Abstract
The bacterial RNA polymerase (RNAP) recognizes promoters through sequence-specific contacts of its promoter-specificity components (σ) with two DNA sequence motifs. Contacts with the upstream (‘−35') promoter motif are made by σ domain 4 attached to the flap domain of the RNAP β subunit. Bacteriophage T4 late promoters consist solely of an extended downstream (‘−10') motif specifically recognized by the T4 gene 55 protein (gp55). Low level basal transcription is sustained by gp55-RNAP holoenzyme. The late transcription coactivator gp33 binds to the β flap and represses this basal transcription. Gp33 can also repress transcription by Escherichia coli σ70-RNAP holoenzyme mutated to allow gp33 access to the β flap. We propose that repression is due to gp33 blocking an upstream sequence-independent DNA-binding site on RNAP (as σ70 domain 4 does) but, unlike σ70 domain 4, providing no new DNA interaction. We show that this upstream interaction is essential only at an early step of transcription initiation, and discuss the role of this interaction in promoter recognition and transcriptional regulation.
Keywords: gene regulation, repression, RNA polymerase, phage T4 gp33, promoter
Introduction
Promoter recognition by RNA polymerase (RNAP) is a key step of gene regulation in bacteria. To begin transcription, the initiating form of RNAP, the holoenzyme, which comprises the catalytic core (subunit composition α2ββ′ω) and one of several promoter-specificity components (sigma subunits), must first recognize a promoter among other DNA sequences to form the initial, closed complex. The closed promoter complex then isomerizes into the open complex, in which ∼13 base pairs (bp) around the transcriptional start site are melted (McClure, 1985; Record et al, 1996). The open promoter complex can begin RNA synthesis, but must pass an additional checkpoint (of abortive initiation) before engaging in productive transcript elongation. Each step of transcription initiation involves multiple reaction intermediates and is subject to extensive and often tight regulation in the cell (reviewed by Lloyd et al, 2001).
A majority of promoters for the most-studied holoenzyme, containing the Escherichia coli primary sigma subunit σ70, are defined by two hexanucleotide motifs located ∼35 and ∼10 bp upstream of the transcriptional start site (+1) (Harley and Reynolds, 1987). These promoters are recognized through sequence-specific interactions of σ70 RNAP holoenzyme with DNA: an upstream interaction with the −35 motif by σ70 domain 4, bound to the RNAP core β subunit's flap (Murakami et al, 2002; Murakami and Darst, 2003); and a downstream interaction with the −10 motif by σ70 domain 2, bound to the RNAP core β′ subunit's coiled coil domain. Some promoters contain an extension of the −10 motif (Kumar et al, 1993; Burns et al, 1999). These ‘−10 extended' promoters do not require the −35 motif, and can be recognized by holoenzymes that lack σ70 domain 4 or the β flap (Kumar et al, 1993; Kuznedelov et al, 2002).
During infection of E. coli with bacteriophage T4, transcription of viral genes occurs in three stages, successively engaging three families of promoters. T4 early and middle promoters require RNAP holoenzyme containing σ70; late promoters require a holoenzyme containing the T4-encoded late promoter-specificity subunit gp55 (Stitt and Hinton, 1994; Williams et al, 1994; Brody et al, 1995). The 185 amino acid gp55 is a highly divergent member of the σ70 family, sharing only a limited similarity with σ70 core-binding segments 2.1 and 2.2 (Gribskov and Burgess, 1986; Helmann and Chamberlin, 1988; Lonetto et al, 1992). Gp55 also lacks a σ domain 4 equivalent. Accordingly, T4 late promoters contain only a −10 motif with the closely adhered-to consensus sequence TATAAATA, and have no additional sequence determinants (Elliott and Geiduschek, 1984; Kassavetis et al, 1986). In vitro, RNAP core and gp55 allow specific, albeit low-level, basal transcription from T4 late promoters. Two additional T4 proteins, the RNAP-bound gp33 and the DNA-loaded sliding clamp, gp45, together greatly enhance this transcription (Herendeen et al, 1990, 1992), facilitating both the formation of the closed promoter complex and its isomerization into the open complex (Kolesky et al, 2002). Gp33 binds the RNAP core at its β flap (Nechaev et al, 2004), and gp45 interacts with gp55 and gp33 (Sanders et al, 1997; Wong and Geiduschek, 1998).
In the absence of the gp45-sliding clamp, gp33 represses basal transcription from T4 late promoters (Williams et al, 1989; Herendeen et al, 1990). Here, we investigate the mechanism of this repression in detail and show the following: (1) Gp33 prevents RNAP binding to internal sites of double-stranded DNA, but does not prevent interaction with DNA ends. (2) Gp33 represses transcription only if present before the T4 late promoter opens; even partial pre-opening of the promoter blocks repression. (3) Gp33 binds to the upstream end of the open promoter complex, and remains with, or can reattach to, the elongating transcription complex. (4) Repression by gp33 is not restricted to the gp55-RNAP holoenzyme; gp33 can also repress transcription by the nonconjugate σ70 holoenzyme when the interaction of σ70 domain 4 with the β flap is weakened by mutation. We suggest that, like σ domain 4, gp33 blocks an upstream, sequence-nonspecific DNA-binding site on RNAP core but, in contrast to σ domain 4, gp33 does not replace it with a new DNA interaction. We also discuss the role of an upstream DNA interaction in promoter recognition and transcriptional regulation.
Results
Gp33 prevents the formation of open complexes, but does not inhibit transcription by preformed open complexes
The first experiment examines the effect of gp33 on the formation and stability of T4 late promoter complexes. E. coli RNAP core and T4 gp55 were incubated with 125-bp linear DNA containing a T4 late promoter (P23) to form open complexes and promoter opening was monitored by KMnO4 probing (Figure 1A). When added to RNAP before DNA, gp33 prevented formation of open complexes (compare lanes 2 and 3 and lanes 5 and 6). In contrast, when added after pre-incubating RNAP with DNA, gp33 did not affect already opened complexes (compare lanes 2 and 4 and lanes 5 and 7), even if these were allowed to form for only 5 min (lanes 2 and 4).
Figure 1.
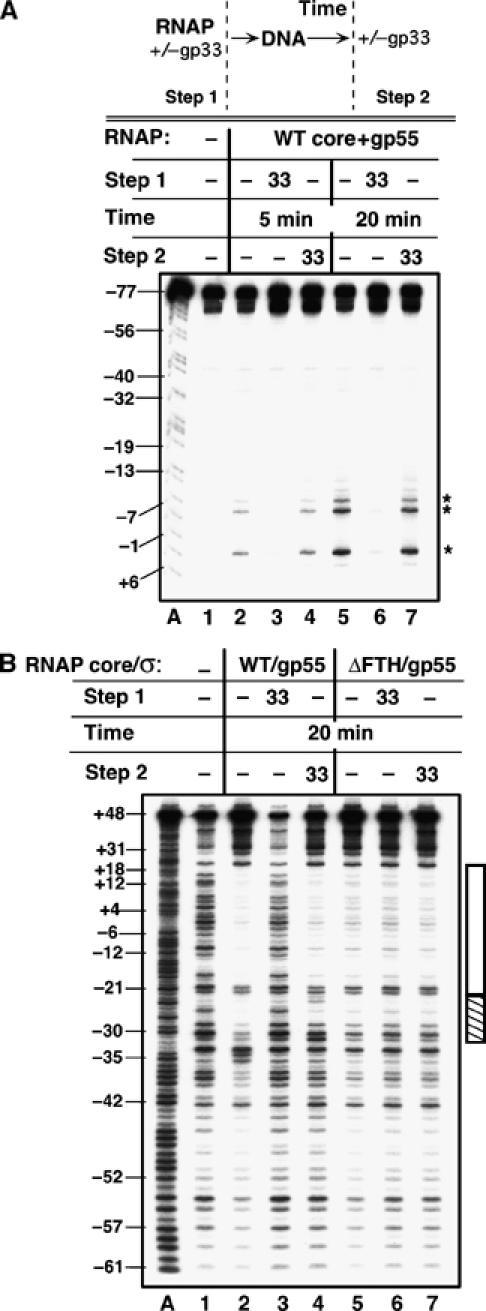
The effect of gp33 on T4 late open promoter complexes. (A) Open promoter complexes monitored by KMnO4 probing. RNAP was combined with gp33, where indicated (Step 1), followed by the addition of P23 promoter DNA, 32P-labeled on the transcribed (template) strand and incubation for 5 or 20 min, as specified. Alternatively, gp33 was added to preformed open promoter complexes (Step 2). DNA cleavage at T +2, −6, and −8, indicative of promoter opening, is shown by asterisks. (B) Open complexes monitored by DNase I footprinting. Complexes containing the indicated RNAP were formed for 20 min, with gp33 added before DNA (Step 1) or after open complex formation (Step 2), as indicated. Heparin was added (to 100 μg/ml) for 30 s prior to DNase addition. Lanes A: A-sequence ladders; lanes 1: DNA without protein. The DNA segment protected by the open promoter complex is indicated by the vertical bar, with gp33-dependent changes highlighted by striping.
Monitoring the formation of open complexes by DNase I footprinting (in the presence of heparin, to selectively visualize open complexes) yielded a similar result: gp33 prevented the formation of open complexes (Figure 1B, compare lanes 1 and 3) but did not destroy already formed open complexes (lane 4). However, gp33 did change the DNase cleavage pattern of the preassembled, open promoter complex both on the nontemplate strand (Figure 1B, compare lanes 2 and 4) and the template strand (Supplementary Figure 1), generating a consistently observed change between bp −23 and −32 and diminishing protection upstream of bp −32. These changes indicate that gp33 interacts with the open promoter complex at its upstream end. This interaction is specific, since gp33 had no effect on the DNase I footprint of the open promoter complex assembled with RNAP core lacking its gp33-binding site, the β flap tip helix (ΔFTH; Figure 1B, lanes 5–7). The result is fully consistent with an earlier finding that gp33 binds to the RNAP flap (Nechaev et al, 2004), and suggests that the RNAP flap domain does not participate in maintaining the open promoter complex.
Gp33 only blocks interaction with internal DNA sites
To determine whether attaching gp33 to the open promoter complex alters its transcriptional output, open T4 late promoter complexes formed as above were supplied with a mixture of NTPs and heparin, enabling a single round of transcription (Figure 2A). Transcription of this 125 bp promoter fragment yielded multiple discrete RNA products, several of which exceeded the length of the template. These transcripts can be divided into two groups (Figure 2A). The lengths of one group of transcripts (estimated by comparison with labelled single-stranded DNA markers that are not shown) correspond to the run-off transcript (P1; 48 nt) and 48 nt plus multiples of the template length (P(n+1)≅(48+125n) nt); these transcripts are presumed to have initiated at the promoter and, for n>0, to have been extended by RNAP ‘switching' from one template end to another without releasing the nascent transcript. The lengths of the second group of transcripts (E) correspond to multiples of the template length (125n), indicating that these transcripts were end-initiated. In the presence of gp33, promoter-originating (P) transcription was repressed, as expected; in sharp contrast, end-initiated (E) transcription was not repressed by gp33 (Figure 2A, lanes 1 and 2). There was no repression of transcription by gp33 in control reactions containing the ΔFTH holoenzyme (lanes 4–6).
Figure 2.
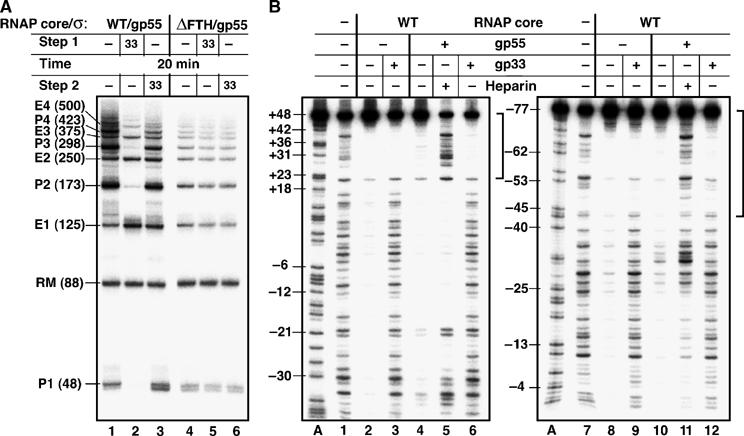
Gp33 prevents RNAP binding to internal DNA sites, but not to DNA ends. (A) Repression of transcription by gp33. Open complexes were formed on a 125 bp template containing a T4 late P23 promoter; where indicated, gp33 was added before DNA (Step 1) or after the formation of the open promoter complex (Step 2). Transcripts originating at the promoter and DNA ends are identified as P and E, respectively, and their predicted lengths are indicated in brackets. P1 and E1 are run-off transcripts; longer transcripts are products of template switching. RM: labelled DNA recovery marker. (B) The effect of gp33 on nonspecific DNA binding, monitored by DNase I footprinting. The same 125 bp P23 DNA was labelled in the nontranscribed (lanes 1–6) or transcribed (lanes 7–12) strands. RNAP core or gp55 holoenzyme was incubated with DNA for 20 min at 25°C and treated with DNase I in the absence of heparin, except for samples for lanes 5 and 11, to which heparin was added 30 s prior to DNase. Where indicated, gp33 was added to RNAP prior to DNA. The extent of DNA end that is protected by RNAP in the presence of gp33 is shown by brackets.
The production of these ‘too-long' P and E transcripts was relatively insensitive to DNA concentration. In particular, serial dilution from the standard assay concentration of 4–0.4 nM DNA progressively reduced the extent of template transfer by a factor of less than 1.4 (versus the expected 10-fold reduction if template switching had been exclusively intermolecular). These observations suggest that the template transfer had a substantial intramolecular component, that is, that RNAP was able to ‘catch' the other end of the already engaged 125 bp template. This is a somewhat unexpected finding for a transcription template shorter than the DNA persistence length (∼150–175 bp) and considerably shorter than optimal DNA lengths for cyclic ligation (Shore et al, 1981; Hagerman, 1988; Crothers et al, 1992; Kahn et al, 1994; Bouchiat et al, 1999). On the other hand, the persistence length of DNA is sensitive to the concentrations of counterions and especially Mg2+ (present in the reaction medium at relatively high concentration), as well as sequence (Crothers et al, 1992; Baumann et al, 1997) (this DNA is A/T-rich and contains partly phased An:Tn tracts); the DNA path in the elongating transcription complex is also sharply bent (Kahn and Crothers, 1993; Rees et al, 1993); each of these specific structural features would be expected to make this relatively short DNA more compliant with cyclic template switching.
We also noted that gp33 reduced the proportion of higher molecular weight P and E transcripts (Figure 2A, compare transcripts P1–E4 in lanes 1 and 3; see also Figure 5). This result indicates that the apparent stimulation of transcripts E1 (lane 2) and P1 (lane 3) by gp33 was due to reduced template switching during transcript elongation, and not to stimulation of initiation (see also Figure 1A). These effects of gp33 were seen with wild-type RNAP (lanes 1–3) but not with ΔFTH RNAP (lanes 4–6), indicating that they require binding of gp33 to the RNAP β flap, and implying that gp33 remains bound to, or is capable of reattaching to, transcript-elongating RNA polymerase through interaction with its β flap.
Figure 5.
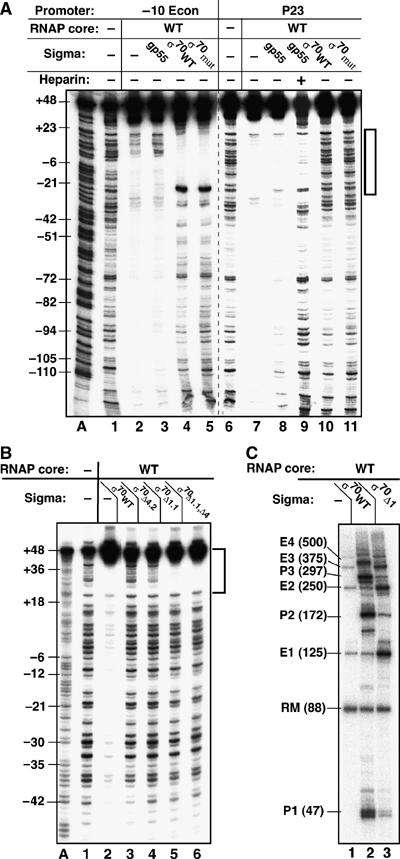
Nonspecific DNA binding by σ70 RNAP holoenzyme derivatives. (A) Comparison of DNase I footprints of gp55- and σ70-RNAP holoenzymes binding DNA probes containing cognate or noncognate promoters in the absence of heparin (except for lane 9). Complexes were assembled on 260 bp DNA (extended upstream by 136 bp relative to the standard ∼125 bp DNA templates) containing a consensus extended −10 (‘−10 Econ') (lanes A and 1–5) or T4 late (‘P23') promoter (lanes 6–11), 5′ end-labelled on the nontranscribed strand. Lane A: an A sequence ladder; lanes 1 and 6: digestion patterns for DNA. σ70mut is the R541C/L607P double mutant. DNA segments protected by RNAP in open promoter complexes are indicated. (B) DNA-end binding by σ70Δ1.1 holoenzyme, monitored on 125 bp P23 promoter DNA labelled in the nontemplate strand. The extent of DNA end protected by RNAP is indicated. (C) Transcription of the 126 bp extended −10 consensus promoter template by RNAP core (lane 1), wild-type σ70 (lane 2), and σ70Δ1.1 (lane 3) holoenzymes. Transcripts originating at the promoter (P) and DNA ends (E) are indicated as in Figure 2A.
When added to the preformed open complex, gp33 increased the production of P1 (promoter-initiated run-off) transcripts that are shorter by 1 nt (Figure 2A, lanes 1 and 3). Primer extension revealed that the start site was not changed (data not shown), and therefore that gp33 must have affected the last step of nucleotide addition to the run-off transcript. The proportion of shorter P1 transcripts produced by the ΔFTH enzyme was not affected by gp33 (lanes 4–6).
That gp33 inhibits promoter-initiated transcription but not end-initiating transcription implies that it only prevents RNAP binding to promoters, and not to DNA ends. To monitor relatively weak, non-promoter DNA binding, DNase I footprinting was carried out in the absence of the heparin competitor (Figure 2B). RNAP core, alone or in the presence of gp55, generated general protection of the 125 bp P23 DNA from DNase I cleavage (Figure 2B, lanes 2 and 4). (The residual cleavage in lane 4 at positions −20, −21, and ∼−35 is due to the gp55-dependent open complex formation, as revealed by the addition of heparin to the reaction (lane 5).) In the presence of gp33 (added before DNA), protection from DNase I cleavage was lost everywhere, except for an approximately 25 bp segment at the labeled DNA end (lane 3). The same result was obtained in the presence of gp55 (conditions under which gp33 represses promoter-specific transcription) (lane 6). Footprinting the other DNA strand (lanes 7–12) showed that gp33 exerts its protective effect at both DNA ends, indicating that DNA end-binding by core or gp55 holoenzyme does not depend on a unique DNA sequence.
That gp33 prevents RNAP binding to internal DNA sites but not to DNA ends is consistent with an observation that gp33 only partially inhibited transcription of promoterless templates such as poly(dG-dC):poly(dG-dC) (Supplementary Figure 2). Partial inhibition can be rationalized as inability to block initiation at DNA ends (and possibly also single-strand breaks, branch junctions, and other structural singularities) while preventing internal initiation.
Gp33 represses transcription by a nonconjugate σ holoenzyme
Domain 4 of σ70 is at a very large advantage in competing with gp33 for occupancy of the RNAP core's β flap because it is tethered to the core by attachment of the rest of σ70, which effectively greatly increases its local concentration. However, gp33 does bind to σ70 holoenzymes with deletions of, or mutations in, σ70 domain 4 that weaken its interaction with the β flap (Nechaev et al, 2004). The ability of gp33 to repress transcription by these holoenzymes was tested with a variant template in which a σ70 consensus extended −10 promoter replaced the P23 promoter (Figure 3A). Gp33 had no effect on transcription by the wild-type σ70 holoenzyme, as expected (Figure 3B, lanes 1–3), but did repress transcription by σ70 holoenzyme with two σ70 region 4 point mutations that weaken its interaction with the β flap (compare lanes 4 and 5), and by holoenzyme deleted for σ70 region 4.2 (compare lanes 7 and 8). As shown above for gp55 holoenzyme, gp33 did not repress promoter-specific transcription by any of the σ70 holoenzymes when added to preformed open complexes (Figure 3B, lanes 3, 6, and 9). Thus, repression of transcription by gp33 is not restricted to its cognate gp55 holoenzyme; it does not depend on any interaction with gp55 and appears to depend only on the accessibility of the β flap on RNAP.
Figure 3.
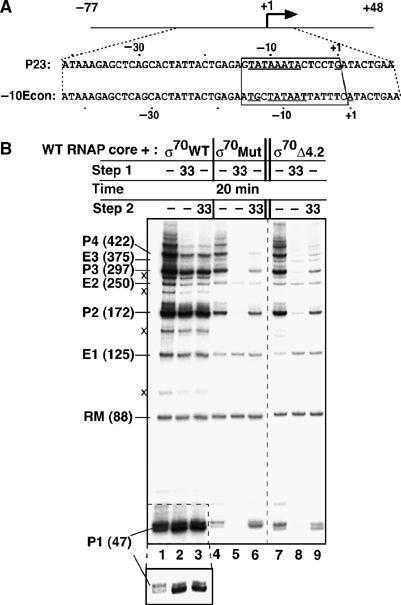
Transcription by σ70 holoenzyme can be repressed by gp33. (A) The sequences of the 126 bp consensus extended −10 (−10 Econ), and 125 bp T4 late (P23) promoter templates differ only within the box-enclosed segment. Only the nontranscribed strands are shown. (B) Single-round transcription on the −10 extended consensus promoter template with wild-type RNAP core and the indicated σ70-derivative holoenzymes was performed as for Figure 2A. The bottom panel shows P1 transcripts in a shorter exposure, to emphasize the 1 bp transcript shortening referred to in the text. Transcripts indicated by (x), obtained with wild-type σ70 holoenzyme, are of unknown origin.
Similar to transcription by gp55 holoenzyme, end-initiated transcription by these σ70 holoenzymes was not blocked by gp33 (E transcripts in Figure 3), and a comparable reduction of template switching efficiency and 1-bp shortening of the run-off product was observed, even for the wild-type σ70 holoenzyme (see the short-exposure inlay for the P1 transcript in Figure 3B). These observations are compatible with σ70 release from the elongation complex (Kapanidis et al, 2005; Mooney et al, 2005; Raffaelle et al, 2005) or with release of σ70 domain 4 from the β flap to make way for gp33 attachment to the RNA chain-elongating transcription complex.
Gp33 does not repress transcription from partially opened DNA templates
The above experiments show that gp33 blocks initiation of transcription at an early step. If binding to double-stranded DNA were the sole step blocked by gp33, then bypassing it by presenting templates containing a preopened promoter should abolish repression. P23-derived templates preopened at the promoter to varying extent (bp −13/−8; −13/−10; −13/−12; −13; −11/−10, and −10), by changing the sequence of the transcribed (bottom) strand, were used to examine transcription in the presence or absence of gp33 (Figure 4). Each template gave rise to two run-off transcripts: the canonical 37 nt ‘R' transcript, and a 70 nt ‘L' transcript. Primer extension mapped the 5′-end of ‘L' transcripts to bp −19 of the template strand (Figure 4, bottom). None of these templates was transcribed by RNAP core in the absence of gp55 (data not shown).
Figure 4.
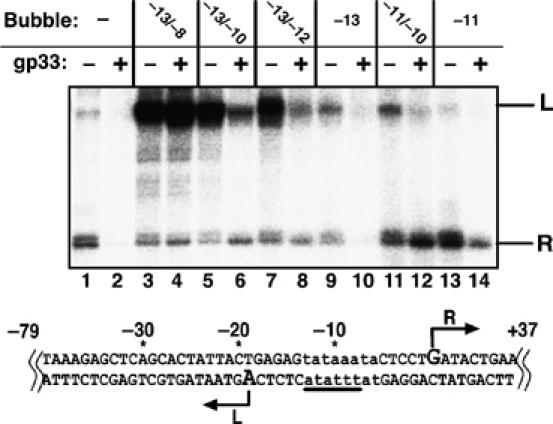
Repression of transcription of heteroduplex DNA by gp33. Top: Single rounds of transcription of 117 bp P23-based heteroduplex templates, with the extent of unpairing indicated above the gel. Gp33 was added to free RNAP for reactions shown in the even-numbered lanes. Leftward (L) and rightward (R) transcripts are identified at the right. Bottom: sequence of the P23 promoter template; nucleotide numbering is relative to the start of the R transcript; the T4 late promoter is written in lower case; the region changed to create mismatches is underlined; initiating nucleotides of the L and R transcripts are in bold and are marked with arrows.
Preopening the transcription bubble strongly affected the ability of gp33 to repress transcription. Even two mismatched bp sufficed to abolish repression of T4 late transcription (R transcript, Figure 4, compare lanes 7 and 8 and lanes 1 and 2). We noted that the production of the L transcript, unlike the R transcript, did not become resistant to gp33 until 6 bp were premelted (Figure 4, lanes 3 and 4). Since the formation of the transcript was not specific to gp55 and also occurred in the presence of σ70 (data not shown), the basis of this difference was not investigated further. We conclude that preopening the promoter abolishes repression by gp33, indicating that the upstream DNA interaction that is abrogated by gp33 is only required at an early step of transcriptional initiation, either before the promoter is melted or at the nucleation of promoter melting.
Gp55- and σ70 holoenzymes differ with respect to non-promoter DNA binding
Although gp33 represses transcription of double-stranded DNA by σ70- and gp55 holoenzymes, the nonspecific DNA-binding properties of σ70- and gp55 holoenzymes are different. This was shown by a DNase I footprinting experiment that examined binding to ∼260 bp DNA probes that were identical to the T4 late and extended −10 (−10 Econ) promoter derivatives used above, except that they contained an extra 136 bp at the upstream end. Footprinting was carried out in the absence of heparin, that is, under conditions allowing detection of unstable nonspecific interaction, except as specified (Figure 5A). Under these conditions, RNAP core (lanes 2 and 7) and gp55 holoenzyme (lanes 3 and 8) protected the entire length of both probes, and heparin exposed the footprint of the gp55-specific T4 late promoter open complex (lane 9), as expected (see also Figure 2B). In contrast, σ70 holoenzymes (both the wild-type and the σ70 domain 4 mutant) only protected the conjugate extended −10 promoter (including its upstream region extending from ∼bp −35 to −65, attributable to the α subunit C-terminal domain), but did not protect the rest of the −10E probe (lanes 4 and 5). The σ70 holoenzymes also did not protect DNA containing the nonconjugate P23 T4 promoter (lanes 10 and 11). Thus, both gp55 and σ70 holoenzymes form open complexes on their conjugate promoters, yet are sharply different in binding to non-promoter DNA. Accordingly, we did not expect gp33 to have any effect on nonspecific DNA binding by σ70Δ4.2 holoenzyme, and this is what was observed, with the exception that, unlike RNAP core or gp55 holoenzyme, σ70Δ4.2 holoenzyme did not protect DNA ends even in the presence of gp33 (Supplementary Figure 3).
Gp55 lacks recognizable amino acid similarity to domains 1, 3, and 4 of σ70, so it was anticipated that deleting one of these domains from σ70 would make the corresponding σ70 holoenzymes resemble gp55 holoenzyme in regard to non-promoter DNA binding. Several σ70 derivatives were tested, including deletions of σ70 domain 4.2 (σ1–565), all of σ70 domain 4 (σ1–516), as well as σ70 domains 3 and 4 (σ1–448), and none of the corresponding σ70 holoenzymes protected non-promoter DNA from DNase I (Figure 5B and data not shown).
Deletion of the 100 N-terminal amino acids of σ70 (domain 1.1), while not affecting the protection of internal DNA sites, generated a holoenzyme that fully protected ∼25 bp at the DNA end (Figure 5B, compare lanes 3 and 5); the DNase I footprint of this holoenzyme was similar to that of RNAP core or gp55 holoenzyme in the presence of gp33 (compare Figure 5B, lane 5 with Figure 2B, lanes 3 and 6). Removing all of domain 4 effected no further change (compare lanes 5 and 6). In accordance with these findings, σ70Δ1.1 holoenzyme was seen to have a much greater propensity for end-initiated transcription (Figure 5C), indicating that region 1.1 of σ70 competes for binding of double-stranded DNA ends. A comparison of lanes 2 and 3 also suggests that DNA end-binding by the σ70Δ1.1 holoenzyme effectively competed for extended −10 promoter utilization.
Discussion
The role of the upstream DNA interaction in initiation of transcription
We have demonstrated that gp33, the small bacteriophage T4 late transcription coactivator that binds to RNAP β flap, represses transcription by preventing RNAP binding to internal sites on DNA. Gp33 also binds to RNAP in the open promoter complex but, in contrast to its action on free RNAP, without diminishing initiation of transcription. The results indicate that the upstream DNA interaction that is blocked by gp33 is essential during early steps of promoter opening (possibly including promoter search) but is not essential for maintaining the open promoter complex. This is consistent with recent studies (Young et al, 2004; Niedziela-Majka and Heyduk, 2005), which show that of the two interactions of RNAP holoenzyme with the promoter, only the downstream interaction (with the −10 motif) is absolutely required for promoter opening.
The fact that gp33 blocks RNAP binding to double-stranded DNA but not to DNA ends argues that RNAP makes at least two separate contacts with DNA, and that only one of these, the upstream contact, is blocked by gp33 (Figure 6A–C). RNAP core binds DNA nonspecifically and with relatively high affinity; binding of σ70 to core reduces the nonspecific general affinity of the resulting holoenzyme for DNA, while dramatically increasing its affinity for promoters (Hinkle and Chamberlin, 1972). σ70 plays a dual role in this process: it blocks nonspecific interactions of RNAP core with DNA, and introduces two new DNA sequence-specific interactions with precisely defined sites on the RNAP holoenzyme surface (Figure 6D). Only a σ domain 2-equivalent domain is present in gp55 holoenzyme; we suggest that the nonspecific, upstream interaction in this holoenzyme is not blocked and, moreover, that it remains essential for promoter binding. The sensitivity of the σ70Δ4.2 holoenzyme to gp33-mediated repression suggests that it also possesses an unblocked upstream DNA-binding determinant. Gp33 binds to the β flap, just as σ domain 4 does, and thus can be viewed as a T4 late analog of σ domain 4 that fills only one of the two domain 4 functions, blocking a nonspecific DNA interaction site in RNAP core but without contributing a new DNA specificity or affinity (Figure 6E and H).
Figure 6.
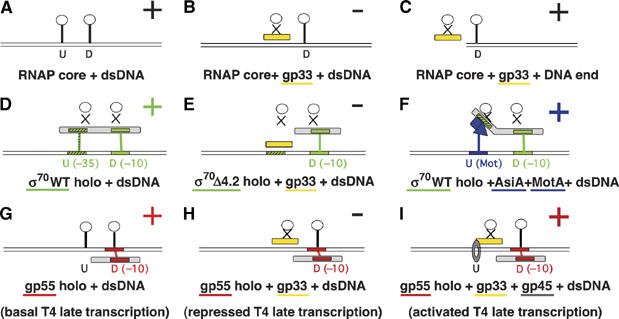
The role of the upstream interaction in initiation of transcription. Binding of RNAP core and various holoenzymes to double-stranded DNA and DNA ends (panel C) is schematized. (+) marks complexes that can bind DNA, (−) marks complexes unable to bind DNA. The horizontal double lines show DNA; white circles symbolize nonspecific DNA-binding sites on RNAP core (RNAP core itself is not shown). U and D designate, respectively, upstream and downstream interactions with DNA that are available to the specified RNAP. Grey rectangles represent promoter-specificity (σ) subunits, with their promoter motifs and the cognate promoter motifs on DNA shown in the same color: red for gp55, green for σ70, blue for σ70 modified by AsiA and MotA, and black for sequence-nonspecific binding. Solid vertical lines denote RNAP–DNA interactions; blocked interactions are shown by X. Gp33 is shown in yellow; AsiA, MotA, and their cognate MotA box in DNA in blue; the grey circle in panel I symbolized the gp45 sliding clamp. (A) RNAP core binds DNA sequence nonspecifically. (B) Gp33 prevents RNAP core binding to DNA by blocking the upstream interaction. (C) The upstream interaction is dispensable for DNA end binding. (D) Wild-type σ70 holoenzyme interacts with its cognate promoter through −35 and −10 promoter motifs. (E) Gp33 prevents σ70Δ4.2 holoenzyme binding to DNA. (F) Changing promoter specificity by modulating the upstream interaction by the accessory proteins AsiA and MotA. Kinking of the sigma symbol represents the remodelling of σ70 domain 4 that is induced by AsiA and MotA. (G) T4 late basal transcription: gp55 holoenzyme binds to DNA through two interactions, one specific and the other nonspecific; unlike σ70, gp55 does not block the downstream DNA-binding site on RNAP core (symbolized by placing the gp55 symbol below the DNA). (H) Repressed T4 late transcription: gp33 blocks the nonspecific upstream interaction. (I) Activated T4 late transcription: DNA-loaded gp45 restores the upstream interaction; tethering of the sliding clamp in the promoter is secured by interactions with the C-termini of gp55 and gp33 that are not indicated.
These observations refine our view of the minimal DNA requirements of the bacterial promoter. We suggest that the sequence-specific interaction at the −10 site, and even the extended −10 site, is not sufficient for establishing the promoter complex, and that an upstream interaction with DNA, perhaps near bp −30 to −40, is also required. The interaction can be sequence-nonspecific in the context of the 9 bp σ70 extended −10 motif (TGxTATAAT) or the 8 bp T4 late motif (TATAAATA), which evidently contain enough sequence information to define a promoter (Figure 6G). σ70 domain 4 and its paralogues impose sequence specificity at this locus, but this is merely a ubiquitous adaptation (which makes it possible to generate promoter diversity through multiple σ paralogues) rather than a necessity imposed by the structure of the multisubunit RNAP and the mechanism of transcriptional initiation. The origin of the upstream interaction in RNAP core remains to be identified, although we note that the β flap tip helix itself is unlikely to contribute to this interaction, since ΔFTH RNAP core binds non-promoter DNA similarly to the wild-type RNAP (data not shown).
Gene regulation in the T4 multiplication cycle hinges on modifications of this upstream interaction. AsiA binds to σ70 domain 4 (Adelman et al, 1997; Colland et al, 1998; Severinova et al, 1998; Urbauer et al, 2001) and interferes with sequence-specific binding of σ70 holoenzyme to its conjugate DNA −35 site; AsiA also serves as the adaptor for attachment of MotA, which recognizes a T4 middle promoter-specific 9 bp site, the Mot box, centered on bp −30 (panel F). AsiA and MotA together determine a switch in promoter recognition by adapting σ70 domain 4 to a new use (Ouhammouch et al, 1995; Hinton et al, 1996, 2005; Lambert et al, 2004). In contrast, gp33 binds to the β flap and represses transcription by occluding a required site for nonspecific interaction with DNA (panel H), and can also interact with the topologically DNA-bound gp45 sliding clamp to restore DNA interaction (Figure 6I). We argue elsewhere that this restoration of upstream DNA confinement by gp45 makes a large contribution to activation of T4 late transcription (NS and EPG, manuscript in preparation).
Promoter finding by different forms of RNAP
σ70-and gp55 holoenzymes differ in their interactions with non-promoter DNA (Figures 5A and 6). In this regard, gp55 holoenzyme is more similar to RNAP core than to σ70 holoenzyme and can be viewed essentially as a core enzyme that is endowed with an extra ability to form open complexes at T4 late promoters. Deletion of σ70 domains 1, 3, or 4 failed to produce a σ70 holoenzyme that binds DNA as promiscuously as does the core enzyme (Figure 5B). We suggest that domain 2 of σ70 is responsible for this difference. It appears possible for RNAP core to bind double-stranded DNA through its main channel (Vassylyev et al, 2002), which is likely to be responsible, at least in part, for its nonspecific downstream DNA interaction (Figure 6A–C). When σ70 domain 2 binds to the β′ coiled coil, it partially blocks the main channel (Vassylyev et al, 2002) and replaces a nonspecific DNA interaction within the channel with a sequence-specific interaction outside the channel (Murakami and Darst, 2003). We speculate that gp55 binds the β′ coiled coil (Wong and Geiduschek, 1998) in such a way that gp55 holoenzyme does not restrict access of DNA to the main channel.
This difference between nonspecific DNA binding by σ70- and gp55 holoenzymes points to two disparate modes of DNA scanning by RNAP. We speculate that while σ70 holoenzyme may scan DNA through interactions with σ domains 2 and 4 outside the channel, gp55 holoenzyme may do so through interactions of its domain 2 with DNA in the channel. In this regard, σ70 holoenzyme has been shown to track along DNA (see also Guthold et al, 1999; Sakata-Sogawa and Shimamoto, 2004). It may be of interest to compare DNA tracking of σ70 holoenzyme and other holoenzymes, especially gp55- and σ54 enzymes.
A role for σ70 domain 1.1 in initiation of transcription
The highly acidic domain 1.1 of σ70 blocks direct σ–DNA interaction (Dombroski et al, 1992, 1993) and has been proposed to act as a DNA mimic that is located in the downstream DNA channel in free σ70 holoenzyme (Mekler et al, 2002). Finding that removing domain 1.1 enhances binding to DNA ends and end-originating transcription is fully consistent with that earlier work. Domain 1.1 is unique to the primary sigma factors such as σ70; it would be of interest to know whether alternative sigma factors contain a functional substitute for σ70 domain 1.1 and/or whether their conjugate holoenzymes interact with DNA ends. One role of domain 1.1 might be to prevent potentially deleterious binding of the primary and most abundant holoenzyme to partially single-stranded DNA, replication forks, and DNA lesions.
A post-initiation role of gp33
Transcription of linear DNA yields RNA molecules whose lengths exceed the span of their templates. This common observation is commonly ignored, but Nudler et al (1996) showed that RNA polymerase can continue to elongate RNA by engaging a new DNA template upon reaching the end of the first one. The efficiency of this process is remarkably high in our experiments, and analysis indicates that intramolecular template recycling contributes to it. Gp33 reduces template switching and recycling by wild-type RNAP but not by ΔFTH RNAP and affects the addition of the last nucleotide at run-off transcription. We propose a single explanation for both effects: gp33 facilitates the relatively slow process of disassembly of the transcription complex at template ends. Our results indicate that gp33 is capable of associating with RNAP that is elongating RNA or transiently stalled during template switching. This result is not at odds with the proposed obligatory release of σ70 domain 4 from the β flap by emerging nascent RNA upon promoter escape (Murakami and Darst, 2003; Nickels et al, 2005) because gp33 and σ70 domain 4 might not occupy the same space on RNAP core. It may also be relevant that gp33 binds to the β flap more strongly than does the isolated σ70 domain 4 (e.g., gp33-core complexes are readily isolated by native gel electrophoresis and σ70 domain 4-core complexes dissociate (Nechaev et al, 2004; unpublished observations)). It remains to be seen whether gp33 binding to elongating RNAP contributes to the shift from σ70-dependent T4 middle, to gp55-dependent late transcription.
Materials and methods
Plasmids
Plasmids for overproduction of untagged E. coli σ70 derivatives Δ1.1 (amino acids 100–613), Δ1.1,4 (amino acids 100–516), and the wild-type protein (amino acids 1–613) were constructed by cloning the corresponding σ70 ORF segments in pET21b using primers providing NdeI and XhoI restriction sites, as described or referenced (Kolesky et al, 1999). An expression plasmid for N-terminally His6-tagged σ70 R541C/L607P (two mutations that impair attachment of σ70 domain 4 to the RNAP core) (Gregory et al, 2004) and the corresponding wild-type σ70 was kindly provided by A Hochschild, and a coexpression plasmid for RNAP core subunits α, β′ (with a C-terminal chitin-binding domain) and β (Δ900–909) (with an N-terminal His6 tag) (Toulokhonov and Landick, 2003) was kindly provided by I Toulokhonov and R Landick.
Protein and DNA
The preparation of gp55, gp33, and E. coli RNAP C-terminally His6-tagged in the β′ subunit has been described or referenced (Kolesky et al, 1999). Untagged σ70 and derivatives were overproduced in E. coli BL21(DE3) and purified from inclusion bodies, dissolved in 7 M urea-containing denaturing buffer and dialyzed against storage buffer (50%(v/v) glycerol/40 mM Tris–HCl, pH 8.0/200 mM NaCl/1 mM DTT/1 mM EDTA) to final protein concentrations of ∼50 μM, as described by Kolesky et al (1999) for preparation of untagged gp55. σ70Δ4.2 (amino acids 1–565) was a generous gift from L Minakhin and K Severinov. Duplex DNA templates for transcription were prepared by PCR amplification using Vent DNA polymerase, with plasmids containing the T4 gene 23 late promoter (P23) or a consensus extended −10 promoter. The 125 bp T4 late promoter fragment (bp −77 to +48, relative to the transcriptional start site as +1) was PCR-amplified out of the previously described plasmid pSK110-rrnB(T1+T2) (Kolesky et al, 2002). A 126 bp extended −10 promoter fragment (bp −79 to +47) was amplified out of a plasmid (pE-10/SK110) constructed by replacing bp −14 to +1 of pSK110-rrnB(T1+T2) as shown in Figure 3A. For the footprinting experiment shown in Figure 5A, longer DNA (bp −213 to +78) was prepared in the same way (with the upstream, nontranscribed strand primer 32P-labeled at its 5′ end).
Partially heteroduplex P23 promoter DNA for transcription was prepared by annealing synthetic oligonucleotides corresponding to bp −79 to +7 of the nontranscribed (nontemplate; top) strand and +37 to −26 on the transcribed (template; bottom) strand of plasmid pSK110-rrnB(T1+T2), and filling in with Klenow fragment DNA polymerase. Mismatches were created by changing the bottom (transcribed) strand to create AA or TT noncomplementarity. DNA was gel-purified as described (Kassavetis et al, 2001), and duplex DNA serving as the control for transcription experiments with these unpaired templates was prepared in the same way.
Transcription
Single-round transcription essentially following Kolesky et al (2002) was carried out in Standard Reaction Buffer (200 mM K acetate/33 mM Tris acetate, pH 7.8/10 mM Mg acetate/1 mM DTT/0.12% (w/v) Tween 20) at 25°C, augmented with 5% (w/v) polyethylene glycol (PEG)3350 for transcription with σ70-RNAP holoenzymes. DNA (100 fmol), RNAP core (1 pmol), and σ70, gp55 and/or gp33, as appropriate (at 12-fold molar excess over RNAP), were incubated in 20 μl volume for 20 min (unless specified otherwise), and single rounds of transcription were initiated by adding nucleotides (to 1 mM ATP and GTP, 100 μM CTP and 100 μM [α-32P]UTP) and heparin (to 100 μg/ml) in 5 μl volume of the same reaction buffer for 5 min. Sample preparation, resolution of transcripts by denaturing gel electrophoresis, phosphor image visualization, and quantification followed standard procedures (e.g. Kolesky et al, 1999).
Footprinting
DNA for footprinting was prepared by PCR as above, except that one primer was 32P-end-labelled. Promoter complexes of RNAP core or holoenzyme (1 pmol) and DNA (50 fmol) in 20 μl Standard Reaction Buffer were formed as above and treated with DNase I (titrated to achieve the desired extent of single-hit DNA cleavage) for 30 s. Standard Reaction Buffer was supplemented with 5% PEG 3350 for experiments with σ70Δ4.2 holoenzyme (Figure 3B) and for the experiment shown in Figure 5A. Where indicated, heparin was added for 30 s, and gp33 for 5 min before DNase I. Digestion was terminated, samples were processed, resolved by denaturing gel electrophoresis and analyzed by phosphorimaging. For probing of promoter opening with KMnO4, specific promoter complexes were formed as above, except that DTT was omitted from the Standard Reaction Buffer. Treatment with 1 mM KMnO4 was for 20 s. Samples were processed following standard procedures.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Data
Acknowledgments
We thank A Hochschild, I Toulokhonov, and K Severinov for generously providing materials, GA Kassavetis for helpful discussions, and M Ouhammouch, GA Kassavetis, and K Severinov for critical and helpful comments on the manuscript. M Kamali-Moghaddam conducted initial experiments on transcription of synthetic homopolymer templates. Support of this research by the NIGMS is gratefully acknowledged.
References
- Adelman K, Orsini G, Kolb A, Graziani L, Brody EN (1997) The interaction between the AsiA protein of bacteriophage T4 and the sigma70 subunit of Escherichia coli RNA polymerase. J Biol Chem 272: 27435–27443 [DOI] [PubMed] [Google Scholar]
- Baumann CG, Smith SB, Bloomfield VA, Bustamante C (1997) Ionic effects on the elasticity of single DNA molecules. Proc Natl Acad Sci USA 94: 6185–6190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchiat C, Wang MD, Allemand J, Strick T, Block SM, Croquette V (1999) Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys J 76: 409–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody EN, Kassavetis GA, Ouhammouch M, Sanders GM, Tinker RL, Geiduschek EP (1995) Old phage, new insights: two recently recognized mechanisms of transcriptional regulation in bacteriophage T4 development. FEMS Microbiol Lett 128: 1–8 [DOI] [PubMed] [Google Scholar]
- Burns HD, Ishihama A, Minchin SD (1999) Open complex formation during transcription initiation at the Escherichia coli galP1 promoter: the role of the RNA polymerase alpha subunit at promoters lacking an UP-element. Nucleic Acids Res 27: 2051–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colland F, Orsini G, Brody EN, Buc H, Kolb A (1998) The bacteriophage T4 AsiA protein: a molecular switch for sigma 70-dependent promoters. Mol Microbiol 27: 819–829 [DOI] [PubMed] [Google Scholar]
- Crothers DM, Drak J, Kahn JD, Levene SD (1992) DNA bending, flexibility, and helical repeat by cyclization kinetics. Methods Enzymol 212: 3–29 [DOI] [PubMed] [Google Scholar]
- Dombroski AJ, Walter WA, Gross CA (1993) Amino-terminal amino acids modulate sigma-factor DNA-binding activity. Genes Dev 7: 2446–2455 [DOI] [PubMed] [Google Scholar]
- Dombroski AJ, Walter WA, Record MT Jr, Siegele DA, Gross CA (1992) Polypeptides containing highly conserved regions of transcription initiation factor sigma 70 exhibit specificity of binding to promoter DNA. Cell 70: 501–512 [DOI] [PubMed] [Google Scholar]
- Elliott T, Geiduschek EP (1984) Defining a bacteriophage T4 late promoter: absence of a ‘−35' region. Cell 36: 211–219 [DOI] [PubMed] [Google Scholar]
- Gregory BD, Nickels BE, Garrity SJ, Severinova E, Minakhin L, Urbauer RJ, Urbauer JL, Heyduk T, Severinov K, Hochschild A (2004) A regulator that inhibits transcription by targeting an intersubunit interaction of the RNA polymerase holoenzyme. Proc Natl Acad Sci USA 101: 4554–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M, Burgess RR (1986) Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res 14: 6745–6763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthold M, Zhu X, Rivetti C, Yang G, Thomson NH, Kasas S, Hansma HG, Smith B, Hansma PK, Bustamante C (1999) Direct observation of one-dimensional diffusion and transcription by Escherichia coli RNA polymerase. Biophys J 77: 2284–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman PJ (1988) Flexibility of DNA. Annu Rev Biophys Biophys Chem 17: 265–286 [DOI] [PubMed] [Google Scholar]
- Harley CB, Reynolds RP (1987) Analysis of E. coli promoter sequences. Nucleic Acids Res 15: 2343–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD, Chamberlin MJ (1988) Structure and function of bacterial sigma factors. Annu Rev Biochem 57: 839–872 [DOI] [PubMed] [Google Scholar]
- Herendeen DR, Kassavetis GA, Geiduschek EP (1992) A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science 256: 1298–1303 [DOI] [PubMed] [Google Scholar]
- Herendeen DR, Williams KP, Kassavetis GA, Geiduschek EP (1990) An RNA polymerase-binding protein that is required for communication between an enhancer and a promoter. Science 248: 573–578 [DOI] [PubMed] [Google Scholar]
- Hinkle DC, Chamberlin MJ (1972) Studies of the binding of Escherichia coli RNA polymerase to DNA. I. The role of sigma subunit in site selection. J Mol Biol 70: 157–185 [DOI] [PubMed] [Google Scholar]
- Hinton DM, March-Amegadzie R, Gerber JS, Sharma M (1996) Characterization of pre-transcription complexes made at a bacteriophage T4 middle promoter: involvement of the T4 MotA activator and the T4 AsiA protein, a sigma 70 binding protein, in the formation of the open complex. J Mol Biol 256: 235–248 [DOI] [PubMed] [Google Scholar]
- Hinton DM, Pande S, Wais N, Johnson XB, Vuthoori M, Makela A, Hook-Barnard I (2005) Transcriptional takeover by sigma appropriation: remodelling of the sigma70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and co-activator AsiA. Microbiology 151: 1729–1740 [DOI] [PubMed] [Google Scholar]
- Kahn JD, Crothers DM (1993) DNA bending in transcription initiation. Cold Spring Harb Symp Quant Biol 58: 115–122 [DOI] [PubMed] [Google Scholar]
- Kahn JD, Yun E, Crothers DM (1994) Detection of localized DNA flexibility. Nature 368: 163–166 [DOI] [PubMed] [Google Scholar]
- Kapanidis AN, Margeat E, Laurence TA, Doose S, Ho SO, Mukhopadhyay J, Kortkhonjia E, Mekler V, Ebright RH, Weiss S (2005) Retention of transcription initiation factor sigma70 in transcription elongation: single-molecule analysis. Mol Cell 20: 347–356 [DOI] [PubMed] [Google Scholar]
- Kassavetis GA, Letts GA, Geiduschek EP (2001) The RNA polymerase III transcription initiation factor TFIIIB participates in two steps of promoter opening. EMBO J 20: 2823–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Zentner PG, Geiduschek EP (1986) Transcription at bacteriophage T4 variant late promoters. An application of a newly devised promoter-mapping method involving RNA chain retraction. J Biol Chem 261: 14256–14265 [PubMed] [Google Scholar]
- Kolesky S, Ouhammouch M, Brody EN, Geiduschek EP (1999) Sigma competition: the contest between bacteriophage T4 middle and late transcription. J Mol Biol 291: 267–281 [DOI] [PubMed] [Google Scholar]
- Kolesky SE, Ouhammouch M, Geiduschek EP (2002) The mechanism of transcriptional activation by the topologically DNA-linked sliding clamp of bacteriophage T4. J Mol Biol 321: 767–784 [DOI] [PubMed] [Google Scholar]
- Kumar A, Malloch RA, Fujita N, Smillie DA, Ishihama A, Hayward RS (1993) The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an ‘extended minus 10' promoter. J Mol Biol 232: 406–418 [DOI] [PubMed] [Google Scholar]
- Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochschild A, Heyduk T, Severinov K (2002) A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science 295: 855–857 [DOI] [PubMed] [Google Scholar]
- Lambert LJ, Wei Y, Schirf V, Demeler B, Werner MH (2004) T4 AsiA blocks DNA recognition by remodeling sigma70 region 4. EMBO J 23: 2952–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd G, Landini P, Busby S (2001) Activation and repression of transcription initiation in bacteria. Essays Biochem 37: 17–31 [DOI] [PubMed] [Google Scholar]
- Lonetto M, Gribskov M, Gross CA (1992) The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol 174: 3843–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure WR (1985) Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem 54: 171–204 [DOI] [PubMed] [Google Scholar]
- Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis AN, Niu W, Ebright YW, Levy R, Ebright RH (2002) Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell 108: 599–614 [DOI] [PubMed] [Google Scholar]
- Mooney RA, Darst SA, Landick R (2005) Sigma and RNA polymerase: an on-again, off-again relationship? Mol Cell 20: 335–345 [DOI] [PubMed] [Google Scholar]
- Murakami KS, Darst SA (2003) Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol 13: 31–39 [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA (2002) Structural basis of transcription initiation: an RNA polymerase holoenzyme–DNA complex. Science 296: 1285–1290 [DOI] [PubMed] [Google Scholar]
- Nechaev S, Kamali-Moghaddam M, Andre E, Leonetti JP, Geiduschek EP (2004) The bacteriophage T4 late-transcription coactivator gp33 binds the flap domain of Escherichia coli RNA polymerase. Proc Natl Acad Sci USA 101: 17365–17370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels BE, Garrity SJ, Mekler V, Minakhin L, Severinov K, Ebright RH, Hochschild A (2005) The interaction between sigma70 and the beta-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc Natl Acad Sci USA 102: 4488–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedziela-Majka A, Heyduk T (2005) Escherichia coli RNA polymerase contacts outside the −10 promoter element are not essential for promoter melting. J Biol Chem 280: 38219–38227 [DOI] [PubMed] [Google Scholar]
- Nudler E, Avetissova E, Markovtsov V, Goldfarb A (1996) Transcription processivity: protein–DNA interactions holding together the elongation complex. Science 273: 211–217 [DOI] [PubMed] [Google Scholar]
- Ouhammouch M, Adelman K, Harvey SR, Orsini G, Brody EN (1995) Bacteriophage T4 MotA and AsiA proteins suffice to direct Escherichia coli RNA polymerase to initiate transcription at T4 middle promoters. Proc Natl Acad Sci USA 92: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaelle M, Kanin EI, Vogt J, Burgess RR, Ansari AZ (2005) Holoenzyme switching and stochastic release of sigma factors from RNA polymerase in vivo. Mol Cell 20: 357–366 [DOI] [PubMed] [Google Scholar]
- Record MT Jr, Reznikoff WS, Craig ML, McQuade KL, Schlax PJ (1996) Escherichia coli RNA polymerase (Es70), promoters, and the kinetics of the steps of transcription initiation. In Escherichia coli and Salmonella, Neidhardt FC (ed) 2nd edn, pp 792–821. Washington, DC: ASM Press [Google Scholar]
- Rees WA, Keller RW, Vesenka JP, Yang G, Bustamante C (1993) Evidence of DNA bending in transcription complexes imaged by scanning force microscopy. Science 260: 1646–1649 [DOI] [PubMed] [Google Scholar]
- Sakata-Sogawa K, Shimamoto N (2004) RNA polymerase can track a DNA groove during promoter search. Proc Natl Acad Sci USA 101: 14731–14735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders GM, Kassavetis GA, Geiduschek EP (1997) Dual targets of a transcriptional activator that tracks on DNA. EMBO J 16: 3124–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severinova E, Severinov K, Darst SA (1998) Inhibition of Escherichia coli RNA polymerase by bacteriophage T4 AsiA. J Mol Biol 279: 9–18 [DOI] [PubMed] [Google Scholar]
- Shore D, Langowski J, Baldwin RL (1981) DNA flexibility studied by covalent closure of short fragments into circles. Proc Natl Acad Sci USA 78: 4833–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt B, Hinton DM (1994) Regulation of middle-mode transcription. In Molecular Biology of Bacteriophage T4, Karam JD (ed) pp 142–160. Washington, DC: American Society for Microbiology [Google Scholar]
- Toulokhonov I, Landick R (2003) The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell 12: 1125–1136 [DOI] [PubMed] [Google Scholar]
- Urbauer JL, Adelman K, Urbauer RJ, Simeonov MF, Gilmore JM, Zolkiewski M, Brody EN (2001) Conserved regions 4.1 and 4.2 of sigma (70) constitute the recognition sites for the anti-sigma factor AsiA, and AsiA is a dimer free in solution. J Biol Chem 276: 41128–41132 [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S (2002) Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 417: 712–719 [DOI] [PubMed] [Google Scholar]
- Williams KP, Kassavetis GA, Herendeen DR, Geiduschek EP (1994) Regulation of late-gene expression. In Molecular Biology of Bacteriophage T4, Karam JD (ed) pp 161–175. Washington, DC: American Society for Microbiology [Google Scholar]
- Williams KP, Muller R, Ruger W, Geiduschek EP (1989) Overproduced bacteriophage T4 gene 33 protein binds RNA polymerase. J Bacteriol 171: 3579–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, Geiduschek EP (1998) Activator–sigma interaction: a hydrophobic segment mediates the interaction of a sigma family promoter recognition protein with a sliding clamp transcription activator. J Mol Biol 284: 195–203 [DOI] [PubMed] [Google Scholar]
- Young BA, Gruber TM, Gross CA (2004) Minimal machinery of RNA polymerase holoenzyme sufficient for promoter melting. Science 303: 1382–1384 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Data


