Abstract
DExH/D proteins catalyze NTP-driven rearrangements of RNA and RNA-protein complexes during most aspects of RNA metabolism. Although the vast majority of DExH/D proteins displays virtually no sequence-specificity when remodeling RNA complexes in vitro, the enzymes clearly distinguish between a large number of RNA and RNP complexes in a physiological context. It is unknown how this discrimination between potential substrates is achieved. Here we show one possible way by which a non-sequence specific DExH/D protein can discriminately remodel similar RNA complexes. We have measured in vitro the disassembly of model RNPs by two distinct DExH/D proteins, DED1 and NPH-II. Both enzymes displace the U1 snRNP from a tightly bound RNA in an active, ATP-dependent fashion. However, DED1 cannot actively displace the protein U1A from its binding site, whereas NPH-II can. The dissociation rate of U1A dictates the rate by which DED1 remodels RNA complexes with U1A bound. We further show that DED1 disassembles RNA complexes with slightly altered U1A binding sites at different rates, but only when U1A is bound to the RNA. These findings suggest that the “inability” to actively displace other proteins from RNA can provide non-sequence specific DExH/D proteins with the capacity to disassemble similar RNA complexes in a discriminatory fashion. In addition, our study illuminates possible mechanisms for protein displacement by DExH/D proteins.
Keywords: helicase, DEAD, DExH/D, RNP, RNPase, RNA
INTRODUCTION
DExH/D proteins are the largest group of enzymes in eukaryotic RNA metabolism (Anantharaman et al. 2002). Proteins from this highly conserved family are essential for numerous ATP-driven conformational changes in ribonucleoprotein (RNP) assemblies, such as the machineries that catalyze pre-mRNA splicing and ribosome biogenesis (Staley and Guthrie 1998; Tanner and Linder 2001). The biological function of many DExH/D proteins correlates with their capacity to unwind purified RNA duplexes in vitro (Tanner and Linder 2001; Schneider et al. 2002). However, while DExH/D proteins act at distinct points in RNA metabolism, the vast majority of enzymes displays virtually no sequence-specificity when unwinding RNA duplexes in vitro (Tanner and Linder 2001). It is unclear how proteins that bind and unwind RNA complexes in a non-sequence specific manner are able to discriminate between the large number of potential RNA or RNP substrates in a cellular context.
Here we present data suggesting that certain DExH/D proteins might function in a discriminatory fashion even in vitro, but only when the enzymes are confronted with RNA-protein complexes, rather than with pure RNA duplexes. DExH/D proteins have recently been shown to directly rearrange RNPs, and it is believed that the ability to remodel RNPs is central to the biological function of DExH/D “helicases” (Linder 2004). To understand scope and mechanism(s) of RNP remodeling by DExH/D enzymes, we have previously started to examine the rearrangement of model RNA-protein complexes in vitro (Jankowsky et al. 2001; Fairman et al. 2004). In the present study we have measured remodeling of additional, non-physiological model RNPs in vitro, using two distinct DExH/D proteins, DED1 from Saccharomyces cerevisiae (Linder 2003) and NPH-II from vaccinia virus (Shuman 1992). We show that both NPH-II and DED1 actively displace the complex U1 snRNP from its target RNA. However, DED1, in contrast to NPH-II, is unable to actively displace the U1A protein from its cognate RNA, which indicates that not all DExH/D proteins are able to actively remodel the same range of RNPs in vitro. Significantly, our data show that DED1 disassembles model RNA complexes bound to U1A at a rate constant that is determined by the dissociation rate constant of the U1A protein. This finding demonstrates that the rate by which DED1 disrupts RNA complexes can be controlled by an RNA binding protein that does not directly interact with DED1. When confronted with a pool of RNA complexes with slightly altered U1A binding sites (that lead to different U1A dissociation rate constants), DED1 disassembles these RNA complexes in a discriminatory fashion, but only in the presence of U1A. These findings suggest that a DExH/D protein can function in a discriminatory manner, provided the enzyme encounters RNA-protein complexes from where it cannot actively displace the protein(s). This observation illuminates one possible means for a non-sequence specific RNA helicase to discriminate between many potential RNA or RNP substrates.
In addition, our study supports the notion that protein displacement by DExH/D proteins is based on ATP-driven function of the enzymes on single-stranded RNA. Our data further suggest that both the architecture of the RNP as well as biochemical properties of the DExH/D protein determine whether a given enzyme can actively disassemble certain RNPs.
RESULTS
Active displacement of the U1 snRNP by NPH-II and DED1
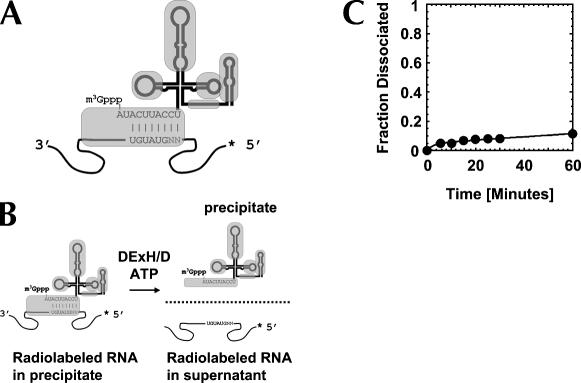
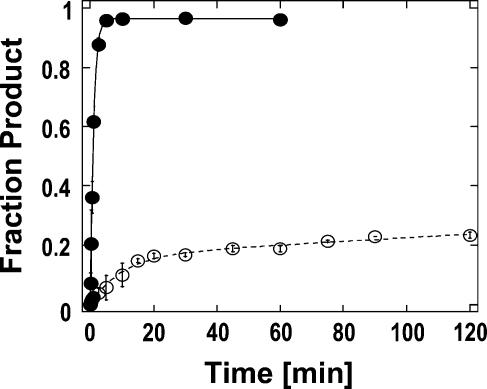
To date, RNP remodeling by DExH/D proteins had been tested only on model substrates where the cognate RNA was bound through RNA-protein interactions (Jankowsky et al. 2001; Fairman et al. 2004). However, in many RNPs, such as in spliceosomal complexes, RNA targets are bound through a combination of both RNA-protein and RNA-RNA interactions (Staley and Guthrie 1998). To examine whether DExH/D proteins are able to disrupt such RNPs, we investigated whether DED1 and NPH-II could displace the U1 snRNP from its target RNA (Fig. 1A,B). The U1 snRNP, which contains 10 proteins and the U1snRNA (Stark et al. 2001), is part of the eukaryotic splicing apparatus where it is involved in the recognition of the 5′ splice site (Will and Lührmann 2001). U1 snRNP binds its cognate RNA at a 5′ splice site through a short helix between the U1 snRNA and through several RNA-protein interactions. Nuclease digestion indicates sequestration of the helix by proteins and extensive protein binding to the RNA substrate 5′ to the helix (P.A. Maroney and T.W. Nilsen, unpubl.). The RNA substrate bound to U1 snRNP with an affinity of Kd ∼4 nM (P.A. Maroney and T.W. Nilsen, unpubl.). The RNA–U1 snRNP complex dissociated with a rate constant of k diss U1 snRNP= (1.2 ± 0.1) × 10−3 min−1 under our reaction conditions (Fig. 1C).
FIGURE 1.
U1 snRNP model system. (A) U1 snRNP bound to RNA containing a 5′ splice site. Lines in the U1 snRNP symbolize the U1 snRNA, gray shapes are U1 snRNP specific proteins on their approximate binding sites (Stark et al. 2001). Substrate RNA is depicted by the curved line; the sequence surrounding the 5′ splice site and the complementary part in the U1 snRNA are indicated. The asterisk shows the radiolabel at the 5′ end of the substrate RNA. (B) Measurement of U1 snRNP dissociation and displacement. U1 snRNP is immunoprecipiated by an antibody against the U1A protein. Thus, radiolabeled substrate RNA bound to U1 snRNP precipitates as well. Radiolabeled substrate RNA released from the U1 snRNP (through spontaneous dissociation or by displacement) is found in the supernatant. (C) Representative time course for spontaneous dissociation of the U1 snRNP from the RNA. Reactions were preformed as described under Materials and Methods and data points were fitted to the integrated rate law for a heterogeneous reaction with two first order components. A fraction of ∼0.05 of the U1 snRNP dissociated faster than experimental accessible, the majority of U1 snRNP (∼95%) dissociated with a rate constant of k D [U1 snRNP] = (1.2 ± 0.1) × 10−3 min−1.
To examine whether NPH-II could displace the U1 snRNP from its RNA target, purified U1 snRNP was bound to a radiolabeled, 75-nucleotide (nt) single-strand RNA containing an authentic 5′ splice site (Fig. 1A). This U1 snRNP–RNA complex was then incubated with NPH-II and ATP. U1 snRNP displacement was monitored by separating released from bound RNA through immunoprecipitation of the U1 snRNP, followed by quantification of the radioactivity in supernatant and precipitate (Fig. 1B). In the presence of NPH-II and ATP, U1 snRNP was displaced with a rate constant of k displ NPH-II ≥ 6 min−1 (Fig. 2A). Without ATP, no displacement beyond spontaneous dissociation of U1 snRNP from the RNA was observed (Fig. 2A). No significant displacement was observed with the non-hydrolysable ATP analog AMPPNP (data not shown; AMPPNP binds to DED1 and promotes RNA binding. In addition, AMPPNP inhibits both ATPase and helicase activities; data not shown).
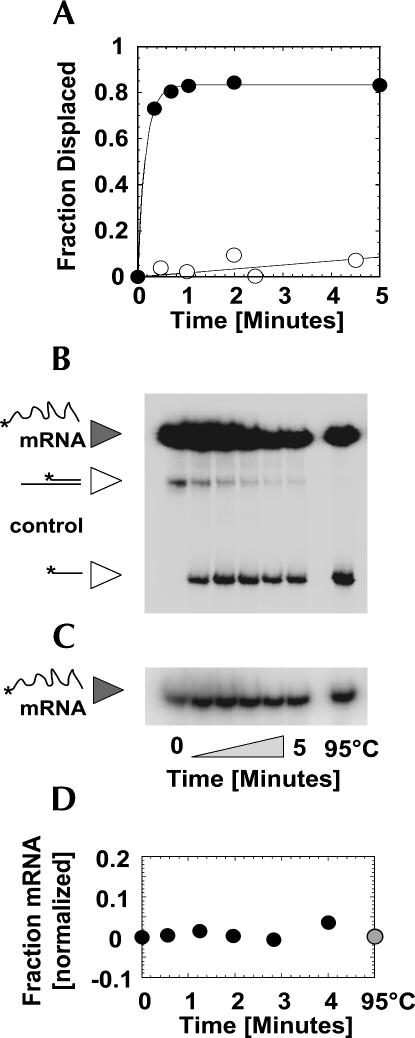
FIGURE 2.
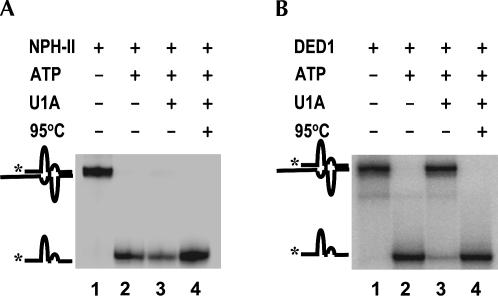
Displacement of the U1 snRNP by NPH-II. (A) Representative time course for U1 snRNP displacement by NPH-II in the presence (filled circles) and in the absence (open circles) of ATP. Data were fitted to the integrated rate law for a first order reaction. With ATP, the displacement rate constant is k displ > 6 min−1 (limit is given since the reaction amplitude has already reached >90% of its final value at the first timepoint). (B) Representative PAGE of an U1 snRNP displacement reaction performed in the presence of a control duplex to test the integrity of the NPH-II helicase activity. The duplex of a control RNA at 0.5 nM is unwound, reaction time is given underneath panel C. No significant degradation of the U1 snRNP substrate RNA (mRNA) is detected. The U1 snRNP substrate RNA is labeled with more radioactivity than the control duplex to minimize the influence of the radiolabeled control RNA on the measurement of U1 snRNP displacement. (C) Representative PAGE of U1 snRNP substrate RNA (mRNA) during the reaction shown in panel B, but at lower detection intensity to illustrate the virtually unchanged level of RNA throughout the reaction. (D) Amount of radioactivity in U1 snRNP substrate RNA normalized to the amount of radioactivity in the control duplex (panel B). The constant value indicates no significant degradation of either RNA during the displacement reaction.
Degradation of the radiolabeled RNA during the displacement reaction was insignificant (Fig. 2B–D), indicating that the increase of radioactivity attributed to free RNA was indeed due to disruption of the RNA–U1 snRNP complex by NPH-II. During the U1 snRNP displacement reaction, the functional integrity of NPH-II was preserved, as evidenced by the full retention of the RNA helicase activity of the enzyme (Fig. 2B). Collectively, these results show that in the presence of ATP, NPH-II increases the rate constant for spontaneous U1 snRNP dissociation by more than three orders of magnitude. That is, NPH-II displaces the U1 snRNP from its target RNA in an active, ATP-dependent fashion.
We then performed an identical displacement reaction with DED1 and ATP (Fig. 3). In the presence of ATP and DED1, the rate constant of U1 snRNP dissociation was also increased by at least three orders of magnitude over the basal dissociation rate constant (Fig. 3). As seen with NPH-II, the fast U1 snRNP displacement by DED1 was strictly ATP-dependent (Fig. 3). DED1 retained its helicase activity under the reaction conditions and no significant hydrolysis of the substrate RNA was detected during the displacement reaction (data not shown). These observations demonstrate that DED1, too, actively dislodged the U1 snRNP from the RNA substrate in an ATP-dependent fashion. We conclude that DExH/D proteins have the capacity to actively remodel RNPs that bind their cognate RNAs through a combination of RNA-RNA and RNA-protein interactions.
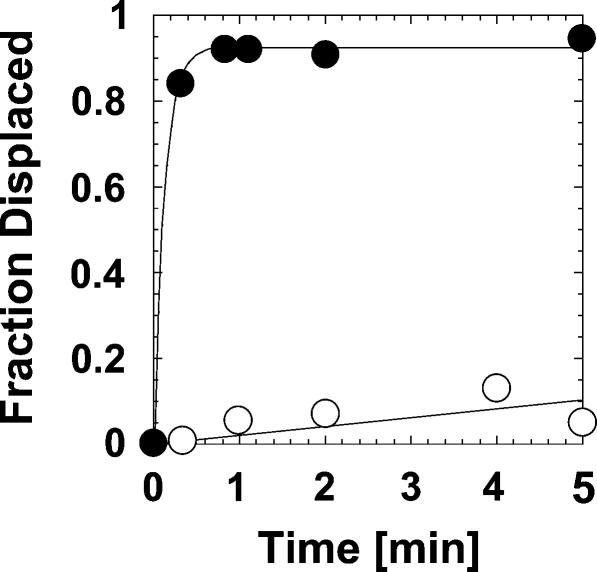
FIGURE 3.
Displacement of the U1 snRNP by DED1. Representative time course for U1 snRNP displacement by DED1 in the presence (filled circles) and in the absence (open circles) of ATP. Data were fitted to the integrated rate law for a first order reaction. With ATP, the displacement rate constant was k displ > 6.4 min−1.
DED1 does not actively displace U1A from RNA
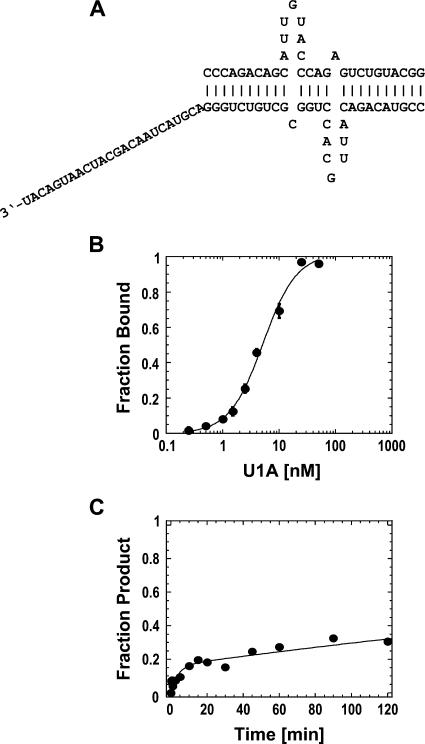
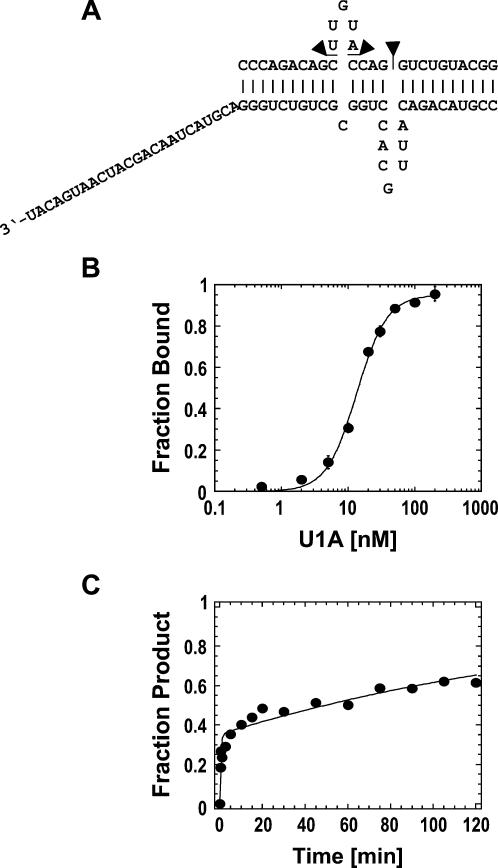
To further investigate the scope of RNP remodeling by different DExH/D proteins, we tested whether DED1 could actively displace the protein U1A from its cognate RNA. NPH-II had been previously shown to actively displace U1A (Jankowsky et al. 2001). In the complex used here, U1A binds its cognate RNA as a homodimer at the single-stranded loops that are embedded into helical structures (Varani et al. 2000). A 24-nt long single-stranded region was appended 3′ end to one helix, in order to provide a binding site for the helicase on the RNP (Fig. 4A). Under our reaction conditions, U1A bound to the RNA with an affinity of K d = 5.1 ± 0.5 nM and dissociated in a biphasic reaction (Fig. 4B,C). The biphasic dissociation kinetics of U1A, which was observed with different U1A preparations, was independent of the U1A concentration, and changes in the reaction conditions did not alter the biphasic shape of the dissociation time course (data not shown). For these reasons, the biphasic dissociation kinetics of U1A is likely to reflect either inherent heterogeneity in the U1A–RNA complex or induced fit binding of U1A to the RNA (Katsamba et al. 2001; Pitici et al. 2002).
FIGURE 4.
U1A-based RNP. (A) RNP design. Sequence of the RNA strands. U1A binds to the single-stranded loops. (B) Equilibrium binding of U1A to the RNA. Data points represent the average of at least three independent measurements. Error bars represent one standard deviation. Data were fit to the Hill-equation (K D = 5.1 ± 0.5 nM, n = 1.4 ± 0.1). (C) Spontaneous dissociation of U1A from the RNA. The representative time course was fit to the sum of two exponentials (dissociation rate constants were, for the first phase: k I d = 0.24 ± 0.1 min−1, and for the second phase: k II d = (1.8 ± 0.2) × 10−3 min−1).
To measure whether DED1 could disassemble the U1A–RNA complex, we followed the separation of the two RNA strands, which, in the presence of U1A, indicates protein removal (Fig. 5; Jankowsky et al. 2001). To ensure the integrity of the U1A–RNA complex, we first conducted the displacement reaction with NPH-II. Consistent with previous results, NPH-II readily unwound the RNA strands with and without U1A bound (Fig. 5A). DED1 readily unwound the RNA complex in the absence of U1A (Fig. 5B). However, in stark contrast to NPH-II, DED1 did not efficiently disassemble the RNA strands with U1A bound (Fig. 5B).
FIGURE 5.
NPH-II but not DED1 actively displaces U1A from the RNA. (A) Displacement of U1A by NPH-II (representative PAGE). Protein displacement is indicated by the ability of the DExH/D protein to separate the RNA strands with U1A bound (lane 3, Jankowsky et al. 2001). Mobilities of the RNA complex and the single-stranded RNA are indicated by cartoons on the left. Reactions were allowed to proceed for 5 min. Lanes are as follows: (1) NPH-II only; (2) NPH-II and ATP; (3) NPH-II and ATP with U1A; (4) boiled control. (B) Inability of DED1 to actively displace U1A from the RNA (representative PAGE). Lanes are as follows: (1) DED1 only; (2) DED1 and ATP; (3) DED1 and ATP with U1A; (4) boiled control.
To further understand these dissimilarities between NPH-II and DED1, we measured the kinetics of the RNA strand separation with and without bound U1A (Fig. 6). Without U1A, DED1 readily unwound the RNA strands with an apparent first order rate constant of k unw = 0.9 ± 0.1 min−1 (Fig. 6). With U1A bound, the time course for strand separation strikingly resembled the biphasic kinetics of the spontaneous U1A dissociation (Fig. 6, cf. Fig. 4C). Thus, DED1 did not accelerate the dissociation of U1A from the RNA. The enzyme could only separate the RNA strands upon spontaneous dissociation of U1A (Fig. 6). Increases in ATP and DED1 concentrations did not change the shape of the time courses or the kinetic parameters for strand separation in the presence of U1A, and we confirmed that DED1 did not dislodge U1A from the RNA without unwinding the RNA strands (data not shown). We also examined U1A displacement from RNA substrates with shortened helices surrounding the U1A binding site and with artificial linkers in one of the RNA strands, all of which had no significant effect on shape and kinetic parameters of the unwinding time course in the presence of U1A (data not shown). Finally, we verified that U1A did not prevent DED1 from unwinding RNA structures in general (data not shown). Taken together, these observations confirmed that DED1, in contrast to NPH-II, is unable to actively displace U1A from the RNA.
FIGURE 6.
Time course of DED1-catalyzed unwinding of the RNA complex with and without U1A bound. RNA strand separation by DED1 with U1A bound (open circles) and in the absence of U1A (filled circles). Data points represent the average of least two independent measurements, error bars indicate one standard deviation. The resulting time course for the reaction without U1A was fit against the integrated rate law for a homogenous first-order process, yielding an unwinding rate constant of k r [RNA] = 0.98 ± 0.02 min−1. The time course for the reaction with U1A was fit to the sum of two exponentials, yielding unwinding rate constants for the first phase of k I r [RNP] = 0.10 ± 0.01 min−1, and for the second phase of k II r [RNP] = (8.0 ± 1.7) × 10−4 min−1.
The inability of DED1 to accelerate U1A dissociation from its RNA binding site contrasts with the capacity of the enzyme to actively disrupt the U1 snRNP–RNA complex. Thus, DED1 remodels only certain RNPs in an active fashion. This finding complements previous results where DED1 actively displaced the EJC but not the TRAP protein from their respective RNA targets (Fairman et al. 2004).
Discriminatory disassembly of RNP complexes by DED1
It occurred to us that the inability of DED1 to actively displace U1A might also provide a straightforward means to enable a non-sequence specific DExH/D protein to remodel similar RNA substrates in a discriminatory fashion. Because the U1A off-rate dictated the velocity by which DED1 could disassemble the RNA strands, an RNA with a slight alteration in the U1A binding site that affected the U1A off-rate should be remodeled by DED1 at a rate determined by this altered U1A off-rate. We, therefore, hypothesized that confronting DED1 with a pool of similar RNAs containing slightly different U1A binding regions should result in a discriminatory remodeling of these RNAs in the presence, but not in the absence, of U1A.
To test this hypothesis, we designed an RNA with a slightly altered U1A binding site (Fig. 7A). We deleted three nucleotides from the U1A cognate site, otherwise the RNA was identical to the complex used above (Fig. 7A). U1A bound to this altered RNA with an affinity of Kd= 13.5 ± 0.7 nM (Fig. 7B), i.e., only slightly weaker than the RNA with the authentic U1A binding site (cf. Fig. 4B). However, U1A dissociated from the altered RNA significantly faster than from the wtRNA (Fig. 7C). As observed for RNAs containing the wild-type U1A binding site, U1A dissociation followed a biphasic time course whose shape did not change upon alterations in the reaction conditions and increases in the U1A concentration (data not shown). The biphasic U1A dissociation kinetics from the altered RNA most likely reflects inherent heterogeneity or an induced-fit binding mode of the U1A–RNA complex (Katsamba et al. 2001; Pitici et al. 2002).
FIGURE 7.
Altered U1A-based RNP. (A) RNP design. Three nucleotides were deleted from the upper RNA strand (indicated by triangles) of the RNA complex with the authentic U1A binding site (Fig. 4A). (B) Equilibrium binding of U1A to the altered RNA complex. Data points represent the average of three independent measurements. Data were fit to the Hill-equation (K D = 13.5 ± 0.7 nM, n = 1.9 ± 0.2). (C) Spontaneous dissociation of U1A from the RNA. The representative time course was fit to the sum of two exponentials. Dissociation rate constants were, for the first phase: k I d = 2.2 ± 0.6 min−1, and for the second phase: k II d = (5.3 ± 0.7) × 10−3 min−1.
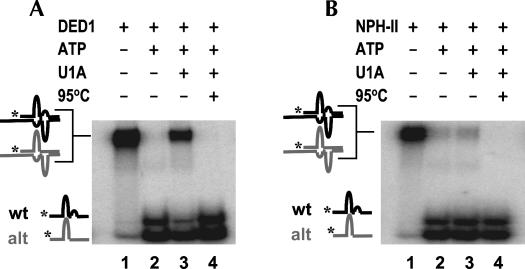
The distinct dissociation kinetics of U1A from wild-type and altered RNAs rendered the altered RNA suitable for testing whether the presence of U1A would indeed enable DED1 to unwind both RNAs in a discriminatory fashion. We combined both RNA complexes in an equimolar ratio and monitored strand separation by DED1 with and without U1A (Fig. 8). Without U1A, both substrates were readily unwound by DED1 to a virtually identical degree (Fig. 8A). With U1A, DED1 unwound both RNAs in a clearly differential fashion. The RNA with the altered U1A binding site was unwound to a significantly greater extent than the RNA with the authentic U1A binding site (Fig. 8A). An identical experiment with NPH-II showed no comparable differences in the unwinding of both RNAs in the presence of U1A, thereby verifying the integrity of the RNAs and the RNPs (Fig. 8B).
FIGURE 8.
DED1 but not NPH-II disassembles the two RNA complexes in a discriminatory fashion. (A) DED1 disassembles the altered RNA complex more efficiently than the complex with the authentic U1A binding site when U1A is bound (lane 3) but not without U1A (lane 2). Reactions were allowed to proceed for 5 min. Mobilities of the RNA complexes and the single-stranded RNAs are indicated by cartoons on the left of the representative PAGE. The altered RNA is in gray, the RNA with the authentic U1A binding site in black. Lanes are as follows: (1) DED1 only; (2) DED1 and ATP; (3) DED1 and ATP with U1A; (4) boiled control. (B) NPH-II disassembles both, altered RNA complex and the complex with the authentic U1A binding site with comparable efficiency. Reactions were allowed to proceed for 5 min. Lanes are as follows: (1) NPH-II only; (2) NPH-II and ATP; (3) NPH-II and ATP with U1A; (4) boiled control.
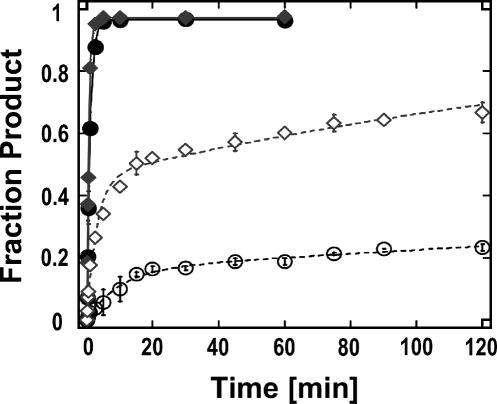
Kinetic analysis of the unwinding time courses with DED1 revealed that the disassembly of both RNA complexes with bound U1A mirrored the dissociation kinetics of U1A from the respective RNA (Fig. 9). Thus, DED1 does not actively displace U1A from either RNA. In the absence of U1A, however, DED1 unwound both RNAs with virtually identical rate constants (Fig. 9). These results demonstrate that U1A binding to distinct binding sites in otherwise similar RNAs enables DED1 to differentially remodel these RNAs. NPH-II, which actively dislodges U1A from both complexes, is unable to differentiate between both RNAs, either with or without U1A bound.
FIGURE 9.
Representative time courses of DED1-catalyzed unwinding of both RNA complexes with and without U1A bound. DED1-catalyzed strand separation of both, the RNA complex with altered and authentic U1A binding site with and without U1A bound (filled circle: wt RNA, no U1A; filled diamond: altered RNA, no U1A; open circle: wt RNA with U1A bound; open diamond: altered RNA with U1A bound). Strand separation of the altered RNA complex without U1A was fit to a single exponential, yielding an unwinding rate constant k r [RNA] = 1.63 ± 0.15 min−1. Strand separation of the altered RNA complex with U1A present was fit to a sum of two exponentials yielding an unwinding rate constant for the first phase: k I r [RNP] = 0.33 ± 0.04 min−1, and for the second phase: k II r [RNP] = (4.8 ± 0.7) × 10−3 min−1. Kinetic data for strand separation of the RNA complex with the authentic U1A binding (with and without U1A) site are reported in Figure 6.
DISCUSSION
In this study, we have shown (1) that two distinct DExH/D proteins, DED1 and NPH-II, can displace the U1 snRNP from a tightly bound RNA in an active, ATP-dependent fashion; (2) that DED1 cannot actively displace the protein U1A from its binding site, whereas NPH-II can; and (3) that the “inability” to actively displace U1A can provide the non-sequence specific DED1 with a means to nonetheless disassemble very similar RNA complexes in a discriminatory fashion.
The different remodeling efficiencies of NPH-II and DED1 toward diverse RNPs reveal possible mechanism(s) for active protein displacement by DExH/D proteins
The active displacement of the U1 snRNP from a tightly bound RNA by both NPH-II and DED1 indicates that DExH/D proteins are able to disrupt complex RNA-protein interfaces in an active, ATP-dependent manner. While illumination of the detailed mechanism by which both enzymes dislodge the U1 snRNP was beyond the scope of this study, our data suggest, nonetheless, that both NPH-II and DED1 may displace the U1 snRNP through an ATP-driven function on the single-stranded substrate RNA (Fairman et al. 2004; Kawaoka et al. 2004; von Hippel 2004). This is mainly because both NPH-II and DED1 require extended stretches of unpaired nucleotides (∼20 nt) in order to bind RNA/RNP substrates with high affinity (Fairman et al. 2004). Such extended stretches of unpaired nucleotides are only present on the substrate RNA but not in the U1 snRNP (Stark et al. 2001). Therefore, both enzymes are more likely to bind and subsequently act on the substrate RNA, rather than directly on the U1 snRNP.
While both DED1 and NPH-II actively dislodged the U1 snRNP from an RNA substrate, only NPH-II, but not DED1, could actively displace U1A. This is an unexpected result because (1) both the U1 snRNP- and the U1A-based RNPs involved a combination of RNA-RNA and RNA-protein interactions; (2) both U1 snRNP and U1A bound their cognate RNAs with similar affinity; and (3) both U1 snRNP and U1A dissociated from the RNA with similar rate constants. The different remodeling efficiencies of NPH-II and DED1 toward both RNPs, however, mirror previous observations with RNPs that involved proteins bound to unstructured RNA (Fairman et al. 2004). There, NPH-II but not DED1 actively displaced the TRAP protein, whereas both enzymes actively dislodged the exon junction complex (EJC), even though TRAP dissociated much faster from the RNA than the EJC (Fairman et al. 2004). Thus, data collected for the rearrangement of four different RNPs (U1A, TRAP, EJC, U1 snRNP) by NPH-II and DED1 indicate that not all DExH/D proteins are able to actively remodel the same range of RNPs. NPH-II actively displaces a greater range of proteins than DED1. It is worth noting that DED1 actively displaces the multicomponent complexes EJC and U1 snRNP, but not the homo-oligomeric proteins TRAP and U1A. These observations indicate that neither stability (the EJC is the most stable RNA-protein complex) nor the rate for spontaneous dissociation of a given RNP determine whether DED1 can actively disrupt an RNA-protein complex. Rather, the architecture of a given RNP may dictate whether enzymes such as DED1 can actively remodel the complex.
However, the data clearly show that both NPH-II and DED1 can displace proteins without unwinding RNA duplexes. The actual protein displacement may be based on the ability of the DExH/D proteins to capture nucleotides that are normally part of the RNA-protein interface. The nucleotide capture could be accomplished either by the DExH/D enzyme exerting force on the other protein or by occupying transiently fraying nucleotides. In any event, nucleotide capture reduces the number of RNA-protein contacts in the RNP, thereby increasing the off-rate for the bound protein, which causes the active protein displacement. If the DExH/D protein dissociates from the RNA before capturing the critical number of nucleotides necessary to accelerate dissociation of the RNP, no active protein displacement is observed. This scenario might explain why DED1, which does not display high processivity during RNA unwinding (protein dissociates from the RNA with higher frequency, Fairman et al. 2004), is unable to actively displace proteins such as U1A or TRAP, whereas the processive NPH-II can dislodge those proteins in an active fashion. In a multicomponent complex such as the EJC or the U1 snRNP, a small decrease in the number of RNA-protein contacts might lead to dissociation of one critical component that in turn unravels the entire RNP. Thus, the transient capture of only a small number of nucleotides from the RNA-protein interface by a less processive enzyme such as DED1 may suffice to accelerate the dissociation of multicomponent RNPs.
We note that tracking on single-stranded nucleic acid and capture of fraying nucleotides are also considered to be important for unwinding of DNA duplexes by DNA helicases (von Hippel and Delagoutte 2001). It is perhaps not surprising that similar mechanisms may underlie both duplex unwinding and protein displacement by “helicase” enzymes.
However, given the limited data for RNP remodeling by DExH/D proteins, alternative mechanisms by which these enzymes cause active protein displacement should not be discounted. For example, instead of capturing fraying nucleotides, DExH/D proteins may be able to force other proteins off the nucleic acid in one step, perhaps through physical clashes between protein domains that are driven by ATP binding and/or hydrolysis in the DExH/D protein.
The inability of DED1 to actively displace other proteins enables discriminatory function in vitro
Although our study has illuminated possible mechanisms by which DExH/D enzymes actively displace other proteins from RNA, the inability of DExH/D proteins to actively dislodge other proteins might be equally significant, most notably for enabling non-sequence specific enzymes to act in a discriminatory fashion. We underscore that the mechanisms for the discriminatory function of DED1 discussed below are based solely on the data collected with artificial model systems in vitro. We do not imply that DED1 targets either of the tested model substrates, U1 snRNP, U1A, EJC, or TRAP in vivo, although we cannot rule out that DED1 might act on the first two substrates. We further do not suggest that DED1 invariably functions in its physiological environment as described in this model study. Finally, we note that the mechanism for discriminatory RNP remodeling does in no way preclude the recruitment and perhaps specific activation of DExH/D proteins by their specific targets. Rather, the proposed discriminatory function of DExH/D proteins may complement their recruitment to specific targets by (1) preventing disassembly of RNPs beyond intended target regions, (2) allowing the timing of RNA remodeling reactions without directly affecting the DExH/D enzyme, and (3) providing one possible way to consider conformational proofreading by RNA helicases in straightforward terms (see discussion below).
The inability of DED1 to actively displace certain other proteins from RNA highlights the possibility for discriminatory RNP remodeling on two levels. The first level of discrimination is based on the ability of DED1 to actively (efficiently) disassemble only certain RNPs. Possible mechanisms for this phenomenon have been discussed above. Thus, confronting an enzyme like DED1 with a pool of different RNPs (e.g., U1A-RNP and U1 snRNP) will result in the remodeling of some but not other RNPs. It is unclear to which degree this situation resembles physiological reactions. Nonetheless, this level of discriminatory RNP remodeling is, despite its simplicity, not a trivial finding. This is (1) since not all DExH/D proteins behave in a similar fashion and (2) because no direct interactions between the DExH/D protein and other co-factors and no modifications of the biochemical activities of the enzymes are involved.
The second level of discrimination is less obvious than the one discussed above. We have observed here that DED1 was not only unable to actively displace U1A from its cognate RNA, but also that the dissociation rate constant of U1A determined how fast DED1 remodeled RNAs with bound U1A. Alterations in the RNA that change the U1A off-rate affect the rate of RNA remodeling by DED1 and thus enable the non-sequence specific DED1 to discriminately remodel RNAs based on only slight sequence differences. By unwinding RNAs containing altered U1A binding sites at a faster rate than RNAs with an authentic U1A binding site, in principle, DED1 could be viewed as “proofreading” for RNAs with an authentic U1A binding site (RNAs with altered U1A binding sites are preferentially disassembled, i.e., “discarded”). Thus, “proofreading” by RNA helicases could be considered in straightforward terms, although physiological ramifications of this possible “proofreading” mechanism are unclear. However, we note that the proposed mode for “proofreading” is consistent with the ATP-dependent kinetic proofreading function of the spliceosomal DExH/D protein Prp16 (Burgess and Guthrie 1993), and with activity modulations of the DExH/D proteins Prp22 and Brr2 by the spliceosomal protein Prp8 (Kuhn et al. 2002; Schneider et al. 2004).
The inability of DED1 to actively displace other proteins from RNA also illuminates a straightforward means to time RNA rearrangement steps by DExH/D enzymes without the need to establish specific protein-protein interactions. Events unrelated to the DExH/D protein function, such as a transesterification step during pre-mRNA splicing, or protein phosphorylation may simply alter the off-rate of a regulatory protein, and thereby time the ATP-driven remodeling function of a DExH/D protein. It is not known whether in a physiological context the timing of some DExH/D protein-catalyzed RNA rearrangements occur according to this mechanism. However, it may be attractive to specifically test whether certain genetic interactions between DExH/D enzymes and RNA binding proteins arise due to such “control” of DExH/D enzymes by RNA binding proteins.
We note that NPH-II, which actively displaces U1A from its cognate RNA, could not be “controlled” by U1A. Consequently, NPH-II could not preferentially remodel any of the RNA complexes tested. Therefore, it may be critical for possible discriminatory functions of DExH/D proteins that the enzymes in question are unable to actively displace certain other proteins from RNA. Consequently, DExH/D proteins such as DED1 may have mechanistic characteristics that result in less potent RNA helicase or RNPase activities in vitro. On the other hand, viral proteins such as NPH-II might have evolved as “cleaners” that indiscriminately remove other proteins and RNAs from a target RNA.
MATERIALS AND METHODS
Protein expression and purification
DED1 was expressed in Escherichia coli as described (Iost et al. 1999), except that bacteria were grown at 28°C. Purification of DED1 was as described (Iost et al. 1999) with an additional purification step in which DED1 was adsorbed to phosphocellulose resin (P11, Whatman) and eluted with 300 mM NaCl. NPH-II was expressed in baculovirus-infected insect cells as described (Fairman et al. 2004). Cells were lysed and NPH-II was purified by adsorption to Ni-agarose (Qiagen) and phosphocellulose resin (P11, Whatman) (Gross and Shuman 1996). Homogeneity (>98%) and concentration of DED1 and NPH-II were assessed by SDS-PAGE and subsequent coomassie staining of the peptide. Purified U1A containing residues 2 through 117 was a gift from Dr. Kyoshi Nagai (Cambridge, UK). U1 snRNP was prepared as described (Stark et al. 2001).
RNA preparation
The 75-nt substrate RNA for U1 snRNP was prepared by in vitro transcription using T7 RNA polymerase (Milligan and Uhlenbeck 1989). All other RNA oligonucleotides were purchased from DHARMACON (Lafayette) and deprotected according to manufacturer's protocols. RNA, where applicable, was labeled with γ32P-ATP (ICN) using T4 Polynucleotide Kinase (New England BioLabs) and purified on denaturing PAGE. To form RNA duplexes, labeled and complementary unlabeled RNA were combined in 50 mM KCl, 10 mM MOPS (pH 6.5), 2 mM EDTA, heated to 95°C for 2 min and cooled to room temperature over 2 h. Bipartite complexes were separated from single-stranded RNA by non-denaturing PAGE, visualized by autoradiography, excised, eluted from the gel, and precipitated with ethanol (Jankowsky et al. 2001). RNA complexes were quantified by measuring incorporated 32P using a scintillation counter.
U1 snRNP dissociation and displacement
Spontaneous U1 snRNP dissociation was measured by pre-forming the U1 snRNP–RNA complex for 10 min at room temperature in a buffer containing 40 mM Tris-HCl pH 8.0, 4 mM MgCl2, 0.01% (v/v) Nonidet P40. The dissociation reaction was initiated by addition of an excess of U1 snRNP scavenger RNA (unlabeled RNA, identical to substrate RNA, 200 nM final concentration) and ATP to a final concentration of 3.5 mM (to provide reaction conditions consistent with the subsequent remodeling reactions). Aliquots were removed at the indicated times and subjected to immunoprecipitation of the U1 snRNP using antibodies against the U1A protein in order to separate bound and free RNA. Aliquots were incubated with the antibody for 60 min on ice (150 mM NaCl, 50 mM Tris/HCl, pH 8.0, 0.1% [v/v] Nonidet P40). Subsequently, aliquots were centrifuged and the supernatant was removed. Radioactivity was measured in precipitate and supernatant by scintillation counting. The fraction of free RNA was calculated from the ratio of radioactivity in precipitate and supernatant. Dissociation rate constants were determined from plots of the fraction of free RNA versus time by least square fitting to the appropriate kinetic model using Kaleidagraph (Synergy software).
To measure U1 snRNP remodeling by NPH-II and DED1, the U1 snRNP complex was pre-formed as described above. NPH-II (20 nM final concentration) or DED1 (600 nM final concentration) were added and incubated for five more minutes. The remodeling reactions were started by adding a mixture of ATP (3.5 mM final concentration) and U1 snRNP scavenger RNA, in order to prevent dissociated U1 snRNP to rebind labeled RNA. Displacement reactions were conducted at room temperature, aliquots were removed at the indicated times and the remodeling reaction was stopped by addition of EDTA (5 mM final) and 5 μM NPH-II scavenger RNA (to prevent further binding of NPH-II to the U1 snRNP–RNA complex) and by placing the aliquot on ice. The amount of radiolabeled RNA bound to the U1 snRNP at the given reaction times was determined by immunoprecipitation as described above. Displacement rate constants were determined from plots of the fraction of free RNA versus time by least square fitting to the appropriate kinetic model.
Equilibrium binding of U1A
U1A equilibrium binding studies were performed in buffer containing 40 mM Tris-HCl, pH 8.0, 50 mM NaCl, 0.01% P40, and 2 mM DTT. Radiolabeled RNA duplex (0.5 nM) in a reaction volume of 20 μL was incubated with increasing concentrations of U1A on ice for 10 min, followed by incubation at 19°C for 5 min. Subsequently, 20 μL loading buffer were added to each individual reaction and the solutions were immediately loaded on non-denaturing PAGE (run at 4°C). Gels were dried and the bands corresponding to single strand and duplex RNA were visualized using a PhosphorImager (Molecular Dynamics). Radioactivity in each band was quantified using the ImageQuant software (Molecular Dynamics). Equilibrium binding constants were calculated according to
Fraction Bound = Kn/(Kn + [U1A]n)
K is the equilibrium binding constant, n is the Hill-coefficient. Curve fitting was performed using Kaleidagraph (software).
U1A dissociation and displacement
RNP/RNA remodeling assays were performed as described (Jankowsky et al. 2001; Fairman et al. 2004) at room temperature in a buffer containing 40 mM Tris-HCl, pH 8.0, 50 mM NaCl, 0.01% (v/v) NP40, and 1 mM DTT with 0.5 nM radiolabeled duplex RNA and U1A, where applicable. U1A–RNA complexes were formed prior to the reaction for 10 min. Spontaneous U1A dissociation was measured by incubating pre-formed U1A–RNA complexes with a large excess of U1A scavenger RNA (1.25 μM, unlabeled RNA based on U1A binding site in U1 snRNA) (Jankowsky et al. 2001), which prevents rebinding of U1A to the radiolabeled RNA complex once U1A has dissociated from this RNA. Aliquots were removed from the reaction, glycerol (10% v/v final) was added and the aliquots were immediately applied to non-denaturing PAGE (run at 4°C). The fraction of disassembled RNA complexes was determined by quantifying radioactivity in U1A-bound and free RNA using a PhosphorImager (Molecular Dynamics). Dissociation rate constants were determined by plotting the fraction free RNA versus time and fitting the resulting time courses with the integrated rate law for a biphasic first order reaction (sum of two exponentials). Curve fitting was performed using Kaleidagraph software.
U1A displacement by DED1 and NPH-II was measured by pre-forming U1A–RNA complexes as described above. Subsequently, DED1 (500 nM) or NPH-II (20 nM) was incubated at room temperature with the complex for an additional 5 min. Longer incubation times or higher protein concentrations (U1A, DED1, NPH-II) did not alter the results. Remodeling reactions were started by adding 5 mM (final) ATP (where applicable), 5 mM (final) MgCl2, and 1.25 μM (final) U1A scavenger RNA to prevent U1A from rebinding to RNA once it has been displaced. Aliquots were removed at appropriate times and stopped with equal volumes of buffer containing 10 mM EDTA, 1% SDS (v/v), 0.1% bromophenol blue, 0.1% xylene cyanol, and 20% glycerol. Subsequently, aliquots were applied to 15% non-denaturing PAGE (Jankowsky et al. 2001). Gels were dried and the bands corresponding to single strand and duplex RNA were visualized using a PhosphorImager (Molecular Dynamics). Radioactivity in each band was quantified using the ImageQuant software (Molecular Dynamics). In the presence of U1A, single-stranded RNA corresponds to the amount of displaced U1A (Jankowsky et al. 2001). We specifically confirmed that DED1 did not displace U1A without separating the RNA strands (data not shown). Rates for strand separation were determined by plotting the fraction single-stranded RNA versus reaction time and fitting the resulting time courses with the integrated rate law for a homogenous first order reaction (without U1A) or with the integrated rate law for a biphasic first order reaction (with U1A bound).
ACKNOWLEDGMENTS
We thank Drs. Kyoshi Nagai and Gabriele Varani for the gift of purified U1A. We are grateful to Drs. Anna Marie Pyle, Pieter deHaseth, and Marc Caprara as well as members of our laboratories for comments on the manuscript. This work was supported by a grant from the NIH to E.J. (GM067700) and by a grant from the NIH to T.W.N. E.J. is a Scholar of the Damon Runyon Cancer Research Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2323406.
REFERENCES
- Anantharaman V., Koonin E.V., Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S., Guthrie C. Beat the clock: Paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem. Sci. 1993;18:381–384. doi: 10.1016/0968-0004(93)90094-4. [DOI] [PubMed] [Google Scholar]
- Fairman M., Maroney P.A., Wang W., Bowers H., Gollnick P., Nilsen T.W., Jankowsky E. Protein displacement by DExH/D RNA helicases without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- Gross C.H., Shuman S. Vaccinia virus RNA helicase: Nucleic acid specificity in duplex unwinding. J. Virol. 1996;70:2615–2619. doi: 10.1128/jvi.70.4.2615-2619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I., Dreyfus M., Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- Jankowsky E., Gross C.H., Shuman S., Pyle A.M. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- Katsamba P.S., Myszka D.G., Laird-Offringa I.A. Two functionally distinct steps mediate high affinity binding of U1A protein to U1 hairpin II RNA. J. Biol. Chem. 2001;276:21476–21481. doi: 10.1074/jbc.M101624200. [DOI] [PubMed] [Google Scholar]
- Kawaoka J., Jankowsky E., Pyle A.M. Backbone tracking by the SF2 helicase NPH-II. Nat. Struct. Mol. Biol. 2004;11:526–530. doi: 10.1038/nsmb771. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Reichl E.M., Brow D.A. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc. Natl. Acad. Sci. 2002;99:9145–9149. doi: 10.1073/pnas.102304299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P. Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol. Cell. 2003;95:157–167. doi: 10.1016/s0248-4900(03)00032-7. [DOI] [PubMed] [Google Scholar]
- Linder P. The life of RNA with proteins. Science. 2004;304:694–695. doi: 10.1126/science.1097850. [DOI] [PubMed] [Google Scholar]
- Milligan J.F., Uhlenbeck O.C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Pitici F., Beveridge D.L., Baranger A.M. Molecular dynamics simulation studies of induced fit and conformational capture in U1A–RNA binding: Do molecular substates code for specificity? Biopolymers. 2002;65:424–432. doi: 10.1002/bip.10251. [DOI] [PubMed] [Google Scholar]
- Schneider S., Hotz H.R., Schwer B. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J. Biol. Chem. 2002;277:15452–15458. doi: 10.1074/jbc.M112473200. [DOI] [PubMed] [Google Scholar]
- Schneider S., Campodonico E., Schwer B. Motifs IV and V in the DEAH box splicing factor Prp22 are important for RNA unwinding, and helicase-defective Prp22 mutants are suppressed by Prp8. J. Biol. Chem. 2004;279:8617–8626. doi: 10.1074/jbc.M312715200. [DOI] [PubMed] [Google Scholar]
- Shuman S. Vaccinia virus RNA helicase: An essential enzyme related to the DE-H family of RNA-dependent NTPases. Proc. Natl. Acad. Sci. 1992;89:10935–10939. doi: 10.1073/pnas.89.22.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J.P., Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Stark H., Dube P., Lührmann R., Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409:539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- Tanner N.K., Linder P. DExD/H box RNA helicases. From generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–261. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Varani L., Gunderson S.I., Mattaj I.W., Kay L.E., Neuhaus D., Varani G. The NMR structure of the 38 kDa U1A protein—PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein. Nat. Struct. Biol. 2000;7:329–335. doi: 10.1038/74101. [DOI] [PubMed] [Google Scholar]
- von Hippel P.H. Helicases become mechanistically simpler and functionally more complex. Nat. Struct. Mol. Biol. 2004;11:494–496. doi: 10.1038/nsmb0604-494. [DOI] [PubMed] [Google Scholar]
- von Hippel P.H., Delagoutte E. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell. 2001;104:177–190. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]