Abstract
EBER 1, a small noncoding viral RNA abundantly expressed in all cells transformed by Epstein-Barr virus (EBV), has been shown to associate with the human ribosomal protein L22. Here we present in vitro binding studies using purified RNAs and recombinant proteins. Electrophoretic mobility-shift assays (EMSAs) show that recombinant L22 (rL22) and maltose-binding protein (MBP)-tagged L22 protein bind EBER 1 in vitro, both forming three specific protein-dependent mobility shifts. Use of a mixture of rL22 and MBP-L22 indicates that these three shifts contain one, two, or three L22 proteins per EBER 1 molecule. EMSAs performed with EBER 1 deletion constructs and EBER 1 stem–loops inserted into a nonbinding RNA, HSUR 3, identify stem–loops I, III, and IV as L22 binding sites. The existence of multiple L22 binding sites on EBER 1 inside cells is demonstrated by in vivo UV cross-linking. Our results are discussed with respect to the function of EBER 1 in EBV-infected human B cells.
Keywords: Epstein-Barr virus, EBER 1, ribosomal protein L22, electrophoretic mobility-shift assay, in vivo UV cross-linking, RNA–protein interaction
INTRODUCTION
Epstein-Barr virus (EBV) is a member of the human γ herpesvirus family that infects >90% of adults worldwide and maintains a lifelong presence. EBV is the causative agent of infectious mononucleosis (Henle et al. 1968) and is associated with Burkitt lymphoma (BL) (Henle and Henle 1966), nasopharyngeal carcinoma (zur Hausen et al. 1970), and other human tumors including Hodgkin's disease (Johansson et al. 1970, 1975) and AIDS-associated lymphoma (Subar et al. 1988; Haluska et al. 1989). While most of the EBV genome is silent in EBV-transformed B-lymphocytes, two genes encoding the small RNAs EBER 1 and EBER 2 (Epstein-Barr encoded RNAs) are actively transcribed by RNA polymerase III (∼106 copies per cell) (Lerner et al. 1981; van Santen et al. 1981; Arrand et al. 1989). Recently, a recombinant EBV with the two EBER genes deleted was shown to exhibit 100-fold lower efficiency in malignant transformation of resting B cells compared with its wild-type counterpart (Yajima et al. 2005), suggesting a role for EBERs in cellular transformation. This confirms earlier reports that expression of EBERs alone promotes the tumorogenic phenotype as assessed by cell growth in soft agar and tumorgenicity in nude mice (Komano et al. 1999; Ruf et al. 2000; Yamamoto et al. 2000). However, the molecular mechanisms underlying the function(s) of the EBERs in the viral life cycle remain unclear.
EBER 1 and EBER 2 (167 and 172 nt, respectively) have considerable secondary structure consisting of several stable stem–loops (Glickman et al. 1988). They contain 3′ terminal polyuridylate stretches that interact stably with the La antigen (Lerner et al. 1981). In addition to La, EBER 1 (Fig. 1A) binds the human ribosomal protein L22 tightly (Toczyski and Steitz 1991) and the interferon-inducible protein kinase R (PKR) more weakly (Clarke et al. 1990, 1991; Clemens et al. 1994). L22 is a 14.8-kDa protein constituent of the large ribosomal subunit, which is not conserved in prokaryotes (Nakao et al. 2004) and does not share RNA binding motifs with any other known protein (http://us.expasy.org/prosite). In addition, L22 associates with human telomerase (Le et al. 2000), the small viral RNA HVP-1 in Herpesvirus papio–infected baboon cells (Toczyski and Steitz 1991), the 3′X region within the 3′-untranslated region (UTR) of the human Hepatitis C virus (HCV) genomic RNA (Wood et al. 2001), poly (ADP-ribose) polymerase in Drosophila melanogaster (Koyama et al. 1999), and the Herpes simplex–infected cell proteins (ICP) 4 and ICP 22 in human cells (Leopardi and Roizman 1996; Leopardi et al. 1997). However, the function of L22 both in the ribosome and in these several RNP complexes remains unknown.
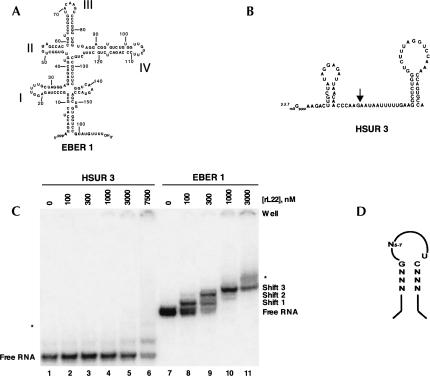
FIGURE 1.
L22 interactions with EBER 1 and HSUR 3 in EMSAs. (A) Secondary structure of wild-type EBER 1 as determined previously (Glickman et al. 1988). Bold roman numerals denote the stem–loops. (B) Secondary structure of HSUR 3 from Herpesvirus saimiri (Lee et al. 1988). The arrow indicates where stem–loops I, II, III, or IV were inserted into the RNA to generate the HSUR 3 chimeras analyzed in Figure 5. (C) EMSAs performed with 5 nM body-labeled HSUR 3 (lanes 1–6) or EBER 1 (lanes 7–11) and increasing concentrations of recombinant L22 protein. EBER 1, which alone migrates as a close doublet, forms three distinct shifts with L22, while HSUR 3 shifts only at protein concentrations ≥3 μM. An asterisk (*) indicates an additional, nonspecific shift that appears at very high L22 concentrations. Note that the free HSUR 3 contains a slower-migrating band even without L22. (D) The generalized RNA consensus motif proposed to mediate interaction between L22 and RNA (adapted from Dobbelstein and Shenk 1995).
In uninfected human B cells, L22 is located in the nucleoli and cytoplasm, whereas following EBV infection, L22 is found in the nucleoplasm as well (Toczyski et al. 1994). In situ hybridization indicates that EBERs are largely nucleoplasmic (Howe and Steitz 1986; Barletta et al. 1993), suggesting that L22 relocalization results from its association with EBER 1 in vivo (Toczyski et al. 1994). In transformed B cells, EBER 1 is quantitatively bound by L22 (>95%) (Toczyski and Steitz 1993), whereas only 30%–50% of the L22 protein associates with this RNA (Toczyski et al. 1994). Previous studies concluded that L22 binds EBER 1 via stem–loop III (Toczyski and Steitz 1993; Fig. 1A), but that additional binding site(s) may also exist (Toczyski and Steitz 1993; Dobbelstein and Shenk 1995). SELEX (systematic evolution of ligands by exponential enrichment) experiments established a generalized RNA motif (Fig. 1D) for L22 binding (Dobbelstein and Shenk 1995). Both stem–loops III and IV of EBER 1 fit this motif, thereby implicating stem–loop IV as an additional binding site for L22 on EBER 1.
To investigate the molecular interactions involved in RNP formation by EBER 1 and L22, we carried out more extensive binding studies using untagged and MBP-tagged L22 protein. EMSAs (electrophoretic mobility-shift assays) performed with wild-type EBER 1 and these recombinant proteins revealed three specific mobility shifts that are dependent on protein concentration and differ in the ratios of protein/RNA in the shifted complexes. Examination of EBER 1 deletion constructs and chimeric RNA molecules containing isolated EBER 1 stem–loops in EMSAs identified three L22 binding sites. These include the above-mentioned stem–loops III and IV, and a previously undescribed site involving stem–loop I. In addition, we demonstrate the existence of multiple L22 binding sites on EBER 1 inside cells by a UV cross-linking assay. Understanding how L22 associates with EBER 1 may help elucidate the contribution of the EBER RNPs to the EBV life cycle.
RESULTS
Stem–loops I, III, and IV are all binding sites for rL22
EMSA was used to study the in vitro interaction of human ribosomal protein L22 with EBER 1 in comparison to an unrelated RNA derived from Herpesvirus saimiri, HSUR 3 (Lee et al. 1988). Increasing concentrations of untagged recombinant L22 (rL22) were incubated with either 5 nM 32P-labeled full-length EBER 1 (Fig. 1A) or HSUR 3 (Fig. 1B). Three distinct protein-dependent mobility shifts were observed for EBER 1 (Fig. 1C, lanes 8–11), suggesting that the dominant species present at 1 μM rL22 is the triply bound RNA. An additional diffuse shift can also be seen at 3 μM rL22. In contrast, HSUR 3 begins to shift at protein concentrations only at or above this level (Fig. 1C, lanes 2–6). Collectively, these data argue that EBER 1 shifts 1, 2, and 3 (Fig. 1C, lanes 8–11) represent specific rL22–EBER 1 complexes, whereas the additional diffuse shift most likely arises from nonspecific electrostatic association with the RNA at high protein concentrations.
The three distinct mobility shifts (Fig. 1C) suggest that rL22 forms at least three distinct complexes with wild-type EBER 1. One interpretation is that wild-type EBER 1 possesses three unique L22 binding sites. However, it cannot be assumed that each shift represents a unique molecular complex; a single shift could contain heterogeneous mixtures of different complexes that travel with the same mobility in native polyacrylamide gels. Immunoprecipitation studies previously indicated that stem–loop III of EBER 1 binds L22 (Toczyski and Steitz 1993), and SELEX and EMSAs (Dobbelstein and Shenk 1995) suggested stem–loop IV as an additional L22 protein binding site. To verify these and identify other RNA elements within EBER 1 important for binding L22, we performed a series of mobility-shift experiments using EBER 1 deletion constructs in which each of the stem–loops was deleted independently (Fig. 2B–E).
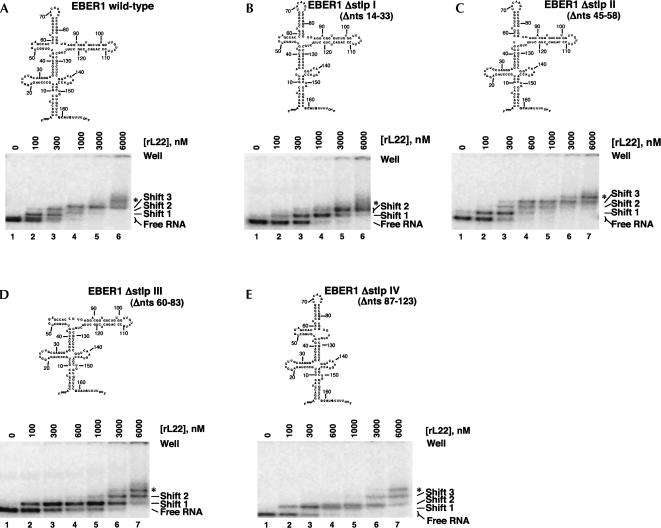
FIGURE 2.
EMSAs performed with EBER 1 deletion constructs identify stem–loops I, III, and IV as binding sites for L22 on EBER 1. Predicted secondary structures of the EBER 1 deletion RNAs tested (top) and EMSAs performed with 5 nM RNA (body-labeled) and increasing concentrations of L22 (bottom). (A) Full-length EBER 1, (B) EBER 1 Δstlp I (Δnts 14–33), (C) EBER 1 Δstlp II (Δnts 45–58), (D) EBER 1 Δstlp III (Δnts 60–83), (E) EBER 1 Δstlp IV (Δnts 87–123). The deletion mutants are numbered according to the full-length RNA. An asterisk (*) indicates an additional, nonspecific shift that only appears at L22 concentrations ≥3 μM.
Interestingly, single deletion of three of the stem–loops of EBER 1 pictured in Figure 1A abolished or greatly hindered the formation of the three specific shifts. These were stem–loop I (EBER 1 Δstlp I, Δnts 14–33; Fig. 2B), stem–loop III (EBER 1 Δstlp III, Δnts 60–83; Fig. 2D), and stem–loop IV (EBER 1 Δstlp IV, Δnts 87–123; Fig. 2E). In contrast, deletion of stem–loop II from EBER 1 (EBER 1 Δstlp II, Δnts 45–58; Fig. 2C) produced an RNA with rL22-binding behavior qualitatively identical to that of wild-type EBER 1 (Fig. 2A). These differences are best exemplified by examination of EBER 1 shifts in the presence of 1 μM rL22. The majority of the signal from full-length (Fig. 2A, lane 4) and Δstlp II RNAs (Fig. 2C, lane 5) is found in shift 3. In contrast, the bulk of the signal from EBER 1 Δstlp I (Fig. 2B, lane 4) or EBER 1 Δstlp III (Fig. 2D, lane 5) is found in shift 1, while that from EBER 1 Δstlp IV (Fig. 2E, lane 5) is found in shift 2.
These results corroborate the previous findings that stem–loops III and IV are essential for full rL22 binding to EBER 1 and suggest an additional, previously undescribed protein binding site in stem–loop I. It was surprising, however, that deletion of stem–loop III did not appear to retard the formation of the first protein shift. In fact, the first protein shift was observed even at the lowest concentration (0.1 μM) shown for all single-loop deletion constructs, suggesting malleability in the rL22–EBER 1 interaction.
EBER 1 binds three protein molecules simultaneously
The protein-concentration dependence of the appearance of the mobility shifts, in concert with the presence of binding sites on stem–loops I, III, and IV, suggests that EBER 1 can bind multiple copies of rL22 simultaneously. To determine the protein/RNA stoichiometry for shifts 1–3, binding studies were performed using a mixture of rL22 (15 kDa) and MBP-L22 fusion protein (56 kDa) (Fig. 3). EBER 1 Δstlp II was substituted for full-length EBER 1, as it exhibited very similar binding behavior with both rL22 (Fig. 2A,C) and MBP-L22 (data not shown). However, the increased resolution with this smaller RNA between bound species greatly facilitated interpretation of the results. In the presence of either rL22 or MBP-L22 alone, three specific concentration-dependent bands appeared (Fig. 3, lanes 2–5 or 9–11, respectively). When EBER 1 Δstlp II was incubated with a mixture of the two proteins (Fig. 3, lanes 6–8), these individual shifts were observed, as well as three distinct intermediate shifts. The formation of a band intermediate to a shift seen with each protein alone would suggest that two (or more) molecules of protein bind.
FIGURE 3.
Shifts 2 and 3 contain multiple L22 molecules. EMSAs performed with 5 nM body-labeled EBER 1 Δstlp II and the indicated concentrations of rL22 and/or MBP-L22 fusion protein. EBER 1 Δstlp II was chosen for the mixing experiment, as it exhibits L22-binding behavior similar to full-length EBER 1 but better separation between protein-bound species. Mixed shift 1 is presumed to comprise one rL22 and one MBP-L22 molecule bound to the RNA, with two rL22 and one MBP-L22 molecules in mixed shift 2, and one rL22 and two MBP-L22 molecules in mixed shift 3. An asterisk (*) indicates a minor multiply shifted species.
The simplest interpretation of the above mixing experiment (Fig. 3) is that protein shifts 1, 2, and 3 contain one, two, and three of the relevant L22 molecules bound to EBER 1, respectively, while the intermediate shifts contain both rL22 and MBP-L22 molecules simultaneously bound to EBER 1. Mixed shift 1 represents EBER 1 bound to two L22 moieties (one molecule of rL22 and one molecule of MBP-L22), while mixed shifts 2 and 3 represent EBER 1 bound to three L22 moieties (two rL22 and one MBP-L22 molecules in mixed shift 2, and one rL22 and two MBP-L22 molecules in mixed shift 3). Although both mixed shifts 2 and 3 contain three protein molecules, the latter migrates significantly more slowly, consistent with its proposed greater bulk.
EBER 1 fragments bind rL22
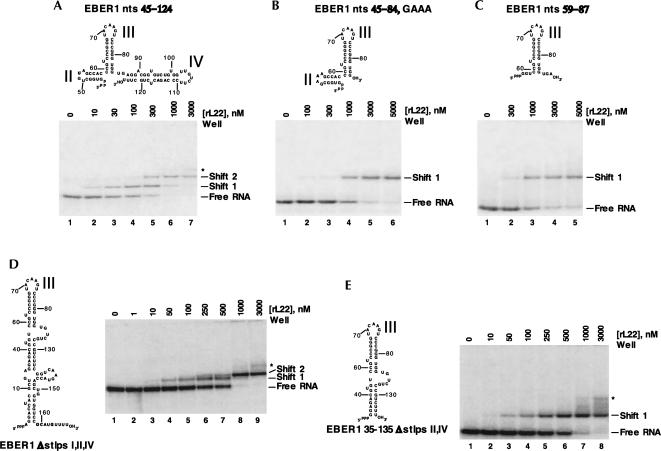
To probe the rL22-EBER 1 interactions further, we performed mobility-shift assays with several EBER 1 fragments. EBER 1 nts 45–124, which comprises stem–loops II–IV of EBER 1, formed two shifts with rL22. These shifts appeared at similar rL22 concentrations to the first two shifts of full-length EBER 1. Shortening stem–loop IV reduced the association of the RNA fragment with the first protein molecule and abrogated the second shift (data not shown), supporting the conclusion that stem–loop IV is an rL22-binding site. Identical results were seen upon substituting the loop of stem–loop II, which was shown above not to be essential for protein binding, with a GAAA tetraloop (data not shown). Our rationale was that a smaller construct comprised of just stem–loops II and III would use this GAAA tetraloop in stem–loop II to promote proper RNA folding. Again, this construct showed only one shift with rL22 (Fig. 4B), but with slightly reduced affinity for rL22. This suggests that elements in the base of stem–loop IV might be necessary either for direct binding to rL22 or for correct presentation of stem–loop III to the rL22-binding surface. Similar results were seen upon removal of stem–loop II entirely (Fig. 4C).
FIGURE 4.
EMSAs investigating the interaction of L22 with EBER 1 fragments. Predicted secondary structures of EBER 1 truncations and EMSAs performed with 5 nM RNA and increasing concentrations of L22 are shown. (A) EBER 1 nts 45–124, (B) EBER 1 nts 45–84, GAAA, (C) EBER 1 nts 59–87, (D) EBER 1 Δstlps I, II, and IV, and (E) EBER 1 35–135 Δstlps II and IV. The deletion mutants are numbered according to the full-length RNA. EMSAs were performed with body-labeled RNAs, with the exception of the constructs in A and E, which were 5′-end labeled. An asterisk (*) indicates an additional, nonspecific shift that only appears at very high L22 concentrations. As stlp II was shown not to be involved in L22 binding, its loop was replaced with a GAAA tetraloop to promote correct folding of the construct studied in B.
Another series of EBER 1 fragments removed stem–loops I, II, and IV from the backbone of the RNA molecule. Unexpectedly, EBER I Δstlps I, II, and IV formed two RNA–protein complexes in mobility-shift assays (Fig. 4D). This behavior may be explained by the presence of two energetically comparable RNA conformers, one of which contains a novel binding site for L22 (see Discussion). Shortening the terminal RNA stem abolished the second shift without affecting the binding affinity of the first shift (Fig. 4E; data not shown).
Stem–loops I, II, and IV can individually bind rL22
Although the data in Figures 2 and 4 argue that stem–loops I, III, and IV are necessary for L22 binding to EBER 1, they do not address whether these regions are sufficient. Because stem–loop III alone binds poorly to rL22 even at 10 μM concentration (data not shown) and decreases in binding affinity were observed when stem–loop IV was deleted (Fig. 4, cf. A and B/C), we reasoned that the stem–loops of EBER 1 might require the context of a larger RNA molecule to fold properly. We therefore performed binding assays on the individual EBER 1 stem–loops inserted (see Fig. 1B for site of insertion) into the nonbinding HSUR 3 molecule to form HSUR 3 stem–loop chimeras.
The insertion of EBER 1 stem–loop I (nts 14–33) (Fig. 5, lanes 6,7) or IV (nts 87–123) (Fig. 5, lanes 14,15) into HSUR 3 each produced a single-shifted complex with rL22. The same amounts (1 and 4 μM) of rL22 did not yield a specific shift with HSUR 3 alone (Fig. 5, lanes 3,4) or with the HSUR 3 stem–loop II chimera (data not shown). We conclude that stem–loops I and IV can independently serve as binding sites for rL22. Interestingly, the HSUR 3 stem–loop III (nts 59–84) chimera consistently yielded two differently migrating forms of the free RNA (Fig. 5, lane 8) in multiple RNA preparations purified from polyacrylamide gels, arguing that it adopts two major conformations (see Discussion). Increasing amounts of rL22 shifted both RNA conformers into specific complexes (Fig. 5, lanes 9–11) but with more background than the profiles of the HSUR 3 stem–loop I or IV chimeras. The diminution of the free HSUR 3 stem–loop III band even at 0.4 μM rL22 (Fig. 5, lanes 9–11) argues that stem–loop III is an L22-binding site. Taken together, these data support the idea that EBER 1 stem–loops I, III, and IV can individually bind L22.
FIGURE 5.
L22 interacts with HSUR 3 chimeras containing individual EBER 1 stem–loop sequences. EMSAs were performed with 5 nM HSUR 3 without an insert (lanes 1–3) and HSUR 3 chimeras containing either stem–loop I (lanes 4–7), stem–loop III (lanes 8–11), or stem–loop IV (lanes 12–15) in the presence of the indicated concentrations of rL22 protein. Note that HSUR 3 stem–loop III contains two forms of the free RNA; their respective shifts are marked with single (*) and double (**) asterisks. The ** shift sometimes, but not always, appears as a doublet.
EBER 1 binds multiple L22 Proteins in vivo
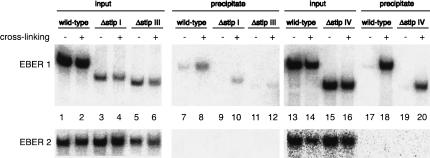
Having established that EBER 1 contains three L22-binding sites in vitro, we wished to determine whether multiple L22 proteins likewise interact with EBER 1 inside transformed cells where other EBER 1-binding proteins could potentially compete. We used a powerful in vivo UV cross-linking method (Cook et al. 2004) in which anti-L22 antibody was used to immunoprecipitate cross-linked EBER 1–L22 complexes under semidenaturing conditions. When a DNA construct expressing wild-type EBER 1 and EBER 2 from their native promoters was transfected into HEK 293 cells, EBER 1 was detected in the anti-L22 precipitate in a cross-linking-dependent manner (Fig. 6, top, cf. lanes 8,18 and 7,17).
FIGURE 6.
In vivo interactions between L22 and EBER 1 deletion constructs. HEK 293 cells were transfected with the following EBER 1 and EBER 2 expression constructs: wild-type EBER 1 (lanes 1,2,7,8,13,14,17,18), Δstlp I (lanes 3,4,9,10), Δstlp III (lanes 5,6,11,12), or Δstlp IV (lanes 15,16,19,20). Intact cells were irradiated with 254-nm UV light to induce cross-linking between protein and RNA in vivo (even-numbered lanes); untreated cells served as negative controls (odd-numbered lanes). Whole-cell lysates were subjected to immunoprecipitation with anti-L22 antibody under denaturing conditions. Precipitated RNAs were released from the antibody by proteinase K digestion and were resolved in an 8% urea polyacrylamide gel. A total of 0.5% of the input and 100% of the precipitates were analyzed, and EBER 1 sequences were detected by Northern blotting (top). The low expression levels of EBER 1 Δstlp I (lanes 3,4) and Δstlp III (lanes 5,6) are due to the disruption of Box A or Box B of the EBER 1 RNA polymerase III promoter, respectively. To control for nonspecific cross-linking and immunoprecipitation, the membranes were stripped and reprobed for EBER 2 sequences (bottom).
If EBER 1 binds multiple copies of the L22 protein in vivo, deletion of any one binding site from the RNA should not abolish precipitation after UV cross-linking. Indeed, upon expression in HEK 293 cells and UV cross-linking, EBER 1 Δstlp I (Fig. 6, top, lane 10), EBER 1 Δstlp III (Fig. 6, top, lane 12), and EBER 1 Δstlp IV (Fig. 6, top, lane 20) were detected in the anti-L22 precipitate. The specificity of these interactions is demonstrated by the absence of EBER 2 in the precipitates (Fig. 6, bottom). We conclude that multiple L22 molecules are bound to EBER 1 inside cells, as in vitro, but we cannot distinguish whether two or more L22-binding sites exist.
DISCUSSION
In vitro binding experiments presented here demonstrate that wild-type EBER 1 forms three distinct mobility shifts with rL22 or MPB-L22 that are protein-concentration dependent (Fig. 1; data not shown). The association is direct since it occurs in the absence of any other viral or host proteins and does not require post-translational modifications. The three distinct RNP complexes observed differ in the total number of bound L22 protein molecules; shifts 1, 2, and 3 appear to contain one, two, and three L22 molecules bound to EBER 1, respectively (Fig. 3). Because the deletion of EBER 1 stem–loop I, III, or IV greatly affects the formation of a third complex (Fig. 2), these three stem–loops appear to comprise the three binding sites for L22 in vitro. Indeed, our data agree with previous observations that stem–loops III and IV are L22 binding sites (Toczyski and Steitz 1993; Dobbelstein and Shenk 1995). Insertion of stem–loop I into HSUR 3 confers binding activity on this unrelated nonbinding RNA, confirming that stem–loop I is a binding site for L22 (Fig. 5). Finally, in vivo cross-linking experiments show that there are also multiple L22-binding sites on EBER 1 inside cells (Fig. 6).
Can stem–loops be assigned to particular shifts?
With the wild-type EBER 1 molecule, we estimate the apparent binding constants (Kd) for the first, second, and third shifts to be 0.1, 0.3, and 1 μM, respectively. In an attempt to assign these shifts to particular binding sites, we first examined fragments of EBER 1. We constructed two small stem–loop III-containing fragments, EBER 1 nts 45–84, GAAA (Fig. 4B) and EBER 1 nts 59–87 (Fig. 4C), and estimated that stem–loop III has a Kd of 1 μM. However, in a molecule that contains stem–loops II, III, and IV (EBER 1 nts 45–124; Fig. 4A) the first and second shifts have Kds of 0.1 and 1 μM, respectively. These affinities suggest that shift 1 with EBER 1 nts 45–124 corresponds primarily to L22 binding at stem–loop IV, with an additional L22 molecule at stem–loop III to form shift 2. Alternatively, additional sequences from stem–loop IV might allow stem–loop III to bind L22 with a higher affinity, perhaps by facilitating proper RNA folding. In this case, shift 1 on EBER 1 nts 45–124 would correspond primarily to L22 binding at stem–loop III.
We used M-fold (Zucker 2003) to predict RNA secondary structures for wild-type EBER 1 and all of the deletion construct RNAs used in Figures 2 and 4. M-fold confirmed the structures shown in all cases except for EBER 1 Δstlps I, II, and IV (Fig. 4D). Here, an alternate structure of comparable predicted stability can form by refolding nts 127–138 to create a hairpin loop with a 4-bp stem closed by GC and a U in the 3′ position of the loop, conforming to the consensus motif for L22 binding (Fig. 1D). L22 binding both to stem–loop III and to this novel stem–loop in the molecule would then explain the two shifts seen in the EMSA profile of this molecule (Fig. 4D). Moreover, this structure would not form with EBER 1 35–135 Δstlps II and IV (Fig. 4E), explaining loss of the second shift.
Because individual stem–loops of EBER 1 may require the context of a larger RNA molecule to fold properly, we also inserted them into a nonspecific RNA, HSUR 3. Curiously, the HSUR 3 stlp III chimera assumed two major conformations (Fig. 5, lane 8), again predicted by M-fold (data not shown). While the two folds are dramatically different, they both retain stem–loop III. Thus, both conformers are shifted (Fig. 5, lanes 9–11) and the estimated Kds, based on the reduction of the free RNA signals, are <1 μM. The estimated Kd for the HSUR 3 stlp IV chimera (whose predicted structure is consistent with M-fold) is 2 μM (Fig. 5). However, for HSUR 3 stlp I, M-fold predicts four isoenergetic structures, three of which lack stem–loop I. Its apparent Kd of >4 μM (Fig. 5, lanes 4–7) is therefore not meaningful. Thus, we can only conclude that, when inserted into HSUR 3, stem–loop III exhibits stronger L22 binding than stem–loop IV. It is striking that the estimated Kds for the individual stem–loop chimeras (Fig. 5) are all at least fourfold lower than when the same stem–loops are part of EBER 1 (Figs. 1C, 2A).
Given the results with both the EBER 1 fragments and with the HSUR 3-EBER 1 chimeras, can we assign the various shifts to L22 interactions with specific stem–loops? Figure 2 shows that the individual deletion of stem–loops I, III, or IV surprisingly has no effect on the appearance of shift 1. Thus, it is impossible to deduce which stem–loop is the strongest binding site for L22 in the context of the full-length RNA. Together our results indicate that any major (Fig. 4) or minor (Fig. 2) deletion of EBER 1 or the isolation of any stem–loop from the rest of the molecule (Fig. 5) negatively affects the interaction between L22 and EBER 1 stem–loops. Therefore, we conclude from our results only that wild-type EBER 1 has three in vitro L22-binding sites, whose relative affinities remain unknown. It is quite possible that each shift represents a mixture with L22 bound to several of its binding sites.
What molecular features does L22 recognize?
SELEX experiments predicted a consensus RNA structure that enables specific interactions between L22 and RNA (Dobbelstein and Shenk 1995; Fig. 1D). The domains of EBER 1 that fit this motif best are stem–loops III and IV. Since stem–loop III was already established as a binding site (Toczyski and Steitz 1993), Dobbelstein and Shenk (1995) proposed stem–loop IV as the additional binding site on EBER 1. EMSAs performed with isolated stem–loop IV in the absence of competitor RNA indeed demonstrated weak binding that could be competed by the addition of isolated stem–loop III (Dobbelstein and Shenk 1995).
Here we have identified stem–loop I of EBER 1 as a third binding site for L22. This assignment is supported by a comparison of the EBER 1 stem–loop I to a sequence from the 3′X sense strand of HCV genomic RNA that is reported to bind L22 (Wood et al. 2001); nt 19–25 within the 3′X sense strand are identical to nt 13–19 in EBER 1 stem–loop I (Wood et al. 2001). Yet, stem–loop I of EBER 1 does not fit the predicted consensus sequence (Dobbelstein and Shenk 1995) entirely (see Fig. 1A,D); it maintains a GC base pair at the apical end, with the G 5′ and the C 3′, but lacks the U residue in the 3′ position of the loop. A common feature is that the stems of both stem–loops I and III consist of 6 canonical bp, with no bulged residues. The identification of more recognition sites for L22, including that in the large subunit of the ribosome, would provide additional insights into the consensus-binding sequence and structure.
Significance of the association between EBER 1 and ribosomal protein L22
The high abundance of EBER 1 and EBER 2 in EBV-infected cells, together with their evolutionary conservation (Arrand and Rymo 1982; Howe and Shu 1988), strongly suggests that these RNAs contribute importantly to the EBV life cycle. The association between EBER 1 and ribosomal protein L22 immediately suggests that the EBERs might modulate translation. Based on the estimate that the intracellular concentration of L22 (∼50 μM) is 10-fold that of EBER 1, nearly all of EBER 1 is predicted to be bound by multiple molecules of L22 in vivo. This is confirmed by the in vivo cross-linking data in Figure 6. Since depletion of L22 from the ribosomes of EBV-transformed cells was not experimentally observed (Toczyski et al. 1994), L22 levels may be selectively up-regulated in EBV-infected cells.
It is also possible that the sequestering of L22 by EBER 1 is not important, but rather that the RNP consisting of EBER 1, L22, and additional host proteins fulfills some specific function in EBV-transformed cells. EBER 1 has been shown to interact stably with the La antigen (Lerner et al. 1981) and PKR (Clarke et al. 1991; Sharp et al. 1993; Clemens et al. 1994; Elia et al. 1996; Clemens and Elia 1997; Vuyisich et al. 2002). La is sufficiently abundant (∼2 × 107 molecules/cell) that binding by EBERs (∼106 molecules/cell) should not sequester La away from other roles in the cell (Wolin and Cedervall 2002). On the other hand, PKR is much less abundant (Clemens and Elia 1997) and its binding by EBER 1 could block its ability to inhibit protein synthesis in infected cells (Clarke et al. 1991; Sharp et al. 1993; Clemens et al. 1994). Recently, this model has been extended to suggest that L22 may exert a protective effect against the transforming potential of EBER 1 in vivo by making EBER 1 unavailable for binding to PKR (Elia et al. 2004). This hypothesis rests on the idea that L22 and PKR compete for a common binding site on EBER 1, namely stem–loop IV (Dobbelstein and Shenk 1995; Vuyisich et al. 2002), while other studies, in contrast (Ruf et al. 2005; Wang et al. 2005), do not find evidence for interactions between EBER 1 and PKR. Clearly, there is much more to be learned concerning the role of the EBER 1 RNP in EBV-transformed cells.
MATERIALS AND METHODS
Plasmid construction
Wild-type EBER 1 was previously cloned into a pUC119 vector (Toczyski and Steitz 1993). This gene fragment (225 nt) was removed from the pUC119 vector and placed into a pUC19 vector. Briefly, the EBER 1 gene (167 nt), preceded by an EcoRI restriction site and a T7 RNA polymerase promoter (23 nt) and followed by a HindIII restriction enzyme site, was excised from the pUC119 vector by cleavage with the enzymes EcoRI and HindIII and inserted into the EcoRI and HindIII sites of pUC19. All EBER 1 stem–loop deletion plasmids were synthesized using the QuikChange Site-Directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. Wild-type EBER 1 plasmid was used as the DNA template in all cases, unless otherwise specified. The following 5′ oligonucleotides were used to create the corresponding deletion constructs (with 3′ oligonucleotides complementary to their respective 5′ oligonucleotides): EBER 1 Δstlp I, 5′-AGGACCTACGCTGAGGAGACGTGTGTGGCTG; EBER 1 Δstlp II, 5′-GCTAGGGAGGAGACGTGTCCGTCCCGGGTACAAGT; EBER 1 Δstlp III, 5′-CGTGTGTGGCTGTAGCCACCGTGAGGACGGTGTCTGTG. EBER 1 Δstlp IV was constructed in two rounds. The first round removed nt 106–123 using 5′-GGACGGTGTCTGTGGTTGTCTGCCGTCTTCGGTC. The second round used this EBER 1 plasmid as the DNA template and removed nt 87–105 using 5′-CAAGTCCCGGGTGGTGTCTGCCGTCTTCGGTC. EBER 1 Δstlps (I, II, and IV) used EBER 1 Δstlp IV plasmid as the DNA template and removed stem–loops I and II sequentially using the corresponding oligonucleotides described above. EBER 1 35–135 Δstlp II, IV was generated by PCR amplification using EBER 1 Δstlps (I, II, and IV) plasmid as the DNA template and the following primers: 5′ primer-GGGAATTCTAATACGACTCACTATAGGAGACGUGUGUGGCUGU, which contains an EcoRI restriction site followed by a T7 RNA polymerase promoter, and 3′ primer-CGCCGGATCCTTTAAAGACGGCAGACACCACC, which contains DraI and BamHI restriction sites. The resulting product was cloned into a pUC19 plasmid using the EcoRI and BamHI restriction sites. EBER 1 45–124 was generated by PCR amplification using wild-type EBER 1 plasmid and the following primers: 5′ primer-GGGAATTCTAATACGACTCACTATAGUGGCUGUAGCCACCC, which contains an EcoRI restriction site, followed by a T7 RNA polymerase promoter, and 3′ primer-CGCCAAGCTTTAAAGCAGAGUCUGGGAAGAC, which contains DraI and HindIII restriction sites. The resultant product was cloned into a pUC19 plasmid using the EcoRI and HindIII restriction sites. For EBER 1 nts 45–84, GAAA and EBER 1 nts 59–87, DNA templates containing a T7 RNA polymerase promoter followed by the relevant sequence were synthesized by the Keck Facility at Yale University and purified on a 15% denaturing polyacrylamide gel. Stem–loops were added to HSUR 3 by ligating two DNA inserts (5′ and 3′ fragments) into the HSUR 3 construct using the EcoRI and SmaI restriction sites. The Herpesvirus saimiri HSUR 3 plasmid was previously constructed (Myer et al. 1992) and was used as PCR template. The 5′ fragments of HSUR 3 stlp I, HSUR 3 stlp III, and HSUR 3 stlp IV were generated by PCR amplification with 5′-CCTCTAGGGCTTGGGTTGTTAATCTCCTA, 5′-TGTACCCGGGACGGCTTGGGTTGTTAATCTCCTA, and 5′-CAACCACAGACACCGTCCTCCTTGGGTTGTTGTTAATCTCCTA, respectively, and T7 primer. The 3′ fragments of HSUR 3 stlp I, HSUR 3 stlp III, and HSUR 3 stlp IV were generated by PCR amplification with 5′-TTTTGCTAGGGAATAATTTTTGAAGGCTCTGG, 5′-AGTCCCGGGTGGAATAATTTTTGAAGGCTCTGG, and 5′-TCTTCCCAGACTCTGCTTTAATAATTTTTGAAGGCTCTGG, respectively, and SP6 primer.
For the in vivo UV cross-linking experiments, EBER 1 and its deletion variants were expressed using their endogenous promoters from a plasmid encoding both EBER 1 and EBER 2. p73EBERs was constructed by ligating a 1.1-kb DNA fragment from the EcoRI J fragment of EBV into pSP73 (Promega) using the SacI and BamHI restriction sites. Stem–loops were deleted from EBER 1 by ligating two DNA inserts (5′ and 3′ fragments) into p73EBERs using the EcoRI and BamHI restriction sites. The 5′ fragments of Δstlp I, Δstlp III, and Δstlp IV were generated by PCR amplification with 5′-AGGAGACGTGTGTGGCTG, 5′-TGAGGACGGTGTCTGTGG, and 5′-TCTGCCGTCTTCGGTCAA, respectively, and an SP6 primer. The 3′ fragments of Δstlp I, Δstlp III, and Δstlp IV were generated by PCR amplification with 5′-GCGTAGGTCCTGAGGTTCA, 5′-GTGGCTACAGCCACACAC, and 5′-CACCACCCGGGACTTGTACC, respectively, and a T7 primer. DNA sequences of all plasmids were verified by the Keck Facility at Yale University.
Protein expression and purification
Overexpression of recombinant L22 (rL22) and MBP-L22 proteins followed a similar procedure. A total of 0.4–1.2 L of LB medium containing 50 μg/mL ampicillin and 34 μg/mL chloramphenicol were inoculated with 0.5–1 mL of an overnight culture of Rosetta (DE3) pLysS cells (Novagen) harboring an L22 expression plasmid. Cells were grown at 37°C for 3.5–5 h to an A595 of 0.6–0.7, and IPTG was added to a final concentration of 1 mM. Protein expression was allowed for 2 h at 37°C. Cells were harvested by centrifugation and resuspended at A595 of 30–60 in 50 mM Tris-HCl at pH 8.0/10% sucrose for rL22 or in column buffer (20 mM Tris-HCl at pH 7.4, 200 mM NaCl, 1 mM EDTA, and 10% glycerol) for MBP-L22 proteins before being stored at −80°C. Frozen cells were thawed in an ice-water bath. All subsequent steps were carried out at 4°C. Protease inhibitor cocktail (Calbiochem) and a final concentration of 1–4 mM EDTA were added to the thawed cells. Lysed cells were cleared by centrifugation at 100,000g for 1 h at 4°C. Both L22 proteins were purified as described below to 90%–95% homogeneity as judged by SDS-PAGE. The calculated extinction coefficients (ε280) for rL22, 1209 M−1 cm−1, and for MBP-L22, 76,810 M−1cm−1, were used to determine protein concentrations.
Purification of recombinant L22 protein
Plasmid pET11a-L22 constructed by PCR amplification of the L22-encoding sequence and ligation into the pET11a vector was used for expression of rL22. The DNA sequence was confirmed by the Keck Facility. Cleared cell lysates were adjusted to Buffer A (50 mM HEPES at pH 7.9, 4 mM EDTA, and 10% glycerol) containing 200 mM NaCl and loaded onto a 1-mL SP-Sepharose Fast Flow column equilibrated with the same buffer. The column was washed with 1 mL Buffer A containing 200 mM NaCl and then 10 mL Buffer A containing 400 mM NaCl. rL22 was eluted in 6 mL Buffer A containing 900 mM NaCl. The purified rL22 protein was dialyzed twice against 25 mM HEPES (pH 7.9), 50 mM NaCl, 2 mM DTT, and 10% glycerol for 4–12 h.
Cloning and purification of MBP-L22 fusion protein
The L22 encoding sequence was PCR amplified from the GST-L22 plasmid (Toczyski and Steitz 1993) using a 5′ primer-GCGGAATTCGCCCCCGTGAAGAAGCTGTG and a 3′ primer-TTTCACCGTCATCACCGAA. The resulting PCR product was restriction digested with EcoRI and SalI enzymes and ligated into pMAL-c2X vector (NEB) cut with the same enzymes. The MBP-L22 sequence was confirmed by DNA sequencing (Keck Facility).
Before centrifugation, thawed cells were sonicated on ice for 10 sec twice, 20-sec apart, with a Branson Sonifier at setting 3 and 100% duty cycle. The soluble lysates were diluted with 20 mL column buffer and then loaded onto a 4-mL amylose column (NEB) equilibrated with column buffer. The column was washed with 60 mL column buffer and the bound MBP-L22 was eluted in 6 mL column buffer containing 10 mM maltose. The purified MBP-L22 protein was dialyzed twice against 25 mM HEPES (pH 7.5), 50 mM NaCl, 2 mM DTT, and 10% glycerol for 4–8 h. A fraction of the MBP-L22 preparation may have been inactive, since rL22 exhibits about threefold higher binding affinity to EBER1 as compared with MBP-L22.
In vitro transcription
RNAs for binding studies were prepared by in vitro run-off transcription. Wild-type or deletion EBER 1 plasmids were linearized with DraI, which yields a 3′ end identical to that of EBER 1. HSUR 3 and HSUR 3 chimeras were linearized with SmaI and PmlI, respectively, resulting in the addition of a G nucleotide at the 3′ end. To obtain unlabeled RNAs, in vitro transcription was performed overnight at 37°C in a final volume of 100 μL containing 40 mM Tris-HCl (pH 7.5), 15 mM MgCl2, 4 mM spermidine, 0.05% Triton X-100, 10 mM DTT, 2.5 mM NTPs, 0.05 μg/μL linearized plasmid or DNA template, and 0.1 μg/μL T7 RNA polymerase. The RNA transcripts were purified on 8% denaturing polyacrylamide gels. Gel slices were eluted overnight in TE buffer (10 mM Tris-HCl at pH 7.5, 1 mM EDTA) containing 1% SDS, and then phenol-extracted (pH 4.5), chloroform-extracted twice, and ethanol-precipitated in 0.3 M NaOAc (pH 4.5). Precipitated RNAs were washed with 70% ethanol and dissolved in water. The RNA concentration was determined by measuring the UV absorbance at 260 nm. A total of 50 ρmols of each transcript were treated for 1 h at 37°C with 0.5 U/μL calf intestinal alkaline phosphatase (Roche) to remove the 5′ triphosphate. The RNA was extracted with phenol (pH 4.5), followed by a chloroform extraction and an ethanol precipitation. It was resuspended in water and 5′ phosphorylated by adding 10 U of T4 polynucleotide kinase (PNK), 1 × T4 PNK buffer (Roche), and 150 μCi of [γ-32P]ATP for 45 min at 37°C. Radiolabeled transcripts were purified by separation on denaturing polyacrylamide gels, elution from gel slices, and precipitation as described above. The pelleted RNAs were dissolved in water.
To produce [α-32P]UTP body-labeled RNAs, in vitro transcription was performed for 90 min at 37°C in a final volume of 10 μL containing 40 mM Tris-HCl (pH 7.5), 6 mM MgCl2, 2 mM spermidine, 10 mM DTT, 0.5 mM ATP and CTP, 0.5 or 1.0 mM GTP, 0.1 or 0.05 mM UTP, 20 or 40 μCi of [α-32P]UTP (400 Ci/mmol), 0.1 μg/μL linearized plasmid or 6 ng/μL DNA template, and 0.1 μg/μL T7 RNA polymerase. RNAs were purified as described above.
Electrophoretic mobility-shift assays
The 5′-end-labeled or [α-32P]UTP body-labeled RNAs (5 nM) were heated at 95°C for 3 min, followed by snap cooling on ice for 3 min. Binding reactions (10 μL) containing the indicated amounts of protein were incubated for 30 min on ice in RNA-binding buffer containing 37.5 mM HEPES (pH 7.9), 75 mM NaCl, 5 mM MgCl2, 8% glycerol, and 1 μg Escherichia coli tRNA. Prior to loading, 2 μL of 50% glycerol were added to each sample. The samples were loaded on a pre-electrophoresed (200V, 30 min), 8% native gel in 1X TBE buffer (100 mM Tris, 100 mM borate, 2 mM EDTA). Electrophoresis was carried out at 200V for 3–6 h at 4°C.
UV cross-linking
To detect in vivo cross-linking between L22 and RNA, 20 μg of p73EBERs or its deletion variants were transfected into HEK293 cells at 40%–60% confluency in a 10 cm-diameter plate using the TransIT 293 reagent (Mirus) according to the manufacturer's protocol. After 48 h, the cells were washed twice with cold phosphate-buffered saline and resuspended in 3 mL of phosphate-buffered saline. Cells were irradiated (or not irradiated as a negative control) on ice with 254-nm UV light at a distance of 4 cm for 4.5 min and collected by scraping and centrifugation in Eppendorf tubes. Cell lysates were prepared, and immunoprecipitation with anti-L22 antibody (Toczyski and Steitz 1993) was carried out as described (Cook et al. 2004). Precipitated RNAs were released from the antibody by proteinase K digestion and resolved on an 8% urea polyacrylamide gel. EBER 1 and EBER 2 sequences were detected by Northern blotting using [α-32P]-labeled DNA oligonucleotide probes, EBER 1R152 (5′-CCAGCTGGTACTTGACCGAAGAC) and EBER2 R134 (5′-ATTAGAGAATCCTGACTTGCAAATGCTCT).
ACKNOWLEDGMENTS
We thank Kazio Tycowski and Nikolay Kolev for comments on the manuscript and Enrique De La Cruz, Robert P. Rambo, and Timothy C. Taylor for their contributions to earlier phases of this study. R.M.M-F. is a fellow of the Jane Coffin Childs Fund for Medical Research. This work was supported by grant CA16038 from the NIH. J.A.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2339606.
REFERENCES
- Arrand J.R., Rymo L. Characterization of the major Epstein-Barr virus-specific RNA in Burkitt lymphoma-derived cells. J. Virol. 1982;41:376–389. doi: 10.1128/jvi.41.2.376-389.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrand J.R., Young L.S., Tugwood J.D. Two families of sequences in the small RNA-encoding region of Epstein-Barr virus (EBV) correlate with EBV types A. B. J. Virol. 1989;63:983–986. doi: 10.1128/jvi.63.2.983-986.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barletta J.M., Kingma D.W., Ling Y., Charache P., Mann R.B., Ambinder R.F. Rapid in situ hybridization for the diagnosis of latent Epstein-Barr virus infection. Mol. Cell. Probes. 1993;7:105–109. doi: 10.1006/mcpr.1993.1014. [DOI] [PubMed] [Google Scholar]
- Clarke P.A., Sharp N.A., Clemens M.J. Translational control by the Epstein-Barr virus small RNA EBER-1. Reversal of the double-stranded RNA-induced inhibition of protein synthesis in reticulocyte lysates. Eur. J. Biochem. 1990;193:635–641. doi: 10.1111/j.1432-1033.1990.tb19381.x. [DOI] [PubMed] [Google Scholar]
- Clarke P.A., Schwemmle M., Schickinger J., Hilse K., Clemens M.J. Binding of Epstein-Barr virus small RNA EBER-1 to the double-stranded RNA-activated protein kinase DAI. Nucleic Acids Res. 1991;19:243–248. doi: 10.1093/nar/19.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens M.J., Elia A. The double-stranded RNA-dependent protein kinase PKR: Structure and function. J. Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- Clemens M.J., Laing K.G., Jeffrey I.W., Schofield A., Sharp T.V., Elia A., Matys V., James M.C., Tilleray V.J. Regulation of the interferon-inducible eIF-2 α protein kinase by small RNAs. Biochimie. 1994;76:770–778. doi: 10.1016/0300-9084(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Cook H.L., Mischo H.E., Steitz J.A. The Herpesvirus saimiri small nuclear RNAs recruit AU-rich element-binding proteins but do not alter host AU-rich element-containing mRNA levels in virally transformed T cells. Mol. Cell. Biol. 2004;24:4522–4533. doi: 10.1128/MCB.24.10.4522-4533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelstein M., Shenk T. In vitro selection of RNA ligands for the ribosomal L22 protein associated with Epstein-Barr virus-expressed RNA by using randomized and cDNA-derived RNA libraries. J. Virol. 1995;69:8027–8034. doi: 10.1128/jvi.69.12.8027-8034.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia A., Laing K.G., Schofield A., Tilleray V.J., Clemens M.J. Regulation of the double-stranded RNA-dependent protein kinase PKR by RNAs encoded by a repeated sequence in the Epstein-Barr virus genome. Nucleic Acids Res. 1996;24:4471–4478. doi: 10.1093/nar/24.22.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia A., Vyas J., Laing K.G., Clemens M.J. Ribosomal protein L22 inhibits regulation of cellular activities by the Epstein-Barr virus small RNA EBER-1. Eur. J. Biochem. 2004;271:1895–1905. doi: 10.1111/j.1432-1033.2004.04099.x. [DOI] [PubMed] [Google Scholar]
- Glickman J.N., Howe J.G., Steitz J.A. Structural analyses of EBER 1 and EBER2 ribonucleoprotein particles present in Epstein-Barr virus-infected cells. J. Virol. 1988;62:902–911. doi: 10.1128/jvi.62.3.902-911.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haluska F.G., Russo G., Kant J., Andreef M., Croce C.M. Molecular resemblance of an AIDS-associated lymphoma and endemic Burkitt lymphomas: Implications for their pathogenesis. Proc. Natl. Acad. Sci. 1989;86:8907–8911. doi: 10.1073/pnas.86.22.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle G., Henle W. Immunofluorescence in cells derived from Burkitt's lymphoma. J. Bacteriol. 1966;91:1248–1256. doi: 10.1128/jb.91.3.1248-1256.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle G., Henle W., Diehl V. Relation of Burkitt's tumor-associated herpes-type virus to infectious mononucleosis. Proc. Natl. Acad. Sci. 1968;59:94–101. doi: 10.1073/pnas.59.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe J.G., Shu M.D. Isolation and characterization of the genes for two small RNAs of herpesvirus papio and their comparison with Epstein-Barr virus-encoded EBER RNAs. J. Virol. 1988;62:2790–2798. doi: 10.1128/jvi.62.8.2790-2798.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe J.G., Steitz J.A. Localization of Epstein-Barr virus-encoded small RNAs by in situ hybridization. Proc. Natl. Acad. Sci. 1986;83:9006–9010. doi: 10.1073/pnas.83.23.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson B., Klein G., Henle W., Henle G. Epstein-Barr virus (EBV)-associated antibody patterns in malignant lymphoma and leukemia. I. Hodgkin's disease. Int. J. Cancer. 1970;6:450–462. doi: 10.1002/ijc.2910060316. [DOI] [PubMed] [Google Scholar]
- Johansson B., Killander D., Holm G., Mellstedt H., Henle G., Henle W., Klein G., Soderberg G. Epstein-Barr virus (EBV)-associated antibody patterns in relation to the deficiency of cell-mediated immunity in patients with Hodgkin's disease. IARC Sci. Publ. 1975;11:237–247. [PubMed] [Google Scholar]
- Komano J., Maruo S., Kurozumi K., Oda T., Takada K. Oncogenic role of Epstein-Barr virus-encoded RNAs in Burkitt's lymphoma cell line Akata. J. Virol. 1999;73:9827–9831. doi: 10.1128/jvi.73.12.9827-9831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama Y., Katagiri S., Hanai S., Uchida K., Miwa M. Poly(ADP-ribose) polymerase interacts with novel Drosophila ribosomal proteins, L22 and l23a, with unique histone-like amino-terminal extensions. Gene. 1999;226:339–345. doi: 10.1016/s0378-1119(98)00529-0. [DOI] [PubMed] [Google Scholar]
- Le S., Sternglanz R., Greider C.W. Identification of two RNA-binding proteins associated with human telomerase RNA. Mol. Biol. Cell. 2000;11:999–1010. doi: 10.1091/mbc.11.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.I., Murthy S.C., Trimble J.J., Desrosiers R.C., Steitz J.A. Four novel U RNAs are encoded by a herpesvirus. Cell. 1988;54:599–607. doi: 10.1016/s0092-8674(88)80004-7. [DOI] [PubMed] [Google Scholar]
- Leopardi R., Roizman B. Functional interaction and colocalization of the herpes simplex virus 1 major regulatory protein ICP4 with EAP, a nucleolar-ribosomal protein. Proc. Natl. Acad. Sci. 1996;93:4572–4576. doi: 10.1073/pnas.93.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopardi R., Ward P.L., Ogle W.O., Roizman B. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the U(L)13 protein kinase. J. Virol. 1997;71:1133–1139. doi: 10.1128/jvi.71.2.1133-1139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M.R., Andrews N.C., Miller G., Steitz J.A. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. 1981;78:805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer V.E., Lee S.I., Steitz J.A. Viral small nuclear ribonucleoproteins bind a protein implicated in messenger RNA destabilization. Proc. Natl. Acad. Sci. 1992;89:1296–1300. doi: 10.1073/pnas.89.4.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao A., Yoshihama M., Kenmochi N. RPG: The ribosomal protein gene database. Nucleic Acids Res. 2004;32:D168–D170. doi: 10.1093/nar/gkh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf I.K., Rhyne P.W., Yang C., Cleveland J.L., Sample J.T. Epstein-Barr virus small RNAs potentiate tumorigenicity of Burkitt lymphoma cells independently of an effect on apoptosis. J. Virol. 2000;74:10223–10228. doi: 10.1128/jvi.74.21.10223-10228.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf I.K., Lackey K.A., Warudkar S., Sample J.T. Protection from interferon-induced apoptosis by Epstein-Barr virus small RNAs is not mediated by inhibition of PKR. J. Virol. 2005;79:14562–14569. doi: 10.1128/JVI.79.23.14562-14569.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp T.V., Schwemmle M., Jeffrey I., Laing K., Mellor H., Proud C.G., Hilse K., Clemens M.J. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993;21:4483–4490. doi: 10.1093/nar/21.19.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subar M., Neri A., Inghirami G., Knowles D.M., Dalla-Favera R. Frequent c-myc oncogene activation and infrequent presence of Epstein-Barr virus genome in AIDS-associated lymphoma. Blood. 1988;72:667–671. [PubMed] [Google Scholar]
- Toczyski D.P., Steitz J.A. EAP, a highly conserved cellular protein associated with Epstein-Barr virus small RNAs (EBERs) EMBO J. 1991;10:459–466. doi: 10.1002/j.1460-2075.1991.tb07968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1993The cellular RNA-binding protein EAP recognizes a conserved stem-loop in the Epstein-Barr virus small RNA EBER 1 Mol. Cell. Biol. 13703–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toczyski D.P., Matera A.G., Ward D.C., Steitz J.A. The Epstein-Barr virus (EBV) small RNA EBER 1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc. Natl. Acad. Sci. 1994;91:3463–3467. doi: 10.1073/pnas.91.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Santen V., Cheung A., Kieff E. Epstein-Barr virus RNA VII: Size and direction of transcription of virus-specified cytoplasmic RNAs in a transformed cell line. Proc. Natl. Acad. Sci. 1981;78:1930–1934. doi: 10.1073/pnas.78.3.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuyisich M., Spanggord R.J., Beal P.A. The binding site of the RNA-dependent protein kinase (PKR) on EBER 1 RNA from Epstein-Barr virus. EMBO Rep. 2002;3:622–627. doi: 10.1093/embo-reports/kvf137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xue S.A., Hallden G., Francis J., Yuan M., Griffin B.E., Lemoine N.R. Virus-associated RNA I-deleted adenovirus, a potential oncolytic agent targeting EBV-associated tumors. Cancer Res. 2005;65:1523–1531. doi: 10.1158/0008-5472.CAN-04-3113. [DOI] [PubMed] [Google Scholar]
- Wolin S.L., Cedervall T. The La protein. Annu. Rev. Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- Wood J., Frederickson R.M., Fields S., Patel A.H. Hepatitis C virus 3′X region interacts with human ribosomal proteins. J. Virol. 2001;75:1348–1358. doi: 10.1128/JVI.75.3.1348-1358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M., Kanda T., Takada K. Critical role of Epstein-Barr Virus (EBV)-encoded RNA in efficient EBV-induced B-lymphocyte growth transformation. J. Virol. 2005;79:4298–4307. doi: 10.1128/JVI.79.7.4298-4307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Takizawa T., Iwanaga Y., Shimizu N. Malignant transformation of B lymphoma cell line BJAB by Epstein-Barr virus-encoded small RNAs. FEBS Lett. 2000;484:153–158. doi: 10.1016/s0014-5793(00)02145-1. [DOI] [PubMed] [Google Scholar]
- Zucker M. M-fold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H., Schulte-Holthausen H., Klein G., Henle W., Henle G., Clifford P., Santesson L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1056–1058. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]