Abstract
The human telomerase ribonucleoprotein particle (RNP) shares with box H/ACA small Cajal body (sca)RNPs and small nucleolar (sno)RNPs the proteins dyskerin, hGar1, hNhp2, and hNop10. How dyskerin, hGar1, hNhp2, and hNop10 assemble with box H/ACA scaRNAs, snoRNAs, and the RNA component of telomerase (hTR) in vivo remains unknown. In yeast, Naf1p interacts with H/ACA snoRNP proteins and may promote assembly of Cbf5p (the yeast ortholog of dyskerin) with nascent pre-snoRNAs. Here we show that the human HsQ96HR8 protein, thereafter termed hNaf1, can functionally replace endogenous Naf1p in yeast. HeLa hNaf1 associates with dyskerin and hNop10 as well as box H/ACA scaRNAs, snoRNAs, and hTR. Reduction of hNaf1 steady-state levels by RNAi significantly lowers accumulation of these components of box H/ACA scaRNP, snoRNP, and telomerase. hNaf1 is found predominantly in numerous discrete foci in the nucleoplasm and fails to accumulate within Cajal bodies or nucleoli. Altogether, these results suggest that hNaf1 intervenes in early assembly steps of human box H/ACA RNPs, including telomerase.
Keywords: H/ACA RNAs, RNPs, telomerase, nucleolus, Cajal bodies
INTRODUCTION
The telomerase holoenzyme catalyzes the addition of simple repeat sequences at telomeres, the ends of chromosomes. In all organisms in which it has been characterized, telomerase is a ribonucleoprotein particle (RNP), constituted of an RNA component associated with several proteins, some of which are species-specific (Cech 2004). The catalytic core of telomerase comprises a protein reverse transcriptase, termed hTERT in humans, and the so-called template sequence of telomerase RNA. This sequence is reverse-transcribed by the telomerase reverse transcriptase using telomere 3′ protruding ends as primers. In addition, the telomerase holoenzyme contains a domain organized around the 3′ terminal part its RNA moiety, which is implicated in the formation, trafficking, and stability of the holoenzyme (Chen and Greider 2004). In human telomerase, this domain is highly related to box H/ACA small RNPs (Mitchell et al. 1999a; Chen et al. 2000; Chen and Greider 2004; Jády et al. 2004). These RNPs function in RNA modification. The first box H/ACA RNPs to be discovered, the box H/ACA small nucleolar (sno)RNPs, accumulate in the nucleolus and are involved in modification of rRNAs and spliceosomal U6 snRNA (Kiss 2002). The box H/ACA small-Cajal-body specific (sca)RNPs, on the other hand, accumulate within Cajal bodies and are involved in nucleotide modification of spliceosomal snRNAs transcribed by RNA polymerase II (Darzacq et al. 2002). Box H/ACA snoRNPs and scaRNPs share four conserved proteins, dyskerin (also called Cbf5p in yeast and Nap57 in rodents), hGar1, hNhp2, and hNop10; and their RNA components contain two conserved box elements, termed the H-box and the ACA-box (Meier 2005). In addition, the RNA components of scaRNPs, scaRNAs, contain the so-called CAB-box that functions in the retention of scaRNPs within Cajal bodies (Richard et al. 2003). The human telomerase holoenzyme also possesses the proteins dyskerin, hGar1, hNhp2, and hNop10 (Cong et al. 2002; Chen and Greider 2004); and its RNA component (called hTR) features the conserved H-, ACA-, and CAB-boxes at its 3′-end (Mitchell et al. 1999a; Chen et al. 2000; Jády et al. 2004). Integrity of the H-box or ACA-box is necessary for normal processing and accumulation of hTR (Mitchell et al. 1999a). Likewise, modifications of dyskerin that cause dyskeratosis congenita induce a drastic drop in hTR levels (Mitchell et al. 1999b). Consistent with it possessing a CAB-box, hTR accumulates within Cajal bodies (Jády et al. 2004, 2006; Tomlinson et al. 2006). Thus human telomerase contains a bona fide box H/ACA RNP domain that is critical for the accumulation and localization of the holoenzyme.
How eukaryotic box H/ACA small RNPs and telomerase are assembled in vivo still remains little understood, although it has recently been shown that a Nap57/Nop10/Nhp2 complex could specifically bind to in vitro synthesized box H/ACA snoRNAs and hTR (Wang and Meier 2004) and that yeast Nop10p may directly contact RNA (Khanna et al. 2006). It has also been recently demonstrated that archaeal aCbf5 can directly and specifically bind in vitro to archaeal box H/ACA small RNAs (Baker et al. 2005; Charpentier et al. 2005). Moreover, recombinant archaeal H/ACA RNP proteins aCbf5, aGar1, L7Ae, and aNop10 can assemble onto the H/ACA domain of telomerase RNA (Hamma et al. 2005). Thus, it is tempting to speculate that dyskerin/Cbf5p also plays a pivotal role in the assembly of eukaryotic box H/ACA snoRNPs, scaRNPs, and telomerase. In yeast, the Naf1p protein might intervene in early steps of yeast box H/ACA snoRNP assembly. Yeast Naf1p is a nucleoplasmic protein that associates with RNA and protein components of box H/ACA snoRNPs (Dez et al. 2002; Fatica et al. 2002; Yang et al. 2002). Yeast Naf1p also interacts with the C-terminal domain of the large subunit of RNA polymerase II (Fatica et al. 2002). Moreover, both Naf1p and Cbf5p are found in close proximity to genes encoding snoRNAs, and such proximity depends on ongoing transcription (Ballarino et al. 2005; Yang et al. 2005). Thus, it is envisaged that Cbf5p is cotranscriptionally delivered by Naf1p to nascent box H/ACA pre-snoRNAs to initiate box H/ACA snoRNP assembly. We wondered whether human cells possess an ortholog of Naf1p and if so, whether it would be required for the accumulation of all RNPs containing a box H/ACA domain, including telomerase. We report here the characterization of human Naf1, hNaf1, and demonstrate that it is, indeed, required for accumulation of all types of box H/ACA RNPs, including telomerase.
RESULTS AND DISCUSSION
HsQ96HR8 is the human ortholog of yeast Naf1p
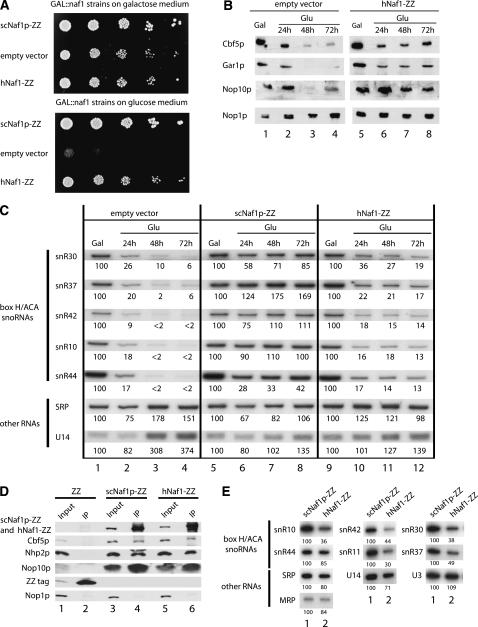
The hypothetical protein HsQ96HR8 was predicted by Fatica and colleagues (Fatica et al. 2002) to be the ortholog of yeast Naf1p. To check whether HsQ96HR8 could partially fulfill the functions of endogenous Naf1p, the gene encoding HsQ96HR8 was expressed constitutively from a plasmid in a yeast strain in which the NAF1 gene is transcribed from the GAL1-10 promoter (Dez et al. 2002); hence, in this strain, Naf1p expression can be turned off by switching cells from a medium containing galactose to one containing glucose as carbon source. As controls, the same strain was transformed with an empty plasmid, or the same plasmid containing the yeast NAF1 gene transcribed from the GAR1 promoter. Expression of HsQ96HR8 corrected to a significant degree the growth defect caused by depletion of endogenous Naf1p (Fig. 1A). Naf1p depletion leads to a drop in the steady-state levels of Cbf5p, Gar1p, Nop10p, and H/ACA snoRNAs (Fig. 1B,C, lanes 1–4; Dez et al. 2002). Expression of HsQ96HR8 significantly reduced the drop in the levels of these proteins and RNAs following depletion of endogenous Naf1p (Fig. 1B, lanes 5–8; Fig. 1C, lanes 9–12), and these effects were correlated with the ability of HsQ96HR8 to interact with yeast H/ACA snoRNP proteins (Fig. 1D). Finally, we showed by plasmid shuffling that haploid yeast cells devoid of the NAF1 gene are viable when they express human HsQ96HR8 from a plasmid (data not shown) and that steady-state levels of H/ACA snoRNAs in these cells range from ∼30% to 85% of wild-type levels (Fig. 1E). From these data, we conclude that HsQ96HR8 is, indeed, the ortholog of yeast Naf1p and is hence thereafter referred to as hNaf1.
FIGURE 1.
HsQ96HR8 (hNaf1) is the human ortholog of yeast Naf1p. (A) Expression of HsQ96HR8 improves growth of yeast following Naf1p depletion. Yeast strain GAL::naf1, in which the NAF1 gene is transcribed from the GAL1-10 promoter, was transformed with an empty expression vector (line empty vector) or derivatives thereof containing wild-type yeast NAF1 (line scNaf1p) or the human HsQ96HR8 gene (line hNaf1) transcribed from the GAR1 promoter and tagged with the ZZ sequence encoding two IgG-binding domains. The resulting transformed strains were grown in galactose-containing medium, washed, and grown for a further 24 h in glucose-containing medium. Serial dilutions of samples from galactose- or glucose-grown cultures were plated on galactose- or glucose-containing plates, respectively. (B) Expression of HsQ96HR8 increases steady-state levels of yeast H/ACA proteins Cbf5p, Gar1p, and Nop10p or (C) steady-state levels of yeast H/ACA snoRNAs, following depletion of endogenous yeast Naf1p. The transformed GAL::naf1 strains described in A were grown in galactose-containing medium, washed, and transferred to glucose-containing medium to repress transcription of chromosomal NAF1. Samples from galactose-grown cultures (GAL, lanes 1,5,9) or from cultures grown for 24 h (lanes 2,6,10), 48 h (lanes 3,7,11), or 72 h (lanes 4,8,12) were collected, from which total proteins (B) or RNAs (C) were extracted. In panel B, total proteins were subjected to SDS-polyacrylamide gel electrophoresis and transferred to a cellulose membrane. Specific proteins were detected by enhanced chemiluminescence using antibodies listed in the “Materials and Methods” section. In panel C, total RNAs were separated by denaturing acrylamide gel electrophoresis and transferred to nylon membranes. Specific RNAs were detected by hybridization with antisense oligonucleotide probes. Signal intensities were measured by PhosphorImager scanning. Values (indicated below each lane) are expressed as percentages of the intensities obtained in the case of galactose-grown samples. (D) HsQ96HR8 interacts with yeast box H/ACA snoRNP proteins. Transformed GAL::naf1 strains expressing the ZZ tag alone (lanes 1,2), tagged yeast Naf1p (lanes 3,4), or tagged human HsQ96HR8 expressed from the TDH3 promoter (lanes 5,6) were grown in galactose-containing medium, washed, and transferred to glucose-containing medium for 24 h. Total extracts were prepared and immunoprecipitation experiments were carried out using IgG-Sepharose beads. After precipitation and washing, beads were resuspended in protein denaturing buffer. Aliquots of the resulting supernatants (IP, lanes 2,4,6) and 1/20th of the corresponding input extract (Input, lanes 1,3,5) were submitted to SDS-PAGE and Western analysis as described in B. The upper band detected in lane 4, anti-Nhp2p Western, probably corresponds to a breakdown product of ZZ-tagged yeast Naf1p. (E) H/ACA snoRNA levels in naf1Δ cells expressing wild-type yeast Naf1p (lane 1) or human HsQ96HR8 (lane 2) from a plasmid. RNAs were analyzed as described for panel C.
hNaf1 interacts with protein and RNA components of HeLa box H/ACA snoRNPs, scaRNPs, and telomerase
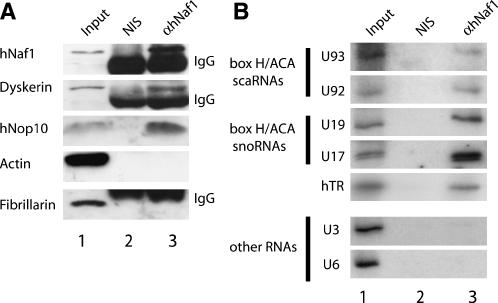
The human family of box H/ACA RNPs includes not only snoRNPs that accumulate in the nucleolus as in Saccharomyces cerevisiae, but also scaRNPs, which accumulate in Cajal bodies, and telomerase. We wondered whether hNaf1 does interact with protein and RNA components of all types of box H/ACA RNPs. Such hypothetical interactions were evaluated by precipitation experiments carried out using purified anti-hNaf1 antibodies (raised in rabbit against hNaf1 peptides) and HeLa cell extracts. hNaf1 interacts with dyskerin and hNop10 but not with fibrillarin (Fig. 2A), consistent with results obtained with yeast cells. Interaction with hGar1 and hNhp2 could not be evaluated for lack of suitable antibodies able to detect these proteins in the Western blot procedure. In addition, not only box H/ACA snoRNAs, but also box H/ACA scaRNAs and hTR were precipitated with anti-hNaf1 antibodies from total HeLa cell extracts (Fig. 2B). In contrast, we could not detect an interaction between hNaf1 and box C/D U3 snoRNA or spliceosomal U6 snRNA (Fig. 2B). We conclude that hNaf1 is specifically associated with protein and RNA components of box H/ACA snoRNPs, scaRNPs, and telomerase in HeLa cells.
FIGURE 2.
hNaf1 interacts with components of box H/ACA snoRNPs, box H/ACA scaRNPs, and telomerase. Immunoprecipitations were carried out using purified anti-hNaf1 antibodies (αhNaf1) or non-immune serum (NIS) and HeLa cell extracts. (A) Western analysis of hNaf1-associated proteins. After precipitation and washing, protein A-agarose beads were resuspended in protein denaturing buffer. Aliquots of the resulting supernatants (lanes 2,3) or 3% of the corresponding input extract (Input, lane 1) were submitted to SDS-PAGE and Western analysis as described in Figure 1B. The strong bands labeled “IgG” correspond to immunoglobulins detached from the beads during pellet resuspension after the precipitation. (B) Detection of small RNAs associated with hNaf1. RNAs extracted from the pellets obtained following precipitation (lanes 2,3) or from an amount of input extract (Input) corresponding to 3% of that used for precipitations were analyzed by Northern blot (to detect snRNAs, snoRNAs, and scaRNAs) as described in the legend of Figure 1C, or RNase protection to detect hTR.
hNaf1 is required to maintain normal cellular levels of box H/ACA RNP and human telomerase components
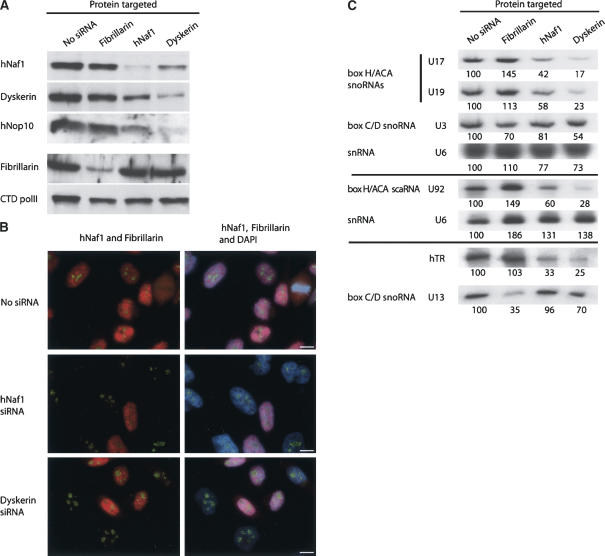
We next sought to determine if hNaf1 is required to maintain normal levels of the box H/ACA RNP and telomerase components with which it interacts. To address this question, we reduced the levels of hNaf1, dyskerin, or fibrillarin as control, by inducing turn-over of the corresponding mRNAs using specific siRNAs (Fig. 3A,B). Reduction of the quantities of hNaf1 induced a mild drop in the levels of dyskerin and hNop10, while levels of fibrillarin remained unchanged (Fig. 3A). Conversely, treatment with siRNAs targeting dyskerin mRNAs reduced the levels of hNaf1 (Fig. 3A,B). In addition, reduction of the quantities of hNaf1 or dyskerin was correlated with a decrease in the steady-state levels of box H/ACA snoRNAs, scaRNAs, and hTR (Fig. 3C). In contrast, levels of box C/D snoRNAs U3 and U13 were unaffected by a reduction in the amounts of hNaf1.
FIGURE 3.
hNaf1 is required for normal accumulation of box H/ACA RNP and telomerase components. (A) Western analysis of the consequences of the reduction of fibrillarin, hNaf1, or dyskerin levels obtained by RNA interference. HeLa cells were mock-transfected (No siRNA) or transfected with double-stranded siRNAs targeting fibrillarin, hNaf1, or dyskerin mRNAs and grown for 60 h. Total proteins were then extracted, separated, and analyzed by the Western blot procedure as described in the legend of Figure 1. (B) Fluorescence in situ immunodetection of hNaf1 (red) and fibrillarin (green) in HeLa cells mock-transfected (No siRNA) or transfected with double-stranded siRNAs targeting hNaf1 or dyskerin mRNAs. Bar, 10 μm. (C) Analysis of small RNA levels following siRNA treatment. HeLa cells were transfected with siRNAs as described in A. After 60 h of incubation, total RNAs were extracted. U17, U19, U92, U3, and U6 (acting as an internal control) were then separated and analyzed by Northern blot, while U13 and hTR were analyzed by RNase protection. U17, U19, and U3, on the one hand, and U92, on the other, were analyzed on different blots. For both procedures, equal amounts of total RNA were used for all samples. Signal intensities were measured by PhosphorImager scanning. Values (indicated below each lane) are expressed as percentages of the intensities obtained in the case of mock-transfected samples (No siRNA).
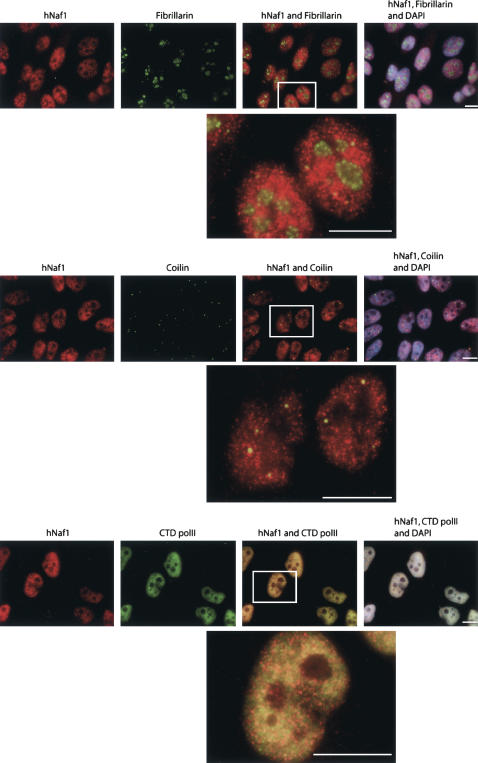
hNaf1 is located in numerous discrete nucleoplasmic foci and fails to accumulate in nucleoli or Cajal bodies
To investigate where hNaf1 might function in the cell, its intracellular distribution was assessed by fluorescence in situ immunodetection using purified anti-hNaf1 antibodies (Fig. 4). hNaf1 is detected primarily in a large number of discrete foci throughout the nucleoplasm that often coincide with, but are less numerous than, nucleoplasmic foci containing the large subunit of RNA polymerase II. Faint diffuse labeling of the cytoplasm with anti-hNaf1 antibodies can also be observed. The same distribution is observed in mammary tumor cells (MCF7), osteocarcinoma cells (U2OS), and in mouse cells (NIH3T3) (data not shown). We failed to detect any hNaf1 within nucleoli, here labeled with anti-fibrillarin antibodies. Likewise, although hNaf1-containing foci are sometimes juxtaposed to Cajal bodies, most of these seem devoid of hNaf1. HA-tagged hNaf1 transiently expressed in transfected HeLa cells is also predominantly detected in the nucleoplasm and seems absent from nucleoli (data not shown). These data demonstrate that hNaf1 is not part of mature box H/ACA snoRNPs and scaRNPs and suggest that it intervenes in early steps of assembly of these particles, as has been proposed for yeast Naf1p (Fatica et al. 2002; Ballarino et al. 2005; Yang et al. 2005).
FIGURE 4.
hNaf1 is located in numerous discrete nucleoplasmic foci. Subcellular localization of hNaf1 in HeLa cells. Bar, 10 μm.
Conclusions and perspectives
Our results establish that hNaf1 is the human ortholog of yeast Naf1p and is required for the accumulation of several box H/ACA snoRNP, scaRNP, and telomerase components with which it physically, directly or indirectly, interacts. The finding that hNaf1 can salvage growth of a yeast strain lacking an NAF1 gene is remarkable given the moderate overall identity between yeast and human Naf1 (28% identity, 59% similarity). This emphasizes the notion that not only the composition and function of mature box H/ACA RNPs have been conserved between lower and higher eukaryotes (Pogačić et al. 2000; Yang et al. 2000), but also at least some aspects of the biogenesis of these particles. Our data also provide further evidence for a kinship between box H/ACA RNPs and telomerase.
Dyskerin has recently been found associated with an unspliced pre-mRNA hosting a box H/ACA snoRNA (Richard et al. 2006). Thus, assembly of human box H/ACA snoRNPs is initiated very early, either immediately following or during transcription of pre-mRNAs hosting box H/ACA snoRNAs. The fact that endogenous hNaf1 is predominantly found in discrete nucleoplasmic foci is consistent with the idea that hNaf1, as was proposed for yeast Naf1p, is involved in early assembly steps of box H/ACA RNPs. Whether hNaf1 acts while box H/ACA RNAs are being transcribed remains an open question: our attempts to assess by chromatin immunoprecipitation assays whether hNaf1, like yeast Naf1p (Ballarino et al. 2005; Yang et al. 2005), is present in the immediate vicinity of box H/ACA snoRNA genes have repeatedly failed, perhaps for trivial technical reasons. The immunoprecipitation experiments presented in Figure 2 demonstrate that hNaf1 interacts with box H/ACA snoRNAs and scaRNAs that have been trimmed to their mature or nearly mature size, as judged from their mobility on acrylamide gels. Hence, we surmise that hNaf1 remains associated with box H/ACA RNAs during most processing steps. At what stage hNaf1 dissociates from maturing particles remains unclear. Box H/ACA scaRNAs and hTR accumulate in Cajal bodies (Darzacq et al. 2002; Jády et al. 2004), and some canonical box H/ACA snoRNAs may transit through Cajal bodies on their way to the nucleolus (Richard et al. 2003). It is possible that hNaf1 dissociates from the particles before they enter Cajal bodies, since we failed to obtain any convincing evidence for the presence of hNaf1 in these nucleoplasmic entities. However, the presence of hNaf1 within Cajal bodies may be too transient to be clearly detectable under our experimental conditions. In that respect, it is interesting to note that only traces of the endogenous PHAX protein, which is required for routing box C/D snoRNPs to Cajal bodies, can be detected in these and only when the box C/D U3 snoRNA is overexpressed does PHAX become concentrated in Cajal bodies (Boulon et al. 2004). hNaf1 could not be seen in nucleoli either. In this case again, we are reluctant to conclude that hNaf1 is totally excluded from nucleoli, given that some yeast Naf1p could be detected in the nucleolus by electron microscopy (Dez et al. 2002). In any case, the observation that hNaf1 fails to accumulate within Cajal bodies or nucleoli is a strong indication that it is not a permanent component of mature box H/ACA scaRNPs and snoRNPs.
The biogenesis of human box H/ACA RNPs and telomerase is likely to be complex, as has recently been shown for the mammalian box C/D U3 snoRNP (Boulon et al. 2004; Watkins et al. 2004). In addition to hNaf1, the Nopp140 protein, which shuttles between cytoplasm, Cajal bodies, and nucleoli, has been shown to transiently interact with box H/ACA RNP components (Meier and Blobel 1992; Isaac et al. 1998; Yang et al. 2000). Moreover, the SMN protein, required for assembly of spliceosomal snRNPs (Yong et al. 2004) and probably box C/D snoRNPs also (Watkins et al. 2004) interacts with hGar1 (Pellizzoni et al. 2001; Whitehead et al. 2002) and with the human telomerase RNP (Bachand et al. 2002). Surprisingly, a recent report has shown that the transport adaptor PHAX interacts with hTR (Boulon et al. 2004). Finally, the Rvb2p and Shq1p proteins are required for box H/ACA snoRNA accumulation in yeast (King et al. 2001; Yang et al. 2002), and both proteins have likely orthologs in human cells. All of these factors are most probably involved in the assembly and/or trafficking of human box H/ACA RNPs and telomerase. Deciphering their roles and determining how hNaf1 fits in the overall picture is the next challenge in the field.
MATERIALS AND METHODS
Strains, media, plasmids, and siRNAs
To assess the ability of hNaf1 to complement growth of a yeast strain depleted of endogenous yeast Naf1p, the GAL::naf1 strain (Dez et al. 2002) was transformed with plasmids pCH32 (Lebaron et al. 2005), pCH31, or pCH32-hNaf1, which constitutively express from the GAR1 promoter the ZZ tag alone or S. cerevisiae Naf1p or hNaf1, respectively. To obtain pCH31, the S. cerevisiae NAF1 open reading frame was PCR-amplified using oligonucleotides NterNaf1p (5′-CCCCCCAGATCTTATGAGCGATGACTTGTTTTCTAAG-3′) and CterNaf1p (5′-CCCCCCAGATCTGCGGTTCTTGGATCTTGCTGATGCTGA-3′). The PCR product was digested with BglII and inserted into BamHI-cut pCH32, creating pCH31. pCH32-hNaf1 was obtained as follows: The open reading frame of hNaf1 was PCR-amplified from the 3,447,276 IMAGE clone (purchased from the Mammalian Gene Collection) using oligonucleotides 5′hNaf1pHA113 (5′-CCCCCCGGATCCTATGGAGGTAGTGGAGGCCG-3′) and 3′hNaf1pHA113 (5′-CCCCCCGGATCCGCATAGTAAGGTCCAAAATGA-3′). The PCR product was digested with BamHI and inserted into BamHI-cut pCH32, creating pCH32-hNaf1. To assess whether hNaf1 can interact with yeast snoRNP proteins, the GAL::naf1 strain was transformed with plasmid pFH144, which directs expression of the open reading frame of hNaf1 from the strong TDH3 promoter. pFH144 was produced in two steps as follows. The GAR1 promoter in pCH32 was removed by BamHI/EcoRI digestion and replaced by the TDH3 promoter, digested by the same enzymes, that was obtained by PCR amplification performed with plasmid pBS2585 (a generous gift of B. Séraphin) and oligonucleotides 5′TDH3pr1 (5′-GGGGGGAATTCAGATTCCTTGATTACGTAAGGGAGTTAGAATC-3′) and 3′TDH3pr (5′-GGGGGGGATCCTTTGTTTGTTTATGTGTGTTTATTCGAAACTAAG-3′). This procedure generated plasmid pFH143. In a second step, the open reading frame of hNaf1, obtained as described above, was digested with BamHI and inserted into pFH143 digested with the same enzyme, generating pFH144. To assess whether hNaf1 could salvage growth of a yeast strain totally devoid of endogenous yeast Naf1p, we proceeded as follows. An naf1::kanMX4 strain possessing a centromeric plasmid containing a URA3 marker gene and the wild-type S. cerevisiae NAF1 gene was transformed with TRP1-containing plasmids pCH9 or pFH146, directing expression of ZZ-tagged S. cerevisiae Naf1p or hNaf1, respectively. pCH9 was produced by amplifying the S. cerevisiae NAF1 open reading frame as described above and cloning the BglII-digested PCR product into the BamHI site of pHA113 (Henras et al. 2001). To produce pFH146, the TRP1-containing 1977-bp HindIII/PciI fragment from pHA113 was ligated to the 5732-bp HindIII/PciI fragment from pFH144. To select cells able to survive in the absence of the URA3-containing plasmid, the transformed naf1::kanMX4 strains were streaked on minimal medium containing fluoroorotic acid.
S. cerevisiae strains were grown either in YP medium (1% yeast extract, 1% peptone) supplemented with either 2% galactose and 2% sucrose or 2% glucose as carbon sources. G418 was added when required at 0.2 mg/mL final concentration. Fluoroorotate-resistant clones were selected on supplemented yeast nitrogen base proline medium containing 0.6 g/L fluoroorotic acid (McCusker and Davis 1991). HeLa cells were grown in DMEM supplemented with antibiotics, 1% sodium pyruvate, and 10% FCS (all from Invitrogen). Transfection of double-stranded siRNAs was performed by electroporation as described in Rouquette et al. (2005). The sequences of the siRNAs (manufactured by Eurogentec) used to target hNaf1 or dyskerin mRNAs are the following: SiRNA-hNaf1-1, 5′-GGAAAGAUAUUCGAGAUAU-3′; SiRNA-hNaf1-2, 5′-UGAACUGCCUUCUGUUGAA-3′; SiRNA-hNaf1-3, 5′-ACAGAUUCAGAUAGUUCAA-3′; SiRNA-Dyskerin-1, 5′-GGACAGGUUUCAUUAAUCU-3′; SiRNA-Dyskerin-2, 5′-CCACAAGGAUGAGAGUUAC-3′; SiRNA-Dyskerin-3, 5′-UCAAACCUGAAUCCAAAGU-3′. The siRNA used to target fibrillarin mRNA was described in Watkins et al. (2004).
Immunoprecipitations
Immunoprecipitations of ZZ-tagged proteins from yeast extracts were performed as described in Lebaron et al. (2005), except for the following modifications: After precipitation, immunoglobulin G (IgG)-Sepharose beads (Amersham Pharmacia Biotech) and associated complexes were washed five times with 1 mL of 20 mM Tris-HCl (pH 8), 5 mM MgAc, 0.2% Triton X-100, 200 mM KCl, and 1 mM dithiothreitol (DTT) and then twice with 1 mL of 20 mM Tris-HCl (pH 8), 5 mM MgAc, 0.2% Triton X-100, 200 mM NaCl, and 1 mM DTT. After washing, the beads were resuspended in 50 μL of 100 mM Tris-HCl (pH 8), 4% SDS, 20% glycerol, 0.04% bromophenol blue, and 200 mM DTT. Immunoprecipitation of human proteins was performed as follows: About 202 × 2106 HeLa cells previously washed twice with ice-cold PBS were sonicated in 50 mM Tris-HCl (pH 7.4), 200 mM NaCl, 0.05% Nonidet P-40 (NET-2 buffer) containing 0.5 unit/μL RNasin (Promega), and protease inhibitors (Roche). The extract was then clarified by centrifugation at 10,000g for 10 min. The supernatant was incubated for 2 h with protein A agarose beads (Sigma) or IP beads (Rabbit TrueBlot Set from eBioscience) previously saturated for 2 h with anti-hNaf1 antibodies or non-immune serum. After immunoprecipitation, the beads were washed seven times with 20 volumes of NET-2 buffer. For protein analysis, beads were resuspended in an appropriate volume of 100 mM Tris-HCl (pH 8), 4% SDS, 20% glycerol, 0.04% bromophenol blue, and 200 mM DTT. For RNA analysis, beads were resuspended in 50 μL of 300 mM NaCl, 40 mM Tris-HCl (pH 8), and 1.7% SDS containing 60 μg of proteinase K (Promega). After 30 min at 37°C, RNAs were extracted with phenol/chloroform and precipitated.
Western analysis
Proteins from total extracts or extracted from immunoprecipitated fractions were separated on 8% or 13% polyacrylamide/SDS gels and transferred to Hybond-C extra membranes (Amersham Pharmacia Biotech). Yeast Gar1p, Nhp2p, Nop10p, Nop1p, ZZ-tagged proteins, and hNop10 were detected as described in Bousquet-Antonelli et al. (2000), Henras et al. (2001), and Dez et al. (2002). Yeast Cbf5p was detected using purified antibodies, diluted 500-fold, raised in rabbits (by Eurogentec) against the recombinant protein lacking the C-terminal KKE repeats. hNaf1 was detected using purified antibodies diluted 1000-fold that had been raised in rabbits (by Eurogentec) against the following hNaf1 peptides: H2N-REFTRGFSRARYPRSC-CONH2 and H2N-CPPSSGDSNSHFGPYY-CONH2. Dyskerin was detected using purified antibodies diluted 1000-fold that had been raised in rabbits (by Eurogentec) against the following dyskerin peptide: H2N-CERDTYPRKWGLGPKASQ-COOH. Fibrillarin was detected using rabbit polyclonal antibodies raised against the protein from Xenopus laevis diluted 200-fold, actin using a mouse monoclonal antibody (Chemicon) diluted 10,000-fold, and CTD Pol II using the CTD4H8 mouse monoclonal antibody (Upstate) diluted 500-fold.
RNA extractions, Northern hybridizations, and RNase protection assay
RNA extractions from yeast cells were performed as described by Tollervey and Mattaj (1987). RNA fractionations by polyacrylamide gel electrophoresis were performed as described by Henras et al. (1998). Small RNAs, except hTR and U13, were analyzed by Northern hybridization using 32P-labeled oligodeoxynucleotide probes. Sequences of antisense oligonucleotides used to detect yeast small RNAs have been reported in Henras et al. (1998, 2001) and Dez et al. (2002) and to detect human small RNAs in Kiss et al. (2002) and Jády et al. (2004). Blots were hybridized with 5′-end-labeled oligodeoxynucleotide probes and washed as described by Henras et al. (1998). hTR and U13 were detected by RNase A/T1 protection assay as described in Goodall et al. (1990).
Fluorescence in situ immunodetection
Cells grown on coverslips were fixed for 15 min at 4°C with 4% paraformaldehyde in PBS, washed with PBS, and permeabilized with 0.4% Triton X-100 in PBS. After two washes with PBS, cells were incubated for 1 h with 1% BSA in PBS, then for 1 h with primary antibodies in PBS containing 1% BSA. Fibrillarin was detected using 72B9 mouse monoclonal antibody diluted 200-fold (provided by J. Steitz), human p80-coilin using a polyclonal rabbit anti-coilin antibody diluted 400-fold (provided by A. Lamond), and hNaf1 and CTD Pol II were detected using the antibodies listed in the “Western Analysis” section above. After three washes with PBS, cells were incubated for 1 h with anti-rabbit IgG TRITC conjugates diluted 2000-fold and anti-mouse IgG FITC conjugates diluted 1000-fold (Sigma). The coverslips were then washed three times with 1× PBS and then mounted in mowiol/DAPI (0.1 μg/mL). Images were captured with a 12-bit CoolSnap ES camera (Roper Scientifics) mounted on a DMRB microscope (Leica) using the Metavue software (Universal Imaging).
ACKNOWLEDGMENTS
We thank S. Aveilla and A. Hochkoeppler for constant support and T. Meier for communicating results prior to publication. We are grateful to A. Lamond, B. Séraphin, and J. Steitz for reagents and C. Chailleux, V. Choesmel, B. Jády, P. Richard, and J. Rouquette for reagents and advice. We are thankful to members of the Ferrer lab for help and numerous discussions. We thank Y. de Préval for synthesis of oligonucleotides, D. Villa for art work, and A. Rivals for expert technical assistance. C.H.-A. is a recipient of a post-graduate fellowship from the Ministère de l′Education Nationale et de la Recherche. M.B. is a recipient of a “Marco Polo” fellowship from the University of Bologna. This work was supported by the CNRS, the Université Paul Sabatier, and a grant from La Ligue Nationale contre le Cancer (“Equipe Labellisée”).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2344106.
REFERENCES
- Bachand F., Boisvert F.M., Cote J., Richard S., Autexier C. The product of the survival of motor neuron (SMN) gene is a human telomerase-associated protein. Mol. Biol. Cell. 2002;13:3192–3202. doi: 10.1091/mbc.E02-04-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.L., Youssef O.A., Chastkofsky M.I., Dy D.A., Terns R.M., Terns M.P. RNA-guided RNA modification: Functional organization of the archaeal H/ACA RNP. Genes & Dev. 2005;19:1238–1248. doi: 10.1101/gad.1309605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballarino M., Morlando M., Pagano F., Fatica A., Bozzoni I. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae . Mol. Cell. Biol. 2005;25:5396–5403. doi: 10.1128/MCB.25.13.5396-5403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S., Verheggen C., Jády B.E., Girard C., Pescia C., Paul C., Ospina J.K., Kiss T., Matera A.G., Bordonné R., et al. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol. Cell. 2004;16:777–787. doi: 10.1016/j.molcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Vanrobays E., Gélugne J.-P., Caizergues-Ferrer M., Henry Y. Rrp8p is a yeast nucleolar protein functionally linked to Gar1p and involved in pre-rRNA cleavage at site A2. RNA. 2000;6:826–843. doi: 10.1017/s1355838200992288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T.R. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- Charpentier B., Muller S., Branlant C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 2005;33:3133–3144. doi: 10.1093/nar/gki630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.L., Greider C.W. Telomerase RNA structure and function: Implications for dyskeratosis congenita. Trends Biochem. Sci. 2004;29:183–192. doi: 10.1016/j.tibs.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Chen J.L., Blasco M.A., Greider C.W. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- Cong Y.S., Wright W.E., Shay J.W. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Jády B.E., Verheggen C., Kiss A.M., Bertrand E., Kiss T. Cajal body-specific small nuclear RNAs: A novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C., Noaillac-Depeyre J., Caizergues-Ferrer M., Henry Y. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 2002;22:7053–7065. doi: 10.1128/MCB.22.20.7053-7065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Dlakić M., Tollervey D. Naf1 p is a box H/ACA snoRNP assembly factor. RNA. 2002;8:1502–1514. [PMC free article] [PubMed] [Google Scholar]
- Goodall G.J., Wiebauer K., Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- Hamma T., Reichow S.L., Varani G., Ferré-D'Amaré A.R. The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nat. Struct. Mol. Biol. 2005;12:1101–1107. doi: 10.1038/nsmb1036. [DOI] [PubMed] [Google Scholar]
- Henras A., Henry Y., Bousquet-Antonelli C., Noaillac-Depeyre J., Gélugne J.-P., Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A., Dez C., Noaillac-Depeyre J., Henry Y., Caizergues-Ferrer M. Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic Acids Res. 2001;29:2733–2746. doi: 10.1093/nar/29.13.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac C., Yang Y., Meier U.T. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády B.E., Bertrand E., Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády B.E., Richard P., Bertrand E., Kiss T. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol. Biol. Cell. 2006;17:944–954. doi: 10.1091/mbc.E05-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna M., Wu H., Johansson C., Caizergues-Ferrer M., Feigon J. Structural study of the H/ACA snoRNP components Nop10p and the 3′ hairpin of U65 snoRNA. RNA. 2006;12:40–52. doi: 10.1261/rna.2221606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King T.H., Decatur W.A., Bertrand E., Maxwell E.S., Fournier M.J. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol. Cell. Biol. 2001;21:7731–7746. doi: 10.1128/MCB.21.22.7731-7746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Kiss A.M., Jády B.E., Darzacq X., Verheggen C., Bertrand E., Kiss T. A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 2002;30:4643–4649. doi: 10.1093/nar/gkf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaron S., Froment C., Fromont-Racine M., Rain J.-C., Monsarrat B., Caizergues-Ferrer M., Henry Y. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker J.H., Davis R.W. The use of proline as a nitrogen source causes hypersensitivity to, and allows more economical use of 5FOA in Saccharomyces cerevisiae . Yeast. 1991;7:607–608. doi: 10.1002/yea.320070608. [DOI] [PubMed] [Google Scholar]
- Meier U.T. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier U.T., Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Mitchell J.R., Cheng J., Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 1999a;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.R., Wood E., Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999b;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Baccon J., Charroux B., Dreyfuss G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 2001;11:1079–1088. doi: 10.1016/s0960-9822(01)00316-5. [DOI] [PubMed] [Google Scholar]
- Pogačić V., Dragon F., Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 2000;20:9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P., Darzacq X., Bertrand E., Jády B.E., Verheggen C., Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22:4283–4293. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P., Kiss A.M., Darzacq X., Kiss T. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol. Cell. Biol. (in press) 2006 doi: 10.1128/MCB.26.7.2540-2549.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette J., Choesmel V., Gleizes P.-E. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Mattaj I.W. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 1987;6:469–476. doi: 10.1002/j.1460-2075.1987.tb04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson R.L., Ziegler T.D., Supakorndej T., Terns R.M., Terns M.P. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Meier U.T. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23:1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N.J., Lemm I., Ingelfinger D., Schneider C., Hossbach M., Urlaub H., Lührmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Whitehead S.E., Jones K.W., Zhang X., Cheng X., Terns R.M., Terns M.P. Determinants of the interaction of the spinal muscular atrophy disease protein SMN with the dimethylarginine-modified box H/ACA small nucleolar ribonucleoprotein GAR1. J. Biol. Chem. 2002;277:48087–48093. doi: 10.1074/jbc.M204551200. [DOI] [PubMed] [Google Scholar]
- Yang Y., Isaac C., Wang C., Dragon F., Pogačić V., Meier U.T. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol. Biol. Cell. 2000;11:567–577. doi: 10.1091/mbc.11.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P.K., Rotondo G., Porras T., Legrain P., Chanfreau G. The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J. Biol. Chem. 2002;277:45235–45242. doi: 10.1074/jbc.M207669200. [DOI] [PubMed] [Google Scholar]
- Yang P.K., Hoareau C., Froment C., Monsarrat B., Henry Y., Chanfreau G. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol. Cell. Biol. 2005;25:3295–3304. doi: 10.1128/MCB.25.8.3295-3304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J., Wan L., Dreyfuss G. Why do cells need an assembly machine for RNA–protein complexes? Trends Cell Biol. 2004;14:226–232. doi: 10.1016/j.tcb.2004.03.010. [DOI] [PubMed] [Google Scholar]