M. Sullivan, M. R. Chance, and M. Brenowitz. 1997. Time-resolved synchrotron x-ray “footprinting”, a new approach to the study of nucleic acid structure and function: application to protein-DNA interactions and RNA folding. J. Mol. Biol. 266:144–159. [DOI] [PubMed] [Google Scholar]
- 58.Rueda, D., K. Wick, S. E. McDowell, and N. G. Walter. 2003. Diffusely bound Mg2+ ions slightly reorient stems I and II of the hammerhead ribozyme to increase the probability of formation of the catalytic core. Biochemistry. 42:9924–9936. [DOI] [PubMed] [Google Scholar]
- 59.Tinsley, R. A., D. A. Harris, and N. G. Walter. 2004. Magnesium dependence of the amplified conformational switch in the trans-acting hepatitis delta virus ribozyme. Biochemistry. 43:8935–8945. [DOI] [PubMed] [Google Scholar]
- 60.Baird, N. J., E. Westhof, H. Qin, T. Pan, and T. R. Sosnick. 2005. Structure of a folding intermediate reveals the interplay between core and peripheral elements in RNA folding. J. Mol. Biol. 352:712–722. [DOI] [PubMed] [Google Scholar]
- 61.Onoa, B., S. Dumont, J. Liphardt, S. B. Smith, I. Tinoco, and C. Bustamante. 2003. Identifying kinetic barriers to mechanical unfolding of the T-thermophila ribozyme. Science. 299:1892–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tinoco, I. 2004. Force as a useful variable in reactions: unfolding RNA. Annu. Rev. Biophys. Biomol. Struct. 33:363–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tewari, R., R. K. Nanda, and G. Govil. 1974. Spatial configuration of single-stranded polynucleotides—calculations of average dimensions and NMR coupling-constants. Biopolymers. 13:2015–2035. [DOI] [PubMed] [Google Scholar]
- 64.Haasnoot, C. A. G., and C. Altona. 1979. Nucleic-acids constitutes. 9. Conformational study of nucleic-acid phosphate ester bonds using P-31 nuclear magnetic-resonance. Nucleic Acids Res. 6:1135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Theimer, C. A., C. A. Blois, and J. Feigon. 2005. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 17:671–682. [DOI] [PubMed] [Google Scholar]
- 66.Record, M. T. Jr., and B. Richey. 1988. Physical chemical analysis of biopolymer self-assembly interactions. In ACS Sourcebook for Physical Chemistry Instructors. T. Lippencott, editor. American Chemical Society, Washington, D.C. 145–159.
- 67.Voss, N. R., and M. Gerstein. 2005. Calculation of standard atomic volumes for RNA and comparison with proteins: RNA is packed more tightly. J. Mol. Biol. 346:477–492. [DOI] [PubMed] [Google Scholar]
- 68.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D‘Souza, Y. S. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. D. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]