Abstract
In a case study on five homologous α-amylases we analyzed the properties of unfolded states as obtained from treatments with GndHCl and with elevated temperatures. In particular the wavelength of the tryptophan fluorescence emission peak (λmax) is a valuable parameter to characterize properties of the unfolded state. In all cases with a typical red shift of the emission spectrum occurring during structural unfolding we observed a larger magnitude of this shift for GndHCl-induced unfolding as compared to thermal unfolding. Although a quantitative relation between aggregation and reduction of the unfolding induced red shifts cannot be given, our data indicate that protein aggregation contributes significantly to smaller magnitudes of red shifts as observed during thermal unfolding. In addition, other properties of the unfolded states, most probable structural compactness or simply differences in the conformational scrambling, also affect the magnitude of red shifts. For the irreversible unfolding α-amylases studied here, transition temperatures and magnitudes of red shifts are strongly depending on heating rates. Lower protein concentrations and smaller heating rates lead to larger red shifts upon thermal unfolding, indicating that under these conditions the protein aggregation is less pronounced.
INTRODUCTION
Most proteins are characterized by well-defined three-dimensional structures, which in general exist only within the limits of specific environmental conditions. Outside of these conditions, proteins exhibit denatured and structurally unfolded states. Depending on the kind of denaturing conditions (high salt concentrations, extreme pH, elevated temperatures, pressure, etc.) the ensemble of unfolded states is characterized by diverse structural and conformational properties. Besides the disappearance of secondary structure elements, in most cases the three-dimensional structures are substantially altered. In particular denatured states of larger multidomain proteins quite often show up as more expanded structures with a strong tendency to form aggregates (1,2). Various techniques have been employed to analyze the diverse aspects of unfolding as well as of (re)folding transitions. Among a large variety of different methods, such as absorption-, circular dichroism spectroscopy, calorimetry, NMR, and light scattering techniques, in particular fluorescence spectroscopy is a powerful technique to monitor unfolding transitions and to characterize properties of the unfolded state (see for example (3,4)). The fluorescence of tryptophan residues is very sensitive to the local environment. In principle mainly two properties of the fluorescence emission exhibit the most pronounced signal changes upon conformational changes of the protein. Due to static and dynamic quenching of solvent molecules or nearby intramolecular subgroups, the intensity of the fluorescently emitted light can vary significantly. If, for example, a tryptophan residue becomes more accessible to the solvent, which quite often is the case for unfolded states, the emission intensity decreases. The other major change in a fluorescence signal upon conformational changes of the protein is related to solvent relaxation, which determines the magnitude of the stokes shift. In less-polar environments of the tryptophan chromophore the stokes shift will be less pronounced as compared to more-polar surroundings. Therefore, the maximum of the fluorescence emission spectrum (λmax) is sensitive for the polarity of the microenvironment of the intrinsic fluorophore. With respect to tryptophan residues, five spectral classes of emission spectra have been identified which exhibit significantly different emission peak maxima. The peak maxima range from ∼308 nm for relatively nonpolar environments inside the protein globule to λmax-values of 350 nm which represent chromophores at the protein surface surrounded by polar water molecules (5,6). For most proteins, which typically bear more than one tryptophan, the observed emission spectrum is composed of multiple contributions of the five spectral classes. As a consequence the observed emission spectra of individual proteins (and of their corresponding conformational states) do not even vary with respect to λmax but also can show different spectral shapes and widths (7,8). However, upon unfolding transitions generally distinct shifts of the emission spectra are reported (see for example (4,9–14)). To derive thermodynamic parameter from spectroscopic data it is crucial whether the signal changes during the unfolding process are proportional to the molar fractions of the folded and unfolded protein. Although the fluorescence intensity signal exhibits this proportionality to the population of the microstates (i.e., folded, unfolded), the use of this signal to measure protein stability often gives rise to difficulties. For example, problems arise from the intrinsic temperature dependence of the tryptophan fluorescence emission and from a pronounced protein concentration dependence, which leads to a significant scattering in the data. Furthermore, in some cases the differences in intensity signals from folded and unfolded states are very small, which makes precise measurements difficult (12). Most of these difficulties are not present if λmax-signals are employed to characterize unfolding transitions, although these signals can give a skewed assignment of the population of microstates, because a priori the λmax-signal is not proportional to the population of the states. However, it has been shown in various cases that (and how) under specific conditions λmax-signals can be used for quantitative analyses of unfolding transitions (4,12,15).
As already mentioned above, the unfolding transition of proteins is not only characterized by the disappearance of the well-defined three-dimensional structure but can also cause distinct changes in the compactness of the unfolded structures or may induce pronounced protein aggregation. In principle all these phenomena can alter the microenvironment of the intrinsic fluorophores and would therefore affect the emission spectra. In this work we report a case study on five homologous α-amylases aiming to elucidate the impact of protein aggregation and conformational scrambling of the unfolded states on fluorescence emission. For this purpose we analyzed the unfolding transitions of the medium-sized multitryptophan protein α-amylase mainly by the use of λmax-signals.
MATERIALS AND METHODS
Enzymes
α-Amylase from Bacillus licheniformis (BLA; purchased from Sigma, Taufkirchen, Germany), from Bacillus amyloliquefaciens (BAA; purchased from Fluka, Buchs SG, Switzerland), from Bacillus subtilis (BSUA; from Fluka), and from Aspergillus oryzae (TAKA; from Sigma) was obtained as lyophilized powder. Pig pancreas α-amylase (PPA, from LaRoche, Mannheim, Germany) was obtained in ammonium sulfate solution. Powders were dissolved in buffer (for details see below) and all enzymes were purified by the use of a desalting column (PD-10, Sephadex G-25, Amersham Biosciences, Freiburg, Germany). For all studies the following buffer was used: 30 mM Mops, 50 mM NaCl, 2 mM CaCl2, pH 7.4. In the case of circular dichroism (CD) spectroscopy, the same buffers was used, but with 10 mM Mops.
Fluorescence spectroscopy
Intrinsic fluorescence was measured using a RF-1501 fluorospectrometer (Shimadzu, Duisburg, Germany) and a LS55 luminescence spectrometer (Perkin-Elmer, Rodgau, Germany). An excitation wavelength of 280 nm (bandwidth 3 nm for the Perkin-Elmer and 10 nm for the Shimadzu spectrometer) was applied while emission spectra were recorded in the range between 300–450 nm (bandwidth 5 nm and 10 nm for the Perkin-Elmer and the Shimadzu spectrometer, respectively). The whole emission spectrum with a wavelength increment of 1 nm was acquired approximately within 30 s. All fluorescence spectra were corrected for background scattering as measured with pure buffer. In some cases (when using the Shimadzu spectrometer) an UG1 filter (Schott, Mainz, Germany) was used to attenuate the exciting beam, to avoid photobleaching and to obtain most reproducibly fluorescence emission spectra. Temperature scans were performed using a constant-temperature cuvette holder connected to an external constant temperature circulator/bath (F25, Julabo, Seelbach, Germany; using a special Julabo bath fluid H10S). Protein solutions (protein concentrations: 7 μg/ml to 1 mg/ml) were filled into quartz-cuvettes with an optical pathlength of 1 cm (104F-QS, Hellma, Muehlheim, Germany). Due to a tight sealing of the quartz cuvettes temperatures of protein solutions up to ∼120°C were applicable without evaporation and boiling.
Light scattering
To estimate protein aggregation occurring upon thermal unfolding, the RF-1501 Fluorospectrometer was employed to measure elastic scattering at 500 nm. For this purpose the same experimental setup (temperature controlling unit, quartz-cuvettes with 1 cm optical pathlength) was used as for fluorescence studies.
Circular dichroism
CD spectra in the far-UV region (200–280 nm) were recorded on a Jasco J-810 equipped with peltier thermostating cuvette holder under constant nitrogen flow. The spectra were recorded in a 0.2-cm cell at a protein concentration of 0.05 mg/ml, averaged over three scans, and finally corrected for the buffer signal. The raw data were corrected for pre- and posttransition slopes. Thermal unfolding transitions were monitored by taking the CD signal at 222 nm as a function of temperature.
RESULTS AND DISCUSSION
Thermal and GndHCl-induced unfolded states
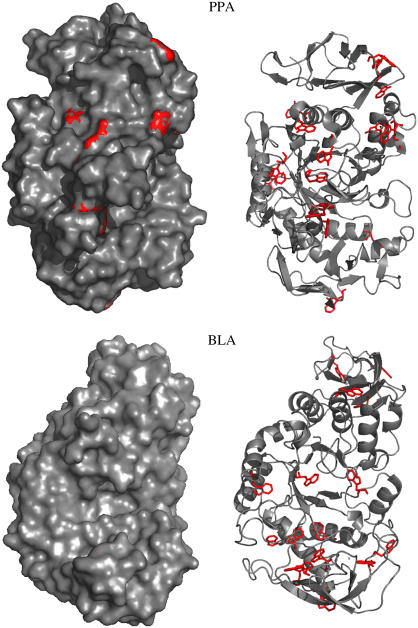
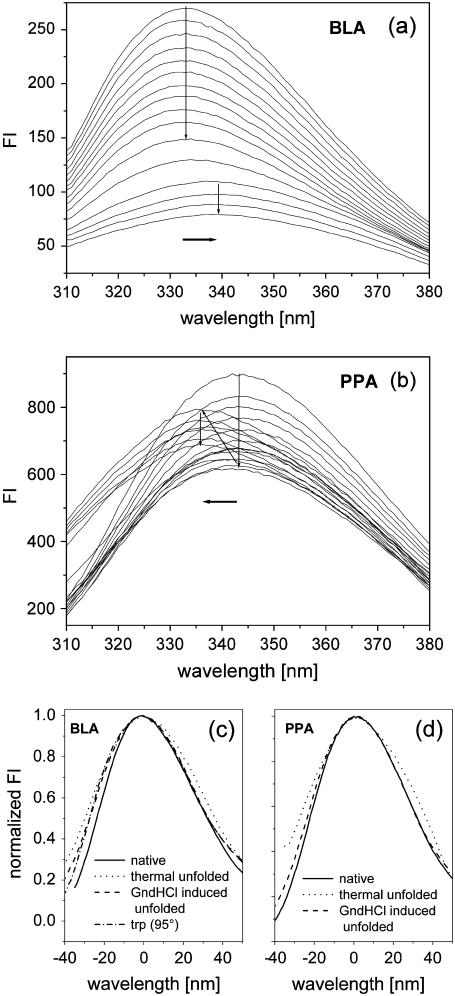
All α-amylases investigated in this study consist of a monomeric structure with three domains. Although the three-dimensional structure of these α-amylases show up rather similar, the corresponding stabilities differ significantly (16,17). It is known from various previous studies that for nearly all α-amylases both thermal and GndHCl-induced unfolding appear as an irreversible process (17–19). In contrast to the GndHCl-induced unfolding, thermal unfolding in most cases is accompanied by strong aggregation (14,17,18). All α-amylase structures bear multiple tryptophan residues more or less equally distributed over the whole structure (see Table 1 and Fig. 1). In the case of multitryptophan proteins, tyrosine fluorescence emission is generally not visible, mainly due to much larger absorption coefficients and quantum yields of the tryptophans, and at least for folded states because of distinct Förster energy transfer from tyrosines to tryptophans (3,14). With respect to the location of tryptophan residues we can roughly distinguish between two different classes of proteins among the α-amylases. One class is represented by PPA, which shows several trytophan residues that are located at the protein surface and have more or less direct contact to the polar solvent. The other class, comprising all other α-amylases, exhibit tryptophan residues that are much more buried in the interior of the protein (see Fig. 1). As a consequence the corresponding emission spectra that were measured during unfolding appear completely different (Fig. 2, a and b). As a representative for the first class, BLA shows a λmax-value for the native state of 333 nm, which increases up to 339 nm upon thermal unfolding. Furthermore the intensity is decreasing during the heating process with an additional decrease in the transition region. A similar behavior, which is typical for most proteins (20), is also observed for TAKA, BSUA, and BAA (see Table 2). The observed red shift is caused by the fact that tryptophan residues are replaced from the less-polar interior of the protein to solvent exposed regions upon unfolding. In contrast PPA shows already in the native state a rather red-shifted emission spectrum (λmax = 343 nm) which first decreases in intensity upon heating. Reaching the transition region the intensity increases accompanied by a significant blue shift of ∼8 nm. Here the process seems to run the other way around. Rather solvent accessible tryptophan residues in the folded state (Fig. 1) reach a more buried and nonpolar microenvironment in the unfolded state. Most probable a well-pronounced aggregation of the thermal unfolded state contributes to the burial of tryptophan residues (discussion of this subject follows in the next section). GndHCl-induced unfolded states show for all α-amylases emission spectra with λmax of ∼348–349 nm which is nearly the value one would expect for fully accessible tryptophan, namely 350 nm. In contrast all thermal unfolded states (with the exception of those obtained from PPA) exhibit λmax values around 340 nm, which correspond to much smaller red shifts upon unfolding. A similar difference in emission peak maxima between thermal and GndHCl-induced unfolding was also observed for other proteins (see for example (10,21)). The shape of emission spectra as obtained from GndHCl-induced and thermal unfolded states also reveal some clear differences (Fig. 2, c and d). On the red edge of the emission spectra a shoulder is visible for thermal unfolded states but not for the native and the GndHCl-induced unfolded states. This shoulder seems to represent a subpopulation of thermal unfolded states which exhibit less buried tryptophan residues, possibly related to unfolded proteins which are not aggregated. The blue edge of the spectra is characterized by larger intensities for the unfolded states as compared to the folded state. This feature can either by related to arising tyrosine emission (often observed around 305 nm for unfolded states when energy transfer to tryptophans disappears during a structural expansion of the protein; see, for example, Schmid (20)) or to elastic scattering caused by aggregation. The shape of the emission spectrum of pure l-tryptophan (see Fig. 2 c) coincides best with the spectrum of the GndHCl-induced unfolded state of α-amylase, indicating that this state represents more or less fully solvent accessible tryptophan residues. Interestingly, the characteristics of spectral shapes as observed for BLA and PPA appear rather similar, although BLA shows a red shift and PPA a blue shift upon thermal unfolding. To summarize the above described observations one can state, that GndHCl-induced unfolding reveals for all α-amylases rather uniform emission spectra with large red shifts, whereas thermal unfolded states appear with more divers and in general with much smaller λmax-values as compared to the former.
TABLE 1.
List of homologous α-amylases used in this study
| Origin α-amylase from | PDB entry | MW (Da) | No. of Trp residues | No. of Tyr residues |
|---|---|---|---|---|
| Pig pancreatic (PPA) | 1DHK | 55,357 | 19 | 19 |
| Aspergillus oryzae (TAKA) | 6TAA | 52,490 | 10 | 34 |
| Bacillus subtilis (BSUA) | 1BAG | 68,421 | 14 | 28 |
| Bacillus amyloliquefaciens (BAA) | 1E43 | 58,843 | 17 | 29 |
| Bacillus licheniformis (BLA) | 1BLI | 58,274 | 17 | 30 |
FIGURE 1.
With respect to the two classes of α-amylases (see text), BLA is shown for the case of buried tryptophan residues whereas PPA represents solvent exposed tryptophan residues in the respective folded states. Tryptophan residues are shown in red color and all presentations, the ribbon model and the surface plot, are shown for the same orientation of the molecules (view into the (β/α)-barrel). With this perspective it is well observable in the surface plot representation, that only PPA shows some red spots, indicative for the fact that only for this α-amylase the corresponding tryptophan residues have direct contact with solvent molecules. In contrast, BLA as a well as all other α-amylases of this class do not show any tryptophan residues in direct contact with the surface. Structure presentations were produced using PyMOL (DeLano, W.L. The PyMOL Molecular Graphics System (2002) DeLano Scientific, San Carlos, CA).
FIGURE 2.
(a, b): Fluorescence emission spectra obtained with an excitation wavelength of 280 nm during thermal unfolding. Both series of spectra were taken with a protein concentration of 50 μg/ml using a heating rate of 1°C/min for BLA between 65°–117°C and for PPA between 25°–78°C. (c, d): Presentations of spectral shapes for three different protein states and for a solution of pure L-tryptophan are shown here. The spectral shapes of emission spectra obtained with L-tryptophan at different temperatures (20 and 95°C, only the latter is shown in c) revealed no differences. To overlay the different spectra, original spectra were normalized and shifted from their respective emission maximum (see Table 2) to a joint reference point (zero).
TABLE 2.
Properties of the emission spectra as obtained under the following conditions (see below)
| Origin α-amylase from | Native λmax (nm) | Thermal unfolded λmax (nm) | T1/2 (°C) | 8 M GndHCl λmax (nm) |
|---|---|---|---|---|
| Pig pancreatic (PPA) | 343 | 335 | 65 | 348 |
| Aspergillus oryzae (TAKA) | 327 | 342 | 70 | 348 |
| Bacillus subtilis (BSUA) | 335 | 341 | 86 | 348 |
| Bacillus amyloliquefaciens (BAA) | 335 | 340 | 86 | 349 |
| Bacillus licheniformis (BLA) | 333 | 339 | 102 | 348 |
Protein concentration, 0.05 mg/ml; thermal unfolding, heating rate of 1°C/min, the respective transition temperatures T1/2 are given for all enzymes; GndHCl-induced unfolding, 8 M GndHCl for 2 weeks. For comparison the emission peak of pure L-tryptophan was observed at 350 nm.
The impact of aggregation on fluorescence emission spectra
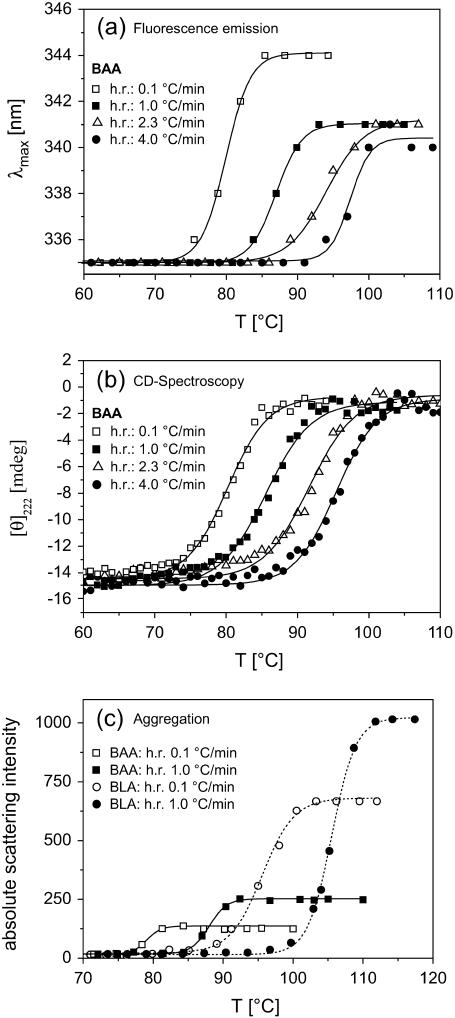
To investigate how fluorescence emission spectra are affected by aggregation we measured the aggregation directly by the use of elastic light scattering and we analyzed unfolding transitions under different conditions which lead to a different extent of protein aggregation. As already mentioned above, there is indication from previous studies on several α-amylases that a structural unfolding is followed by an irreversible process (14,22–25). Due to a rather slow structural unfolding but a fast irreversible step we observe an unfolding process which is under kinetic control. As a consequence the obtained transition temperatures are strongly depending on the heating rates (for a more extended discussion on this subject see (17)). In Fig. 3 this behavior is shown for BAA. Besides the shift of transition temperatures caused by different heating rates, pronounced differences in the λmax-values for the unfolded states are visible in Fig. 3 a. Smaller heating rates are related to larger red shifts. In contrast, CD spectroscopy revealed no differences for the unfolded states at the respective heating rates (Fig. 3 b). For all heating rates the secondary structure elements (mainly α-helices) fully disappear and compared to GndHCl-induced unfolding (data not shown here) the CD signals are identical. In addition, the extent of aggregation as measured by elastic light scattering (Fig. 3 c) shows a distinct dependence on heating rates. Smaller heating rates are accompanied by less-pronounced aggregation. Our data therefore indicate that unfolded states exhibit a larger red shift of the fluorescence emission spectrum when theses states under the respective conditions are less susceptible for aggregation. Furthermore, Fig. 3 c reveals that the native state exhibits no affinity for aggregation which is also true for all other α-amylases within limits of protein concentrations used here.
FIGURE 3.
(a): Thermal unfolding transitions of BAA as measured with 50 μg/ml protein concentration at different heating rates. To obtain most reliable λmax-values, individual emission spectra were fitted with a sum of three Gaussians which fitted all measured emission spectra satisfactorily well. The λ-value at the peak of the resulting fitting curve was taken for λmax (b) Corresponding CD data obtained with the same samples as described in (a). (c) Elastic light scattering as measured at 500 nm for samples with a protein concentration of 50 μg/ml. Similar scattering profiles during thermal unfolding have been observed for BSUA but are not shown here.
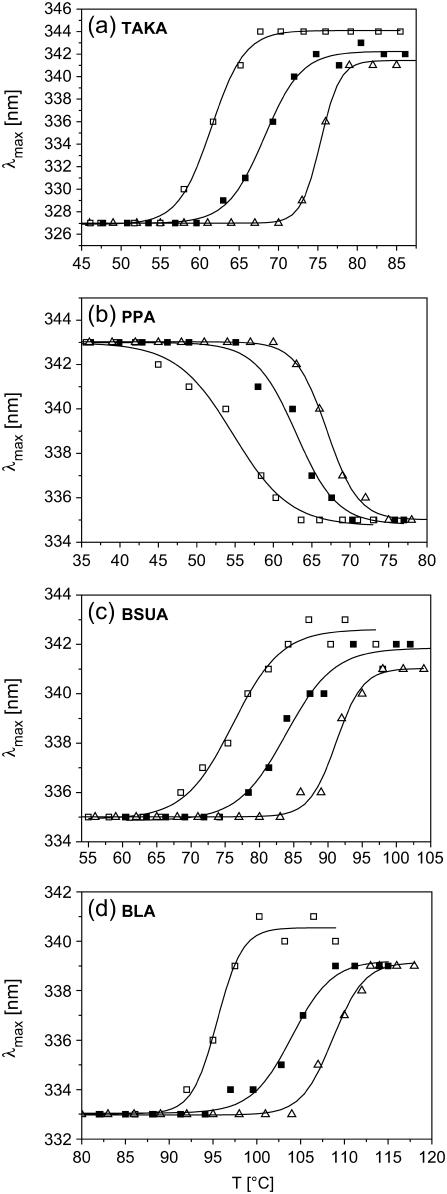
With the exception of TAKA and PPA qualitatively a similar behavior was observed for the other α-amylases, namely for BLA and BSUA (Fig. 4). For PPA, which already exposes tryptophan residues to the protein surface in the folded state, a blue shift of the emission spectrum occurs upon thermal unfolding (see Fig. 2 b). Furthermore, the characteristic shift of transition temperatures with increasing heating rates is as well visible for PPA (see Fig. 4 b). Although PPA exhibits a strong aggregation for the thermal unfolded state, we do not observe any dependence of the magnitude of the blue shift on the heating rate. All thermal unfolded states of PPA are characterized by emission spectra peaked around 335 nm. With respect to all α-amylases investigated here, TAKA is the only protein which does not show a visible aggregation upon thermal unfolding (17). However, with respect to the heating rate dependence of the respective λmax-values obtained for unfolded states, TAKA exhibits the same behavior as compared to the other α-amylases (Fig. 4 a). Another noticeable feature is related to the fact that TAKA shows the largest red shift upon thermal unfolding. The fluorescence emission of the folded state is already rather blue shifted (λmax = 327 nm), but also rather large λmax-values (341–344 nm) for the unfolded states contribute significantly to the large red shift. A possible explanation for this large red shift during thermal unfolding, which is almost as large as those obtained with GndHCl-induced unfolding (see Table 2), may be given by the fact that the absent aggregation gives rise to only a weak burial of tryptophan residues.
FIGURE 4.
(a–d): Thermal unfolding transitions of further α-amylases as measured with 50 μg/ml protein concentration at different heating rates (□, 0.1°C/min; ▪, 1°C/min; ▵, 3°C/min).
Although we observe a qualitative proportionality between smaller red shifts and a significant aggregation of thermal unfolded states (at least for BAA, BLA, and BSUA), we can not exclude other and maybe additional mechanisms responsible for smaller red shifts during thermal unfolding as compared to those obtained with GndHCl-induced unfolding. In principle, unfolded states are rather heterogeneous (26–28) and in addition the mechanisms leading to unfolded states are rather different for thermal and GndHCl-induced unfolding (1). Therefore, the respective unfolded states can have rather different structures (21). For example, more compact structures of the thermal unfolded states as compared to chemical unfolded states would also explain more buried tryptophan residues which exhibit smaller red shifts. Furthermore, the conformational scrambling of both unfolded states might be different due to different mechanisms of denaturing the proteins. It is obvious that this also can have different effects on the microenvironment of tryptophan residues. However, our data indicate that aggregation and in addition at least one of these other mechanisms seems to play a role for the α-amylases with red shifts upon thermal unfolding (i.e., with the exception of PPA). Even for TAKA, which does not show any aggregation, we observed slightly smaller red shift for thermal unfolding, but due to missing aggregation still larger as compared to BLA, BAA, and BSUA.
For thermal and GndHCl-induced unfolded states of BLA, BAA, and of BSUA we made an attempt to initiate a potential refolding process. For this purpose, either a cooling back to room temperature or a removal of GndHCl (by dialysis) has been carried out to measure properties of the enzymes after the denaturing impact was eliminated. With respect to corresponding CD spectra we observed for both types of samples (i.e., cooled and GndHCl removed) no or only very weak secondary structure elements. Furthermore both types of samples exhibit associated fluorescence emission spectra that are rather similar. The obtained peak maxima around 336–338 nm indicate rather small red shifts (∼2–3 nm) with respect to the native state (see also Fitter and Haber-Pohlmeier (14)). Interestingly, the sample with removed GndHCl exhibits now a strong elastic scattering related to pronounced aggregation, which was not observed under unfolding conditions with 8 M GndHCl. For the thermal unfolded proteins we observe more or less the same extent of aggregation after cooling, which was observed at temperatures above the transition temperature. From these results the following scenario about the underlying processes emerges: Elevated temperatures as well as high concentrations of chemical denaturants dissolve the secondary structure elements and the three-dimensional integrity of the protein structures. In contrast to high temperatures the presence of GndHCl efficiently suppresses aggregation. After eliminating the denaturing force the proteins are not able to fold back into their native (and functional) structure and the still unfolded proteins form (or just stay on with) aggregates. For this final state we do not watch a significant difference between formerly thermal or GndHCl-induced unfolded states. For both cases we observe only a rather small magnitude of red shift upon unfolding, most probable due to the strong tendency to form aggregates.
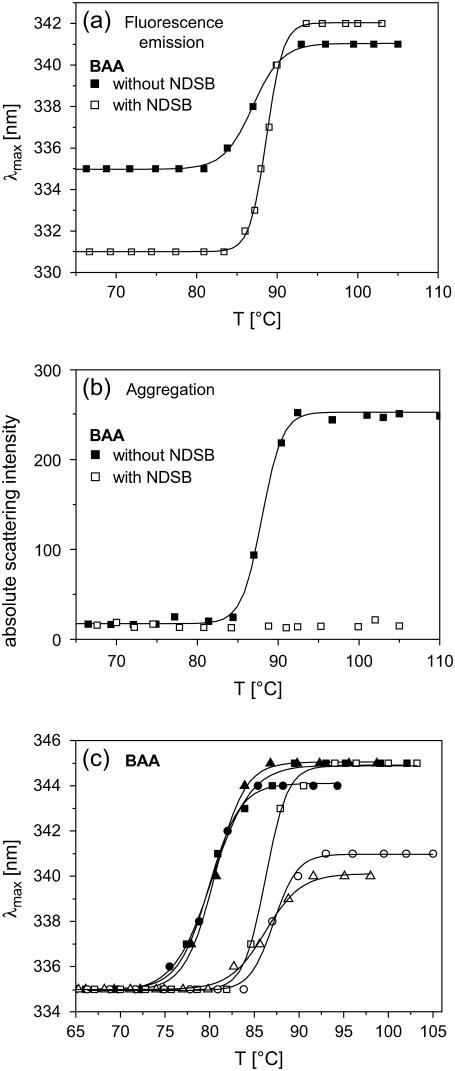
Further and more detailed insights in the mechanisms of how aggregation affects properties of the unfolded state can be gained from studies where aggregation, in particular of thermal unfolded states, is suppressed. Encapsulation techniques, either using liposomes (29) or sol-gel glasses (30,31), have recently been used to trap individual proteins in small molecular voids. Under specific conditions, single proteins can be encapsulated in small volumes (e.g., 10–100 nm) and therefore aggregation does not occur (see for example (29)). Furthermore, studies with solubilizing agents such as detergents, reversed micelles, or nondetergent sulfobetaines (NDSB) proved that aggregation, which typically occurs during thermal unfolding in pure buffers, is strongly suppressed when such agents are added to the solution (29,32). As a first step in this direction, we measured the thermal unfolding transition of α-amylases in the presence of NDSB, which is known to be an effective agent to prevent aggregation of thermal unfolded proteins (32–34). In contrast to samples with pure buffer solutions, samples with NDSB do not show significant aggregation of thermal unfolded states. This result is shown for BAA in Fig. 5 b, but was also observed for all other α-amylases which otherwise exhibit aggregation upon thermal unfolding. As shown in Fig. 5 a, the transition temperature is nearly identical for samples with and without NDSB. With respect to the fluorescence emission spectra α-amylases in NDSB solutions show peak maxima for the folded state which are significantly blue shifted as compared to pure buffer samples. The λmax-values of the unfolded states in NDSB solutions were slightly larger (see Fig. 5 a for BAA) or identical within the limits of error as compared to samples without NDSB. In addition, all thermal transitions in NDSB solutions exhibit also the missing reversibility as observed for pure buffer solutions. The pronounced blue shift for native proteins in NDSB solutions is most probably related to the fact that NDSB interacts with the surface of the proteins which causes a more hydrophobic nonpolar environment of surface exposed tryptophan residues. It is obvious from the data shown in Fig. 5 a that the λmax-values obtained for the native and most probably also for the nonaggregated thermal unfolded state are strongly biased by direct interaction with NDSB. The fact that for NDSB samples the obtained Δλmax-value between the folded and the unfolded state is much larger as compared to that measured with pure buffer samples is a further indication that aggregates reduce the magnitude of red shifts. However, more meaningful data might be obtained by employing encapsulation techniques, which in principle can ensure studies without interfering interactions of tryptophan residues with the encapsulating material.
FIGURE 5.
(a, b) Thermal unfolding transitions and aggregation as measured with a 0.5 M NDSB (NDSB-201: 3-(1-Pyridino)-1-propane Sulfonate; Fluka) buffer solution and in pure buffer solutions. All measurements were performed with BAA 50 μg/ml protein and with a heating rate of 1 °C/min. (c) Thermal unfolding transitions of BAA (in pure buffer) as measured with heating rates of 0.1 °C/min (solid symbols) and of 1 °C/min (open symbols). In addition three different protein concentrations were used (squares: 7 μg/ml; circles: 50 μg/ml; triangles: 1 mg/ml).
The effect of protein concentration and heating rates
It is well known that the kinetics and the extent of aggregation often depend not only on heating rates and temperatures but also on protein concentration (9,35). As shown in Fig. 5 c, and as known for all α-amylases studied here, the protein concentration has no effect on the transition temperatures (17). However, the fluorescence emission peak maxima obtained with a heating rate of 1°C/min exhibit a pronounced dependence on the protein concentration. Higher protein concentrations lead to smaller red shifts upon thermal unfolding. The lowest protein concentration of 7 μg/ml reveals a large red shift with λmax of ∼345 nm, whereas protein concentrations of 50 μg/ml and 1 mg/ml result in λmax-values ∼340 nm for the unfolded states. In contrast, the slowest heating rate of 0.1°C/min reveals for all protein concentrations λmax-values around 345 nm for the unfolded state. Assuming at least a qualitative relation between the absence of aggregation and larger red shifts for the unfolded states of BSUA, BLA, and BAA we can state the following: (1), the smallest heating rate (0.1°C/min) gives rise to less pronounced aggregation during the thermal unfolding transition as compared to higher heating rates; and (2), the lowest protein concentration (7μg/ml) reveals a less-pronounced aggregation also for a higher heating rate (1°C/min), whereas higher protein concentrations studied with the higher heating rate exhibit more pronounced aggregation.
CONCLUSION
It has been demonstrated in this study that the λmax-values of tryptophan fluorescence emission are meaningful parameter to characterize properties of the unfolded state. Due to the high sensitivity of tryptophan fluorescence with respect to changes in the microenvironment of the intrinsic chromophores, various properties of the unfolded state can have a distinct effect on the λmax-values. In particular differences in the extent of protein aggregation, in structural compactness, or simply in the structural arrangement of tryptophan residues in unfolded states can change the magnitude of the red shift observed during the unfolding transition significantly. Due to the complexity of the investigated proteins (multitryptophan proteins in an ensemble of structurally heterogeneous unfolded states) a quantitative relation between λmax-values and the extent of aggregation or the compactness of the unfolded state cannot be given. However, at least on a qualitative level in particular differences between several unfolded states can be detected easily. Furthermore it was demonstrated here in the case of homologous α-amylases, that accompanying studies with additional techniques (CD, static, and dynamic light scattering, see for example (14,36) for analyzing the compactness in terms of radii of gyration of unfolded states) can help to obtain a more meaningful interpretation of the tryptophan fluorescence data. A promising approach to elucidate the effect of aggregation in more detail and maybe on a quantitative level would be the use of encapsulation techniques. To perform studies in this direction the conformational stability of encapsulating structures at rather elevated temperatures (above 80°C) needs to be ensured.
Acknowledgments
J. F. thanks G. Büldt (Forschungszentrum Jülich) for continuous support in his institute.
References
- 1.Tanford, C. 1968. Protein denaturation. Adv. Protein Chem. 23:121–282. [DOI] [PubMed] [Google Scholar]
- 2.Buchner, J., and T. Kiefhaber. 2005. Protein Folding Handbook. Vol. 1–5. Wiley-VCH Verlag, Weinheim, Germany.
- 3.Lakowicz, J. R. 1999. Principles of Fluorescence Spectroscopy. Kluwer Academic/Plenum Press, New York.
- 4.Eftink, M. R. 1994. The use of fluorescence methods to monitor unfolding transitions in proteins. Biophys. J. 66:482–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burstein, E. A. 1976. Luminescence of protein chromophores: model studies. In Advances in Science and Technology, Ser. Biophysics, Vol. 6, VINITI. Moscow.
- 6.Vivian, J. T., and P. R. Callis. 2001. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 80:2093–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burstein, E. A., S. M. Abornev, and Y. K. Reshetnyak. 2001. Decomposition of protein tryptophan fluorescence spectra into log-normal components. I. Decomposition algorithms. Biophys. J. 81:1699–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reshetnyak, Y. K., and E. A. Burstein. 2001. Decomposition of protein tryptophan fluorescence spectra into log-normal components. II. The statistical proof of discreteness of tryptophan classes in proteins. Biophys. J. 81:1710–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Young, L. R., K. A. Dill, and A. L. Fink. 1993. Aggregation and denaturation of apomyoglobin in aqueous urea solutions. Biochemistry. 32:3877–3886. [DOI] [PubMed] [Google Scholar]
- 10.Kreimer, D. I., V. L. Shnyrov, E. Villar, I. Silman, and L. Weiner. 1995. Irreversible thermal denaturation of Torpedo californica acetylcholinesterase. Protein Sci. 4:2349–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lasagna, M., E. Gratton, D. M. Jameson, and J. E. Brunet. 1999. Apohorseradish peroxidase unfolding and refolding: intrinsic tryptophan fluorescence studies. Biophys. J. 76:443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewert, S., A. Honegger, and A. Pluckthun. 2003. Structure-based improvement of the biophysical properties of immunoglobulin VH domains with a generalizable approach. Biochemistry. 42:1517–1528. [DOI] [PubMed] [Google Scholar]
- 13.Chattopadhyay, A., S. S. Rawat, D. A. Kelkar, S. Ray, and A. Chakrabarti. 2003. Organization and dynamics of tryptophan residues in erythroid spectrin: novel structural features of denatured spectrin revealed by the wavelength-selective fluorescence approach. Protein Sci. 12:2389–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitter, J., and S. Haber-Pohlmeier. 2004. Structural stability and unfolding properties of thermostable bacterial alpha-amylases: a comparative study of homologous enzymes. Biochemistry. 43:9589–9599. [DOI] [PubMed] [Google Scholar]
- 15.Monsellier, E., and H. Bedouelle. 2005. Quantitative measurement of protein stability from unfolding equilibria monitored with the fluorescence maximum wavelength. Protein Eng. Des. Sel. 18:445–456. [DOI] [PubMed] [Google Scholar]
- 16.Fitter, J. 2005. Structural and dynamical features contributing to thermostability in alpha-amylases. Cell. Mol. Life Sci. 62:1925–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duy, C., and J. Fitter. 2005. Thermostability of irreversible unfolding alpha-amylases analyzed by unfolding kinetics. J. Biol. Chem. 280:37360–37365. [DOI] [PubMed] [Google Scholar]
- 18.Tomazic, S. J., and A. M. Klibanov. 1988. Mechanisms of irreversible thermal inactivation of Bacillus alpha-amylases. J. Biol. Chem. 263:3086–3091. [PubMed] [Google Scholar]
- 19.D'Amico, S., J. C. Marx, C. Gerday, and G. Feller. 2003. Activity-stability relationships in extremophilic enzymes. J. Biol. Chem. 278:7891–7896. [DOI] [PubMed] [Google Scholar]
- 20.Schmid, F. X. 1989. Optical spectroscopy to characterize protein conformation and conformational changes. In Protein Structure: A Practical Approach. T. E. Creighton, editor. 261–297. IRL Press, Oxford, UK.
- 21.Dubey, V. K., and M. V. Jagannadham. 2003. Differences in the unfolding of procerain induced by pH, guanidine hydrochloride, urea, and temperature. Biochemistry. 42:12287–12297. [DOI] [PubMed] [Google Scholar]
- 22.Fukada, H., K. Takahashi, and J. M. Sturtevant. 1987. Differential scanning calorimetric study of the thermal unfolding of Taka-amylase A from Aspergillus oryzae. Biochemistry. 26:4063–4068. [DOI] [PubMed] [Google Scholar]
- 23.Tomazic, S. J., and A. M. Klibanov. 1988. Why is one Bacillus alpha-amylase more resistant against irreversible thermoinactivation than another? J. Biol. Chem. 263:3092–3096. [PubMed] [Google Scholar]
- 24.Violet, M., and J. C. Meunier. 1989. Kinetic study of the irreversible thermal denaturation of Bacillus licheniformis alpha-amylase. Biochem. J. 263:665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Declerck, N., M. Machius, R. Chambert, G. Wiegand, R. Huber, and C. Gaillardin. 1997. Hyperthermostable mutants of Bacillus licheniformis alpha-amylase: thermodynamic studies and structural interpretation. Protein Eng. 10:541–549. [DOI] [PubMed] [Google Scholar]
- 26.Dyson, H. J., and P. E. Wright. 1998. Equilibrium NMR studies of unfolded and partially folded proteins. Nat. Struct. Biol. 5(Suppl):499–503. [DOI] [PubMed] [Google Scholar]
- 27.Onuchic, J. N., Z. Luthey-Schulten, and P. G. Wolynes. 1997. Theory of protein folding: the energy landscape perspective. Annu. Rev. Phys. Chem. 48:545–600. [DOI] [PubMed] [Google Scholar]
- 28.Fitter, J. 2003. A measure of conformational entropy change during thermal protein unfolding using neutron spectroscopy. Biophys. J. 84:3924–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin, I., E. Wachtel, E. Roth, C. Bon, I. Silman, and L. Weiner. 2002. Thermal denaturation of Bungarus fasciatus acetylcholinesterase: is aggregation a driving force in protein unfolding? Protein Sci. 11:2022–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brennan, J. D. 1999. Using intrinsic fluorescence to investigate proteins entrapped in sol-gel derived materials. Appl. Spectrosc. 53:106A–121A. [Google Scholar]
- 31.Eggers, D. K., and J. S. Valentine. 2001. Molecular confinement influences protein structure and enhances thermal protein stability. Protein Sci. 10:250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vuillard, L., T. Rabilloud, and M. E. Goldberg. 1998. Interactions of non-detergent sulfobetaines with early folding intermediates facilitate in vitro protein renaturation. Eur. J. Biochem. 256:128–135. [DOI] [PubMed] [Google Scholar]
- 33.Goldberg, M. E., N. Expert-Bezancon, L. Vuillard, and T. Rabilloud. 1996. Non-detergent sulphobetaines: a new class of molecules that facilitate in vitro protein renaturation. Fold. Des. 1:21–27. [DOI] [PubMed] [Google Scholar]
- 34.Vuillard, L., C. Braun-Breton, and T. Rabilloud. 1995. Non-detergent sulphobetaines: a new class of mild solubilization agents for protein purification. Biochem. J. 305:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Bon, C., T. Nicolai, and D. Durand. 1999. Kinetics of aggregation and gelation of globular proteins after heat-induced denaturation. Macromolecules. 32:6120–6127. [Google Scholar]
- 36.Wilkins, D. K., S. B. Grimshaw, V. Receveur, C. M. Dobson, J. A. Jones, and L. J. Smith. 1999. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 38:16424–16431. [DOI] [PubMed] [Google Scholar]