Abstract
MHC class I molecules display peptides from endogenous and viral proteins for immunosurveillance by cytotoxic T lymphocytes (CTL). The importance of the class I pathway is emphasised by the remarkable strategies employed by different viruses to downregulate surface class I and avoid CTL recognition. The K3 gene product from Kaposi's sarcoma-associated herpesvirus (KSHV) is a viral ubiquitin E3 ligase which ubiquitinates and degrades cell surface MHC class I molecules. We now show that modification of K3-associated class I by lysine-63-linked polyubiquitin chains is necessary for their efficient endocytosis and endolysosomal degradation and present three lines of evidence that monoubiquitination of class I molecules provides an inefficient internalisation signal. This lysine-63-linked polyubiquitination requires both UbcH5b/c and Ubc13-conjugating enzymes for initiating mono- and subsequent polyubiquitination of class I, and the clathrin-dependent internalisation is mediated by the epsin endocytic adaptor. Our results explain how lysine-63-linked polyubiquitination leads to degradation by an endolysosomal pathway and demonstrate a novel mechanism for endocytosis and endolysosomal degradation of class I, which may be applicable to other receptors.
Keywords: antigen presentation, MHC class I molecules, ubiquitin, viral evasion
Introduction
MHC class I molecules display peptides from endogenous and viral proteins for immunosurveillance by cytotoxic T lymphocytes (CTL) (Cresswell, 2000). The importance of the class I pathway is emphasised by the remarkable strategies employed by many different viruses to inhibit cell surface MHC class I expression and thus avoid CTL recognition (Yewdell and Hill, 2002). The K3 family of viral genes, first identified in gammaherpesviruses (Coscoy and Ganem, 2000; Haque et al, 2000; Ishido et al, 2000b; Stevenson et al, 2000) and subsequently found in some poxvirus family members (Guerin et al, 2002; Mansouri et al, 2003) provides an unusual example of viral evasion (Lehner et al, 2005). The K3 and K5 gene products of KSHV and the mK3 gene product from murine γ-herpesvirus-68 (mHV68) were initially shown to target MHC class I molecules for degradation. K5 was subsequently found to downregulate a range of host immunoreceptors, including ICAM-1, CD86 and CD1d (Ishido et al, 2000a; Coscoy and Ganem, 2001; Sanchez et al, 2005), perhaps allowing increased flexibility by evading different effector arms of the immune response. The key to understanding the mechanism of action of these viral proteins was the recognition of a shared N-terminal RING-CH domain (Boname and Stevenson, 2001; Coscoy et al, 2001; Hewitt et al, 2002), a variant of the classical RING domain found in ubiquitin E3 ligases (Dodd et al, 2004). K3, K5 and mK3 act as membrane-associated viral ubiquitin E3 ligases, which associate with, ubiquitinate and degrade their target proteins (Boname and Stevenson, 2001; Coscoy et al, 2001; Hewitt et al, 2002).
The intracellular site of ubiquitination can affect the fate of the targeted substrate. For the murine gammaherpesvirus-68 (MHV-g68) mK3 gene product, association with immature ER-resident class I molecules in the peptide-loading complex leads to 26S proteasome-mediated class I degradation (Boname et al, 2004; Wang et al, 2004). In contrast, in KSHV-encoded K3-expressing cells, class I molecules are loaded with peptide in the ER and acquire normal Endo H resistance. In the late secretory pathway, their association with K3 leads to class I ubiquitination, endocytosis, sorting through the endosomal pathway and lysosomal degradation (Hewitt et al, 2002). By acting as a viral ubiquitin E3 ligase, K3 therefore exploits both the host's ubiquitination machinery as well as the ubiquitin-dependent endosomal sorting pathway to degrade MHC class I molecules (Hewitt et al, 2002).
The three steps of the ubiquitin reaction involve (i) ubiquitin activation via an E1 enzyme, (ii) transfer of the ubiquitin via a cysteine residue to an E2 ubiquitin-conjugating enzyme and (iii) in the case of RING finger proteins, the targeting of the activated ubiquitin from the E2 to the lysine residue of the target protein. This latter reaction is catalysed by the ubiquitin E3 ligase, which associates with the substrate and thereby confers specificity to the ubiquitination reaction. Proteomic approaches to understanding ubiquitination have identified detectable in vivo levels of ubiquitin conjugates linked at Lys-11, -33, -27 and -6 (Peng et al, 2003), and the chain structure of polyubiquitin influences the fate of a ubiquitinated substrate (Pickart and Fushman, 2004). 26S proteasome-mediated degradation requires the recognition of a polyubiquitinated substrate containing a minimum of four ubiquitin molecules linked via the lysine residue at position 48 of ubiquitin, commonly referred to as lysine-48-linked (Lys-48) polyubiquitination (Thrower et al, 2000). Unlike Lys-48 chains, Lys-63 ubiquitin chains (ubiquitin molecules linked via the lysine residue at position 63 of ubiquitin) act as nonproteolytic signals in DNA repair, kinase signalling pathways and receptor endocytosis in yeast (Pickart and Fushman, 2004).
Monoubiquitination is also known to play a critical role in regulating the expression of many cell surface receptors (Hicke and Dunn, 2003). This is best characterised for the downregulation of activated growth factor receptor tyrosine kinases (RTKs), where recruitment of Cbl mediates receptor ubiquitination, and is required for internalisation and endosomal sorting (Hicke and Dunn, 2003). Ubiquitination is also required for the downregulation of surface immunoreceptors, including the T-cell receptor (Naramura et al, 2002), Fc receptors (Booth et al, 2002) as well as cytokine receptors (Gesbert et al, 2005). In the case of the epidermal growth factor receptor (EGFR), monoubiquitination appears sufficient to direct receptor sorting for lysosomal degradation (Haglund et al, 2003b; Mosesson et al, 2003). The conjugation of single ubiquitin molecules to multiple lysine residues in the cytoplasmic tail, ‘multiple monoubiquitination' rather than polyubiquitination, was proposed to be the principal signal responsible for movement of RTKs from the plasma membrane to the lysosome for degradation.
In the present study, we sought to explain how polyubiquitination of class I molecules by K3 leads to endocytosis and sorting of class I for lysosomal degradation. We find that monoubiquitin is an insufficient signal for internalisation of MHC class I molecules. Instead, the recruitment of host E2 ubiquitin-conjugating enzymes, in particular Ubc13, leads to modification by Lys-63- rather than Lys-48-linked polyubiquitin chains. Endocytosis of these Lys-63-polyubiquitinated class I molecules is clathrin-dependent and requires the epsin endocytic adaptor for internalisation and subsequent sorting through the endosomal pathway.
Results
MHC class I molecules are polyubiquitinated and degraded in a lysosomal compartment
The K3-induced downregulation of MHC class I molecules (Figure 1A) is dependent on lysines in the cytoplasmic tail of class I (Hewitt et al, 2002). Following ubiquitination, the class I molecules are internalised and sorted via the endosomal pathway for lysosomal degradation (Hewitt et al, 2002). Classical class I molecules have two to three lysines in their cytoplasmic tails. We previously showed that the conserved lysine at position 340 of HLA-A*0201 is the dominant ubiquitin acceptor for K3-mediated ubiquitination (Hewitt et al, 2002). However, K3-associated class I molecules are tagged with two or more ubiquitin adducts (Figure 1B), suggesting that polyubiquitinated class I may be endocytosed and degraded in a bafilomycin-sensitive, vacuolar compartment (Figure 1B). To examine whether the class I molecules are indeed polyubiquitinated (as opposed to multiubiquitinated with monoubiquitin), we employed the FK1 antibody which recognises poly- but not monoubiquitin chains (Fujimuro et al, 1994; Haglund et al, 2003b). Metabolic [35S]methionine radiolabelling followed by immunoprecipitation of K3-associated class I molecules showed a ladder of ubiquitinated class I species (Figure 1C). The FK1 antibody is unable to recognise the initial monoubiquitinated class I band, but readily visualises the higher molecular weight ubiquitinated class I species (Figure 1C), confirming that the K3-associated class I molecules are indeed polyubiquitinated. A K3(W41A) RING mutant, containing a tryptophan–alanine substitution, is unable to recruit a cellular Ubc E2 enzyme. In vivo K3(W41A) was previously shown to associate with class I molecules, but failure to recruit any Ubc enzyme prevents ubiquitination or downregulation of MHC class I molecules (Hewitt et al, 2002) (Figure 1A).
Figure 1.
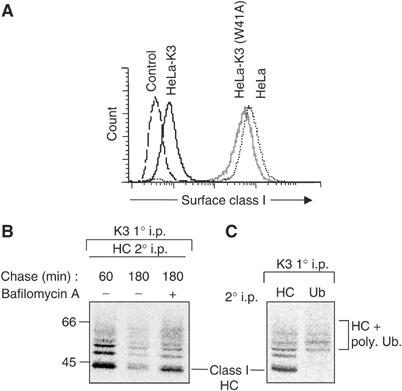
The viral protein K3 promotes the polyubiquitination and endolysosomal degradation of class I molecules. (A) K3 downregulates MHC class I molecules. Cytofluorometric analysis of MHC class I expression in HeLa, HeLa-K3 and HeLa-K3(W41A) cells stained with mAb w6/32 and an FITC-conjugated secondary antibody. (B) K3-associated class I molecules are degraded in an endosomal compartment. HeLa-K3 cells were pulsed for 15 min in [35S]methionine and [35S]cysteine and chased for the times indicated, in the presence or absence of bafilomycin A. Digitonin lysates of K3-transfected HeLas were immunoprecipitated with the anti-FLAG M2 mAb (immunoprecipitates FLAG-tagged K3), dissociated (1% SDS for 30 min) and reprecipitated with the HC10 (class I heavy chain (HC)) mAb and protein A sepharose beads overnight. Washed samples were heated to 70°C in 2 × SDS–PAGE sample buffer and analysed by SDS–PAGE and autoradiography. (C) K3-associated class I molecules are polyubiquitinated. HeLa-K3 cells were labelled and the K3 complex immunoprecipitated as in (B). Following SDS dissociation, lysates were reprecipitated with HC10 (for class I heavy chain) or FK1 (ubiquitin).
Class I molecules are polyubiquitinated through a lysine-63 ubiquitin chain linkage
To determine why a polyubiquitinated receptor is directed to a lysosomal compartment and not recognised by the proteasome, we sought the identity of the ubiquitin E2-conjugating enzyme(s) (Ubc) recruited by K3. RING-containing ubiquitin E3 ligases do not themselves bind ubiqutin, but recruit a ubiquitin-charged Ubc enzyme and promote transfer of ubiquitin from the Ubc to the substrate. There are 40 known Ubcs and in a yeast two-hybrid (Y2H) assay we previously showed that the wild-type K3 RING bound four Ubc proteins, Ubc13 and members of the UbcH5 family (UbcH5A/B/C) (Dodd et al, 2004). No Ubc interactions were seen with the negative control K3(W41A) mutant (Dodd et al, 2004), and in vitro binding assays using recombinantly expressed K3 RING-CH domain protein confirmed the findings of the Y2H analysis (Dodd et al, 2004).
The identification of Ubc13 as a potential K3-binding partner was of particular interest as Ubc13 catalyses the formation of noncanonical Lys-63-linked polyubiquitin chains (Hofmann and Pickart, 1999). To determine whether Ubc13 affects K3 function in vivo, we examined cell surface class I expression in HeLa and HeLa-K3-expressing cells following depletion of Ubc13 by short interfering RNA (siRNA). An effective reduction in Ubc13 protein levels (Figure 2A), but not a control calreticulin protein, was consistently associated with an increase in cell surface class I in HeLa-K3 cells (Figure 2B), with no effect on class I expression in Ubc13-depleted control HeLa cells. Metabolic [35S]Methionine radiolabelling followed by pulse-chase analysis of MHC class I molecules showed that siRNA-mediated depletion of Ubc13 prevented the degradation of class I molecules seen at the 3 h time point in HeLa-K3-expressing cells (Figure 2C). The effect of siRNA-mediated Ubc13 depletion on class I downregulation by K3 was assessed using four separate Ubc13-specific siRNA oligos (Supplementary Table 1). Three out of four oligos increased class I expression in K3-expressing cells, with the degree of class I rescue correlating with the effectiveness of Ubc13 depletion. Furthermore, we did not find evidence of ‘off-target' effects, in terms of an IFN-γ-induced increase in MHC class I expression in control HeLa cells treated with Ubc13-specific siRNAs (see Supplementary Figure 1 online).
Figure 2.
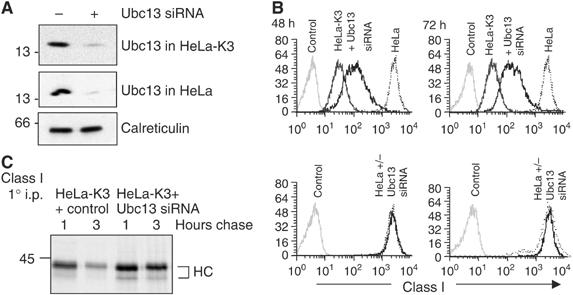
SiRNA-mediated depletion of the Ubc13 ubiquitin E2-conjugating enzyme rescues class I expression in K3-expressing cells. (A) Triton X-100 lysates from HeLa and HeLa-K3 cells targeted with Ubc13 siRNA were probed for Ubc13 (upper 2 panels) or calreticulin control (lower panel). (B) HeLa and HeLa-K3 cells were analysed for MHC class I expression at 48 or 72 h following control or Ubc13 siRNA depletion (as Figure 1A—except that the secondary antibody was Cy5 conjugated). The control HeLa and HeLa-K3 cells were fixed at 48 h and the same control cells were therefore used for the 72 h time point (C) HeLa-K3 cells (+/− Ubc13 siRNA) were pulse-labelled for 10 min, chased for 1 and 3 h and class I molecules immunoprecipitated with the w6/32 mAb.
These results suggest that recruitment of Ubc13 by K3 promotes the assembly of Lys-63-linked ubiquitin chains on class I molecules. To further characterise the class I ubiquitin chain linkage, we expressed either wild-type or mutant GFP-tagged ubiquitin, with lysine to arginine mutations at positions 48 (Lys-48R) or 63 (Lys-63R) (Figure 3A). These mutant ubiquitins will allow formation of a monoubiquitinated substrate but not a polyubiquitinated Lys-48- or Lys-63-linked ubiquitin chain, respectively. Following expression of these ubiquitin genes, the C-terminal GFP tag is proteolytically cleaved and expression of GFP therefore acts as an independent, quantitative marker of wild-type or mutant ubiquitin gene expression (Tsirigotis et al, 2001). Since the introduced ubiquitin must compete with endogenous ubiquitin, the level of GFP expression in the cell provides a useful surrogate marker for the amount of mutant ubiquitin expressed. As shown (Figure 3B), expression of neither the wild-type nor mutant Lys-48R ubiquitin affects the low level of class I expression seen in HeLa-K3 cells. In contrast, expression of high levels of the mutant Lys-63R ubiquitin (Figure 3B, far right panel—x-axis) leads to an increase in cell surface class I (y-axis). These results imply that in those cells expressing high levels of the mutant Lys-63R ubiquitin, a failure to generate Lys-63 polyubiquitin chains promotes the rescue of class I molecules to the cell surface.
Figure 3.
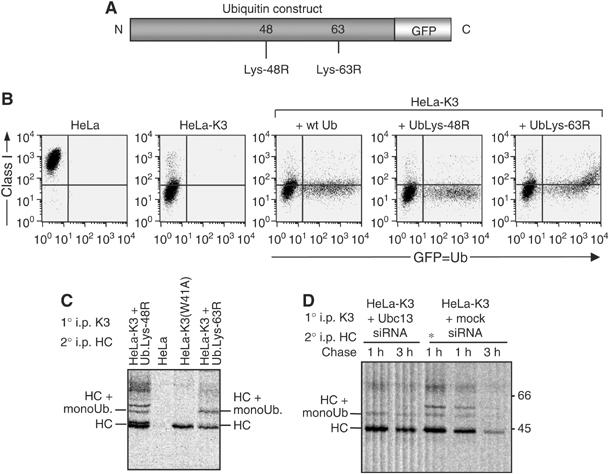
Ubc13 mediates Lys-63 polyubiquitination of class I molecules in K3-expressing cells. (A–D) Conjugation of Lys-63-linked chains is required for class I downregulation. (A) Schematic showing the GFP-tagged ubiquitin constructs with lysine–arginine mutations at either position 48 or 63 of ubiquitin. (B) Cytofluorometric analysis of MHC class I expression in HeLa cells and HeLa-K3 cells following transfection with GFP-tagged wild-type-, Lys-48R- or Lys-63R-ubiquitin. (C) Radiolabelled HeLa-K3 cells (as Figure 1B), transfected with GFP-tagged Lys-48R- or Lys-63R-ubiquitin, HeLa cells expressing mutant K3(W41A) and HeLa cells were chased for 40 min and immunoprecipitated with the FLAG mAb M2, dissociated in 1% SDS and reprecipitated with the heavy chain (HC)-specific mAb. HC10 prior to SDS–PAGE and autoradiography. (D) Ubc13 siRNA depletion of HeLa-K3 cells leaves predominantly monoubiquitinated class I molecules. Pulse-labelled HeLa-K3 cells (+/− Ubc13 siRNA) were labelled for 15 min, chased for the times indicated and immunoprecipitated (as Figure 1B) with the FLAG mAb M2, dissociated in 1% SDS and reprecipitated with the HC-specific mAb HC10 prior to SDS–PAGE and autoradiography. In lane 3, the * represents a 2.5 times increased cell number of mock siRNA-treated control HeLa-K3 cells.
To determine the biochemical consequences of mutant ubiquitin gene expression, we examined the K3-associated class I molecules from radiolabelled cells. The polyubiquitination pattern of K3-associated class I species was not affected by expression of the wild-type (not shown) or mutant Lys-48R ubiquitin (Figure 3C, lane 1). However, following expression of the mutant Lys-63R ubiquitin gene, a change in ubiquitination pattern was observed, with a predominantly monoubiquitinated class I species seen in association with K3 (Figure 3C, lane 4). These results show that formation of Lys-63 polyubiquitin chains on class I molecules are required for efficient downregulation by K3, and that monoubiquitination provides an insufficient signal for efficient class I downregulation.
Two ubiquitin E2-conjugating enzymes are required for formation of Lys-63-linked polyubiquitinated class I molecules
We previously showed that ubiquitination is the signal for dissociation of class I from K3 (Hewitt et al, 2002). We therefore predicted that following siRNA-mediated Ubc13 depletion, we would (i) see an increase in the proportion of K3-associated class I molecules, and (ii) these class I molecules would not be ubiquitinated. Only the first prediction was correct. Analysis of radiolabelled HeLa-K3 cells following Ubc13 depletion did indeed lead to a modest increase in K3-associated, nonubiquitinated class I chains, best seen at the 3 h chase point (Figure 3D, lane 2 versus lane 5). More significantly however, rather than inhibiting class I ubiquitination completely, Ubc13 depletion resulted in a predominantly monoubiquitinated class I species in association with K3 (Figure 3D, lanes 1 and 2). This pattern of class I ubiquitin conjugates is reminiscent of that seen in the presence of the mutant Lys-63R ubiquitin gene (Figure 3C). This unexpected result has two implications. First, the Ubc13 enzyme does not initiate substrate ubiquitination, but requires the prior generation of a monoubiquitin substrate to catalyse formation of Lys-63 polyubiquitin chains and second, that a separate Ubc is required to monoubiquitinate class I molecules. In addition to Ubc13, the Y2H assay identified binding of the UbcH5a/b/c series to the K3 RING-CH domain (Dodd et al, 2004). These three UbcH5 family members share >90% homology and we used the UbcH5-specific siRNA sequences (Saville et al, 2004) to deplete Hela-K3 and control cells of the UbcH5b/c enzymes. Effective UbcH5b/c depletion (Figure 4B) led to an increase in class I expression in HeLa-K3 cells, with up to 28% of depleted cells showing a complete rescue of class I expression (Figure 4A, middle panel), an effect not seen following UbcH5a depletion (see Supplementary Figure 2 online). Depletion of UbcH5b/c together with Ubc13 in HeLa-K3 cells (Figure 4B) showed a synergistic effect, with an almost complete rescue of cell surface class I molecules in cells depleted of both UbcH5b/c and Ubc13 (Figure 4A, right panel). Control siRNA depletions of an additional five Ubc enzymes (see Supplementary Figure 2 online) showed no increase in class I expression in HeLa-K3 cells, suggesting that in vivo, UbcH5b/c together with Ubc13 are the Ubc enzymes responsible for K3-mediated class I ubiquitination.
Figure 4.
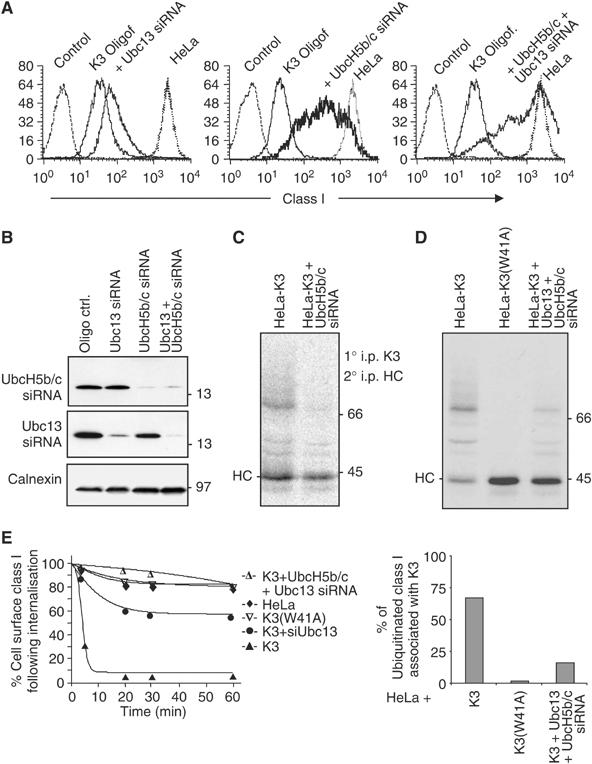
UbcH5 and Ubc13 act synergistically to mediate Lys-63 polyubiquitination of class I molecules. (A–D) Class I expression in HeLa-K3 cells is rescued following cellular depletion of UbcH5b/c+Ubc13. (A, B) HeLa and HeLa-K3 cells were analysed for class I expression (as Figure 1A) following Ubc13 and/or UbcH5b/c siRNA. (C) Cellular depletion of UbcH5b/c inhibits class I ubiquitination. HeLa-K3 following control or UbcH5b/c siRNA depletion were pulsed for 15 min, chased for 1 h and immunoprecipitated (as Figure 1B) with the FLAG mAb M2, dissociated in 1% SDS and reprecipitated with the heavy chain (HC)-specific mAb HC10. (D) Cellular depletion of UbcH5b/c+Ubc13 inhibits class I ubiquitination. (D upper panel) HeLa-K3 or HeLa-K3(W41A) cells were pulsed for 15 min, chased for 1 h and immunoprecipitated (as Figure 1B) with the FLAG mAb M2, dissociated in 1% SDS and reprecipitated with the HC-specific mAb HC10, following control or UbcH5b/c+Ubc13 siRNA depletion. (D lower panel) Phosphorimager analysis of Figure 4D upper panel. Ubiquitinated class I are expressed as a percentage of total class I molecules associated with K3. (E) Ubc13 depletion decreases class I internalisation in the presence of K3. Class I internalisation was determined in a cytofluorometric-based internalisation assay (see Materials and methods).
If the UbcH5b/c enzymes are responsible for monoubiquitination, then UbcH5b/c depletion alone should rescue class I expression and completely inhibit class I polyubiquitination and degradation. However, no more than 28% of UbcH5b/c-depleted cells showed a complete class I rescue (Figure 4A, middle panel). Analysis of radiolabelled HeLa-K3 cells following UbcH5b/c depletion did indeed lead to a marked reduction in K3-associated, polyubiquitinated class I chains (Figure 4C), although our inability to completely rescue class I expression and prevent polyubiquitination may be due to either incomplete UbcH5b/c depletion or redundancy in E2 enzymes able to monoubiqutinate class I molecules.
We next compared the proportion of K3-associated class I molecules in control HeLa-K3 cells, mutant HeLa-K3(W41A) cells and UbcH5b/c+Ubc13 siRNA-depleted HeLa-K3 cells following pulse chase analysis (Figure 4D, upper panel), and used phosphorimager analysis to determine the percentage of K3-associated class I molecules that are ubiquitinated (Figure 4D, lower panel). The K3(W41A) genetic mutant is unable to recruit any Ubc enzyme, leads to a rescue in cell surface class I (Figure 1A), and therefore none of the K3(W41A)-associated class I molecules are ubiquitinated (Figure 4D, lane 2). This mutant therefore represents the maximal amount of class I which can associate with K3 in the absence of ubiquitination. Following UbcH5b/c+Ubc13 depletion, the majority of K3-associated class I molecules are no longer ubiquitinated (Figure 4D, lane 3), and the proportion of ubiquitinated class I molecules, expressed as a percentage of total K3-associated class I, decreases from 69% in the presence of wild-type K3 to 16.5% in the Ubc-depleted cells (Figure 4D, lower panel)—presumably representing cells not effectively depleted of UbcH5b/c and Ubc13 by the siRNAs.
To understand why Lys-63-polyubiquitinated class I chains are required for class I downregulation, we initially examined how K3 and Ubc siRNA depletions from K3-expressing cells affected class I internalisation. Using a cytofluorometric-based internalisation assay, the rapid internalisation of class I molecules seen in K3-expressing cells (t1/2=7 min) was reduced following depletion of UbcH5b/c+Ubc13 to internalisation rates comparable to those seen in HeLas (t1/2>4 h) and HeLa K3(W41A)-expressing cells (t1/2>4 h) (Figure 4E), and a similar profile was seen following depletion of UbcH5b/c (data not shown). However, depletion of Ubc13 alone gave an intermediate phenotype (t1/2=2 h) suggesting that monoubiquitinated class I molecules internalise slowly and are recycled to the cell surface.
An MHC class I/ubiquitin fusion protein is not efficiently endocytosed
To further confirm whether a single ubiquitin is sufficient for class I downregulation, we made a class I/ubiquitin fusion protein (Figure 5A). The HLA-A*0201 gene, with its three cytoplasmic tail lysines mutated to arginine, was fused to a mutant, lysine-less ubiquitin (HLA-A2K0UbK0) (Li et al, 2003). The cytoplasmic tail of this protein therefore contains no lysine residues. HLA-A2K0UbK0 expression at the cell surface of HeLa cells was not as high as wild-type or lysine-less HLA-A2 (Figure 5B), and internalisation studies also suggested an intermediate phenotype, with internalisation rates similar to those seen in HeLa-K3 cells following Ubc13 depletion (Figure 5C). These results imply that monoubiquitination provides class I with a weak signal for endocytosis that is insufficient for degradation.
Figure 5.
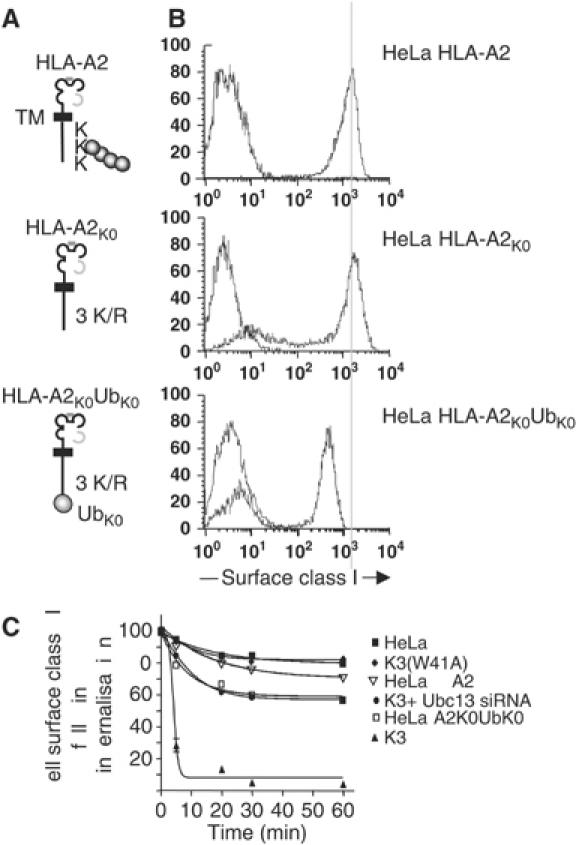
A class I monoubiquitin fusion protein is inefficiently internalised. (A) Schematic of HLA-A*0201 fusion proteins; TM=transmembrane region. (B) Cytofluorometric analysis of HLA-A*0201 constructs expressed in HeLa cells. (C) HLA-A2K0UbK0-expressing cells and Ubc13-depleted HeLa-K3 cells show similar rates of internalisation. Cytofluorometric-based internalisation assay for the HLA-A2K0UbK0-expressing cells (as Figure 4E).
The epsin 1 adaptor is required for clathrin-mediated downregulation of Lys-63-polyubiquitinated class I molecules
To identify which proteins recognise and promote internalisation of Lys-63-polyubiquitinated class I molecules, we initially investigated the role of clathrin. An absolute requirement for clathrin was demonstrated as both overexpression of the C-terminus of AP180 (Figure 6A), an inhibitor of clathrin-mediated endocytosis (Ford et al, 2001), and siRNA-mediated clathrin depletion (Figure 6B–D) prevented K3-dependent class I downregulation. However, for ubiquitin to act as an endocytic signal, additional ubiquitin-binding adaptor proteins must be required for clathrin recruitment and internalisation of ubiquitinated proteins. Binding of ubiquitin to the ubiquitin-interacting motif (UIM) of adaptor proteins requires a hydrophobic binding patch that includes the conserved isoleucine at position 44 of ubiquitin (Ile44). We generated a mutant ubiquitin (Ile44Ala) previously shown not to bind UIMs (Miller et al, 2004). At high expression levels of this ubiquitin (Ile44Ala) mutant in HeLa-K3 cells, the class I molecules are rescued back to the cell surface (Figure 6E), with no effect seen in HeLa cells. There is therefore a requirement for the Ile44 residue of ubiquitin, and by inference the interaction of Lys-63-polyubiquitinated class I molecules with a ubiquitin-binding protein, for internalisation of ubiquitinated class I molecules. The epsin and Eps15 endocytic adaptors contain multiple UIMs (Polo et al, 2002), are localised to the plasma membrane (Benmerah et al, 1998; Chen et al, 1998), and have been implicated in the downregulation of ubiquitinated receptors in yeast (Shih et al, 2002). To determine a requirement for these adaptors in Lys-63-polyubiquitinated class I downregulation, we used siRNA to deplete epsin 1, Eps15 and AP-2 from K3-expressing cells. Depletion of epsin 1, as assessed by immunoblot analysis (Figure 6F), led to a partial, but highly reproducible class I rescue in K3-expressing HeLa cells with no effect seen on class I expression in control HeLas (Figure 6H). This partial rescue was seen with an epsin 1 smartpool as well as two individual epsin 1-specific siRNA oligonucleotides. In contrast neither Eps15 nor depletion of the μ2 subunit of AP-2 affected class I downregulation (Figure 6F–H), and a similar negative result was also seen with the β2 subunit of AP-2 (data not shown). These results imply that internalisation of Lys-63-polyubiquitinated class I molecules requires the epsin 1 endocytic adaptor in a clathrin-dependent, but AP-2-independent pathway.
Figure 6.
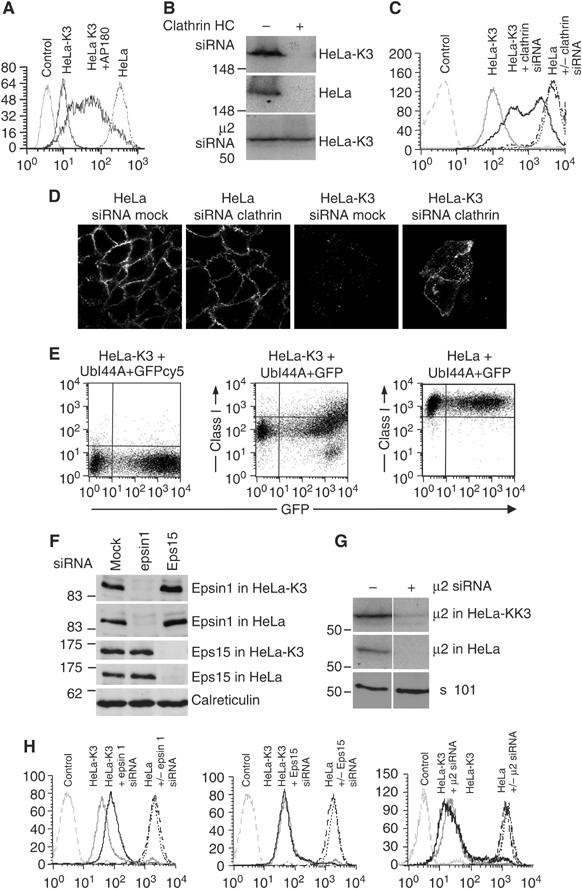
Downregulation of Lys-63-polyubiquitinated class I molecules is clathrin and epsin 1 dependent and AP-2 independent. (A–D) Class I downregulation in HeLa-K3 cells is clathrin dependent. (A) Cytofluorometric analysis of MHC class I expression in HeLa and HeLa-K3 cells transfected with cDNA encoding the C-terminus of AP180 and stained with mAb w6/32 and FITC-conjugated secondary antibody. (B) Immunoblots of clathrin heavy chain (HC) following siRNA depletion. Triton X-100 lysates from HeLa and HeLa-K3 cells were targeted with clathrin HC siRNA and probed with clathrin-specific antisera. (C, D) Cytofluorometric analysis (C) and confocal immunofluorescence microscopy (D) of MHC class I expression in HeLa and HeLa-K3 cells following siRNA-mediated depletion of clathrin HC. For (C), cells were stained with mAb w6/32 and Alexa Fluor 647-conjugated secondary antibody, and for (D) with mAb w6/32 and Alexa Fluor 555-conjugated secondary antibody. (E) Class I downregulation in HeLa-K3 cells is dependent on the UIM binding face of ubiquitin. Cytofluorometric analysis of HeLa or HeLa-K3 cells transfected with the ubiquitin isoleucine–alanine mutant (UbI44A)+GFP, and stained with mAb w6/32 and cy5-conjugated secondary antibody (middle and right panel) or cy5-conjugated secondary antibody alone (left panel). (F, G) Immunoblot analysis following siRNA depletion of epsin 1, Eps15 and AP-2. Triton X-100 lysates from HeLa and HeLa-K3 cells were targeted with epsin 1 (oligo. #1) and Eps15 siRNA (F) and the μ2 subunit of AP-2 (G) and probed with the appropriate antibodies (Materials and methods). (H) Cytofluorometric analysis of MHC class I expression in HeLa and HeLa-K3 cells following siRNA-mediated depletion of epsin 1 (oligo. #1), Eps15 and the μ2 subunit of AP-2. Cells were stained with mAb w6/32 and Alexa Fluor 647-conjugated secondary antibody.
Discussion
Apart from its established role in post-translational control of protein turnover, ubiquitin is the major endocytic signal in yeast and regulates both the internalisation and sorting of an increasing number of mammalian membrane receptors (Hicke and Dunn, 2003; Dupre et al, 2004). Our results (Figure 7) suggest that K3 associates with class I molecules in the late secretory pathway and recruits two Ubc enzymes. UbcH5b/c initiates class I ubiquitination, but a single ubiquitin is inefficient at internalisation of class I molecules for degradation. Additional recruitment of Ubc13 by K3 promotes the generation of Lys-63-polyubiquitinated class I molecules which uses the epsin 1 adaptor for clathrin-dependent endocytosis and sorting via multivesicular bodies for lysosomal degradation.
Figure 7.
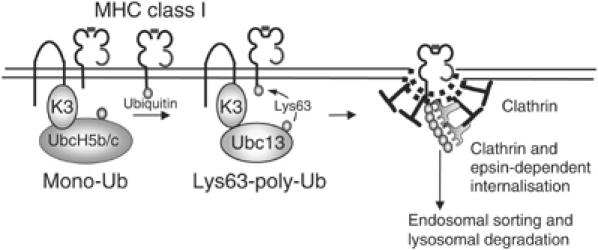
Schematic of Lys-63-linked polyubiquitination of MHC class I molecules by K3. MHC class I molecules associate with K3 in the late secretory pathway. Recruitment of Ubc enzymes of initially the UbcH5 series, followed by Ubc13, leads to mono- followed by lysine-63-linked polyubiquitination. Lys-63-linked polyubiqutinated class I molecules are endocytosed via a clathrin- and epsin 1-dependent pathway, leading to sorting through the endosomal pathway and lysosomal degradation.
Conjugation of monoubiquitin is sufficient for downregulation of several yeast proteins (Hicke and Dunn, 2003; Dupre et al, 2004), and in mammalian cells monoubiquitination of RTKs appears sufficient for endocytosis and lysosomal sorting (Haglund et al, 2003b; Mosesson et al, 2003). We have presented three independent experiments which show that monoubiquitinated class I provides an inefficient signal for class I internalisation and degradation. First, expression of the Lys-63R ubiquitin mutant in K3-expressing cells resulted in a monoubiquitinated class I species which was not efficiently internalised and led to a rescue of class I molecules at the cell surface (Figure 3B and C). Second, Ubc13 depletion in K3-expressing cells caused a partial rescue of class I molecules back to the cell surface (Figure 2B) and left a predominantly monoubiquitinated class I species (Figure 3D). Finally, a class I ubiquitin fusion protein was not efficiently internalised (Figure 5). These findings appear at odds with published data on ligand-induced downregulation of receptor tyrosine kinases. However, analysis of two yeast transporters, Fur4p and Gap1p, in mutants lacking the Doa4p Ub-isopeptidase, show a requirement for Lys-63 ubiquitin chains for efficient internalisation (Galan and Haguenauer-Tsapis, 1997; Blondel et al, 2004). Indeed, the extent of ubiquitin chain topology in vivo may be underestimated, with Lys-63 chains more abundant than previously thought (Peng et al, 2003). Furthermore, a recent study identified UbcH7 as the E2 recruited by the TRAF6 E3 ligase for noncanonical Lys-63 polyubiquitination of the nerve growth factor receptor TrkA, directing its internalisation and signalling (Geetha et al, 2005).
The affinity of interactions between ubiquitin and ubiquitin-binding domain (UBD)-containing receptors along the endocytic pathway is low (Fisher et al, 2003). However, ubiquitin binding is cooperative, and multiple ubiquitins may increase the avidity and stabilise the interaction between ubiquitin and its corresponding UBD (Haglund et al, 2003a). This could occur through either multiple monoubiquitination or, for those receptors with a limited number of available cytoplasmic lysine residues, Lys-63 polyubiquitin conjugates offer an alternative mechanism. Structural analysis shows Lys-48 ubiquitin chains to adopt a closed conformation, while Lys-63 ubiquitin chains are in an extended conformation (Varadan et al, 2004), with a topology similar to adjacent linear ubiquitin chains which, unlike the Lys-48 linkage, have their hydrophobic surfaces available for binding (Pickart and Fushman, 2004). This open conformation of Lys-63 polyubiquitin conjugates may, as with multiple monoubiquitins, increase the avidity for UBDs. Alternatively, the presence of Lys-63 chains may allow specificity for selective UBDs (Raasi et al, 2005). We therefore investigated the internalisation pathway used by class I molecules. Class I downregulation by K3 was clathrin dependent as expression of the AP180 C-terminus, as well as clathrin siRNA depletion, rescued class I downregulation. A similar mechanism has been suggested for K5-mediated class I downregulation which was inhibited by a mutant of the large GTPase dynamin (Coscoy and Ganem, 2001). A functional requirement for ubiquitin to interact with a UIM-containing protein in the downregulation of Lys-63-polyubiquitinated class I molecules was shown by the increase in cell surface class I in the presence of the ubiquitin I44A mutant. Additional ubiquitin-binding adaptors must therefore recruit the ubiquitinated class I to the clathrin-coated vesicle (Hicke and Dunn, 2003; Dupre et al, 2004).
Epsin and Eps15 are clathrin adaptors implicated in the recognition of ubiquitinated plasma membrane proteins through their ubiquitin-interacting motifs (Hicke and Dunn, 2003). Epsin has three UIMs while Eps15 has two C-terminal UIMs (Hofmann and Falquet, 2001). The ability of these proteins to bind ubiquitin and associate with both clathrin and AP-2 makes them attractive candidate adaptors for the internalisation of polyubiquitin chains from the plasma membrane. The partial rescue of class I downregulation following epsin 1 depletion provides supporting evidence for the role of this endocytic adaptor in the clathrin-dependent, but AP-2-independent downregulation of Lys-63-polyubiquitinated class I molecules. However, the intermediate phenotype with only partial rescue of class I downregulation suggests a role for other, unidentified endocytic adaptors, possibly the related epsin 2 and 3 adaptors. The lack of effect of AP-2 depletion is not especially surprising as internalisation of other cell surface receptors following AP-2 depletion has been described, and AP-2 contains no recognised ubiqutin-binding motifs (Motley et al, 2003; Traub, 2005).
Our data suggest a requirement for at least two E2 enzymes for K3 activity. The UbcH5b/c enzymes initiate conjugation of the first ubiquitin, while Ubc13 is responsible for subsequent Lys-63 polyubiquitination. The four members of the UbcH5 family, UbcH5a/b/c and HBUCE1 share more than 90% homology, and our difficulty in demonstrating a complete rescue following UbcH5b/c depletion could be due to an incomplete loss of the UbcH5b/c enzymes. Alternatively, redundancy within this E2 family or other E2s may promote monoubiquitination. However, the requirement for UbcH5b/c and Ubc13 for K3 activity emphasises a role for Ubc E2 enzymes in determining specificity and outcome of the ubiquitination reaction. Recent data examining Lys-48-linked ubiquitin chain synthesis by the cullin-RING ubiquitin ligase SCF, together with its cognate Ubc E2 enzyme Cdc34 (Ubc3), showed that ubiquitin targeting can be separated into two steps (Petroski and Deshaies, 2005). Attachment of the first ‘initiator' ubiquitin is the rate-limiting step and is followed by a dramatic change with rapid elongation of the subsequent growing polyubiquitin chain. The UbcH5 family do not typically form high molecular weight polyubiquitin conjugates and are intrinsically less processive than Cdc34 (Ubc3) in the synthesis of polyubiquitin chains (Petroski and Deshaies, 2005). An alternative strategy to promote chain elongation following attachment of the initial ubiquitin may therefore be to exchange Ubc E2 enzyme. Ubc13 is a unique Ubc that together with Mms2 generates Lys-63 ubiquitin conjugates. Mechanistically, the requirement by K3 for more than one Ubc E2-conjugating enzyme to build a Lys-63-polyubiquitinated class I chain has similarities to the yeast DNA polymerase PCNA (proliferating cell nuclear antigen) involved in DNA repair (Hoege et al, 2002). PCNA is initially monoubiquitinated on a single lysine residue via the RAD18 E3 ligase with Ubc RAD6, and requires a second E3 ligase RAD5 together with Ubc13 to attach Lys-63 ubiquitin conjugates (Hoege et al, 2002). Thus in vivo, Ubc13 is required for the generation of Lys-63 ubiquitin conjugates but may be unable to initiate substrate ubiquitination. This initial ubiquitin conjugation requires an additional Ubc, and in the case of PCNA, an additional E3 ligase to generate a monoubiquitin platform which can be elongated by Ubc13/Mms2. It will be interesting to determine whether this requirement is characteristic of the generation of all Lys-63, or other polyubiquitin conjugates.
Although K3 encodes a viral ubiquitin E3 ligase, viral genes are far more likely to subvert existing cellular pathways than invent new ones. The identification of K3-related gene products from different viral families suggested the appropriation of these genes from vertebrate hosts. A total of 10 human gene products—the Membrane-associated RING-CH (MARCH) proteins—with similar structural organisation to the K3 family, have now been identified and their overexpression leads to downregulation of cell surface immunoreceptors, including MHC class I molecules, CD4, CD86, Fas and the transferrin receptor (Bartee et al, 2004; Lehner et al, 2005). Although the physiological function of these MARCH gene products remains unclear, it will be interesting to determine whether these cellular orthologues of K3 also use Lys-63-linked polyubiquitin chains to endocytose and degrade their target immunoreceptors. We suggest that Lys-63-mediated polyubiquitination may provide a more general mechanism to promote internalisation, endolysosomal sorting and degradation of cell surface receptors.
Materials and methods
Antibodies
The following antibodies were used: mAb HC10 (anti-class I heavy chain), w6/32 (anti-MHC class I), mAb M2 (anti-FLAG) (Sigma), mAb FK1 (anti-ubiquitin) (Affiniti), mAb (anti-Ubc13) (Affiniti), rabbit anti-UbcH5b/c (Saville et al, 2004), mAb BB7.2 (anti-HLA-A2) (Serotec), mAb AP50 (anti-μ2) (Transduction Laboratories); R-20 (goat anti-epsin 1) (Santa Cruz); K-15 (goat anti-Eps15) (Santa Cruz), PA3–900 (rabbit anti-calreticulin) (Affinity BioReagents), mAb A410 (anti-Tsg101) (Genetex), rabbit anti-clathrin heavy chain (a kind gift from M.S. Robinson) and anti-IgG2b isotype-control (eBioscience); Fluorescent secondary antibodies were from Molecular Probes.
Constructs
The constructs and cell lines used were as described (Hewitt et al, 2002) except pCDNA3.1(−)Pac-HLA-A2K0 was generated by site-directed mutagenesis of pCDNA3.1(−)Pac-HLA-A2-K340A/K364A using the QuickChange kit (Stratagene). The sense oligonucleotide used for the K335R mutation was A2-K0-5′. pCDNA3.1(−)Pac-HLA-A2K0UbK0: HLA-A2K0 was fused to a lysine-less form of ubiquitin (a kind gift of Wei Gu) to form the chimera HLA-A2K0UbK0.
The GFP-tagged wild-type and mutant ubiquitin constructs (wtUb, UbK48R and UbK63R) were used as described (Tsirigotis et al, 2001) (a kind gift of Doug Gray). The UbI44A construct was generated by site-directed mutagenesis of HA-wtUb-pcDNA3 (a kind gift from Ivan Dikic) using the QuickChange kit (Stratagene). The sense oligonucleotide used for I44A mutation was UbI44A-5′.
Oligonucleotides:
GATGTGGAGGAGGAGGAGCTCAGATAGAGCA2K0-5′,
TGCGGCCGCCAGATCTTCGTCAGAACGTTAACCNotI UbK0 5′,
TGGATCCTCAACCACCTCTTAGTCUbK0 BamHI 3′,
CAGCTTGTGCAGTGAGCGGCCGCCGAGCTCGGTACCAAGA2K0 NotI SDM5′
ATCAGCAGAGACTGGCCTTTGCTGGCAAGCAGUbI44A-5′
Ubc depletion using siRNA
HeLa-M and HeLa-K3 cells were transfected twice at 24 h intervals with 50–200 pmol of siRNA per well with Oligofectamine (Invitrogen) and analysed 48 h after the second transfection, exactly as described (Hewitt et al, 2002). For clathrin heavy chain depletion, HeLa-M and HeLa-K3 cells were transfected twice at 48 h intervals with 20 pmol of siRNA per well with Oligofectamine as described (Hewitt et al, 2002) and analysed 48 h after the second transfection. For the μ2 subunit of AP-2, epsin 1 and Eps15 depletions, the same protocol as for clathrin heavy chain depletion was implemented, with an additional transfection, making three transfections at 24 h intervals with 50–200 pmol of siRNA per well. All siRNA sequences and references are shown as Table 1 in Supplementary data.
HeLa and HeLa-K3 transient transfections
HeLa and HeLa-K3 cells were transfected at 50% confluency in six well plates with 2 μg of vector per well using TransIT-HeLaMONSTER transfection Kit (Mirus). For the UbI44A transfections, cells were cotransfected with 1.8 μg of UbI44A-pcDNA3 and 0.2 μg pmaxFP-Green-C GFP control vector per well. Transfected cells were analysed by flow cytometry 48 h following transfection.
Radiolabelling and immunoprecipitations
Cells were starved for 1 h in methionine, cysteine-free medium and labelled with [35S]methionine and [35S]cysteine for 15 min and chased in media containing excess cold methionine and cysteine for the indicated time periods. Immunoprecipitations were performed as described (Hewitt et al, 2002). Samples were analysed by SDS–PAGE (8 or 10%) and autoradiography. Visualisation of ubiquitin bands required autoradiography for 7–21 days.
Flow cytometric analysis was performed as described (Hewitt et al, 2002) with the mAbs w6/32 or BB7.2 and anti-mouse secondary antibody as indicated in figure legends.
Immunofluorescence
At 24 h after the second siRNA transfection, cells were transferred to glass coverslips and grown for a further 24 h. Cells were fixed with 4% paraformaldehyde for 15 min. w6/32 primary and anti-mouse Alexa Fluor 555 secondary incubations were carried out in 0.2% BSA. Cells were mounted and visualised on a Zeiss Axiovert 200 M inverted microscope with an LSM 510 META confocal laser scanning attachment.
Internalisation assay
Cells were stained with w6/32 or BB7.2 for 45 min at 4°C, washed twice with cold PBS/0.1% BSA and returned to 37°C. Surface class I molecules were internalised for 0, 5, 20, 30 and 60 min, and cells stained with a goat anti-mouse Cy5 antibody for 45 min at 4°C prior to fixation and FACS analysis. Relative mean fluorescence intensities (MFI) at each time point were calculated by dividing the mean fluorescence intensity of a sample by the mean fluorescence of cells labelled with negative control mAb only. The percentage class I remaining at the cell surface following internalisation was calculated as (MFI tX−MFI control cells/(MFI t0−MFI control cells) × 100.
Supplementary Material
Supplementary Table 1
Supplementary Figure 1
Supplementary Figure 2
Acknowledgments
We thank ZJ Chen (University of Texas Southwestern) for initial Ubc13 siRNA sequences, D Gray (University of Ottawa, Ontario) for mutant ubiquitin/GFP constructs, W Gu (Columbia University) for the lysine-less ubiquitin construct, I Dikic (Goethe University Medical School) for the ubiquitin construct, H McMahon (LMB-MRC, Cambridge) for the AP180 cDNA and MS Robinson (Cambridge University) for the clathrin antisera, P Kozik and E Hutchinson for experimental support. This work was supported by grants from the Wellcome Trust (PJL, SCP) the MRC (CMS, JPL) and CRUK (MKS).
References
- Bartee E, Mansouri M, Hovey Nerenberg BT, Gouveia K, Fruh K (2004) Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J Virol 78: 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerah A, Lamaze C, Begue B, Schmid SL, Dautry-Varsat A, Cerf-Bensussan N (1998) AP-2/Eps15 interaction is required for receptor-mediated endocytosis. J Cell Biol 140: 1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel MO, Morvan J, Dupre S, Urban-Grimal D, Haguenauer-Tsapis R, Volland C (2004) Direct sorting of the yeast uracil permease to the endosomal system is controlled by uracil binding and Rsp5p-dependent ubiquitylation. Mol Biol Cell 15: 883–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boname JM, de Lima BD, Lehner PJ, Stevenson PG (2004) Viral degradation of the MHC class I peptide loading complex. Immunity 20: 305–317 [DOI] [PubMed] [Google Scholar]
- Boname JM, Stevenson PG (2001) MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15: 627–636 [DOI] [PubMed] [Google Scholar]
- Booth JW, Kim MK, Jankowski A, Schreiber AD, Grinstein S (2002) Contrasting requirements for ubiquitylation during Fc receptor-mediated endocytosis and phagocytosis. EMBO J 21: 251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH, Di Fiore PP, De Camilli P (1998) Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature 394: 793–797 [DOI] [PubMed] [Google Scholar]
- Coscoy L, Ganem D (2000) Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc Natl Acad Sci USA 97: 8051–8056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy L, Ganem D (2001) A viral protein that selectively downregulates ICAM-1 and B7-2 and modulates T cell costimulation. J Clin Invest 107: 1599–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy L, Sanchez DJ, Ganem D (2001) A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J Cell Biol 155: 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell P (2000) Intracellular surveillance: controlling the assembly of MHC class I-peptide complexes. Traffic 1: 301–305 [DOI] [PubMed] [Google Scholar]
- Dodd RB, Allen MD, Brown SE, Sanderson CM, Duncan LM, Lehner PJ, Bycroft M, Read RJ (2004) Solution structure of the Kaposi's sarcoma-associated herpesvirus K3 N-terminal domain reveals a Novel E2-binding C4HC3-type RING domain. J Biol Chem 279: 53840–53847 [DOI] [PubMed] [Google Scholar]
- Dupre S, Urban-Grimal D, Haguenauer-Tsapis R (2004) Ubiquitin and endocytic internalization in yeast and animal cells. Biochim Biophys Acta 1695: 89–111 [DOI] [PubMed] [Google Scholar]
- Fisher RD, Wang B, Alam SL, Higginson DS, Robinson H, Sundquist WI, Hill CP (2003) Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem 278: 28976–28984 [DOI] [PubMed] [Google Scholar]
- Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT (2001) Simultaneous binding of PtdIns (4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291: 1051–1055 [DOI] [PubMed] [Google Scholar]
- Fujimuro M, Sawada H, Yokosawa H (1994) Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett 349: 173–180 [DOI] [PubMed] [Google Scholar]
- Galan JM, Haguenauer-Tsapis R (1997) Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J 16: 5847–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha T, Jiang J, Wooten MW (2005) Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol Cell 20: 301–312 [DOI] [PubMed] [Google Scholar]
- Gesbert F, Malarde V, Dautry-Varsat A (2005) Ubiquitination of the common cytokine receptor gammac and regulation of expression by an ubiquitination/deubiquitination machinery. Biochem Biophys Res Commun 334: 474–480 [DOI] [PubMed] [Google Scholar]
- Guerin JL, Gelfi J, Boullier S, Delverdier M, Bellanger FA, Bertagnoli S, Drexler I, Sutter G, Messud-Petit F (2002) Myxoma virus leukemia-associated protein is responsible for major histocompatibility complex class I and Fas-CD95 down-regulation and defines scrapins, a new group of surface cellular receptor abductor proteins. J Virol 76: 2912–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Di Fiore PP, Dikic I (2003a) Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem Sci 28: 598–603 [DOI] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I (2003b) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol 5: 461–466 [DOI] [PubMed] [Google Scholar]
- Haque M, Chen J, Ueda K, Mori Y, Nakano K, Hirata Y, Kanamori S, Uchiyama Y, Inagi R, Okuno T, Yamanishi K (2000) Identification and analysis of the K5 gene of Kaposi's sarcoma-associated herpesvirus. J Virol 74: 2867–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt EW, Duncan L, Mufti D, Baker J, Stevenson PG, Lehner PJ (2002) Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J 21: 2418–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Dunn R (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19: 141–172 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Falquet L (2001) A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci 26: 347–350 [DOI] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96: 645–653 [DOI] [PubMed] [Google Scholar]
- Ishido S, Choi JK, Lee BS, Wang C, DeMaria M, Johnson RP, Cohen GB, Jung JU (2000a) Inhibition of natural killer cell-mediated cytotoxicity by Kaposi's sarcoma-associated herpesvirus K5 protein. Immunity 13: 365–374 [DOI] [PubMed] [Google Scholar]
- Ishido S, Wang C, Lee BS, Cohen GB, Jung JU (2000b) Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J Virol 74: 5300–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner PJ, Hoer S, Dodd R, Duncan LM (2005) Downregulation of cell surface receptors by the K3 family of viral and cellular ubiquitin E3 ligases. Immunol Rev 207: 112–125 [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W (2003) Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302: 1972–1975 [DOI] [PubMed] [Google Scholar]
- Mansouri M, Bartee E, Gouveia K, Hovey Nerenberg BT, Barrett J, Thomas L, Thomas G, McFadden G, Fruh K (2003) The PHD/LAP-domain protein M153R of myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J Virol 77: 1427–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SL, Malotky E, O'Bryan JP (2004) Analysis of the role of ubiquitin-interacting motifs in ubiquitin binding and ubiquitylation. J Biol Chem 279: 33528–33537 [DOI] [PubMed] [Google Scholar]
- Mosesson Y, Shtiegman K, Katz M, Zwang Y, Vereb G, Szollosi J, Yarden Y (2003) Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J Biol Chem 278: 21323–21326 [DOI] [PubMed] [Google Scholar]
- Motley A, Bright NA, Seaman MN, Robinson MS (2003) Clathrin-mediated endocytosis in AP-2-depleted cells. J Cell Biol 162: 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramura M, Jang IK, Kole H, Huang F, Haines D, Gu H (2002) c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR down-modulation. Nat Immunol 3: 1192–1199 [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2005) Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123: 1107–1120 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D (2004) Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 8: 610–616 [DOI] [PubMed] [Google Scholar]
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP (2002) A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416: 451–455 [DOI] [PubMed] [Google Scholar]
- Raasi S, Varadan R, Fushman D, Pickart CM (2005) Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol 12: 708–714 [DOI] [PubMed] [Google Scholar]
- Sanchez DJ, Gumperz JE, Ganem D (2005) Regulation of CD1d expression and function by a herpesvirus infection. J Clin Invest 115: 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville MK, Sparks A, Xirodimas DP, Wardrop J, Stevenson LF, Bourdon JC, Woods YL, Lane DP (2004) Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J Biol Chem 279: 42169–42181 [DOI] [PubMed] [Google Scholar]
- Shih SC, Katzmann DJ, Schnell JD, Sutanto M, Emr SD, Hicke L (2002) Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol 4: 389–393 [DOI] [PubMed] [Google Scholar]
- Stevenson PG, Efstathiou S, Doherty PC, Lehner PJ (2000) Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc Natl Acad Sci USA 97: 8455–8460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J 19: 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM (2005) Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim Biophys Acta 1744: 415–437 [DOI] [PubMed] [Google Scholar]
- Tsirigotis M, Zhang M, Chiu RK, Wouters BG, Gray DA (2001) Sensitivity of mammalian cells expressing mutant ubiquitin to protein-damaging agents. J Biol Chem 276: 46073–46078 [DOI] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D (2004) Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem 279: 7055–7063 [DOI] [PubMed] [Google Scholar]
- Wang X, Lybarger L, Connors R, Harris MR, Hansen TH (2004) Model for the interaction of gammaherpesvirus 68 RING-CH finger protein mK3 with major histocompatibility complex class I and the peptide-loading complex. J Virol 78: 8673–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW, Hill AB (2002) Viral interference with antigen presentation. Nat Immunol 3: 1019–1025 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Supplementary Figure 1
Supplementary Figure 2


