Abstract
The Prp19-associated complex, consisting of at least eight protein components, is involved in spliceosome activation by specifying the interaction of U5 and U6 with pre-mRNA for their stable association with the spliceosome after U4 dissociation. We show here that yeast cells depleted of one or two of the Prp19-associated components, accumulate the free form of U4. In NTC25-deleted cells, the level of U6 was also reduced. Extracts prepared from NTC25-deleted cells contained neither free U4 nor U6 and were ineffective in spliceosome recycling in the in vitro splicing reaction. Overexpression of U6 partially rescued the temperature-sensitive growth defect and decreased the relative amount of free U4 in NTC25-deleted cells, indicating that the accumulation of free U4 was a consequence of insufficient amounts of U6 snRNA. Extracts prepared from U6-overproducing NTC25-deleted cells containing free-form U6 were capable of spliceosome recycling, suggesting a role of free U6 RNP in spliceosome recycling. Our results demonstrate that in addition to direct participation in spliceosome activation, the Prp19-associated complex has an indirect role in spliceosome recycling through affecting the biogenesis of U4/U6 snRNP in the in vivo splicing reaction.
Keywords: Prp19, U4, U6, spliceosome recycling
INTRODUCTION
Splicing of pre-mRNA takes place on a large dynamic ribonucleoprotein complex, the spliceosome, which assembles through ordered interactions of four small nuclear ribonucleoprotein particles (snRNPs), U1, U2, U4/U6, and U5, and numerous protein factors (Will and Lührmann 1997; Staley and Guthrie 1998; Burge et al. 1999; Brow 2002; Hartmuth et al. 2002; Jurica et al. 2002; Zhou et al. 2002). During spliceosome assembly, U1 first binds to the 5′-splice site of the pre-mRNA, followed by binding of U2 to the branch site. The pre-formed U4/U6.U5 tri-snRNP is then recruited to the spliceosome to form a complex containing all five snRNAs. Subsequently, a large conformational rearrangement in the spliceosome occurs in which U1 and U4 are released, accompanied by formation of new base-pairing between U6 and U2, and U6 and the 5′-splice site of the pre-mRNA. This leads to the activation of the spliceosome, on which catalytic reactions can then take place (Brow 2002).
Activation of the spliceosome requires unwinding of base-pairing between U1 and the 5′-splice site and between U4 and U6 as the first step, freeing the 5′-splice site sequence and U6 for new base-pair formation. Factors that mediate unwinding of these base-pairings have not been directly demonstrated, although Brr2 has been implicated in the unwinding of U4/U6 (Raghunathan and Guthrie 1998a), and Prp28 in displacing U1 from the 5′-splice site (Staley and Guthrie 1999; Chen et al. 2001b). U4 snRNA is presumed to function as the repressor of U6 prior to the activation of the spliceosome and does not participate in catalytic reactions (Yean and Lin 1991). After release from the spliceosome, U4 appears to reassociate with U6 immediately to form the di-snRNP complex, and the free form is normally not detected, or only in a very small amount in splicing extracts or during the splicing reaction. The RNA-binding protein Prp24 has been shown to catalyze annealing of U4 and U6, and functions in regeneration of U4/U6 di-snRNP for recycling of the spliceosome (Ghetti et al. 1995; Raghunathan and Guthrie 1998b; Bell et al. 2002; Medenbach et al. 2004). In splicing extracts, U6 is present in excess of U4, and free U6 RNP exists aside from those base-paired with U4 in the U4/U6 di-snRNP or U4/U6.U5 tri-snRNP complexes (Cheng and Abelson 1987). Prp24 has been shown to be associated with U6 in wild-type extracts, but also associated with U4/U6 in U4-G14C mutant extracts, or upon incubation of wild-type extracts with ATP (Shannon and Guthrie 1991; Jandrositz and Guthrie 1995; Raghunathan and Guthrie 1998b). It has therefore been proposed that Prp24 functions to anneal U4 and U6 by binding to U6 first, and is dissociated from U4/U6 upon binding of U5 on formation of tri-snRNP (Raghunathan and Guthrie 1998b). The presence of free U6 is presumed to be important, although never directly demonstrated, for efficient formation of U4/U6 di-snRNP when U4 is dissociated from the spliceosome, since U6 remains associated with the spliceosome until the splicing reaction is complete.
After U4 and U1 are dissociated from the spliceosome, a protein complex associated with Prp19 (named NTC, standing for nineteen complex), identified in the budding yeast Saccharomyces cerevisiae, is required to stabilize binding of U5 and U6 on the spliceosome (Chan et al. 2003). Stabilization of U6 is achieved by confining interactions of U5 and U6 with pre-mRNA to specific positions from a dynamic state, and destabilizing Sm-like (Lsm) complex from U6 to expose the Lsm-binding site of U6 for more interactions with the intron sequence (Chan et al. 2003; Chan and Cheng 2005). NTC consists of at least eight protein components (Tarn et al. 1994; Chen et al. 1998; Tsai et al. 1999; Chen et al. 2001a, 2002), many of which are evolutionarily conserved (Ohi et al. 1998; Dix et al. 1999). Proteomic analysis has revealed a similar protein complex in mammals and in the fission yeast Schizosaccharomyces pombe (McDonald et al. 1999; Ohi et al. 2002). All of the identified NTC components are also found associated with the yeast penta-snRNP, which has been proposed as the preassembled functional spliceosome (Stevens et al. 2002).
In this report, we show that free U4 accumulates in cells depleted of NTC components. Analysis of NTC25 (SNT309)-deleted cells revealed that the amount of U6 snRNA was lower in these cells than in wild-type cells, suggesting that NTC has a secondary effect on the biogenesis of U4 and U6 snRNAs. Splicing extracts prepared from NTC25-deleted cells contained neither free U4 nor U6 and exhibited a defect in recycling of the spliceosome, possibly due to failure in regeneration of U4/U6 di-snRNP during the splicing reaction. Our results suggest a functional link between NTC, U4/U6 biogenesis, and spliceosome recycling.
RESULTS
Accumulation of free U4 in NTC-deficient cells
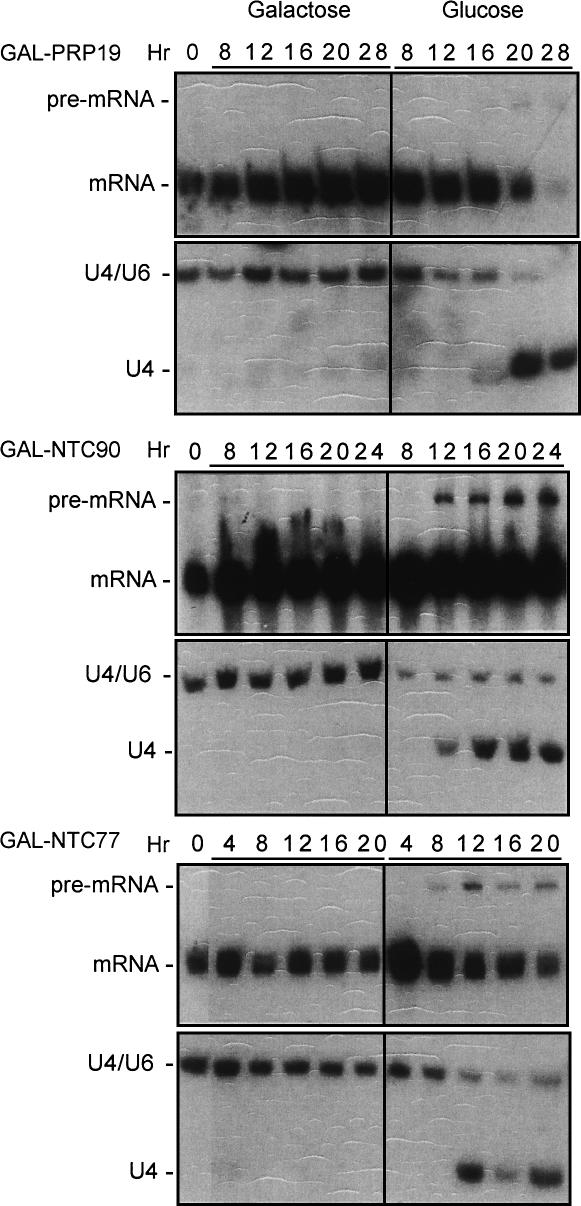
A novel yeast splicing factor, Sad1, was identified in a genetic screen for snRNP assembly factors (Lygerou et al. 1999). The temperature-sensitive sad1-1 mutant accumulates free U4 when shifted to restrictive temperatures. Interestingly, prp19-1 and prp17-1 mutants were shown to exhibit the same phenotype of accumulating free U4 (Lygerou et al. 1999). Since the Prp19-associated complex functions after U4 is dissociated from the spliceosome (Chan et al. 2003), the free U4 accumulation phenotype of prp19-1 might be a result of incomplete spliceosome assembly after U4 dissociation due to malfunction of NTC. In such a case, U4 accumulation may be a general phenotype associated with NTC deficiency rather than a consequence specific to prp19 defect. In order to determine whether U4 accumulation is due to NTC deficiency or a prp19-specific defect, we created several constructs carrying the conditional alleles of genes encoding NTC components under the control of the GAL promoter to regulate their expression. The PRP19, NTC90 (SYF1), and NTC77 (CLF1) genes tested are all essential for cellular growth. Cells were grown in media supplemented with galactose then shifted to glucose-containing media. RNA was isolated from these cells and fractionated on both denaturing and nondenaturing polyacrylamide gels for Northern blotting. As shown in Figure 1, Northern blots probed with the intron-containing gene CRY1 revealed accumulation of pre-mRNA ∼12–16 h after shifting to glucose in all three strains, indicative of a splicing defect (Fig. 1, upper panels). Although the amount of mRNA decreased on prolonged growth in glucose for GAL-PRP19 and GAL-NTC77, no significant reduction in the mRNA level was seen for GAL-NTC90. Western blot analysis revealed the presence of a residual amount of Ntc90, but not Ntc77, even after prolonged growth in glucose (data not shown). It is not known whether residual Ntc90 was able to support a low level of splicing to yield more mRNA in GAL-NTC90. Nevertheless, a splicing defect was observed in all three depletion strains. Correspondingly, free U4 accumulated upon prolonged incubation in glucose media (Fig. 1, lower panels). Cells grown in galactose-containing media did not accumulate pre-mRNA or free U4. These results suggest a link between free U4 accumulation and splicing defect resulting from NTC deficiency.
FIGURE 1.
Free U4 accumulated in cells metabolically depleted of Prp19, Ntc90, or Ntc77. Cells with the PRP19, NTC90, or NTC77 gene under the control of a GAL promoter were grown in galactose or glucose. RNA was extracted from cells retrieved at various times, and fractionated on polyacrylamide denaturing (upper panels) or nondenaturing gels (lower panels) for Northern blotting and probed with CRY1 and U4, respectively.
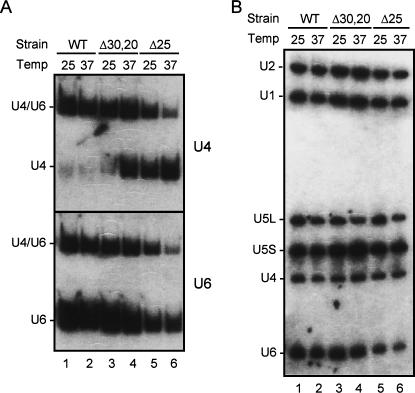
A more prominent effect was seen from the analysis of null mutants of NTC components. Ntc30 (Isy1), Ntc25, and Ntc20 are components of the Prp19-associated complex, and encoded by genes not essential for cell viability. Cells deleted of NTC25 (Δ25) or of both NTC30 and NTC20 (Δ30,20) are viable but temperature-sensitive and slow growing (Chen et al. 1998, 2001a). Total RNA was isolated from Δ25 or Δ30,20 cells grown only at 25°C or at 25°C then shifted to 37°C for 2 h and analyzed by Northern blotting. Figure 2A shows that in wild-type cells, only trace amounts of free U4 were present (Fig. 2A, lanes 1,2). In Δ30,20 cells, a significant amount of free U4 accumulated at 37°C but not at 25°C (Fig. 2A, lanes 3,4), whereas in Δ25 cells, free U4 accumulated at 25°C, and even more conspicuously at 37°C (Fig. 2A, lanes 5,6) accompanied by a reduction in U4/U6. This further confirms that free U4 accumulation is associated with NTC deficiency.
FIGURE 2.
Free U4 accumulated in Δ25 and Δ30,20 cells. Total RNA was isolated from wild-type, Δ25, or Δ30,20 cells grown at 25°C or at 25°C, then shifted to 37°C for 2 h. (A) RNA was fractionated on a 5% nondenaturing polyacrylamide gel for Northern blotting probed sequentially with U4 and U6. (B) RNA was fractionated on a 5% polyacrylamide/8 M urea gel for Northern blotting and probed with U1, U2, U4, U5, and U6.
When the same blot was probed with U6, notably the total amount of U6 was also low in Δ25 cells (Fig. 2A, lanes 5,6). This was better revealed when total RNA was analyzed on denaturing gels for comparison with other snRNAs as shown in Figure 2B. While the overall level of other snRNAs did not vary significantly, the amount of U6 was lower in Δ25 cells at both 25°C and 37°C (Fig. 2B, lanes 5,6).
Overexpression of U6 partially rescues the growth defect of NTC-deficient cells
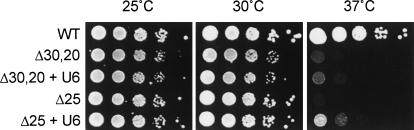
The fact that the total amount of U6 was lower in Δ25 cells raised a question as to whether the growth defect of these cells was a consequence of defective NTC or of U6 inadequacy. This was examined by testing whether overexpression of U6 could complement the growth defect observed in Δ25 and Δ30,20 cells. A multicopy plasmid containing the U6 gene, pRS424-U6, was transformed into Δ25 and Δ30,20 cells, and growth was examined by spot assays. As shown in Figure 3, both Δ25 and Δ30,20 were slower growing than wild-type cells at 37°C, with Δ25 being slightly more adverse than Δ30,20. Overexpression of U6 partially complemented the growth defect of Δ25, but did not significantly improve the growth observed in Δ30,20 cells. In contrast, overexpression of U4 had no detectable effect on the growth of these cells (data not shown). This suggests that the growth defect of Δ25 and Δ30,20 cells might be partly, but not entirely, due to U6 deficiency. The correlation between NTC and U4/U6 biogenesis was further analyzed in Δ25 cells, which gave more drastic effects on free U4 accumulation and reduction in U6.
FIGURE 3.
Spot assays on YPD of Δ30,20 and Δ25 cells carrying plasmids pRS426 or pRS426-U6 (+U6). Cells were grown at 25°C until mid-log phase, normalized to the same density, and diluted in series by 10-folds.
Overexpression of U6 partially complements the free U4 accumulation phenotype of Δ25 cells
In both yeast and mammalian cells, U4 and U6 snRNAs form a di-snRNP complex, on which the two RNA are associated through extensive base-pairing, which further associates with U5 to form a tri-snRNP particle. Nearly all U4 snRNA is associated with U6 snRNA, whereas a large fraction of U6 exists in an independent RNP particle since U6 is more abundant than U4 (Brow and Guthrie 1988; Fig. 2A). The function of the free U6 particle is not known, but is speculated to play a role in facilitating U4/U6 recycling during the splicing reaction. When U6 is underexpressed, U4 snRNP might accumulate as a free form if it is unable to immediately reassociate with U6 to form U4/U6 di-snRNP following dissociation from the spliceosome. This may account for the phenotype of free U4 accumulation in NTC-defective cells and partial complementation of growth defect in these cells by overexpression of U6. Indeed, Northern blotting revealed that although overexpression of U6 did not completely relieve accumulation of free U4 in Δ25 cells, the relative amount of U4 to U4/U6 was much lower at both 25°C and 37°C as shown in Figure 4. These results indicate that free U4 accumulation is the consequence of U6 inadequacy, which is intimately linked to NTC deficiency.
FIGURE 4.
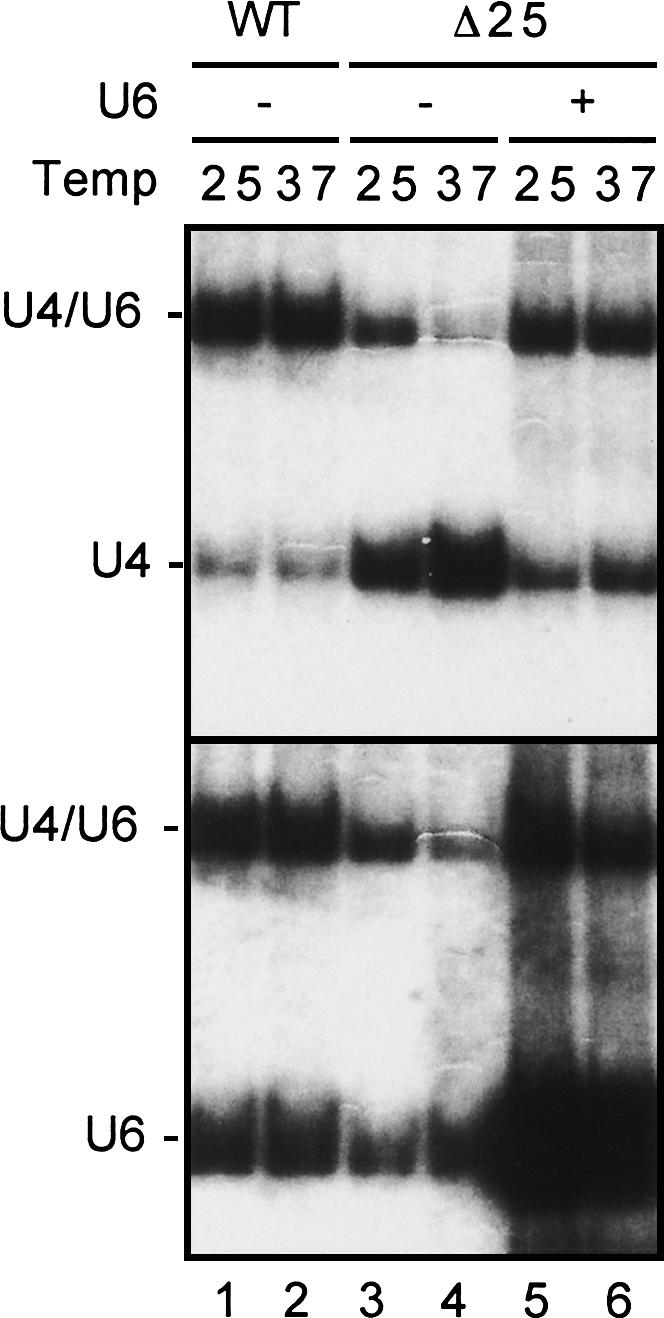
Northern blotting of total RNA isolated from Δ25 cells carrying plasmids pRS426 or pRS426-U6 (+U6). Cells were grown at 25°C, then shifted to 37°C for 2 h. Total RNA was isolated and fractionated on a 5% nondenaturing polyacrylamide gel for Northern blotting and probed sequentially with U4, then U6.
Spliceosome recycling in Δ25 cells
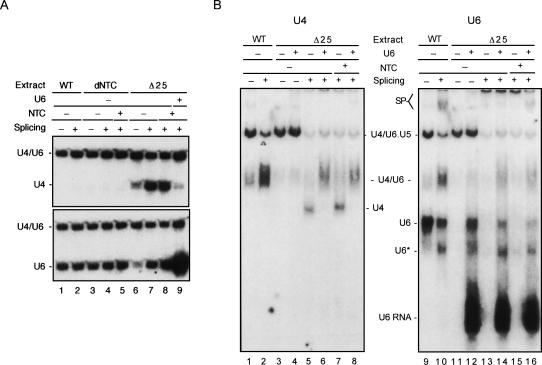
Although free U4 accumulated in Δ25 cells, we found that splicing extracts prepared from such cells did not contain free U4, as shown in Figure 5 (upper panel, cf. lanes 2 and 4), suggesting loss of free U4 during extract preparation. Interestingly, there was also a complete lack of free U6 in the extract from Δ25 cells (Fig. 5, lower panel, cf. lanes 2 and 4) as opposed to that from wild-type cells (Fig. 5, lower panel, cf. lanes 1 and 3). This raises a possibility that there might exist two forms of free U6, one of which was isolated in the wild-type extract and the other was not. Δ25 cells may contain only the free-form U6 that is excluded from the splicing extract. Consistent with this notion, the relative amount of free U6 to U4/U6 was lower in wild-type splicing extracts than in total RNA (Fig. 5, lower panel, cf. lanes 1 and 3), suggesting that a fraction of free U6 was not isolated in the wild-type splicing extract.
FIGURE 5.
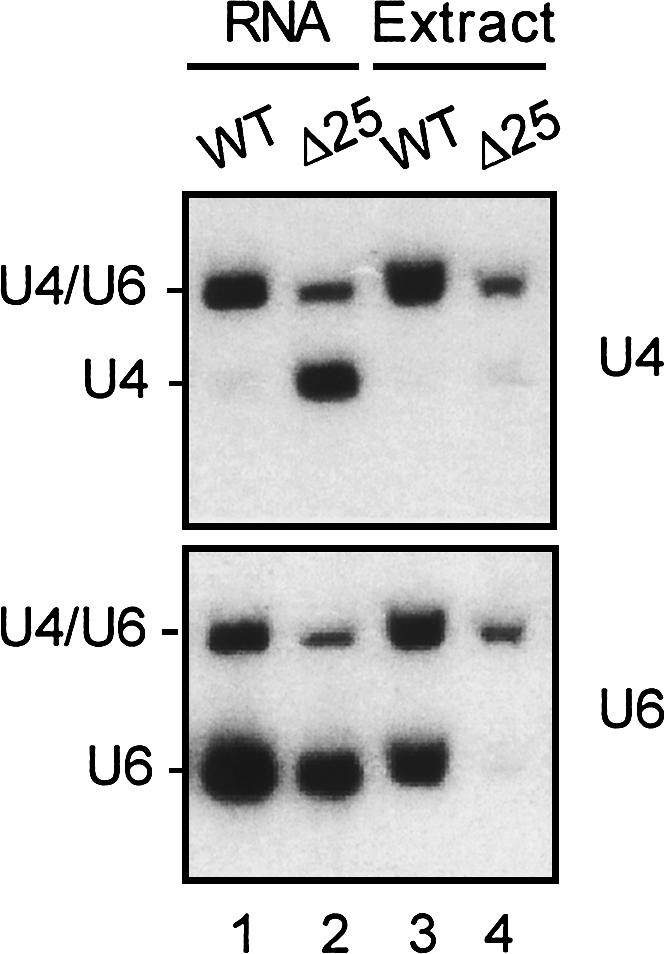
Free U4 and U6 in Δ25 cells were not isolated in splicing extracts. RNA was extracted from wild-type or Δ25 total cell lysates or splicing extracts and fractionated on a 5% polyacrylamide gel for Northern blotting probed sequentially with U4, then U6.
Extracts prepared from Δ25 cells normally have low splicing activity, but can be complemented by the addition of purified NTC (Chen et al. 1999). When total RNA was isolated from splicing reaction mixtures, fractionated on nondenaturing polyacrylamide gels, and analyzed by Northern blotting, free U4 was found to accumulate upon splicing in the presence or absence of added NTC, as shown in Figure 6A (upper panel, lanes 6–8). In contrast, splicing in in vitro NTC-depleted extracts (dNTC) did not result in accumulation of free U4 with or without prior addition of NTC (Fig. 6A, lanes 3–5), as in wild-type extracts (Fig. 6A, lanes 1,2). This indicates that free U4 was generated from the splicing reaction independent of NTC, consistent with the notion of U4 dissociation prior to NTC function (Chan et al. 2003). When the same blot was probed with U6, it was found that a great amount of free U6 also appeared after splicing in Δ25 extracts (Fig. 6A, lower panel, lanes 6–8). Nevertheless, the splicing-generated free U4 and U6 did not anneal to form U4/U6 di-snRNP. Interestingly, when in vitro synthesized U6 RNA was added to the Δ25 extract, free U4 no longer accumulated after splicing (Fig. 6A, lane 9), suggesting that the exogenously added U6 RNA could anneal with the released U4.
FIGURE 6.
Free U4 accumulated in Δ25 extracts after the splicing reaction. (A) Splicing reactions were carried out in wild-type, NTC-depleted, or Δ25 extracts, with or without addition of affinity-purified NTC at a concentration of ∼5 nM. The in vitro synthesized U6 was also added to one reaction in the Δ25 extract (lane 9). RNA was extracted from reaction mixtures and fractionated on a 5% polyacrylamide gel for Northern blotting and probed sequentially with U4, then U6. (B) Splicing reactions were carried out in wild-type or Δ25 extracts, with or without addition of purified NTC. To some reactions carried out in the Δ25 extracts, the in vitro synthesized U6 was also added prior to the reaction (lanes 12,14,16). Reaction mixtures were fractionated on a 4% nondenaturing polyacrylamide gels for Northern blotting and probed sequentially with U4, then U6.
To examine the nature of the free U4 and U6 generated from the splicing reaction in Δ25 extracts, total splicing reaction mixtures were analyzed on nondenaturing polyacrylamide gels for RNP as shown in the Northern blots of Figure 6B. In Δ25 extracts, nearly all U4 and U6 were present in the form of tri-snRNP (Fig. 6B, lanes 3,11). After splicing, U4 became free in the presence or absence of NTC (Fig. 6B, lanes 5,7). When the in vitro synthesized U6 RNA was added to Δ25 extracts, free U4 no longer appeared, but was replaced by the paired U4/U6 form (Fig. 6B, lanes 6,8). This indicates that despite lacking free U6, sufficient amounts of the protein components of U6 and Prp24 were present in the Δ25 extract for formation of U4/U6 after the splicing reaction. Consistently, the blot probed with U6 revealed formation of U4/U6, U6, and U6*, a form of U6 RNP containing no Prp24 (Raghunathan and Guthrie 1998b), when the in vitro synthesized U6 RNA was added to Δ25 extracts (Fig. 6B, lane 12). However, U6 was not seen in its free form after splicing in the absence of exogenously applied U6 in Δ25 extracts, but was associated with the spliceosome (Fig. 6B, lanes 13,15), indicating that the free U6 RNA seen on the RNA gel (Fig. 6A, lower panel, lanes 7,8) was the spliceosomal form of U6 that became free after deproteinization. These results suggest that the accumulation of free U4 in Δ25 extracts upon splicing was a consequence of a lack of free U6 for regeneration of U4/U6.
It has been shown that Prp24 mediates formation of U4/U6 di-snRNP and is required for spliceosome recycling (Raghunathan and Guthrie 1998b; Bell et al. 2002). Spliceosome recycling is inhibited by depletion of Prp24 from splicing extracts, and can be restored upon addition of recombinant Prp24. Since Prp24 is associated with U6 snRNP, depletion of Prp24 concurrently depletes free U6 from splicing extracts. However, the addition of recombinant Prp24 to the depleted extract promotes formation of free U6 snRNP (Raghunathan and Guthrie 1998b). This raises the question as to whether free U6 snRNP is also required for spliceosome recycling, since in its absence reannealing of U4/U6 needs to await completion of the splicing reaction for disassembly of the spliceosome to release U6.
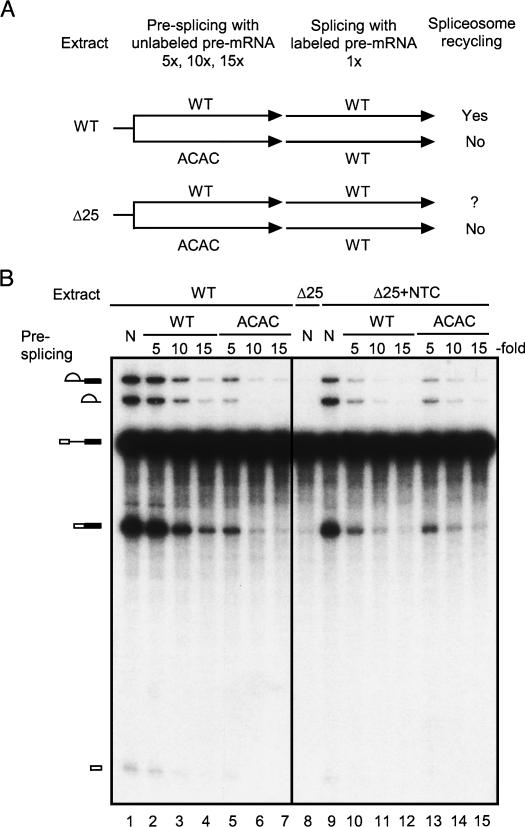
To address whether free U6 contributes to efficient recycling of the spliceosome, we performed a splicing assay designed to detect spliceosome recycling as described by Raghunathan and Guthrie (1998b) using Δ25 extracts. A schematic presentation of such an assay is shown in Figure 7A. Splicing was carried out with increasing amounts of unlabeled pre-mRNA in a presplicing reaction to force at least one round of splicing, then standard amounts of labeled pre-mRNA were added for the splicing assay. A 3′-splice site mutant pre-mRNA ACAC (Vijayraghavan et al. 1986) was used in comparison with the wild-type pre-mRNA in presplicing to reveal recycling of the spliceosome. The ACAC pre-mRNA is able to assemble the spliceosome and undergo the first catalytic reaction, but is blocked for the second step of the splicing reaction. As a result, the spliceosome containing partially spliced mRNA accumulates and is unable to release splicing factors for recycling. Splicing of subsequently added labeled pre-mRNA will be prohibited if enough unlabeled pre-mRNA is used in presplicing to sequester spliceosomal components. In contrast, presplicing using wild-type pre-mRNA allows recycling of spliceosomal components, and splicing of labeled pre-mRNA is not prohibited unless a large excessive amount of unlabeled pre-mRNA is used in presplicing. Whether spliceosome recycles can be judged by comparison of the level of splicing in response to the amount of unlabeled wild-type and ACAC pre-mRNA used in presplicing. Figure 7B shows that in wild-type extracts, splicing of labeled pre-mRNA decreased dramatically with increasing amounts of ACAC used in presplicing (Fig. 7B, lanes 1,5–7). A fivefold excess of unlabeled ACAC pre-mRNA in presplicing resulted in reduction of splicing activity by several folds (Fig. 7B, lane 5). A similar pattern was seen in Δ25 extracts supplemented with affinity-purified NTC (Δ25+NTC) (Fig. 7B, lanes 13–15), suggesting comparable concentrations of spliceosomal components in wild-type and Δ25 extracts. In wild-type extracts, splicing was affected to a lesser extent after presplicing with unlabeled wild-type pre-mRNA (Fig. 7B, lanes 1–4) with no significant effect at fivefold excess (Fig. 7B, lane 2). This indicates that spliceosome recycling did occur with wild-type pre-mRNA. In contrast, presplicing with wild-type and ACAC pre-mRNA affected splicing to the same level in Δ25+NTC extracts (Fig. 7B, cf. lanes 9–12 and lanes 9,13–15), indicating no recycling in Δ25 extracts using either wild-type or ACAC pre-mRNA.
FIGURE 7.
Spliceosome recycling assays. (A) A scheme for spliceosome recycling assays. (B) Splicing reactions were carried out in wild-type (lanes 1–7) or Δ25 extracts supplemented with affinity-purified NTC (∼5 nM) (lanes 9–15) for 30 min using 0.4 nM labeled actin pre-mRNA. Presplicing was carried out without (lanes 1,8,9) or with fivefold (2 nM), 10-fold (4 nM), or 15-fold (6 nM) excessive amounts of unlabeled wild-type (lanes 2–4,10–12) or ACAC (lanes 5–7,13–15) over labeled pre-mRNA for 30 min prior to the addition of labeled pre-mRNA. (WT) Wild type, (N) no presplicing.
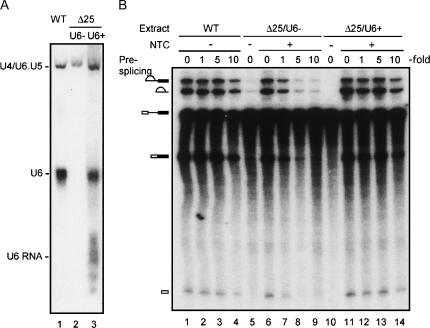
To confirm that failure in spliceosome recycling in Δ25 extracts was due to U6 deficiency, U6 was overexpressed in Δ25 cells and extracts prepared from the U6-overproducing strain were assayed for spliceosome recycling as described above, in comparison with wild-type and Δ25 extracts. Analysis by electrophoresis on nondenaturing polyacrylamide gels for ribonucleoprotein complexes revealed that the U6-overproducing Δ25 extract contained free U6 RNP and U4/U6.U5 tri-snRNP as shown in Figure 8A (lane 3) as opposed to standard Δ25 extracts, which contained only the tri-snRNP (Fig. 8A, lane 2). In addition, some faster migrating species, speculated to be free U6 RNA, were also detected. Figure 8B shows that in spliceosome recycling assays, while Δ25 extracts failed in spliceosome recycling (Fig. 8B, lanes 6–9), the U6-overproducing Δ25 extract, when supplemented with NTC, showed recycling activity with up to five- to 10-fold excess of pre-mRNA in the presplicing reaction (Fig. 8B, lanes 11–14) as in the wild-type extract (Fig. 8B, lanes 1–4). This indicates that free U6 RNP may play an important role in spliceosome recycling.
FIGURE 8.
Spliceosome recycling in U6-overproducing Δ25 extracts. (A) Wild-type (lane 1), Δ25 (lane 2), and U6-overproducing Δ25 (lane 3) extracts were subjected to electrophoresis on native polyacrylamide gels, and RNA was analyzed by Northern blotting. (U6−) Not overproducing U6, (U6+) overproducing U6. (B) Spliceosome recycling assays were carried out as in Figure 7 with onefold (0.4 nM), fivefold (2 nM), or 10-fold (4 nM) excessive amounts of unlabeled wild-type over labeled pre-mRNA in wild-type (lanes 1–4), Δ25 (lanes 6–10), or U6-overproducing Δ25 extracts (lanes 11–14) supplemented with affinity-purified NTC at a concentration of ∼5 nM. (WT) Wild type, (Δ25/U6−) Δ25 not overproducing U6, (Δ25/U6+) U6-overproducing Δ25.
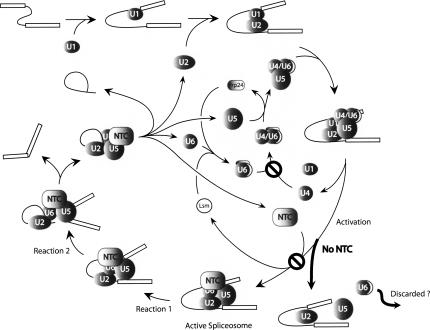
Figure 9 shows a diagram of the pathway for assembly and cycling of the yeast spliceosome illustrating our results. In the absence of NTC, formation of the active spliceosome is blocked and spliceosome assembly undergoes a discard pathway, as shown by the thick arrows, leading to U6 instability. As a consequence, cells defective in NTC function have lower levels of U6 snRNA and fail to form U4/U6, resulting in the accumulation of free U4 snRNA. Altogether, our results suggest that NTC malfunction can result in deficiencies to U4/U6 biogenesis and consequently affects spliceosome recycling.
FIGURE 9.
Schematic representation of the spliceosome cycle in yeast showing ordered interactions of snRNAs and NTC, and recycling of Prp24 and Lsm complex. Thick arrows represent the discard pathway in the absence of NTC that might lead to discard of U6. Ø indicates blockage of the pathway.
DISCUSSION
In this study, we demonstrated that free U4 accumulation was a common phenotype of cells defective in the function of NTC. Having extensively analyzed Δ25 cells, which showed a more drastic phenotype, we found that although the total amount of U4 was not affected in NTC-deficient cells, the level of U6 snRNA was reduced. Reduced levels of U6 have previously been reported in mutants of genes encoding U6, U4/U6, or tri-snRNP components, including PRP3, PRP4, PRP6, PRP24, PRP38, and LSM genes (Blanton et al. 1992; Xie et al. 1998; Mayes et al. 1999). Since these proteins are all involved in the biogenesis of U6-related RNP particles, it is not surprising that their deficiency results in reduced levels of U6 in the cell. In fact, Blanton et al. (1992) also observed a reduction in U6 level by more than threefold in a prp19-1 mutant shifted to restrictive temperatures, although the function of Prp19 was not known at the time. Our finding was consistent with this earlier observation and further extended this phenotype to defects in other NTC components. Distinct from other U6-related components, NTC is not tightly associated with any of the five spliceosomal snRNAs, and only functions in spliceosome activation after U4 is dissociated. Given that NTC is required for stable association of U5 and U6 with the spliceosome after U4 is dissociated, it is possible that destabilization of U6 from the spliceosome in NTC-deficient cells might result in U6 instability, subjecting it to a futile pathway.
The 3′-terminal U-tract of U6, recognized by the Lsm complex, is shown to interact with the intron sequence near the 5′-splice site in the activated spliceosome. This interaction requires prior release of Lsm proteins in an NTC-dependent manner (Chan et al. 2003). Thus, the U6 snRNA destabilized from the premature spliceosome due to the lack of NTC may still associate with the Lsm complex. The U6-associated Lsm complex, which contains Lsm2–Lsm8, shares six subunits with another Lsm complex involved in mRNA degradation (Bouveret et al. 2000). These two heptameric complexes differ only by one subunit yet have distinct functions. While the Lsm2–Lsm8 complex plays a role in stabilizing U6 and facilitating U4/U6 assembly, the Lsm1–Lsm7 complex is involved in mRNA degradation. Phylogenetic analysis reveals that among Lsm proteins, Lsm1 and Lsm8 are both related to SmB of the Sm protein complex (Salgado-Garrido et al. 1999). How these two closely related Lsm complexes distinguish their substrates and function differently is currently unknown. Considering its functional analogy to the Lsm1–Lsm7 complex, it is tempting to think that the Lsm2–Lsm8 protein complex might have an additional function in disposing of U6 snRNA when the normal U4/U6 assembly pathway is interfered with.
Interestingly, free U4 and U6, found in total RNA in Δ25 cells, were not found in the splicing extract prepared from these cells. Since U4 does not normally exist in its free form, this abnormally accumulated free U4 particle in Δ25 cells might be targeted to or associated with distinct structural units to escape extraction. In contrast, free U6 RNP is normally present in the cell and in splicing extracts. The fact that free U6 can be isolated in wild-type but not in Δ25 splicing extracts implies that the nature of free U6 in these cells might be different. Comparison of U6 in total RNA and splicing extracts reveals that the relative amount of U6 to U4/U6 in the wild-type splicing extract is lower than that in total RNA (Fig. 5, cf. lanes 1 and 3), indicating that a fraction of free U6 was not isolated in the splicing extract from wild-type cells. This suggests a possibility that the free U6 seen in total RNA may represent two different forms, among which only one was isolated in the splicing extract, and Δ25 cells contained only the form resistant to extraction. Since U6 accumulated in the spliceosome after splicing in Δ25 extracts but was revealed as free RNA upon extraction of RNA from the splicing reaction mixture (Fig. 6A, lanes 7,8), we speculate that in vivo these extraction-resistant U4 and U6 might be associated with the spliceosome such that they cannot be isolated in the splicing extract. This is consistent with previous observations that yeast splicing extracts contain only negligible amounts of endogenous spliceosome (Cheng and Abelson 1987).
It is interesting that Δ25 extracts were incapable of spliceosome recycling and that overexpression of U6 antagonized this block, indicating that a certain amount of U6 was required for recycling. Although the mechanism by which U6 was down-regulated in Δ25 cells is not known, overexpression of U6 by transforming a multicopy U6-containing plasmid was sufficient to overcome the defect caused by down-regulation of U6 in Δ25 cells, including partial rescue of the growth defect, regeneration of free U6 snRNP, and recovery of spliceosome recycling activity. This points to the possibility that the growth defect of Δ25 cells was in part attributed to failure in sustaining a sufficient amount of U6 snRNP, consequently blocking spliceosome recycling. Prp24, previously demonstrated to catalyze annealing of U4 and U6 to form U4/U6 di-snRNP for recycling of the spliceosome (Raghunathan and Guthrie 1998b), is associated with U6 as a component of U6 snRNP, but is not associated with the U4/U6.U5 tri-snRNP particle (Shannon and Guthrie 1991; Raghunathan and Guthrie 1998b). Immunoblot analysis in this study revealed that the amount of Prp24 was also lower in Δ25 extracts (data not shown), indicating that the steady-state level of Prp24 was also affected by the function of NTC. Nevertheless, overexpression of PRP24 did not rescue the growth defect of Δ25 cells, and overexpression of both U6 and PRP24 rescued the growth defect to the same level as overexpression of U6 alone (data not shown). In this context, Prp24 was probably not the primary factor affecting spliceosome recycling in Δ25 cells.
Although in vivo overproduction of U6 could recover spliceosome recycling activity in Δ25 extracts, addition of in vitro synthesized U6 RNA to Δ25 extracts failed to restore recycling activity (data not shown), despite the fact that U6 RNA could assemble into U6 snRNP and U4/U6 di-snRNP (Fig. 6B). Further attempts with the purified U6 snRNP particle, isolated by affinity chromatography on an anti-HA antibody column of Prp24-HA extracts, also failed to recover spliceosome recycling activity in Δ25 extracts. Since the thus purified U6 snRNP fraction contained Prp24 both in association with U6 and as a free protein, it is unlikely that failure in complementing Δ25 extracts in spliceosome recycling was due to insufficient amounts U6 snRNP or Prp24. This suggests that additional factors, besides U6 snRNP and Prp24, required for spliceosome recycling might be lacking in Δ25 extracts.
Our results demonstrate that NTC-deficient mutants exhibited a common phenotype of accumulating free U4 in the cell, likely as a result of down-regulation of U6 in response to a block in spliceosome activation after dissociation of U4. The cause of free U4 accumulation in sad1-1 and prp17-1 mutants is an interesting question. Although SAD1 was identified in a screen designed to isolate mutations affecting U4/U6 assembly, the sad1-1 mutant has a pre-mRNA accumulation phenotype (Lygerou et al. 1999), and the mechanistic role of Sad1 in U4/U6 assembly or in the pre-mRNA splicing reaction has not been determined. While the human Sad1 homolog was shown to be associated with the tri-snRNP (Makarova et al. 2001), yeast Sad1 was found associated with the yeast penta-snRNP particle like all known NTC components (Stevens et al. 2002). It will be interesting to see whether Sad1 has a similar function to NTC in spliceosome activation to account for the U4 accumulation phenotype in the sad1-1 mutant. Prp17 is one of several protein splicing factors involved in the second step of splicing catalysis (Jones et al. 1995). An allele of prp17 (slu4-1) was found to be synthetically lethal with specific mutations in U5 snRNA (Frank et al. 1992), although physical association of Prp17 with snRNA has not been demonstrated. Whether Prp17 plays a direct role in U4/U6 assembly is unclear. It is unlikely that the free U4 accumulation phenotype of prp17-1 is directly related to a block in the second catalytic reaction since other mutants affecting the second step do not exhibit the same phenotype (Lygerou et al. 1999), although the possibility of allele specificity cannot be excluded.
Taken together, our finding that deficiency in NTC function resulted in down-regulation of U6 and consequent accumulation of free U4 in these cells indicates that NTC not only directly participates in spliceosome activation, but also influences the biogenesis of U4/U6 snRNP in the in vivo splicing reaction. Our in vitro analysis of Δ25 extracts suggests that splicing might not efficiently recycle if NTC does not function properly. This establishes a functional link between NTC, spliceosome recycling, and U4/U6 biogenesis, and suggests that splicing factors involved in a distinct step of the splicing reaction may show pleiotropic phenotypes along the pathway.
MATERIALS AND METHODS
Yeast strains
BJ2168: Mata prc1 prb1 pep4 leu2 trp1 ura3
YSCC1: Mata prc1 prb1 pep4 leu2 trp1 ura3 PRP19HA
YSCC6: Mata leu2 his3 trp1 ade2 lys2 ura3 suc2 URA3::GAL1-PRP19
YSCC11: Mata prc1 prb1 pep4 trp1 ura3 PRP19HA NTC25::LEU2
YSCC14: Mata prc1 prb1 pep4 trp1 PRP19HA NTC20::LEU2 NTC30::URA3
YSCC773: Mata prc1 prb1 pep4 leu2 trp1 ura3 URA3::GAL1-NTC77
YSCC903: Mata prc1 prb1 pep4 leu2 trp1 ura3 URA3::GAL1-NTC90
YSCC24: Mata prc1 prb1 pep4 leu2 trp1 ura3 PRP24HA
Plasmids
pRS424-U6
A 1.2-kb DNA fragment containing the yeast U6 gene, plus 539 bp upstream and 517 bp downstream of the gene-coding sequence, was retrieved from the yeast genome by PCR and inserted into the BamHI site of plasmid vector pRS424.
pG1-PRP24
A 1.8-kb DNA fragment containing the ORF of the yeast PRP24 gene and 485 bp of the downstream region sequence was retrieved from the yeast genome by PCR and inserted into the BamHI site of plasmid vector pG-1.
Preparation of total RNA and splicing extracts
Yeast total RNA was isolated according to Lygerou et al. (1999). Splicing extracts were prepared either by dounce homogenization (Cheng et al. 1990) or by grinding with mortar and pestle (Ansari and Schwer 1995). NTC-depleted extracts were prepared by incubation of PRP19-HA extracts with anti-HA antibody as described (Chan et al. 2003). For metabolic depletion of essential NTC components, YSCC6, YSCC773, or YSCC903 cells were grown in YP-galactose medium until mid-log phase. To half of the culture was then added glucose to a final concentration of 2%, and both cultures were further incubated. Portions of cultures were removed at different periods of time of incubation for isolation of total RNA. For Δ25 and Δ30,20 strains, cells were grown in YPD at 25°C to mid-log phase, then shifted to 37°C or left at 25°C, and allowed to grow for 2 h prior to harvest for isolation of RNA or preparation of splicing extract. For overexpression of U6, strain BJ2168 carrying plasmid pRS424-U6 was grown in YPD until mid-log phase and harvested for preparation of splicing extracts.
Northern blotting
Total RNA from whole cells or from splicing extracts was fractionated by electrophoresis on 5% denaturing or nondenaturing polyacrylamide (29:1) gels and electroblotted onto GeneScreen membranes for Northern blotting probed with CRY1, U4, U6, or five snRNAs. For RNP analysis, splicing reactions were fractionated on 4% nondenaturing polyacrylamide (80:1) gels according to Raghunathan and Guthrie (1998b), followed by Northern blotting probed with U4 or U6 snRNA.
Spliceosome recycling assays
Splicing reactions were carried out with various amounts of unlabeled actin or ACAC pre-mRNA at 25°C for 30 min. Labeled pre-mRNA was then added to the reaction mixtures to a final concentration of 0.4 nM and further incubated for 30 min.
ACKNOWLEDGMENTS
We thank S. Rader and C. Guthrie for kindly providing antibodies against Prp24. We also thank P. Lin and members of Cheng's laboratory for helpful discussions and critical reading of the manuscript, and K.J. Deen for English editing. This work was supported by a grant from the Academia Sinica and National Science Council (Taiwan), NSC92-2321-B-001-017.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2292106.
REFERENCES
- Ansari A., Schwer B. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 1995;14:4001–4009. doi: 10.1002/j.1460-2075.1995.tb00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M., Schreiner S., Damianov A., Reddy R., Bindereif A. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 2002;21:2724–2735. doi: 10.1093/emboj/21.11.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton S., Srinivasan A., Rymond B.C. PRP38 encodes a yeast protein required for pre-mRNA splicing and maintenance of stable U6 small nuclear RNA levels. Mol. Cell. Biol. 1992;12:3939–3947. doi: 10.1128/mcb.12.9.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E., Rigaut G., Shevchenko A., Wilm M., Séraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Brow D.A., Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Tuschl T.H., Sharp P.A. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland R.F., et al., editors. The RNA world. 2d ed., Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. pp. 525–560. [Google Scholar]
- Chan S.-P., Cheng S.-C. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J. Biol. Chem. 2005;280:31190–31199. doi: 10.1074/jbc.M505060200. [DOI] [PubMed] [Google Scholar]
- Chan S.-P., Kao D.-I., Tsai W.-Y., Cheng S.-C. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- Chen H.-R., Jan S.-P., Tsao T.Y., Sheu Y.-J., Banroques J., Cheng S.-C. Snt309p, a component of the Prp19p-associated complex that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19p. Mol. Cell. Biol. 1998;18:2196–2204. doi: 10.1128/mcb.18.4.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-R., Tsao T.Y., Chen C.-H., Tsai W.-Y., Her L.-S., Hsu M.-T., Cheng S.-C. Snt309p modulates interactions of Prp19p with its associated components to stabilize the Prp19p-associated complex essential for pre-mRNA splicing. Proc. Natl. Acad. Sci. 1999;96:5406–5411. doi: 10.1073/pnas.96.10.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-H., Tsai W.-Y., Chen H.-R., Wang C.-H., Cheng S.-C. Identification and characterization of two novel components of the Prp19p-associated complex, Ntc30p and Ntc20p. J. Biol. Chem. 2001a;276:488–494. doi: 10.1074/jbc.M006958200. [DOI] [PubMed] [Google Scholar]
- Chen J.Y-F., Stands L., Staley J.P., Jackups R.R., Jr., Latus L.J., Chang T.-H. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell. 2001b;7:227–232. doi: 10.1016/s1097-2765(01)00170-8. [DOI] [PubMed] [Google Scholar]
- Chen C.-H., Yu W.-C., Tsao T.Y., Wang L.-Y., Chen H.-R., Lin J.-Y., Tsai W.-Y., Cheng S.-C. Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res. 2002;30:1029–1037. doi: 10.1093/nar/30.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.-C., Abelson J. Spliceosome assembly in yeast. Genes & Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Cheng S.-C., Newman A., Lin R.-J., McFarland G.D., Abelson J.N. Preparation and fractionation of yeast splicing extract. Methods Enzymol. 1990;181:89–96. doi: 10.1016/0076-6879(90)81114-a. [DOI] [PubMed] [Google Scholar]
- Dix I., Russell C.S., Ben-Yehuda S., Kupiec M., Beggs J.D. The identification and characterization of a novel splicing protein, Isy1p, of Saccharomyces cerevisiae . RNA. 1999;5:360–368. doi: 10.1017/s1355838299981396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D., Patterson B., Guthrie C. Synthetic lethal mutations suggest interactions between U5 small nuclear RNA and four proteins required for the second step of splicing. Mol. Cell. Biol. 1992;12:5197–5205. doi: 10.1128/mcb.12.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti A., Company M., Abelson J. Specificity of Prp24 binding to RNA: A role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA. 1995;1:132–145. [PMC free article] [PubMed] [Google Scholar]
- Hartmuth K., Urlaub H., Vornlocher H.P., Will C.L., Gentzel M., Wilm M., Lührmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandrositz A., Guthrie C. Evidence for a Prp24 binding site in U6 snRNA and in a putative intermediate in the annealing of U6 and U4 snRNAs. EMBO J. 1995;14:820–832. doi: 10.1002/j.1460-2075.1995.tb07060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.H., Frank D.N., Guthrie C. Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc. Natl. Acad. Sci. 1995;92:9687–9691. doi: 10.1073/pnas.92.21.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M.S., Licklider L.J., Gygi S.R., Grigorieff N., Moore M.J. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z., Christophides G., Séraphin B. A novel genetic screen for snRNP assembly factors in yeast identifies a conserved protein, Sad1p, also required for pre-mRNA splicing. Mol. Cell. Biol. 1999;19:2008–2020. doi: 10.1128/mcb.19.3.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O.V., Makarov E.M., Lührmann R. The 65 and 110 kDa SR-related proteins of the U4/U6 · U5 tri-snRNP are essential for the assembly of mature spliceosome. EMBO J. 2001;20:2553–2563. doi: 10.1093/emboj/20.10.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayes A.E., Verdone L., Legrain P., Beggs J.D. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald W.H., Ohi R., Smelkova N., Frendewey D., Gould K.L. Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol. 1999;19:5352–5362. doi: 10.1128/mcb.19.8.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medenbach J., Schreiner S., Liu S., Lührmann R., Bindereif A. Human U4/U6 snRNP recycling factor p110: Mutational analysis reveals the function of the tetratricopeptide repeat domain in recycling. Mol. Cell. Biol. 2004;24:7392–7401. doi: 10.1128/MCB.24.17.7392-7401.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R., Feoktistova A., McCann S., Valentine V., Look A.T., Lipsick J., Gould K.L. Myb-related Schizosaccharomyces pombe cdc5p is structurally and functionally conserved in eukaryotes. Mol. Cell. Biol. 1998;18:4097–4108. doi: 10.1128/mcb.18.7.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M.D., Link A.J., Ren L., Jennings J.L., McDonald W.H., Gould K.L. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 2002;22:2011–2024. doi: 10.1128/MCB.22.7.2011-2024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan P.L., Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 1998a;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- Raghunathan P.L., Guthrie C. A spliceosomal recycling factor that reanneals U4 and U6 snRNPs. Science. 1998b;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- Salgado-Garrido J., Bragado-Nilsson E., Kandels-Lewis S., Séraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon K., Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes & Dev. 1991;5:773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- Staley J.P., Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Staley J.P., Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- Stevens S.W., Ryan D.E., Ge H.Y., Moore R.E., Young M.K., Lee T.D., Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;8:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- Tarn W.-Y., Hsu C.-H., Huang K.-T., Chen H.-R., Kao H.-Y., Lee K.-R., Cheng S.-C. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 1994;13:2421–2431. doi: 10.1002/j.1460-2075.1994.tb06527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W.-Y., Chow Y.-T., Chen H.-R., Huang K.-T., Hong R.-I., Jan S.-P., Kuo N.-Y., Tsao T.Y., Chen C.-H., Cheng S.-C. Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing. J. Biol. Chem. 1999;274:9455–9462. doi: 10.1074/jbc.274.14.9455. [DOI] [PubMed] [Google Scholar]
- Vijayraghavan U., Parker R., Tamm J., Iimura Y., Rossi J., Abelson J., Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- Xie J., Beickman K., Otte E., Rymond B.C. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J. 1998;17:2938–2946. doi: 10.1093/emboj/17.10.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean S.-L., Lin R.-J. U4 small nuclear RNA dissociates from a yeast spliceosome and does not participate in the subsequent splicing reaction. Mol. Cell. Biol. 1991;11:5571–5577. doi: 10.1128/mcb.11.11.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Licklider L.J., Gygi S.P., Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]