Abstract
Many metalloproteins have the capacity to bind diverse metals, but in living cells connect only with their cognate metal cofactor. In eukaryotes, this metal specificity can be achieved through metal-specific metallochaperone proteins. Herein, we describe a mechanism whereby Saccharomyces cerevisiae manganese superoxide dismutase (SOD2) preferentially binds manganese over iron based on the differential bioavailability of these ions within mitochondria. The bulk of mitochondrial iron is normally unavailable to SOD2, but when mitochondrial iron homeostasis is disrupted, for example, by mutations in S. cerevisiae mtm1, ssq1 and grx5, iron accumulates in a reactive form that potently competes with manganese for binding to SOD2, inactivating the enzyme. Studies in mtm1 mutants indicate that iron inactivation of SOD2 involves the Mrs3p/Mrs4p mitochondrial carriers and iron-binding frataxin (Yfh1p). A small pool of SOD2-reactive iron also exists under normal iron homeostasis conditions and binds SOD2 when mitochondrial manganese is low. The ability to control this reactive pool of iron is critical to maintaining SOD2 activity and has important potential implications for oxidative stress in disorders of iron overload.
Keywords: Aft1p, iron, manganese, mitochondria, SOD2
Introduction
Heavy metals such as zinc, copper, iron and manganese are widely used in biology as enzymatic cofactors and structural determinants in proteins. These ions all accumulate in cells at concentrations well above the extracellular mileu (Outten and O'Halloran, 2001; Finney and O'Halloran, 2003), and all bind polypeptides via coordination to thiol, imidazole and carboxylic side chains. In spite of the seemingly diverse choice of candidate cofactors, metalloproteins generally bind only their cognate metal in vivo. In some cases, metal specificity is inherent to the polypeptide sequence, where the protein only accommodates a metal with a particular binding geometry (Finney and O'Halloran, 2003). However, most metalloproteins seem more flexible, for example, a copper-requiring enzyme may also bind zinc or cobalt in vitro, but is only active when copper-bound. Metal ion selectivity in this case may be achieved in vivo through the action of accessory proteins known as metallochaperones (Pufahl et al, 1997). Metallochaperones capture a specific metal ion in spite of the absence of free intracellular metals (Rae et al, 1999; Outten and O'Halloran, 2001; Finney and O'Halloran, 2003), and directly transfer their cargo to metalloprotein targets. Metallochaperones have been well characterized for copper (O'Halloran and Culotta, 2000; Bartnikas and Gitlin, 2001; Finney and O'Halloran, 2003) and while the metallochaperone concept has been expanded to include other metals (Colpas and Hausinger, 2000; O'Halloran and Culotta, 2000; Anderson et al, 2005), information on chaperones for metals other than copper is quite sparse.
In bacteria, there are rare instances of metal ion mis-incorporation. The manganese-containing superoxide dismutase (MnSOD) of Escherichia coli can also bind iron, and iron binds to MnSOD with similar metal binding geometries and affinities as manganese (Beyer and Fridovich, 1991; Privalle and Fridovich, 1992; Whittaker, 2003; Mizuno et al, 2004). Isolates of MnSOD from E. coli contain a mixture of iron- and manganese-bound molecules, and under anaerobic conditions, the enzyme becomes virtually all iron-bound (Beyer and Fridovich, 1991; Privalle and Fridovich, 1992). Iron binding inactivates the SOD enzyme, due to an aberrant redox potential at the active site (Vance and Miller, 1998) and a possible block in substrate access (Whittaker, 2003).
Eukaryotes express a highly homologous MnSOD (SOD2) in the mitochondrial matrix that is predicted to exhibit similar inactivation by iron based on comparative structural analyses (Borgstahl et al, 1992; Wintjens et al, 2004). However, there has been no documentation of iron misincorporation in SOD2, other than a trace amount of iron-association with the yeast enzyme (Ravindranath and Fridovich, 1975). Eukaryotes have therefore evolved with means for ensuring cofactor specificity in MnSOD.
We have been employing bakers yeast as a model system to explore the mechanism by which Sod2p specifically acquires manganese. (In this article, Sod2p refers to the Saccharomyces cerevisiae polypeptide; all other eukaryotic versions of the enzyme are denoted SOD2). Manganese activation requires a mitochondrial localization for Sod2p, and the protein unfolding step associated with mitochondrial import is thought to drive metal insertion (Luk et al, 2005). In our search for proteins that facilitate manganese insertion into Sod2p, we identified S. cerevisiae Mtm1p, a member of the mitochondrial carrier family of transporters (Luk et al, 2003). Yeast mtm1 mutants accumulate inactive Sod2p in the mitochondrial matrix and enzyme activity can be restored by growing cells in the presence of very high concentrations of manganese (Luk et al, 2003). However, Mtm1p is not a manganese transporter, as mtm1Δ mitochondria are not deprived of manganese (Luk et al, 2003). Interestingly, our analysis of mitochondrial metals revealed elevated iron levels in mtm1Δ mitochondria (Luk et al, 2003). This uncovering of high iron in mtm1 mutants prompted our recent investigations into the manganese versus iron selectivity of Sod2p in vivo.
Herein, we demonstrate that loss of Sod2p activity in mtm1 mutants is due to misincorporation of iron into Sod2p rather than manganese. Such iron inactivation of Sod2p is not unique to mtm1 mutants, and was observed in other yeast mutants affected in mitochondrial iron metabolism. Our studies provide evidence for the existence of at least two pools of mitochondrial iron: an ‘SOD2-inert' pool that predominates with normal iron homeostasis, and an ‘SOD2-reactive' iron form that effectively competes with mitochondrial manganese for binding to SOD2. A small pool of SOD2-reactive iron also exists in the absence of iron-related defects, and this iron pool readily associates with Sod2p when mitochondrial manganese is low. Overall, these studies demonstrate that metallation of mitochondrial SOD2 is not as manganese-specific as originally thought. The differential bioavailability of manganese versus iron in the mitochondria plays an important role in determining the metal ion specificity of this enzyme.
Results
Sod2p associates with iron in mtm1 mutants
We explored the relationship between the high mitochondrial iron and low Sod2p activity in S. cerevisiae mtm1 mutants. Loss of Sod2p activity per se does not lead to high mitochondrial iron since sod2Δ mutants do not hyperaccumulate mitochondrial iron (Yang and Culotta, unpublished). We therefore tested whether the reverse was true and whether mitochondrial iron was interacting with, and inhibiting Sod2p.
The metal that associates with Sod2p in wild-type (WT) versus mtm1 mutants was analyzed in a mitochondrial fractionation study. Soluble components of gradient-purified mitochondria were subjected to Mono Q anion exchange and size exclusion chromatography. Sod2p fractions were identified by immunoblot, and manganese and iron profiles analyzed by ICP-OES (Inductively Coupled Plasma Optical Emission Spectroscopy). Sod2p elutes from Mono Q in a single peak over two fractions with both WT cells (immunoblot of Figure 1A) and mtm1 mutants (immunoblot of Figure 1B). Metal analysis of WT mitochondria revealed a single manganese-containing peak that coeluted with Sod2p during anionic exchange (Figure 1A top). Manganese and Sod2p continued to co-elute during subsequent chromatography by size exclusion (not shown). With WT cells, the bulk of soluble manganese in the mitochondria is bound to Sod2p.
Figure 1.

Sod2p associates with iron in mtm1 mutants. The soluble fraction from gradient-purified mitochondria (see Materials and methods) was loaded onto a Mono Q anion exchange column. Fractions 1–4 represent flow-through fractions containing positive and uncharged molecules that do not bind to the anion exchange resin under these conditions. Fractions 5–20 represent bound molecules eluted with a 0–100% gradient of 1 M NaCl. Fractions were analyzed by ICP-OES for metals. Concentrations of manganese (A, top) and iron (A, bottom; B) are shown. Fractions were also analyzed for Sod2p by immunoblot. (A) Immunoblots represent fractions 1–20 of the WT strain (middle strip) and fractions 11–13 of the mtm1 mutant (inset). With the same column injection volume, Sod2p from WT and mtm1 cells elute at the identical fraction number. (B) Immunoblot represents fractions 1–20 of the mtm1 mutant. The injection volume was smaller (12 versus 15 ml) for the column in (B) compared to (A) explaining the earlier fractionation of Sod2p in case. Strains utilized: WT, BY4741; mtm1Δ, MY019; mtm1Δ sod2Δ, MY020.
Unlike WT cells, there was no anionic peak of soluble manganese with mtm1 mutants (Figure 1A, top). Instead mtm1Δ mitochondria have two major iron-containing components that were absent in WT cells (Figure 1A bottom and 1B top). The first represents soluble material that did not bind Mono Q (fractions ≈1–4) and the second represents an anionic bound fraction. In all cases, this second iron-peak of mtm1 mutants co-eluted with Sod2p as monitored by immunoblot (Figure 1A, bottom and Figure 1B top). Iron and Sod2p continued to co-elute during subsequent size exclusion chromatography (not shown). Among the soluble iron-containing proteins of yeast mitochondria, only the iron–Sod2p peak was visible. Other mitochondrial iron proteins possess high pI values or are of lower abundance (e.g., Yhb1p, Cyc1p, Bio2p and Lys4p) and could easily escape detection by anionic exchange. Furthermore, the labile 4Fe–4S cluster of aconitase may be disrupted during the aerobic processing of mitochondria. In any case, the anionic iron peak of mtm1 mutants clearly represents Sod2p, as this peak is absent in mtm1Δ sod2Δ double mutants lacking Sod2p (Figure 1B).
Iron inactivation of Sod2p
To test whether iron inactivates Sod2p in mtm1Δ mutants, we sought to reduce mitochondrial iron levels through mutations in AFT1, encoding an iron-sensing transcription factor (Yamaguchi-Iwai et al, 1995). A number of iron transporters rely on Aft1p for expression and aft1Δ mutants of yeast accumulate low iron (Yamaguchi-Iwai et al, 1996; Yun et al, 2000; Rutherford et al, 2003). As seen in Figure 2A top, the high mitochondrial iron of mtm1Δ cells was not observed in an mtm1Δ aft1Δ double mutant. Effects on Sod2p activity were monitored by the native gel assay. In these assays, active Sod2p often migrates as two distinct bands (for description, see legend to Figure 2). Mutants of mtm1 exhibit low Sod2p activity (Figure 2A, lane 3) and this was reversed in the double mtm1Δ aft1Δ mutant (lane 4). These results indicate that mitochondrial iron of mtm1 mutants inactivates Sod2p.
Figure 2.

Low mitochondrial iron rescues the Sod2p defect in mtm1 mutants. The designated strains were grown in YPD media containing where indicated (B–E), the specified concentrations of the iron chelator BPS (bathophenanthrolinedisulfonate). (A–C): Bar graphs represent mitochondrial levels of iron and manganese as determined by atomic absorption spectrometry. Data represent averages of four readings obtained from two independent mitochondrial preps; error bars indicate standard deviation. (A–B): Whole-cell lysates were analyzed for SOD activity by native gel electrophoresis and nitroblue tetrazolium staining (top gel), and for Sod2p polypeptide levels by immunoblot (bottom gel). ‘SOD2' and ‘SOD1' indicate activity of Mn Sod2p and Cu/Zn Sod1p. On native gels, Sod2p activity typically resolves into 2–3 distinct bands, especially with BPS treatment. These bands all represent Sod2p, as determined by native gel immunoblot (not shown) and may correspond to different metal occupation states of the tetrameric enzyme. Combined analysis of Sod2p activity and protein levels for the various experimental trials of part (A) can be found in Supplementary Figure S1. (D, E) Anion exchange chromatography analysis of mitochondrial fractions was conducted as in Figure 1 for mtm1Δ(D) or WT (E) cells grown in the absence (solid lines) or presence (dashed lines) of 100 μM BPS. Insets are the immunoblots of Sod2p from fraction 10 of these experiments. (E) The Sod2p of BPS-treated cells exhibits a ≈30–50% increase in manganese association that was reproducible over three experimental trials (see Supplementary Figure S2). Strains utilized: WT, BY4741; mtm1Δ, MY019; mtm1Δ sod2Δ, MY020; aft1Δ, 4438; mtm1Δ aft1Δ, MY030.
To further monitor the effects of mitochondrial iron, we used the impermeable iron chelator, bathophenanthrolinedisulfonate (BPS). As seen in Figure 2B top, the level of mitochondrial iron in mtm1 mutants was reduced in accordance with increasing concentrations of BPS in the growth medium. There was no toxicity to the cell up to 100 μM BPS (not shown) and BPS did not greatly alter mitochondrial levels of manganese (Figure 2C). In the native gel assay, Sod2p activity in mtm1 mutants increased as mitochondrial iron levels dropped (Figure 2B, lanes 3–6). By analysis of mitochondrial anionic fractions, manganese, but not iron, was seen to associate with Sod2p when mtm1 mutants were treated with BPS (Figure 2D). It is important to note that this manganese binding is not due to an exchange of metals in pre-existing Fe–Sod2p, but rather reflects manganese insertion into newly synthesized Sod2p molecules. Metal insertion only occurs with newly synthesized Sod2p molecules that are freshly imported into mitochondria (Luk et al, 2005).
Lowering of mitochondrial iron by BPS not only rescued Sod2p in mtm1 mutants but also slightly improved Sod2p activity in WT cells. WT cells consistently exhibited an ≈30–50% increase in manganese association with Sod2p during BPS treatment (Figure 2E, also see Supplementary Figure S2). These results indicate that a small fraction of Sod2p associates with iron even in WT cells.
The mitochondrial carriers Mrs3/Mrs4p contribute to iron inactivation of Sod2p
We explored the mitochondrial factors that may contribute to iron inactivation of Sod2p in mtm1 mutants. Mrs3p and Mrs4p are transporters of the mitochondrial carrier family that localize to the mitochondrial inner membrane (Wiesenberger et al, 1991). Although the precise substrate of transport by Mrs3p/Mrs4p has not been clarified, these transporters contribute to mitochondrial iron levels (Foury and Roganti, 2002; Muhlenhoff et al, 2003b; Li and Kaplan, 2004) and facilitate synthesis of iron sulfur clusters and heme (Muhlenhoff et al, 2003b; Zhang et al, 2005). As seen in Figure 3A, a double mrs3 mrs4 deletion resulted in a partial lowering of mitochondrial iron in mtm1 mutants. In spite of this seemingly small effect on iron, Sod2p activity increased (Figure 3A, lane 3; also see Supplementary, Figure S1). Mitochondrial iron is also controlled by yeast MMT1 and MMT2, encoding members of the cation diffusion facilitator family that localize to the mitochondria (Li and Kaplan, 1997; Jensen et al, 2004). Compared to the effects of mrs3Δ mrs4Δ mutations, there was no restoration of Sod2p activity with mmt1Δ mmt2Δ mutations (Figure 3B, lane 3; also see Supplementary, Figure S1). The iron relevant to Mrs3/4p seems more bioavailable to inactivate Sod2p.
Figure 3.

Mitochondrial proteins that contribute to iron inactivation of Sod2p. The indicated yeast strains were tested for mitochondrial iron and for Sod2p activity and polypeptide levels as described in Figure 2A. Analysis of the various experimental trials can be found in Supplementary Figure S1. Where indicated in (D), cells were cultured in the presence of 80 μM BPS. Yeast strains utilized—(A, C, D): WT, BY4741; mtm1Δ, MY019; mtm1Δ mrs3-4Δ, the triple mtm1Δ mrs3Δ mrs4Δ strain VC111; ssq1Δ, 5278; grx5Δ, 2769; (B): WT, 1783; mtm1Δmmt1-2Δ, the triple mtm1Δ mmt1Δ mmt2Δ strain VC112.
Effects of mitochondrial iron overload on Sod2p activity
Do elevations in mitochondrial iron always inactivate Sod2p or is this a special case of mtm1 mutants? A number of yeast mutants have been identified that accumulate high mitochondrial iron and many of these are defective in Fe/S cluster biogenesis. SSQ1 and GRX5 encode a molecular chaperone and glutaredoxin, respectively, that are required to assemble Fe/S clusters on target proteins (Knight et al, 1998; Rodriguez-Manzaneque et al, 2002; Muhlenhoff et al, 2003a). In the experiment of Figure 3C, ssq1 and grx5 mutants accumulated elevated levels of mitochondrial iron and also exhibited an impairment in Sod2p activity (Figure 3C). Sod2p activity was restored by BPS treatment (Figure 3D, also see Supplementary Figure S1), demonstrating a role for iron in Sod2p inactivation. Therefore, iron inactivation of Sod2p is not unique to mtm1 mutants and can be observed with other defects in mitochondrial iron homeostasis.
However, we observed two instances where elevated mitochondrial iron did not correlate with Sod2p inactivation. First, culturing WT yeast cells in the presence of high extracellular iron effectively increased mitochondrial iron, yet Sod2p activity was normal (Figure 4A, lane 2). The enzyme only associated with manganese in iron-treated WT cells (Figure 4C). We also tested the effects of mitochondrial iron overload in yfh1 mutants. YFH1 encodes the iron-binding frataxin homologue defective in Friedreich's ataxia (Babcock et al, 1997; Foury and Cazzalini, 1997). In S. cerevisiae, loss of yfh1 has been associated with high mitochondrial iron and defects in Fe/S cluster biogenesis (Babcock et al, 1997; Foury and Cazzalini, 1997; Chen et al, 2002; Muhlenhoff et al, 2002, 2003a). We observed that in spite of high mitochondrial iron, yfh1Δ mutants exhibit normal Sod2p activity (Figure 4B, lane 3). The differential Sod2p activity results obtained with yfh1Δ versus ssq1Δ and grx5Δ mutants seemed surprising since all three have been implicated in Fe/S cluster biogenesis. As one possibility, Yfh1p itself may participate in iron inactivation of Sod2p. To test this, we created an mtm1Δ yfh1Δ double mutant. As seen in Figure 4C lane 3, Sod2p activity was normal in this mutant in spite of the very high levels of mitochondrial iron (also see Supplementary Figure S1). Yfh1p contributes to iron inactivation of Sod2p in mtm1 mutants.
Figure 4.

Sod2p activity in WT and yfh1Δ strains is not inhibited by mitochondrial iron. (A, B) The indicated yeast strains were analyzed for Sod2p activity and protein levels and for mitochondrial iron as in Figure 2. Where indicated (Fe: +), cells were cultured in the presence of 1.0 mM ammonium iron (III) citrate. Analysis of the various experimental trials can be found in Supplementary Figure S1. (C) Mitochondrial anion exchange fractions from WT cells cultured in 100 μM iron (II) chloride (which effects an ∼5-fold increase in total mitochondrial iron; not shown) were analyzed for iron (solid lines) and manganese (dashed lines) as in Figure 1. Inset is immunoblot analysis of fractions 8–10 showing elution of Sod2p in fractions 8–9 of this experiment. Strains used: WT, BY4741; yfh1Δ, MY036; mtm1Δ, MY019; mtm1Δ yfh1Δ, MY038.
Effects of mtm1 mutations on iron homeostasis
Yeast mutants defective in Fe/S cluster biogenesis (e.g., yfh1, ssq1, grx5) are known to activate Aft1p, the iron regulatory transcription factor (Foury and Talibi, 2001; Belli et al, 2004; Chen et al, 2004; Rutherford et al, 2005). Since mtm1 mutations phenocopy grx5 and ssq1 mutants with regard to mitochondrial iron and Sod2p, we addressed whether mtm1 mutations likewise induce Aft1p and block Fe/S cluster synthesis.
To test for Aft1p activation, an FET3-lacZ reporter was used (Jensen and Culotta, 2002). In WT cells, FET3-lacZ was induced by Aft1p during iron starvation with BPS, and repressed somewhat by iron supplements (Figure 5A). Yet in mtm1 mutants, FET3-lacZ was expressed at high levels even with iron supplements, and BPS treatment only moderately increased expression (Figure 5A). Aft1p appears constitutively active in mtm1Δ cells.
Figure 5.
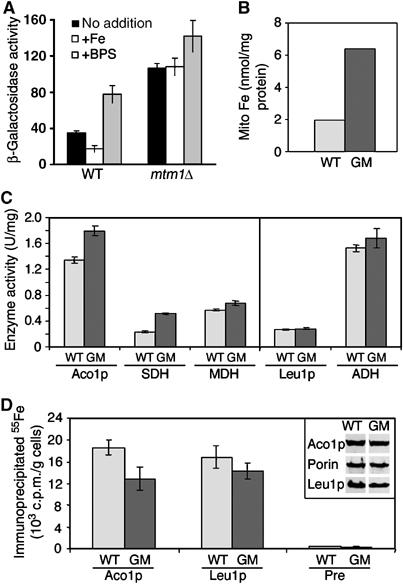
Aft1p activity and Fe/S cluster assembly in mtm1 mutants. (A) The WT BY4741 or mtm1Δ mutant MY019 were transformed with the FET3-lacZ reporter plasmid pAR1 and were cultured in LF-SD medium that was supplemented where indicated with 1.6 mM ammonium iron (III) citrate (‘+Fe') or 100 μM of the iron chelator BPS (‘+BPS'). β-Galactosidase activity was measured as described in Materials and methods. Results represent the averages of four readings obtained from cultures of two independent single colonies, and error bars represent standard deviation. (B–D) The WT (strain W303) and Gal-MTM1 (GM) cells were cultured for 4 days in glucose-containing medium to deplete Mtm1p. (B) The non-heme, non-Fe/S iron content of isolated mitochondria was measured by the bathophenantroline method (Li et al, 1999). (C) Mitochondria were isolated and analyzed for the enzyme activities of the Fe/S proteins aconitase (Aco1p) and succinate dehydrogenase (SDH) as well as the non-Fe/S protein malate dehydrogenase (MDH) as a control. The activity of the cytosolic Fe/S protein isopropylmalate isomerase (Leu1p) and the control enzyme alcohol dehydrogenase (ADH) was measured in total cell lysates. (D) Cells were cultured in LF-SD medium supplemented with glucose for 16 h prior to radiolabeling with 55Fe for 2 h. Whole-cell extracts were used for immunoprecipitation with specific antibodies against Aco1p and Leu1p as well as with a preimmune serum (Pre). The amount of 55Fe that was co-precipitated was quantified by liquid scintillation counting. The inset shows the amounts of Aco1p and Leu1p and as a control, porin detected in WT and Gal-MTM1 cells by immunostaining of the corresponding cell extracts.
We next addressed whether Mtm1p is needed for Fe/S cluster synthesis. As with mutants known to affect Fe/S biogenesis (e.g., yfh1, ssq1 and grx5 (Lill and Kispal, 2000)), mtm1Δ cells accumulate mitochondrial DNA mutations (Luk et al, 2003) which can potentially interfere with Fe/S enzyme activities (Kaut et al, 2000). As such, an Mtm1p depletion study was conducted in which MTM1 under the GAL1-10 promoter was repressed by glucose, and immediate effects on mitochondrial iron and Fe/S proteins was monitored. This identical GAL1 depletion approach has been used to study the roles of various mitochondrial ISC (iron sulfur cluster) components in Fe/S cluster biogenesis (Muhlenhoff et al, 2003a). In the case of Mtm1p, GAL-MTM1 cells exhibited a substantial rise in mitochondrial iron following 4 days of Mtm1p depletion by growth in glucose (Figure 5B). This effect on mitochondrial iron is similar to what is seen with null mutations in mtm1 (Figure 2A and B). However, unlike mtm1Δ cells, the Mtm1p-depleted cells did not loose mitochondrial DNA over this time frame (not shown). In spite of the rise in mitochondrial iron, Mtm1p-depeleted cells exhibited no major defect in Fe/S proteins. Aco1p and Leu1p are mitochondrial and cytosolic 4Fe–4S proteins and the activity of these enzymes remained normal in Mtm1p-depleted cells (Figure 5C). The Fe/S succinate dehydrogenase (SDH) also remained enzymatically active with Mtm1p depletion (Figure 5C). We additionally monitored the de novo assembly of Aco1p and Leu1p by an 55Fe radiolabeling assay (Kispal et al, 1999). The rate of 55Fe incorporation into the apoproteins was only slightly affected by Mtm1p depletion (Figure 5D). By comparison, the analogous depletions of Yfh1p, Ssq1p, Grx5p (and other members of the Fe/S cluster machinery) are known to dramatically inhibit (⩾4-fold) 55Fe labeling of Fe/S proteins as well as activities of Fe/S enzymes (Kaut et al, 2000; Lange et al, 2000; Muhlenhoff et al, 2003a). If Mtm1p has any role in Fe/S biogenesis, this is minor in comparison to known factors for Fe/S biogenesis.
Mitochondrial manganese and iron inactivation of Sod2p
Our studies clearly show that Sod2p activity can be influenced by mitochondrial iron homeostasis. Can mitochondrial manganese counteract the effects of iron? We have previously shown that Sod2p activity of mtm1 mutants can be recovered by supplementing the growth medium with manganese (Luk et al, 2003). However, the levels of manganese required were quite high (>100 μM) and approached toxic quantities (Luk et al, 2003). We now show that this translates to a ∼200–400-fold increase in mitochondrial manganese with no change in mitochondrial iron (Figure 6A, lanes 4–5). In mtm1 cells treated with just 10 μM manganese, mitochondrial manganese levels rise by nearly 10-fold; however, this is not sufficient to restore Sod2p activity (Figure 6A, lane 3 and Luk et al, 2003). The iron of mtm1 mutants is remarkably bioavailable and competes effectively with mitochondrial manganese for binding to Sod2p.
Figure 6.
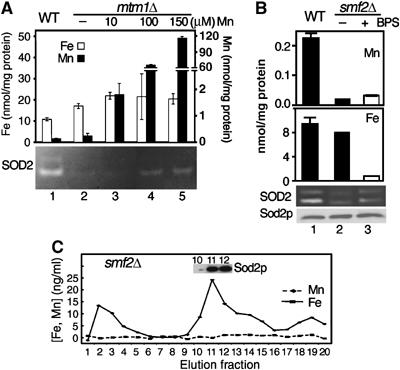
Competition between iron and manganese for binding to Sod2p in vivo. (A, B) The indicated yeast strains were grown in YPD medium supplemented where indicated with 10, 100 or 150 μM MnCl2 (A) or 80 μM BPS (B). Mitochondrial iron and manganese contents and Sod2p activity (A, B) and Sod2 protein (B) were assayed as in Figure 2. Note the different scales for iron and manganese, including the broken scale for manganese in (A). Analysis of the various experimental trials can be found in Supplementary Figure S1. (C) Soluble mitochondrial components from smf2Δ mutant yeast were fractionated by anion exchange, and metal profiles determined (iron (solid) and manganese (dashed)) as in Figure 1. Inset is immunoblot of fractions 10–12 for Sod2p. Strains utilized: WT, BY4741; mtm1Δ, MY019; smf2Δ, 1878.
We also explored the effects of manganese starvation on iron–Sod2p interactions. Manganese starvation can be achieved in yeast cells through a deletion in SMF2, encoding an Nramp transporter for manganese (Luk and Culotta, 2001). There is a cell wide manganese deficiency in smf2 mutants, and mitochondrial manganese is very low (Figure 6B top) (Luk and Culotta, 2001). By comparison, smf2Δ cells exhibit no change in mitochondrial iron (Figure 6B). Sod2p has low activity in smf2 mutants (Figure 6B, lane 2 and Luk and Culotta, 2001) and as expected, there is an absence of detectable manganese association with Sod2p (Figure 6C). However, rather than being apo (no metal bound to Sod2p), the Sod2p of these manganese-deficient cells was found to be associated with iron (Figure 6C). Iron binding to Sod2p inhibits the enzyme, because activity was increased upon lowering mitochondrial iron with BPS (Figure 6B, lane 3; also see Supplementary Figure S1). Hence, when manganese is low, Sod2p becomes vulnerable to iron inactivation. Even in the absence of an iron homeostasis defect, there exists a pool of mitochondrial iron that is capable of inactivating Sod2p.
Discussion
Using bakers yeast as a model system, we provide evidence that eukaryotic manganese SOD2 can associate with iron in vivo. Iron binding precludes insertion of the correct manganese ion cofactor and destroys SOD2 enzymatic activity. Iron inactivation of yeast Sod2p can be promoted when mitochondrial iron homeostasis is disrupted or when cells are starved for manganese.
This is the first report of eukaryotic SOD2 inactivation by mitochondrial iron. While iron inactivation of SOD2 may be rare, iron commonly inhibits bacterial MnSOD in vivo (Beyer and Fridovich, 1991; Privalle and Fridovich, 1992; Mizuno et al, 2004). Bacteria generally express a second, non-manganese SOD (i.e., FeSOD) in the same compartment, but eukaryotic SOD2 is the only SOD of the mitochondrial matrix. SOD2 is an extremely important antioxidant enzyme and its loss can lead to neonatal lethality (Li et al, 1995; Lebovitz et al, 1996) and reduced lifespan (Duttaroy et al, 2003), making it critical to prevent iron interactions with the enzyme.
Some, but not all, cases of mitochondrial iron overload lead to Sod2p inactivation in yeast. Our studies have differentiated mitochondrial iron into two classes in terms of bioavailability to Sod2p. A portion of the mitochondrial iron in mtm1, grx5 and ssq1 yeast mutants is bioavailable to yeast Sod2p (‘SOD2-reactive'), but mitochondrial iron in WT cells and in yfh1 frataxin mutants is largely ‘SOD2-inert'. Grx5p, Ssq1p and Yfh1p all participate in a sequential pathway of Fe/S cluster assembly (Lill and Muhlenhoff, 2005; Rouault and Tong, 2005). Yfh1p acts upstream to donate iron to Fe/S scaffolds, while Ssq1p and Grx5p act downstream to transfer the clusters from these scaffolds to Fe/S proteins (Muhlenhoff et al, 2003a; Lill and Muhlenhoff, 2005). The frataxin-derived iron that is normally employed for Fe/S proteins may be shunted towards Sod2p when downstream components of Fe/S cluster assembly are blocked. In fact, our studies implicate a role for frataxin in iron inactivation of Sod2p, since the mtm1 defect in Sod2p is not seen in a double mtm1 yfh1 mutant (Figure 4B). Frataxin may help to increase the bioavailability of iron not only for Fe/S clusters but inadvertently for Sod2p as well.
Frataxin works in an iron pathway also involving the mitochondrial carriers Mrs3p and Mrs4p (Muhlenhoff et al, 2003b; Lill and Muhlenhoff, 2005; Rouault and Tong, 2005; Zhang et al, 2005). We show here that these same two carriers contribute to iron inactivation of Sod2p in mtm1 mutants (Figure 3A). By comparison, the Mmt1p/Mmt2p diffusion cation transporters of the mitochondria do not affect Sod2p activity (Figure 3B). This differential effect of Mmt1p/Mmt2p versus Mrs3p/Mrs4p indicates that SOD2-inert and SOD2-reactive iron pools may utilize distinct mitochondrial transport systems. Alternatively, distinct mitochondrial localizations of Mmt1p/Mmt2p and Mrs3p/Mrs4p could account for the differential effects of these transporters. Currently, the precise nature of SOD2-reactive versus SOD2-inert iron is not clear, but may represent different oxidation states or ligand-binding properties of the metal.
SOD2-reactive iron is not unique to disruptions in mitochondrial iron homeostasis. A small fraction of Sod2p from S. cerevisiae is normally found iron-bound (Ravindranath and Fridovich, 1975), and treating WT cells with iron chelators increases manganese binding to Sod2p (Figure 2E). Normally, this small pool of SOD-reactive iron cannot compete well with mitochondrial manganese, but when mitochondrial manganese levels drop, the pool of reactive iron gains access to Sod2p (Figure 6B and C). These results also indicate that mitochondrial Sod2p cannot readily accumulate as an ‘apo' enzyme totally devoid of metals, but prefers to exist in either the manganese or iron bound state.
Although Mtm1p was originally proposed to act in manganese activation of Sod2p (Luk et al, 2003), our more recent studies suggest either that Mtm1p has a primary function in mitochondrial iron metabolism or alternatively, mtm1 mutations alter levels of a particular metabolite within the mitochondrial matrix that stabilizes iron in a bioavailable form. As a transport carrier for mitochondria, Mtm1p may exchange a solute that directly or indirectly affects iron homeostasis. We show here that cells respond to a loss in mtm1 through activation of the iron regulator Aft1p. However, Aft1p activation by itself is not responsible for the high mitochondrial iron of mtm1 mutants. Expression of a constitutively active AFT1-1up allele in a WT cell does not cause high mitochondrial iron (Babcock et al, 1997). We also find that AFT1-1up expression does not lead to a loss of Sod2p activity in WT cells (Supplementary Figure S3). Activation of Aft1p, as seen in mtm1 mutants, has been a hallmark of mutants defective in Fe/S cluster biogenesis (Chen et al, 2004; Lesuisse et al, 2005; Rutherford et al, 2005). However, mtm1 mutants exhibit no major loss of either mitochondrial or cytosolic Fe/S clusters (Figure 5). Therefore, in mtm1 mutants, Aft1p is sensing a perturbation in mitochondrial iron homeostasis independent of the mitochondrial ISC assembly and export systems (Lill and Muhlenhoff, 2005).
How does SOD2 normally choose manganese over iron? SOD2 generally binds manganese, but with certain defects in mitochondrial iron homeostasis (e.g., mtm1, ssq1 and grx5 mutants), there is an expansion in the pool of SOD2-reactive iron. This iron is so reactive with yeast Sod2p that it takes a rise in mitochondrial manganese levels of >200-fold to restore Sod2p activity (Figure 6A). We therefore propose a model of ‘differential metal bioavailability' for metal ion selectivity in SOD2. SOD2 normally binds manganese simply because available manganese levels out-compete the small amount of SOD2-reactive iron. Under conditions where SOD2-reactive iron increases or when mitochondrial manganese levels drop, SOD2 shifts to the Fe-bound state. If SOD2 does require a metallochaperone, this factor is expected to bind either iron or manganese depending on which ion is more bioavailable. Alternatively, separate chaperone-like molecules may be involved in delivery of iron versus manganese.
In human disorders of iron overload, it is believed that iron can cause oxidative stress to cells mainly through Fenton chemistry, but this study suggests a new aspect of iron toxicity: inactivation of an important mitochondrial antioxidant enzyme. Although iron may not be largely SOD2-reactive in cases of Friedreich's ataxia where frataxin is defective (Anderson et al, 2005; Seznec et al, 2005), other disorders of iron overload should be considered, for example, neuroferritinopathy and sideroblastic anemia (reviewed in Rouault, 2001; Napier et al, 2005). In this regard, it is noteworthy that SOD2 deficiency itself leads to a sideroblastic anemia-like disorder in mice (Friedman et al, 2004).
Materials and methods
Yeast strains, plasmids and culture conditions
Most of the yeast strains used in this study are isogenic to BY4741 (MATa, leu2Δ0, met15Δ0, ura3Δ0, his3Δ1), including the sod2Δ∷kanMX4 (6605), aft1Δ∷kanMX4 (4438), ssq1Δ∷kanMX4 (5278), grx5Δ∷kanMX4 (2769) and smf2Δ∷kanMX4 (1878) mutants purchased from Research Genetics (Huntsville, AL). The double mrs3Δ∷kanMX4 mrs4Δ∷kanMX4 mutant (781-1A) was a gift from Dr Andrew Dancis (University of Pennsylvania). Disruptions of MTM1 in BY4741 and the corresponding sod2Δ, aft1Δ and mrs3Δmrs4Δ strains were generated using the mtm1Δ∷LEU2 plasmid pVC257Δ (Luk et al, 2003), resulting in strains MY019 (mtm1Δ), MY020 (mtm1Δ sod2Δ), MY030 (mtm1Δ aft1Δ) and VC111 (mtm1Δ mrs3Δ mrs4Δ). Disruption of YFH1 in BY4741 and in the mtm1Δ strain MY019, generating MY036 and MY038, was accomplished using the yfh1Δ∷URA3 plasmid pJS406 (Strain et al, 1998). The yfh1Δ mtm1Δ double mutant was cultured on iron-limited plates, prepared by mixing agarose (GIBCO) with iron-limited selecting minimal synthetic medium (Jensen and Culotta, 2002). The mmt1Δ mmt2Δ double mutant LJ224 was derived from parental strain 1783 (MATa, leu2-3, 112, ura3-52, trp1-1, his4 and can1r) (Jensen et al, 2004). Disruptions of MTM1 in 1783 and LJ224 were generated using the mtm1Δ∷TRP1 plasmid pVC257Δ-T, resulting in strains VC110 (from 1783) and VC112 (mtm1Δ mmt1Δ mmt2Δ). The pVC257Δ-T plasmid was created by mobilizing the MTM1 containing disruption insert from pVC257 by BamHI and SalI digestion, and inserting the cassette into the integrating TRP1 vector pRS404 digested with the same enzymes. To test for effects of active Aft1p expression, the AFT1-1up plasmid, AFT1-1up-313 (Jensen and Culotta, 2002), was used. Creation of the Gal-MTM1 strain for depleting Mtm1p involved strain W303-1A (MATa, ura3-1, ade2-1, trp1-1, his3-11,15 and leu2-3,112). The promoter of MTM1 was exchanged for the galactose-inducible GAL1 promoter by PCR-mediated DNA replacement (http://mips.gsf.de/proj/yeast/CYGD/db/index.html). In brief, PCR fragments carrying the HIS3 gene and the GAL1-10 promoter region of vector pTL26 flanked by region −230 to −180 and +1 to +50 of the MTM1 gene were used to transform strain W303-1A. All gene replacements and gene deletions were verified by PCR.
Yeast cells were propagated at 30°C either in enriched yeast-peptone-based medium with 2% (w/v) glucose (YPD) or in an iron-limited selecting minimal synthetic medium (LF-SD) (Jensen and Culotta, 2002). Gal-MTM1 cells were depleted of Mtm1p to physiologically critical levels by preincubating for 4 days on glucose-containing solid-rich medium.
Chromotographic mitochondrial fractionations
Purified mitochondria were prepared by loading crude mitochondria onto a discontinuous Nycodenz gradient (16% on 22%). Intact mitochondria were recovered from the gradient interface after centrifugation at 150 000 g for 1 h. The intact mitochondria were washed by dilution into isotonic buffer followed by centrifugation at 12 000 g. Mitochondria were lysed by three 30 s pulses of sonication at 50% output of a microtip (Ultrasonic, W-380). The soluble fraction was separated from the insoluble fraction by centrifugation at 15 000 g. The soluble fraction was diluted into Buffer A (20 mM Tris–HCl pH 7.2) and filtered with a 0.45 μm syringe filter. The sample was then loaded onto a HR10/10 MonoQ column (Amersham Biosciences) equilibrated in 10 column volumes (CV) of Buffer A. The unbound proteins were washed from the column with the injection volume and an additional 5 CV of Buffer A and collected as fractions 1–4 (5 ml per fraction). A 25 CV gradient (0–100%) of Buffer B (20 mM Tris–HCl pH 7.2, 1 M NaCl) was then initiated. Fractions (1 ml) were collected for the gradient and all fractions were then analyzed by ICP-OES (Perkin Elmer, Optima 3100XL) versus buffer blanks after dilution in 10% Nitric acid. Concentrations were determined from a standard curve constructed with serial dilutions of commercially available mixed metal standards. Two metal mixes were used: one contained Ca, Cd, Co, Cu, Fe, Ni and Zn and a second mix contained Mg and Mn (Optima). Blanks of Nitric acid or buffer samples with and without ‘metal-spikes' were analyzed to ensure reproducibility.
Analysis of Sod2p activity and polypeptide levels
Cultures (50 ml) of S. cerevisiae strains were typically grown without shaking at 30°C for 17–18 h to a final OD600 of 2–4.0 as described (Luk et al, 2003). In experiments using the slow-growing yfh1Δ mtm1Δ mutant, all cultures including the WT and mtm1Δ controls were grown to a final OD600 of 0.5–0.7. Cell lysates were prepared by glass bead homogenization (Luk and Culotta, 2001) and 60.0 μg of lysate protein was analyzed for both SOD activity by native gel electrophoresis and nitroblue tetrazolium staining (Luk et al, 2003) and for SDS–PAGE and immunoblotting as described (Luk and Culotta, 2001; Luk et al, 2003). To prevent SDS-mediated precipitation of Sod2p, samples for electrophoresis were not boiled.
Miscellaneous biochemical analyses
Atomic absorption analysis of mitochondrial metals employed crude mitochondria preparations obtained from 200 ml cultures according to published methods (Luk and Culotta, 2001). To monitor activation of Aft1p, yeast cells transformed with a FET3-lacZ reporter plasmid (Jensen and Culotta, 2002) were grown to an A600 nm of ∼1.0 in LF-SD medium supplemented as needed with 100 μM BPS or 1.6 mM ammonium iron (III) citrate. Cell extracts prepared by glass bead homogenization were then assayed for β-galactosidase as previously described (Jensen and Culotta, 2002). In vivo labeling of yeast cells with radioactive iron (55FeCl3) and measurement of 55Fe incorporation into mitochondrial or cytosolic Fe/S proteins by immunoprecipitation and liquid scintillation counting was carried out as described previously (Kispal et al, 1999). The anti-Aco1p antibody was raised against purified preparations of Aco1 protein. For analysis of mitochondrial Fe/S enzyme activities, mitochondria were isolated as described (Daum et al, 1982) and enzyme activities of malate dehydrogenase, aconitase, alcohol dehydrogenase, isopropylmalate isomerase (Leu1p) (Kispal et al, 1997) and succinate dehydrogenase were carried out according to published procedures (Robinson et al, 1991; Robinson and Lemire, 1995). The standard error of the determination of enzyme activities was between 5 and 15%.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Andrew Dancis for the 781-1A strain, Matt Hall for critical review of this manuscript and Laran Jensen for the LJ224 strain and for assistance with atomic absorption spectrophotometry and lacZ reporter assays. This research was supported by the JHU NIEHS center and by NIH grants ES 08996 (awarded to VCC) and CA61286 (awarded to DRW) and by grants of the Deutsche Forschungsgemeinschaft (SFB 593 and Gottfried-Wilhelm Leibniz program) and Fonds der Chemischen Industrie (awarded to RL). MY is supported by NIH post-doctoral fellowship F32 GM 074402 and by NIEHS training grant ES 07141.
References
- Anderson PR, Kirby K, Hilliker AJ, Phillips JP (2005) RNAi-mediated suppression of the mitochondrial iron chaperone, frataxin, in Drosophila. Hum Mol Genet 14: 3397–3405 [DOI] [PubMed] [Google Scholar]
- Babcock M, Silva Dd, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J (1997) Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276: 1709–1712 [DOI] [PubMed] [Google Scholar]
- Bartnikas TB, Gitlin JD (2001) How to make a metalloprotein. Nat Struct Biol 8: 733–734 [DOI] [PubMed] [Google Scholar]
- Belli G, Molina MM, Garcia-Martinez J, Perez-Ortin JE, Herrero E (2004) Saccharomyces cerevisiae glutaredoxin 5-deficient cells subjected to continuous oxidizing conditions are affected in the expression of specific sets of genes. J Biol Chem 279: 12386–12395 [DOI] [PubMed] [Google Scholar]
- Beyer WF, Fridovich I (1991) In vivo competition between iron and manganese for occupancy of the active site region of the manganese-superoxide dismutase of Escherichia coli. J Biol Chem 266: 303–308 [PubMed] [Google Scholar]
- Borgstahl GE, Parge HE, Hickey MJ, Beyer WF Jr, Hallewell RA, Tainer JA (1992) The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell 71: 107–118 [DOI] [PubMed] [Google Scholar]
- Chen OS, Crisp RJ, Valachovic M, Bard M, Winge DR, Kaplan J (2004) Transcription of the yeast iron regulon does not respond directly to iron but rather to iron–sulfur cluster biosynthesis. J Biol Chem 279: 29513–29518 [DOI] [PubMed] [Google Scholar]
- Chen OS, Hemenway S, Kaplan J (2002) Inhibition of Fe–S cluster biosynthesis decreases mitochondrial iron export: evidence that Yfh1p affects Fe–S cluster synthesis. Proc Natl Acad Sci USA 99: 12321–12326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpas GJ, Hausinger RP (2000) In vivo and in vitro kinetics of metal transfer by the Klebsiella aerogenes urease nickel metallochaperone, UreE. J Biol Chem 275: 10731–10737 [DOI] [PubMed] [Google Scholar]
- Daum G, Bohni PC, Schatz G (1982) Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem 257: 13028–13033 [PubMed] [Google Scholar]
- Duttaroy A, Paul A, Kundu M, Belton A (2003) A Sod2 null mutation confers severely reduced adult life span in Drosophila. Genetics 165: 2295–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney LA, O'Halloran TV (2003) Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300: 931–936 [DOI] [PubMed] [Google Scholar]
- Foury F, Cazzalini O (1997) Deletion of the yeast homologue of the human gene associated with Friedreich's ataxia elicits iron accumulation in mitochondria. FEBS Lett 411: 373–377 [DOI] [PubMed] [Google Scholar]
- Foury F, Roganti T (2002) Deletion of the mitochondrial carrier genes MRS3 and MRS4 suppresses mitochondrial iron accumulation in a yeast frataxin-deficient strain. J Biol Chem 277: 24475–24483 [DOI] [PubMed] [Google Scholar]
- Foury F, Talibi D (2001) Mitochondrial control of iron homeostasis: a genome wide analysis of gene expression in a yeast frataxin deficient strain. J Biol Chem 276: 7762–7768 [DOI] [PubMed] [Google Scholar]
- Friedman JS, Lopez MF, Fleming MD, Rivera A, Martin FM, Welsh ML, Boyd A, Doctrow SR, Burakoff SJ (2004) SOD2-deficiency anemia: protein oxidation and altered protein expression reveal targets of damage, stress response, and antioxidant responsiveness. Blood 104: 2565–2573 [DOI] [PubMed] [Google Scholar]
- Jensen LT, Culotta VC (2002) Regulation of S. cerevisiae FET4 gene expression by iron and oxygen. J Mol Biol 318: 251–260 [DOI] [PubMed] [Google Scholar]
- Jensen LT, Sanchez RJ, Srinivasan C, Valentine JS, Culotta VC (2004) Mutations in Saccharomyces cerevisiae iron–sulfur cluster assembly genes and oxidative stress relevant to Cu, Zn superoxide dismutase. J Biol Chem 279: 29938–29943 [DOI] [PubMed] [Google Scholar]
- Kaut A, Lange H, Diekert K, Kispal G, Lill R (2000) Isa1p is a component of the mitochondrial machinery for maturation of cellular iron–sulfur proteins and requires conserves cysteine residues for function. J Biol Chem 275: 15955–15961 [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Guiard B, Lill R (1997) The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett 418: 346–350 [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R (1999) The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J 18: 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SAB, Sepuri NBV, Pain D, Dancis A (1998) Mt-Hsp70 Homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J Biol Chem 273: 18389–18393 [DOI] [PubMed] [Google Scholar]
- Lange H, Kaut A, Kispal G, Lill R (2000) A mitochondrial ferredoxin is essential for biogenesis of cellular iron–sulfur proteins. Proc Natl Acad Sci USA 97: 1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebovitz RM, Zhang H, Vogel H, Cartwright J Jr, Dionne L, Lu N, Huang S, Matzuk MM (1996) Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci USA 93: 9782–9787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesuisse E, Knight SA, Courel M, Santos R, Camadro JM, Dancis A (2005) Genome-wide screen for genes with effects on distinct iron uptake activities in Saccharomyces cerevisiae. Genetics 169: 107–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kogan M, Knight SA, Pain D, Dancis A (1999) Yeast mitochondrial protein, Nfs1p, coordinately regulates iron-sulfur cluster proteins, cellular iron uptake, and iron distribution. J Biol Chem 274: 33025–33034 [DOI] [PubMed] [Google Scholar]
- Li L, Kaplan J (1997) Characterization of two homologous yeast genes that encode mitochondrial iron transporters. J Biol Chem 272: 28485–28493 [DOI] [PubMed] [Google Scholar]
- Li L, Kaplan J (2004) A mitochondrial–vacuolar signaling pathway in yeast that affects iron and copper metabolism. J Biol Chem 279: 33653–33661 [DOI] [PubMed] [Google Scholar]
- Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, Wallace DC, Epstein CJ (1995) Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismtuase. Nat Genet 11: 376–381 [DOI] [PubMed] [Google Scholar]
- Lill R, Kispal G (2000) Maturation of cellular Fe–S proteins: an essential function of mitochondria. Trends Biochem Sci 25: 352–356 [DOI] [PubMed] [Google Scholar]
- Lill R, Muhlenhoff U (2005) Iron–sulfur–protein biogenesis in eukaryotes. Trends Biochem Sci 30: 133–141 [DOI] [PubMed] [Google Scholar]
- Luk E, Carroll M, Baker M, Culotta VC (2003) Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proc Natl Acad Sci USA 100: 10353–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk E, Culotta VC (2001) Manganese superoxide dismutase in S. cerevisiae acquires its metal co-factor through a pathway involving the Nramp metal transproter, Smf2p. J Biol Chem 276: 47556–47562 [DOI] [PubMed] [Google Scholar]
- Luk E, Yang M, Jensen LT, Bourbonnais Y, Culotta VC (2005) Manganese activation of superoxide dismutase 2 in the mitochondria of Saccharomyces cerevisiae. J Biol Chem 280: 22715–22720 [DOI] [PubMed] [Google Scholar]
- Mizuno K, Whittaker MM, Bachinger HP, Whittaker JW (2004) Calorimetric studies on the tight-binding metal interactions of Escherichia coli manganese superoxide dismutase. J Biol Chem 279: 27339–27344 [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U, Gerber J, Richhardt N, Lill R (2003a) Components involved in assembly and dislocation of iron–sulfur clusters on the scaffold protein Isu1p. EMBO J 22: 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenhoff U, Richhardt N, Ristow M, Kispal G, Lill R (2002) The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum Mol Genet 11: 2025–2036 [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U, Stadler JA, Richhardt N, Seubert A, Eickhorst T, Schweyen RJ, Lill R, Wiesenberger G (2003b) A specific role of the yeast mitochondrial carriers MRS3/4p in mitochondrial iron acquisition under iron-limiting conditions. J Biol Chem 278: 40612–40620 [DOI] [PubMed] [Google Scholar]
- Napier I, Ponka P, Richardson DR (2005) Iron trafficking in the mitochondrion: novel pathways revealed by disease. Blood 105: 1867–1874 [DOI] [PubMed] [Google Scholar]
- O'Halloran TV, Culotta VC (2000) Metallochaperones: an intracellular shuttle service for metal ions. J Biol Chem 275: 25057–25060 [DOI] [PubMed] [Google Scholar]
- Outten CE, O'Halloran TV (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292: 2488–2492 [DOI] [PubMed] [Google Scholar]
- Privalle CT, Fridovich I (1992) Transcriptional and maturation effects of manganese and iron on the biosynthesis of manganese-superoxide dismutase in Escherichia coli. J Biol Chem 267: 9140–9145 [PubMed] [Google Scholar]
- Pufahl R, Singer C, Peariso KL, Lin SJ, Schmidt P, Fahrni C, Culotta VC, Penner-Hahn JE, O'Halloran TV (1997) Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science 278: 853–856 [DOI] [PubMed] [Google Scholar]
- Rae TD, Schmidt PJ, Pufhal RA, Culotta VC, O'Halloran TV (1999) Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 284: 805–808 [DOI] [PubMed] [Google Scholar]
- Ravindranath SD, Fridovich I (1975) Isolation and characterization of a manganese-containing superoxide dismutase from yeast. J Biol Chem 250: 6107–6112 [PubMed] [Google Scholar]
- Robinson KM, Lemire BD (1995) Flavinylation of succinate: ubiquinone oxidoreductase from Saccharomyces cerevisiae. Methods Enzymol 260: 34–51 [DOI] [PubMed] [Google Scholar]
- Robinson KM, von Kieckebusch-Guck A, Lemire BD (1991) Isolation and characterization of a Saccharomyces cerevisiae mutant disrupted for the succinate dehydrogenase flavoprotein subunit. J Biol Chem 266: 21347–21350 [PubMed] [Google Scholar]
- Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, Herrero E (2002) Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell 13: 1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA (2001) Iron on the brain. Nat Genet 28: 299–300 [DOI] [PubMed] [Google Scholar]
- Rouault TA, Tong WH (2005) Iron–sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat Rev Mol Cell Biol 6: 345–351 [DOI] [PubMed] [Google Scholar]
- Rutherford JC, Jaron S, Winge DR (2003) Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J Biol Chem 278: 27636–27643 [DOI] [PubMed] [Google Scholar]
- Rutherford JC, Ojeda L, Balk J, Muhlenhoff U, Lill R, Winge DR (2005) Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron–sulfur protein biogenesis. J Biol Chem 280: 10135–10140 [DOI] [PubMed] [Google Scholar]
- Seznec H, Simon D, Bouton C, Reutenauer L, Hertzog A, Golik P, Procaccio V, Patel M, Drapier JC, Koenig M, Puccio H (2005) Friedreich ataxia: the oxidative stress paradox. Hum Mol Genet 14: 463–474 [DOI] [PubMed] [Google Scholar]
- Strain J, Lorenz CR, Bode J, Smolen GA, Garland SA, Vickery LE, Culotta VC (1998) Suppressors of superoxide dismutase (SOD1) deficiency in Saccharomyces cerevisiae: identification of proteins predicted to mediate iron–sulfur cluster assembly. J Biol Chem 273: 31138–31144 [DOI] [PubMed] [Google Scholar]
- Vance CK, Miller AF (1998) A simple proposal that can explain the inactivity of metal-substituted superoxide dismutases. J Am Chem Soc 120: 461–467 [Google Scholar]
- Whittaker JW (2003) The irony of manganese superoxide dismutase. Biochem Soc Trans 31: 11318–11321 [DOI] [PubMed] [Google Scholar]
- Wiesenberger G, Link TA, Ahsen U, Waldherr M, Schweyen RJ (1991) MRS3 and MRS4, two suppressors of mtRNA splicing defects in yeast, are new members of the mitochondrial carrier family. J Mol Biol 217: 23–37 [DOI] [PubMed] [Google Scholar]
- Wintjens R, Noel C, May AC, Gerbod D, Dufernez F, Capron M, Viscogliosi E, Rooman M (2004) Specificity and phenetic relationships of iron- and manganese-containing superoxide dismutases on the basis of structure and sequence comparisons. J Biol Chem 279: 9248–9254 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Dancis A, Klausner R (1995) AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J 14: 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner RD (1996) Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J 15: 3377–3384 [PMC free article] [PubMed] [Google Scholar]
- Yun CW, Ferea T, Rashford J, Ardon O, Brown PO, Botstein D, Kaplan J, Philpott CC (2000) Desferrioxamine-mediated iron uptake in Saccharomyces cerevisiae. Evidence for two pathways of iron uptake. J Biol Chem 275: 10709–10715 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lyver ER, Knight SA, Lesuisse E, Dancis A (2005) Frataxin and mitochondrial carrier proteins, Mrs3p and Mrs4p, cooperate in providing iron for heme synthesis. J Biol Chem 280: 19794–19807 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


