Abstract
RNase MRP is a eukaryote-specific endoribonuclease that generates RNA primers for mitochondrial DNA replication and processes precursor rRNA. RNase P is a ubiquitous endoribonuclease that cleaves precursor tRNA transcripts to produce their mature 5′ termini. We found extensive sequence homology of catalytic domains and specificity domains between their RNA subunits in many organisms. In Candida glabrata, the internal loop of helix P3 is 100% conserved between MRP and P RNAs. The helix P8 of MRP RNA from microsporidia Encephalitozoon cuniculi is identical to that of P RNA. Sequence homology can be widely spread over the whole molecule of MRP RNA and P RNA, such as those from Dictyostelium discoideum. These conserved nucleotides between the MRP and P RNAs strongly support the hypothesis that the MRP RNA is derived from the P RNA molecule in early eukaryote evolution.
Keywords: specificity domain, catalytic domain, RNase MRP RNA
RNase P and RNase MRP are ribonucleoprotein (RNP) complexes participating in cellular RNA processing. RNase P is a ubiquitous endoribonuclease that cleaves precursor tRNA (pre-tRNA) transcripts to produce their mature 5′ termini (Altman 1990; Altman and Kirsebom 1999). Bacterial RNase P consists of a catalytic RNA subunit and a protein cofactor, while archaeal and eukaryotic enzymes have a single RNA subunit and more than four proteins (Hartmann and Hartmann 2003). RNase MRP is a eukaryote-specific endoribonuclease that has at least two roles: one in the mitochondrial compartment where it generates RNA primers for the initiation of mitochondrial DNA replication (Chang and Clayton 1989), and a second in the nucleolus where it participates in precursor rRNA processing (Lygerou et al. 1996). Hemiascomycetes, such as the budding yeast Saccharomyces cerevisiae, have been extensively used to study the structure and function of eukaryotic RNase P and MRP. The S. cerevisiae RNase P enzyme possesses one RNA (P RNA) and nine integral protein subunits, while RNase MRP has one RNA (MRP RNA) and 10 proteins (Chamberlain et al. 1998; Salinas et al. 2005). Previously, the catalytic domains of these two RNAs have been demonstrated to have similar secondary structures (Li et al. 2002). Moreover, eight of the associated protein components are identical (Chamberlain et al. 1998), indicating that these two enzyme complexes are structurally and evolutionally related.
Despite structural similarity in the catalytic domains, little sequence homology beyond the P3 and P4 helices of P and MRP RNA has been demonstrated, partially due to limited availability of sequences, especially the MRP RNA sequences. A recent report by Samuelsson and colleagues identified more than 100 new MRP and P RNA sequences from eukaryotes with complete genome sequences or whole-genome-shotgun sequences (Piccinelli et al. 2005). Some of these predicted MRP and P RNA genes from Candida species are demonstrated by biochemical methods in our laboratory to be expressed (data not shown). These sequences allowed us to perform a comprehensive analysis of MRP RNAs and their homology with the corresponding P RNAs. Our analysis reveals unprecedented structural conservation between MRP RNA and P RNA, lending strong support to the hypothesis that the MRP RNA originated from P RNA during earlier evolution in eukaryotes rather than directly from the “RNA world.”
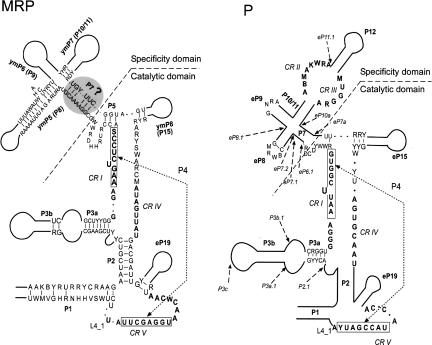
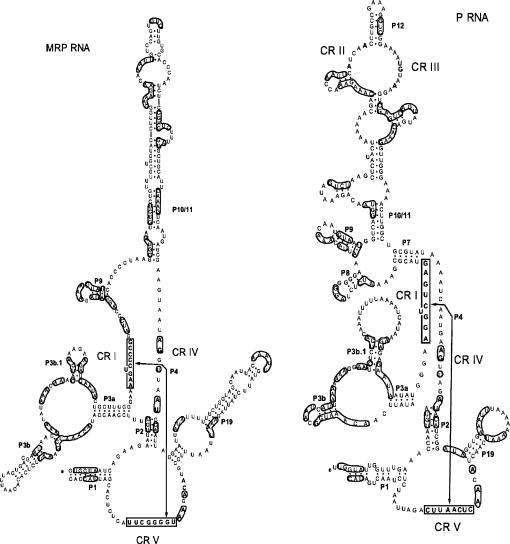
The secondary structure model of MRP RNA from hemiascomycetes
Following the modeling of a core structure for P RNA from hemiascomycetes (Kachouri et al. 2005), we compared and analyzed the secondary structure of their corresponding MRP RNAs. We aligned the RNA sequences to recognize most of the homologous helices, in comparison to the known secondary structures, especially that of S. cerevisiae and others from the Saccharomycetaceae family (Li et al. 2002). The alignment (Supplemental Fig. S1) recognized the helices generated by compensatory changes as reported previously (Li et al. 2002) and was used to derive the secondary structure of MRP RNA (Fig. 1). Some unusually large sequences, like the putative extremely large MRP RNAs from Candida albicans and Candida dubliniensis (Piccinelli et al. 2005) and the large P RNA from Candida glabrata still under investigation (Kachouri et al. 2005), as well as MRP RNA from Candida tropicalis, Debaryomyces hansenii, Pichia guilliermondii, and Yarrowia lipolytica, were not included in Figure 1. Beyond the conserved regions (CR I, CR IV, CR V), the MRP RNAs within hemiascomycetes are conserved in P1, P2, P3a, and ymP5 helices (Fig. 1).
FIGURE 1.
Comparison of the secondary structures of RNase MRP and RNase P RNAs from hemiascomycetes. Boldface letters represent the universally conserved nucleotides for P RNA and MRP RNA, respectively. The degenerate nucleotides are shown according to (Cornish-Bowden 1985). A dot represents any nucleotide. Two parallel straight lines represent a helix, and a curved line represents a loop containing a variable number of nucleotides. The P7 helix with a question mark in MRP RNA indicates it is not defined (see main text). A P7.2 helix insertion in the MRP structure of S. paradoxus is not included. The nomenclature for these conserved regions (CRs) is that of (Chen and Pace 1997). Helix numbering follows their putative homology with bacterial structures (Haas et al. 1994). The P RNA structure model is derived from (Kachouri et al. 2005), while the insertions are shown with arrows.
It has been proposed that ymP5 and ymP6 are equivalent to eP8 and eP9 of P RNA in two subsets of MRP RNA structures: one for human MRP RNA, another for yeast MRP RNA (Walker and Avis 2005). If the ymP7 stem is equivalent to the P10/P11 stem of P RNA, and if there is a P7-like stem in MRP RNA, both MRP RNA and P RNA will possess the cruciform structure (P7, P8, P9, and P10/11 in P RNA; and P7-like, ymP5, ymP6, and ymP7 in MRP RNA), which is well defined as the specificity domain of P RNA (Chen and Pace 1997; Massire et al. 1998). Subsequently, the two-domain structure model of bacterial P RNA (Chen and Pace 1997; Massire et al. 1998) could be well applied to that of MRP RNA in addition to the eukaryotic P RNA (Kachouri et al. 2005). However, the formation of a P7-like structure is not required for the essential function of RNase MRP and may not be in the active complex (Walker et al. 2005). Thus, further concrete evidence is needed before designating the P7 stem in MRP RNA.
We further notice that the deletion of the entire helix of P15 (Shadel et al. 2000; Li et al. 2004) or P10/11 (Li et al. 2004) of S. cerevisiae MRP RNA results in slow-growing yet viable cells at lower temperatures, while the complete removal of P8, P9, or P19 (Li et al. 2004) is lethal. Thus, it appears that, for the helices of MRP RNA, there is no direct association between structural conservation (with corresponding P RNA or other MRP RNA) and their indispensability (to cell viability).
It should be noted that there is less variation in the secondary structure model of MRP RNA compared with that of P RNA (Fig. 1), a tendency also observed previously (Li et al. 2002). We noticed that the ITS1 regions (the substrate for the MRP enzyme) of hemiascomycetes precursor rRNA are highly conserved (Supplemental Fig. S2). Thus, the sequence conservation of MRP RNA may be related to the substrate recognition.
Catalytic domain of MRP RNA
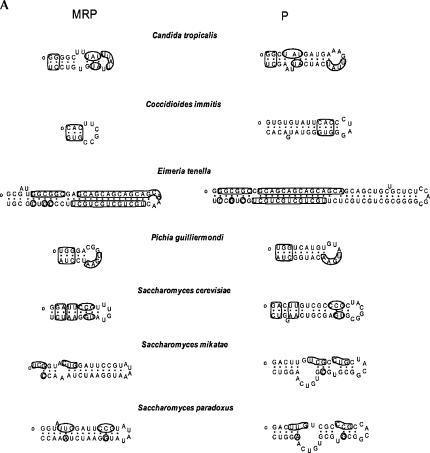
We sought to expand the analysis of MRP RNA to all eukaryotic domains. Wide-range conservation in pairs of the catalytic domain of P and MRP RNAs was observed. The conservation is variable from species to species, beyond the P3 and P4 regions (Fig. 2). Conservation is defined only when three or more conserved nucleotides are found in the corresponding helix in P and MRP RNAs unless they are at the beginning of the helix or have its pairing partner conserved as well.
FIGURE 2.
(Continued on next page)
FIGURE 2.
Sequence conservation in the catalytic domains of RNase P RNA and MRP RNA. Conserved nucleotides between MRP and P RNA are marked. The 5′ ends of the sequences are denoted with a small circle. (A) The P15 helix; (B) the P19 helix; (C) the internal loop of P3 helix is 100% conserved between P and MRP RNAs from C. glabrata, as indicated in boldface letters.
Sequence conservation in the P15 helices, previously identified as ymP8 in MRP RNA (Li et al. 2002), of P and MRP RNAs was observed in hemiascomycete and pezizomycotina (Fig. 2A). In P. guilliermondii, the beginning three base pairs and the UGAA in the terminal loop of P15 are conserved (Fig. 2A). In some coccidia species this homology can be quite extensive. For example, in Eimeria tenella, 37 out of 56 nucleotides are conserved in the P15 helix (Fig. 2A).
The conservation between the eP19 of P and MRP RNA is widely observed (Fig. 2B). P and MRP RNAs have conserved nucleotides in the eP19 in all pezizomycotina and some basidiomycota and hemiascomycete, and rarely in an invertebrata (Anopheles gambiae). These nucleotides may pair with corresponding ones, such as those in Aspergillus nidulans. In certain cases, such as P19 of Podospora anserine, the conserved nucleotides may not base-pair correspondingly (Fig. 2B).
The internal loop of P3 helix has been implicated previously to be conserved among all P and MRP RNA, and it is also true in all newly identified sequences (data not shown). We found an extreme case in C. glabrata where the P3 internal loops of both P and MRP RNA were 100% identical (Fig. 2C).
The specificity domain of MRP RNA
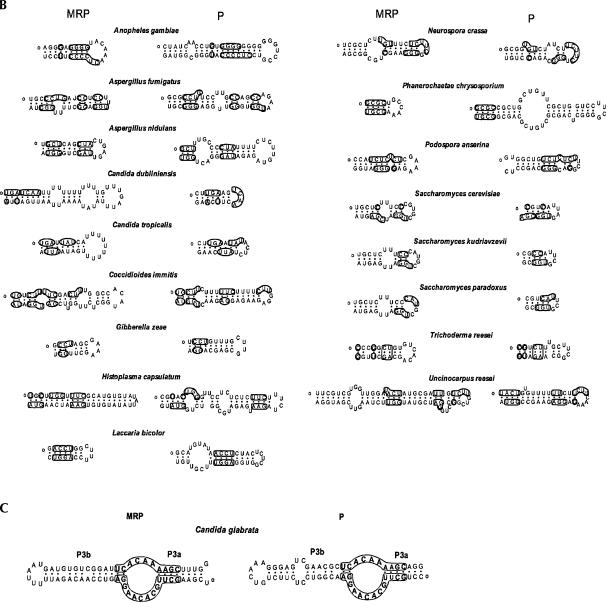
Although it is generally agreed that P RNA and MRP RNA are structurally similar and evolutionarily share a common origin in the catalytic domain (Li et al. 2002), little sequence conservation between their specificity domains has been found. Our results, however, may shed light on the structural similarities of the specificity domains of P and MRP RNA. The specificity domain consists of P7 (undefined in MRP RNA), P8, P9, and P10/11 (or P10/11-like in MRP RNA) in both P and MRP RNAs, as proposed in Figure 1.
The eP8 and eP9 helices are ubiquitous components of eukaryotic P RNAs (Frank et al. 2000; Kachouri et al. 2005; Marquez et al. 2005; Piccinelli et al. 2005). Based on sequence analysis, an equivalent helix in the MRP RNA is the helix that preserves a significant number of nucleotides from its counterpart in the P RNA. This criterion led to the identification of the helices P8 and P9 in the MRP RNA of Encephalitozoon cuniculi (Fig. 3A), where the P8 in the MRP RNA is 100% identical to the P8 in the P RNA, and the P9s have four nucleotides conserved in the terminal loop (Fig. 3A). In the same way, we identified the equivalents to eP8 (Fig. 3A) in some pezizomycotina and some basidiomycota species, and those to eP9 (Fig. 3B) in pezizomycotina, hemiascomycete, and a coccidia species (E. tenella).
FIGURE 3.
Sequence conservation in the specificity domain of RNase P RNA and MRP RNA. Marked are the conserved nucleotides for P RNA and MRP RNA from the same species. The 5′ ends of the sequences are denoted with a small circle. (A) The P8 helix. Boldface letters in E. cuniculi show that the P8 of P RNA is identical to that of MRP RNA; (B) the P9 helix; (C) the P7.2 helix.
P7.2 has been identified as a helix between P7 and P8 in eukaryotic (hemiascomycete) P RNA (Kachouri et al. 2005). The establishment of P8 facilitates the identification of the P7.2 helix in MRP RNA. No P7.2 helix was observed in hemiascomycete MRP RNA except in S. paradoxus. However, pezizomycotina species widely possess the P7.2 helix, and some of them are conserved with that of P RNA (Fig. 3C).
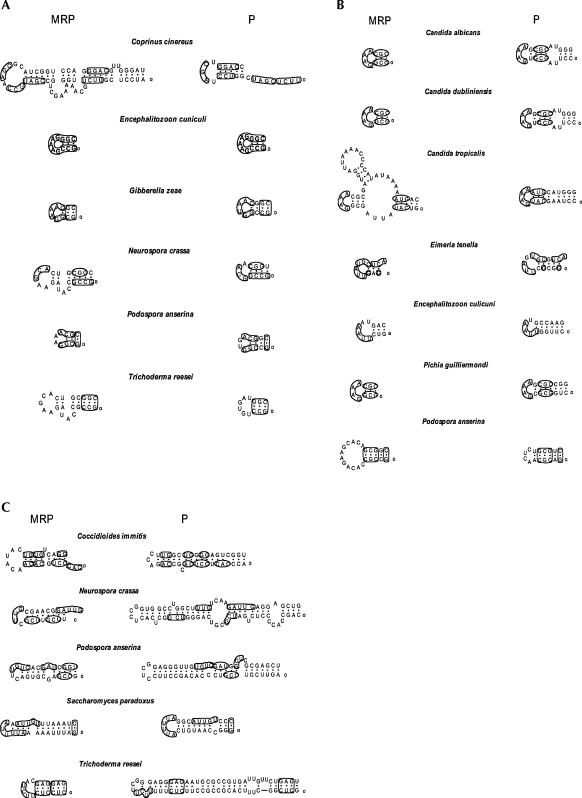
Sequence conservation between P RNA and MRP RNA is not limited to the regions described above. It can also occur sporadically in the helices of P1, P2, and P10/11, etc. For example, in the protist Dictyostelium discoideum, conserved nucleotides between MRP and P RNA were found in nearly every helix (Fig. 4). The conservation can be short or as long as eight nucleotides. Some conserved nucleotides form helices in P RNA but may dissolve into loops in MRP RNA, like the P8 helix (Fig. 4). It should be noted that the RNA folding in Figure 4 is from (Piccinelli et al. 2005), which is solely based on the MFold program version 3.1 (Zuker 2003) and did not contain any phylogenetic information; thus, it may not represent the proper secondary structure, especially in the P10/11 helix. Yet the conservation is too extensive to be coincidental.
FIGURE 4.
The secondary structure of P and MRP RNAs from D. discoideum. Conserved nucleotides are found along the entire sequence of P RNA and MRP RNA. The 5′ ends of the sequences are denoted with a small circle. The boldface letters represent the universally conserved regions (CR I to CR V). The nomenclature for these conserved regions (CRs) is that of (Chen and Pace 1997). Helix numbering follows their putative homology with bacterial structures (Haas et al. 1994).
Three hypotheses
RNase P and RNase MRP are two closely related endoribonucleases. Both enzymes consist of one RNA subunit and about nine proteins in yeast and human cells (Chamberlain et al. 1998; van Eenennaam et al. 2000). Their RNA subunits can be folded into a similar secondary structure, and they share most protein subunits with each other (Chamberlain et al. 1998; van Eenennaam et al. 2000). There have been three hypotheses proposed for the relatedness of P and MRP RNA (Reddy and Shimba 1995; Collins et al. 2000). Hypothesis I is that MRP RNA is derived from P RNA in early eukaryotes, which predicts that MRP RNA is more similar to eukaryotic P RNA than to prokaryotic P RNA (archaeal or bacterial). Hypothesis II is that MRP RNA evolved from an endosymbiont P RNA, which predicts that the secondary structure of MRP RNA would be closer to prokaryotic or organellar P RNA than to eukaryotic P RNA. Hypothesis III is that MRP RNA evolved directly from the “RNA world” (Altman and Kirsebom 1999) and the need for MRP has since been lost in prokaryotes. The secondary structure of MRP RNA is more similar to eukaryotic P RNA than prokaryotic or organellar (mitochondrial or chloroplast) P RNA (Li et al. 2002). Therefore, the possibility of an organellar origin of MRP RNA is unlikely (Collins et al. 2000). Hypothesis I is the most commonly supported in previous studies (Morrissey and Tollervey 1995; Reddy and Shimba 1995) because of the structural similarities of these two RNAs and the fact that RNase P and MRP share most of their protein subunits (Chamberlain et al. 1998). However, Hypothesis III cannot be excluded due to little sequence similarity of P and MRP RNA beyond the P4 and P3 helices. One could argue that MRP RNA, like P RNA, directly evolved from the “RNA world” and that the P3 helix was added to both RNA molecules for recruiting common protein sets, while the related but distinctive P4 helices from P and MRP RNA (Piccinelli et al. 2005) performed similar catalytic reactions.
Our analysis of MRP RNA and P RNA demonstrates that they display extensive structural similarity not only in the catalytic domain but also in the specificity domain in various species. Of note is that there is more sequence conservation between P and MRP RNA in lower eukaryotes such as protists and yeasts (and microsporidia) than in higher eukaryotes such as plants and animals, though there are fewer identified plant P and MRP RNA genes. In most yeast, the conserved CR IV is AUAGUUAU (Fig. 1). However, AGNNNNAU is conserved as CR IV in other lower eukaryotes such as E. cuniculi, T. vaginalis, D. discoideum, (Fig. 4) and Plasmodium species such as P. falciparum (Piccinelli et al. 2005). It is possible the CR IV evolved and diversified from the CR IV (AGNNNNAU) of P RNA. At a molecular level, the P and MRP RNA from D. discoideum share extensive sequence conservation in their P1, P2, P3, P4, P8, P9, P10/P11, and P19 helices, underscoring that MRP RNA is unlikely to have evolved directly from the “RNA world,” but rather from the P RNA.
It has been proposed that RNase MRP may have arisen from early eukaryotes as a form of RNase P specialized for pre-rRNA processing (Morrissey and Tollervey 1995), as tRNA genes are found in the Internal Transcribed Spacer 1 (ITS1) regions of most bacterial and archaeal rRNA genes. To support Hypothesis I, the P RNA should be able to process the ITS1 region even without an embedded tRNA, at least in some cases. In actinobacteria (including the genus of Mycobacterium), a tRNA is not generally present in the ITS1 region. Yet RNase P enzyme from Mycobacterium smegmatis is able to process the tRNA-less ITS1 (Y. Li, unpubl.), supporting that RNase P functions in tRNA-less pre-rRNA processing before RNase MRP arises. The structural similarity between MRP and P RNA may drift apart as a result of the involvement of MRP RNA in mitochondrial DNA replication. We noticed that the exemplary 100% homology in the P8 helices of P and MRP RNA come from E. cuniculi, a species without mitochondria. It is plausible that MRP RNA evolved from eukaryotic P RNA by gene duplication before the hypothetical endosymbiotic fusion that formed the mitochondrion. The diversification of the specificity domain of MRP RNA is dependent on the need for its function in mitochondrial DNA replication.
It is interesting that this interpretation may support that microsporidia is a deeply branched earlier eukaryote (Vossbrinck et al. 1987) rather than a yeast that loses mitochondria, a hypothesis that was supported by small subunit rRNA analysis and cytology, yet negated by multiple protein analysis (Baldauf et al. 2000). We noticed that the multiple protein analysis, no matter how sophisticated, was subject to which proteins are chosen. For instance, the elongation factor 1α (EF-1α) analysis supports that of rRNA phylogeny (Baldauf et al. 2000). Regarding MRP RNA, it is unlikely its sequence would evolve back to the one similar to RNase P RNA after microsporidia loses mitochondria (aerobic respiration). Thus, a more plausible explanation based on the origin of MRP RNA is that microsporidia may be an early offshoot of the eukaryotic line of descent.
The role of protein subunits in the evolution of RNase MRP
RNase MRP and RNase P have been found to extensively share protein subunits. Eight (Pop1p, Pop3p, Pop4p, Pop5p, Pop6p, Pop7p, Pop8p, and Rpp1p) of the 10 protein components of S. cerevisiae RNase MRP are also subunits of its RNase P complex (Chamberlain et al. 1998), while Pop1p binds to both P3 helices of RNase MRP and RNase P (Ziehler et al. 2001). Human RNase MRP and RNase P share at least six (hPop1, Rpp38, Rpp30, Rpp29, Rpp25, Rpp20, hPop5) of their protein components (van Eenennaam et al. 2000). Three proteins (Rpp20, Rpp25, Rpp38) bind the P3 helix of MRP RNA (van Eenennaam et al. 2000), while Rpp25, Rpp38, Rpp40, hPop1, and hPop5 bind that of P RNA (Jiang et al. 2001). The P3 helices of RNase MRP and RNase P from S. cerevisiae are interchangeable without loss of function or specificity (Lindahl et al. 2000), indicating that P3 helix in RNase MRP/P performs similar functions. It is suggested that because the two RNA subunits must interact with a common set of proteins, a given protein and its binding site (such as P3) on the MRP/P RNA, they must co-evolve and remain constrained within each species to constitute a matched set for the functionality of both enzymes (Lindahl et al. 2000).
Regarding the origination of MRP RNA, one could argue that MRP RNA, like P RNA, was directly evolved from the “RNA world” (Hypothesis III) and the P3 helices in both RNAs were added to recruit the same set of proteins. However, it is unlikely that the protein subunits and their interactions with RNA dictate the origination of MRP RNA for two reasons: First, the interacting proteins for both P3 helices are different in yeast (Pop1p) and human (Rpp25 and Rpp38); and second, there is extensive structural similarity in both the catalytic domains and specificity domains of various MRP RNA and P RNA (Figs. 2, 3). It is more plausible that MRP RNA evolved from P RNA by gene duplication after the formation of the eukaryotic RNase P complex and that MRP RNA, while preserving most RNase P proteins, was then derived to perform its distinct functions with diversified specificity domain and acquired MRP-specific proteins such as Snm1 and Rmp1 in S. cerevisiae (Chamberlain et al. 1998; Salinas et al. 2005). The protein components of RNase P/MRP may play a pivotal role in maintaining certain structural similarity, such as the P3 helices after the origination of MRP RNA from P RNA.
To summarize, our analysis of MRP RNA strongly supports the hypothesis that MRP RNA originated from P RNA in earlier eukaryotic evolution. Upon mitochondrion endosymbiosis, MRP RNA became involved in mitochondrial DNA replication with altered structure (especially the specificity domain), while it lost the P RNA-specific CR II and CR III that are required for pre-tRNA binding (Loria and Pan 1997). Additionally, common protein subunits of RNase MRP and RNase P contributed to the RNA structural conservation through protein–RNA interactions.
ACKNOWLEDGMENTS
This project is partially supported by a grant from the Center for Genetics and Molecular Medicine, University of Louisville.
Footnotes
SUPPLEMENTAL MATERIALS Supplementary materials are available upon request from Yong Li, at yong.li@louisville.edu.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2284906.
REFERENCES
- Altman S. Nobel lecture. Enzymatic cleavage of RNA by RNA. Biosci. Rep. 1990;10:317–337. doi: 10.1007/BF01117232. [DOI] [PubMed] [Google Scholar]
- Altman S., Kirsebom L. Ribonuclease P. In: Gesteland R., et al., editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. pp. 351–380. [Google Scholar]
- Baldauf S.L., Roger A.J., Wenk-Siefert I., Doolittle W.F. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- Chamberlain J.R., Lee Y., Lane W.S., Engelke D.R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes & Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.D., Clayton D.A. Mouse RNAase MRP RNA is encoded by a nuclear gene and contains a decamer sequence complementary to a conserved region of mitochondrial RNA substrate. Cell. 1989;56:131–139. doi: 10.1016/0092-8674(89)90991-4. [DOI] [PubMed] [Google Scholar]
- Chen J.L., Pace N.R. Identification of the universally conserved core of ribonuclease P RNA. RNA. 1997;3:557–560. [PMC free article] [PubMed] [Google Scholar]
- Collins L.J., Moulton V., Penny D. Use of RNA secondary structure for studying the evolution of RNase P and RNase MRP. J. Mol. Evol. 2000;51:194–204. doi: 10.1007/s002390010081. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: Recommendations 1984. Nucleic Acids Res. 1985;13:3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D.N., Adamidi C., Ehringer M.A., Pitulle C., Pace N.R. Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA. RNA. 2000;6:1895–1904. doi: 10.1017/s1355838200001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas E.S., Brown J.W., Pitulle C., Pace N.R. Further perspective on the catalytic core and secondary structure of ribonuclease P RNA. Proc. Natl. Acad. Sci. 1994;91:2527–2531. doi: 10.1073/pnas.91.7.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E., Hartmann R.K. The enigma of ribonuclease P evolution. Trends Genet. 2003;19:561–569. doi: 10.1016/j.tig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Jiang T., Guerrier-Takada C., Altman S. Protein–RNA interactions in the subunits of human nuclear RNase P. RNA. 2001;7:937–941. doi: 10.1017/s1355838201010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachouri R., Stribinskis V., Zhu Y., Ramos K.S., Westhof E., Li Y. A surprisingly large RNase P RNA in Candida glabrata . RNA. 2005;11:1064–1072. doi: 10.1261/rna.2130705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Frank D.N., Pace N., Zengel J.M., Lindahl L. Phylogenetic analysis of the structure of RNase MRP RNA in yeasts. RNA. 2002;8:740–751. doi: 10.1017/s1355838202022082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zaman S., Langdon Y., Zengel J.M., Lindahl L. Identification of a functional core in the RNA component of RNase MRP of budding yeasts. Nucleic Acids Res. 2004;32:3703–3711. doi: 10.1093/nar/gkh689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L., Fretz S., Epps N., Zengel J.M. Functional equivalence of hairpins in the RNA subunits of RNase MRP and RNase P in Saccharomyces cerevisiae . RNA. 2000;6:653–658. doi: 10.1017/s1355838200992574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria A., Pan T. Recognition of the T stem-loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1997;36:6317–6325. doi: 10.1021/bi970115o. [DOI] [PubMed] [Google Scholar]
- Lygerou Z., Allmang C., Tollervey D., Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- Marquez S.M., Harris J.K., Kelley S.T., Brown J.W., Dawson S.C., Roberts E.C., Pace N.R. Structural implications of novel diversity in eucaryal RNase P RNA. RNA. 2005;11:739–751. doi: 10.1261/rna.7211705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massire C., Jaeger L., Westhof E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 1998;279:773–793. doi: 10.1006/jmbi.1998.1797. [DOI] [PubMed] [Google Scholar]
- Morrissey J.P., Tollervey D. Birth of the snoRNPs: The evolution of RNase MRP and the eukaryotic pre-rRNA-processing system. Trends Biochem. Sci. 1995;20:78–82. doi: 10.1016/s0968-0004(00)88962-8. [DOI] [PubMed] [Google Scholar]
- Piccinelli P., Rosenblad M.A., Samuelsson T. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res. 2005;33:4485–4495. doi: 10.1093/nar/gki756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R., Shimba S. Structural and functional similarities between MRP and RNase P. Mol. Biol. Rep. 1995;22:81–85. doi: 10.1007/BF00988710. [DOI] [PubMed] [Google Scholar]
- Salinas K., Wierzbicki S., Zhou L., Schmitt M.E. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J. Biol. Chem. 2005;280:11352–11360. doi: 10.1074/jbc.M409568200. [DOI] [PubMed] [Google Scholar]
- Shadel G.S., Buckenmeyer G.A., Clayton D.A., Schmitt M.E. Mutational analysis of the RNA component of Saccharomyces cerevisiae RNase MRP reveals distinct nuclear phenotypes. Gene. 2000;245:175–184. doi: 10.1016/s0378-1119(00)00013-5. [DOI] [PubMed] [Google Scholar]
- van Eenennaam H., Jarrous N., van Venrooij W.J., Pruijn G.J. Architecture and function of the human endonucleases RNase P and RNase MRP. IUBMB Life. 2000;49:265–272. doi: 10.1080/15216540050033113. [DOI] [PubMed] [Google Scholar]
- Vossbrinck C.R., Maddox J.V., Friedman S., Debrunner-Vossbrinck B.A., Woese C.R. Ribosomal RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature. 1987;326:411–414. doi: 10.1038/326411a0. [DOI] [PubMed] [Google Scholar]
- Walker S.C., Avis J.M. Secondary structure probing of the human RNase MRP RNA reveals the potential for MRP RNA subsets. Biochem. Biophys. Res. Commun. 2005;335:314–321. doi: 10.1016/j.bbrc.2005.07.074. [DOI] [PubMed] [Google Scholar]
- Walker S.C., Aspinall T.V., Gordon J.M., Avis J.M. Probing the structure of Saccharomyces cerevisiae RNase MRP. Biochem. Soc. Trans. 2005;33:479–481. doi: 10.1042/BST0330479. [DOI] [PubMed] [Google Scholar]
- Ziehler W.A., Morris J., Scott F.H., Millikin C., Engelke D.R. An essential protein-binding domain of nuclear RNase P RNA. RNA. 2001;7:565–575. doi: 10.1017/s1355838201001996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]