Abstract
All three kingdoms of life employ two methionine tRNAs, one for translation initiation and the other for insertion of methionines at internal positions within growing polypeptide chains. We have used a reconstituted yeast translation initiation system to explore the interactions of the initiator tRNA with the translation initiation machinery. Our data indicate that in addition to its previously characterized role in binding of the initiator tRNA to eukaryotic initiation factor 2 (eIF2), the initiator-specific A1:U72 base pair at the top of the acceptor stem is important for the binding of the eIF2•GTP•Met-tRNAi ternary complex to the 40S ribosomal subunit. We have also shown that the initiator-specific G:C base pairs in the anticodon stem of the initiator tRNA are required for the strong thermodynamic coupling between binding of the ternary complex and mRNA to the ribosome. This coupling reflects interactions that occur within the complex upon recognition of the start codon, suggesting that these initiator-specific G:C pairs influence this step. The effect of these anticodon stem identity elements is influenced by bases in the T loop of the tRNA, suggesting that conformational coupling between the D-loop–T-loop substructure and the anticodon stem of the initiator tRNA may occur during AUG codon selection in the ribosomal P-site, similar to the conformational coupling that occurs in A-site tRNAs engaged in mRNA decoding during the elongation phase of protein synthesis.
Keywords: protein synthesis, translation, initiator tRNA, T loop, mechanism, identity elements
INTRODUCTION
A central goal of translation initiation is to set the reading frame of the mRNA by selecting the correct initiation (AUG) codon at which to begin protein synthesis. The assembly of a ternary complex of the eukaryotic translation initiation factor 2 (eIF2), GTP, and the methionyl initiator tRNA (Met-tRNAi) is the first step in this process in eukaryotes (for review, see Kapp and Lorsch 2004b). The ternary complex and mRNA bind to the 40S ribosomal subunit with the aid of several other initiation factors to form the 43S•mRNA pre-initiation complex. The 43S pre-initiation complex is thought to scan along the message beginning near its 5′ end until the identification of the start codon, at which point irreversible hydrolysis of GTP by eIF2 takes place, in a reaction requiring the GTPase activating protein eIF5 (Chakrabarti and Maitra 1991; Huang et al. 1997; Algire et al. 2005). In solution, the resulting eIF2•GDP complex has an approximately 20-fold lower affinity for the Met-tRNAi than does eIF2•GTP (Kapp and Lorsch 2004a), and so eIF2 is thought to release the tRNA into the P-site of the 40S ribosomal subunit upon GTP hydrolysis and inorganic phosphate dissociation. eIF2•GDP then dissociates from the 40S subunit, allowing the joining of the 60S ribosomal subunit in a step facilitated by a second GTPase, eIF5B (Pestova et al. 2000). The resulting 80S initiation complex can begin making a protein once a second aminoacyl tRNA is carried into the A-site of the ribosome by the eukaryotic elongation factor 1A (eEF1A), a GTPase that is homologous to the γ-subunit of eIF2 (Gaspar et al. 1994; Schmitt et al. 2002).
Although all known organisms (with the exception of mammalian mitochondria) contain two methionine-accepting tRNAs (Sprinzl and Vassilenko 2005), the initiator methionine tRNA (tRNAi), and the elongator methionine tRNA (tRNAe), only the initiator tRNA functions in translation initiation. The combination of pro-initiator identity elements with anti-elongator determinants that serve to block the binding of eEF1A restricts the initiator tRNA to functioning exclusively during initiation. Because the elongator tRNA lacks any pro-initiator elements but contains the binding determinants for eEF1A, it functions exclusively for the insertion of methionine at internal positions of proteins (RajBhandary 1994; Mayer et al. 2001). The identity elements of eukaryotic initiator tRNAs consist of the following: an A1:U72 base pair in the acceptor stem, as opposed to a G1:C72 base pair found in elongator tRNAs; the sequence GAUC in the T loop, as opposed to GrTΨC in elongator tRNAs; an adenosine at position 60 instead of a pyrimidine as is found in elongator tRNAs; and three universally conserved consecutive G:C base pairs in the anticodon stem of cytoplasmic initiator tRNAs (one of which is also present in Saccharomyces cerevisiae elongator methionine tRNA [Sprinzl and Vassilenko 2005]) (Fig. 1). In addition, the initiator tRNA of S. cerevisiae lacks the adenosine found at position 17 in the D-loop of the elongator tRNA and contains a 2′-O-phosphoribosyl adenosine at position 64 that protrudes into the minor groove of the T-stem and blocks the binding of eEF1A (Simsek and RajBhandary 1972; Desgres et al. 1989; Kiesewetter et al. 1990; Basavappa and Sigler 1991; Forster et al. 1993). It has been suggested that separate methionyl tRNAs are required for initiation and elongation to prevent potentially dangerous competition for the same tRNA by the initiation and elongation machinery (Forster et al. 1993; Guillon et al. 1996; Astrom et al. 1999). In addition, the initiator tRNA apparently must take part in decoding of mRNAs in the P-site of the ribosome rather than in the usual A-site decoding center, thus perhaps necessitating the maintenance of identity elements in the tRNA that allow it to perform this unique function.
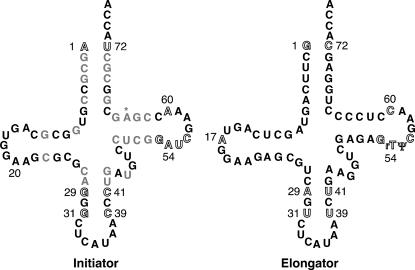
FIGURE 1.
Secondary structures of the yeast initiator methionyl tRNA (left) and elongator methionyl tRNA (right). Numbered positions correspond to bases that were previously found to be important for distinguishing the initiator from the elongator tRNA. Gray nucleotides indicate positions that differ between the two tRNAs and may also be involved in allowing discrimination between the initiator and elongator tRNAs but have not been as thoroughly studied as the other positions. Base modifications are not shown, except those implicated in the discrimination of the initiator from the elongator tRNA. The asterisk marks the position of the 2′-O-phosphoribosyl modification on A64 present in plant and fungal initiator tRNAs that serves as an anti-elongator determinant.
Although there have been a number of studies to determine how the identity elements in the initiator tRNA differentiate it from the elongator tRNA, our understanding of the molecular details of initiator-elongator discrimination is still incomplete. We have recapitulated many of the fundamental steps of eukaryotic translation initiation in vitro using a reconstituted system of S. cerevisiae components (Algire et al. 2002) and have now used this system to examine the roles of various initiator tRNA identity elements in key events in the initiation pathway.
We have found that in addition to the previously characterized key role of the A1:U72 base pair in eIF2-binding, other elements within the initiator tRNA body also play smaller but still significant roles in ternary complex formation. The A1:U72 base pair also influences the binding of the ternary complex to the 40S subunit, a previously unrecognized role for this identity element.
It was previously reported that the G:C base pairs in the anticodon stem of the initiator tRNA are important for binding of the tRNA to the ribosome (Seong and RajBhandary 1987a). We have now refined this conclusion, and show that these identity elements are required for the strong (>1000-fold) thermodynamic coupling between the binding of the ternary complex and mRNA to the 40S ribosomal subunit. This coupling reflects interactions within the complex that occur upon start codon recognition, and thus it is possible that it is a readout of events that are required for accurate response to identification of the AUG codon. Based on these data, we suggest that the anticodon stem G:C base pairs may play a role in mediating the response to start site identification.
Finally, our data suggest functionally important communication between the T-loop and the anticodon stem of the initiator tRNA. This communication may be part of a conformational change in the initiator tRNA upon start codon recognition that is involved in triggering events downstream in the initiation pathway.
RESULTS
Using an in vitro reconstituted system of yeast ribosomes and translation factors, it is possible to monitor key steps of the initiation pathway: ternary complex formation, 43S and 43S•mRNA complex formation, and 80S initiation complex formation. We are also able to assess the ability of the 80S initiation complexes to catalyze the formation of the first peptide bond. Although not all known steps are recapitulated in this system, the steps that can be monitored are dependent on the inclusion of various key components (factors, etc.) in ways consistent with the current model of the initiation process, indicating that the system reflects core events in the initiation pathway (Algire et al. 2002; Maag et al. 2005). Thus the effects on the steps in this system arising from the mutation of identity elements in the initiator tRNA should provide information about the roles of these elements in translation initiation. To study the roles of these elements, we have examined both the effects of replacing them in the initiator tRNA body with the corresponding elongator tRNA sequences, and the effects of adding initiator tRNA elements to the elongator tRNA body. For this work we have used tRNAs transcribed in vitro, which allowed us to generate and study a large number of variant initiator and elongator tRNAs, but did not allow us to probe the roles of base modifications in them. However, previous work has failed to find any detectable differences between the unmodified and native initiator tRNA in a variety of in vitro assays (Pestova and Hellen 2001; Kapp and Lorsch 2004a).
We have focused our work here on three sets of identity elements (Fig. 1): the acceptor stem 1:72 base pair (A1:U72 and G1:C72 in initiator and elongator tRNAs, respectively); the initiator-specific anticodon stem G:C pairs (G29:C41, G31:C39 in the initiator and A29:U41, U31:U39 in the elongator); and the initiator-specific T-loop identities (A54, A60 in the initiator and U54 [rT], C60 in the elongator). Note that for clarity of comparisons the numbering used for both tRNAs refers to the elongator tRNA sequence.
Initiator identities within the tRNA body in addition to the A1:U72 base pair are important for efficient ternary complex formation
Mutant initiator tRNAs
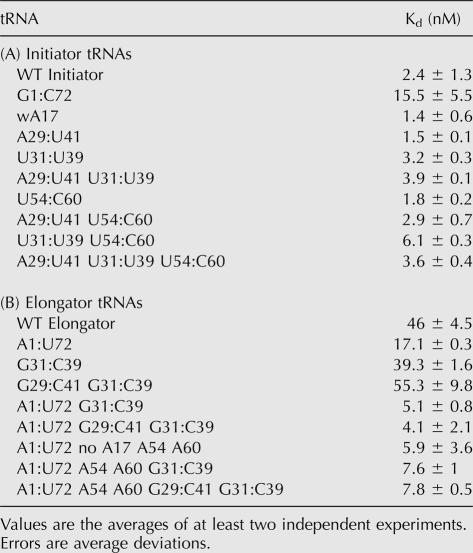
Consistent with previous work demonstrating that the primary eIF2-binding determinant of a eukaryotic tRNAi is the A1:U72 base pair (Farruggio et al. 1996; Kapp and Lorsch 2004a), the mutation that produced the greatest effect (approximately sevenfold; Table 1A) on the affinity of Met-tRNAi for eIF2•GTP was the change of the A1:U72 base pair to its corresponding elongator identity G1:C72.
TABLE 1.
Kds for eIF2·GTP and Met-tRNA
The alteration of either one or both of the initiator-specific G:C base pairs at positions 29:41 and 31:39 in the anticodon stem of tRNAi or alteration of positions 54 and 60 in the T-loop to their corresponding elongator identities had virtually no effect on the affinity of the resultant tRNAis for eIF2•GTP (Table 1A). However, the combination of the elongator U54 and C60 T-loop identities with the elongator U31:U39 anticodon stem base pair in an initiator tRNA resulted in an approximately threefold lower affinity for eIF2•GTP compared with the WT initiator (Kds of 6.1 and 2.4 nM, respectively). In contrast, combining elongator identities in the T-loop with elongator identities in both anticodon stem base pairs (Table 1A, A29:U41 U31:U39 U54 C60) did not result in any significant change to the binding affinity of the resultant tRNA compared with the WT initiator. These data suggest that the anticodon stem and T-loop identities may functionally interact with each other, as discussed in more detail in the following sections.
Mutant elongator tRNAs
WT Met-tRNAe binds approximately 20-fold more weakly to eIF2•GTP than does WT Met-tRNAi (Table 1B; Fig. 2). Alteration of only one or both anticodon stem base pairs at positions 29:41 and 31:39 to G:C base pairs did not produce an increase in the binding affinity relative to WT Met-tRNAe. However, the addition of only the initiator-specific A1:U72 base pair to tRNAe yielded a nearly threefold increase in its affinity for eIF2•GTP, and the combination of the A1:U72 base pair with either G31:C39 alone or G31:C39 along with G29:C41 in the anticodon stem resulted in affinities for eIF2•GTP that were nearly the same as that of tRNAi. Combining the A1:U72 base pair in the acceptor stem with the initiator identities A54 and A60 in the T-loop (in conjunction with the removal of base A17 in the D-loop) resulted in an affinity for eIF2•GTP within twofold of that of WT tRNAi. Thus both the initiator-specific G:C base pairs in the anticodon stem and the adenine bases at positions 54 and 60 in the T-loop influence the ability of Met-tRNAs to bind to eIF2. The dependence of the effects of these elements on the presence of the A1:U72 acceptor stem base pair suggests that sequences distant from one another in space cooperate to tune the structure of the tRNA.
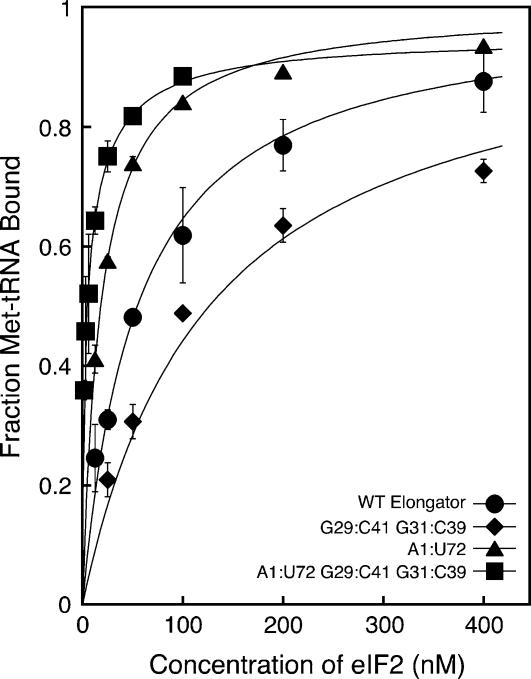
FIGURE 2.
The initiator-specific G:C base pairs in the anticodon stem, G29:C41 and G31:C39, enhance binding of mutant elongator tRNAs containing the A1:U72 initiator identity element to eIF2•GTP. Points are averages of at least two independent experiments. Errors are average deviations. Curve fits were performed as described in the methods section. The concentration ranges are 1.56–100 nM for the A1:U72 G29:C41 G31:C39 mutant elongator tRNA and 12.5–800 nM for the other tRNAs. eIF2 concentrations above 400 nM have been omitted from the figure for clarity.
The conserved G:C base pairs in the anticodon stem of tRNAi are required for thermodynamic coupling between the binding of ternary complex and mRNA to the 40S subunit
40S subunit binding by mutant initiator tRNAs in the absence of mRNA
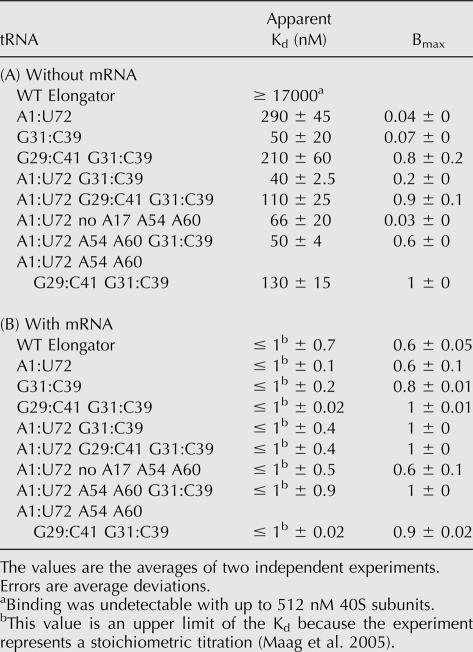
We next investigated the binding of mutant tRNAs to the 40S ribosomal subunit as part of the eIF2•GTP•Met-tRNA ternary complex in the presence of eIFs 1 and 1A and the absence of mRNA. Experiments using a native gel assay (Lorsch and Herschlag 1999; Algire et al. 2002) to examine the binding of Met-tRNAs to the 40S subunit in the absence of mRNA give two distinct parameters, an apparent Kd and an endpoint (Bmax), that can be used to assess the ability of ternary complexes containing mutant tRNAs to form 43S complexes (Table 2A,B). Bmax is the maximum fraction of Met-tRNA that is observed in the 43S complex as a function of 40S subunit concentration. With WT Met-tRNAi in the presence of a model mRNA containing an AUG codon, this value is close to 1, as would be expected; at high concentrations of 40S subunits all the ternary complex is in 43S complexes. In contrast, with WT Met-tRNAi in the absence of mRNA, Bmax is ∼0.4, indicating that even at very high 40S concentrations, it is not possible to observe all of the Met-tRNAi bound in the complex. Other data from our laboratory indicate that there are two conformational states of the 43S complex in the absence of mRNA, only one of which is stable in the native gel, explaining why the apparent endpoint of 43S complex formation with the WT Met-tRNAi is <100%. (D. Applefield, D. Maag, and J. Lorsch, in prep.). If the inverse experiment is done, fixing the concentration of 40S subunits and varying ternary complex, Bmax (now in terms of fraction of 40S subunits in 43S complexes) is still ∼0.4, indicating that the low Bmax is not the result of damaged ternary complex. Increasing the concentrations of eIF1, eIF1A, and mRNA also does not increase Bmax, arguing against a low specific activity of one of these components being responsible for the effect. The fact that addition of mRNA allows for nearly 100% of the ternary complex to bind 40S subunits also argues against one population of complexes being nonfunctional. Therefore, we take Bmax to be a measure of the equilibrium between two states of the 43S complex, one more stable than the other. Consistent with this, we have recently shown that two inter-convertible forms of the 43S complex exist with respect to the stability of eIF1A within the complex, and the equilibrium between these two states shifts dramatically toward the more stable form upon AUG codon recognition (Maag et al. 2006). These two states of the 43S complex may be the scanning-competent “open” and scanning-incompetent “closed” complexes originally proposed by Pestova and colleagues (Pestova et al. 1998; Pestova and Kolupaeva 2002).
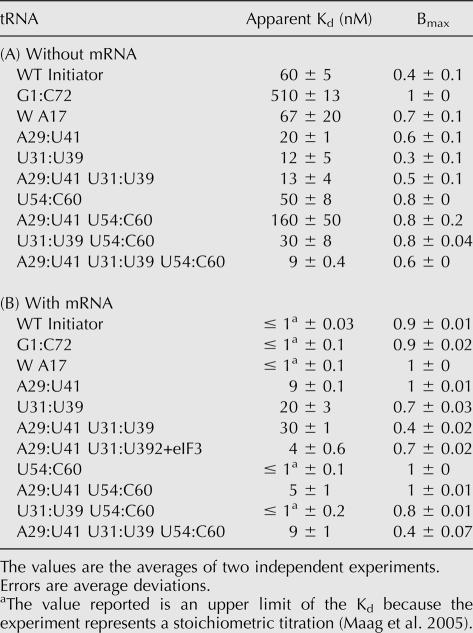
TABLE 2.
40S subunit binding by ternary complexes containing mutant Met-tRNAis
The concentration of eIF2•GTP used in all experiments measuring ternary complex binding to the 40S subunit was at least fourfold in excess of the Kd for eIF2•GTP binding to the Met-tRNAs, and thus defects observed in these experiments were not due to incomplete ternary complex formation. In all cases, observable binding of tRNA to the 40S subunit was dependent on the presence of eIF2•GTP (data not shown).
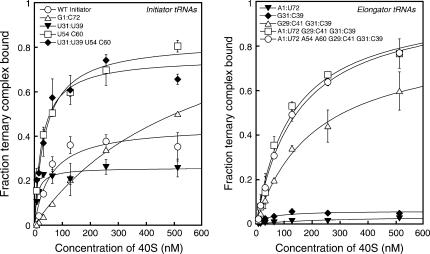
The most significant effect on binding of ternary complex to the 40S subunit came from changing the acceptor stem A1:U72 base pair in tRNAi to the elongator sequence G1:C72, which increased the apparent Kd from 60 to 510 nM and the Bmax from 0.4 to 1.0 (Table 2A; Fig. 3). Changing the anticodon stem G:C base pairs to the elongator sequences (A29:U41, U31:U39) actually increased the affinity of the ternary complex for the ribosome by three- to fivefold. These data indicate that both the acceptor stem 1:72 pair and the anticodon stem initiator specific G:C pairs influence how the ternary complex interacts with the 40S ribosomal subunit.
FIGURE 3.
40S subunit binding by ternary complexes containing mutant initiator tRNAs is enhanced by elongator T-loop identities (U54:C60) but weakened by the elongator acceptor stem identity (G1:C72) (left), while the binding of ternary complexes containing mutant elongator tRNAs is progressively enhanced by increasing initiator identity content (right). No binding to 40S subunits could be detected for ternary complexes containing the WT elongator tRNA. Points are averages of at least two independent experiments. Errors are average deviations. Curve fits were done as described in the methods section. Reactions contained eIFs 1 and 1A in all cases. Ternary complexes were formed using sufficient eIF2 to drive binding of all tRNAs tested (see Materials and Methods). No binding was detected in the absence of eIF2 (data not shown).
Replacement of the initiator T-loop elements A54 and A60 with the elongator bases (U54 and C60) did not affect the affinity of ternary complex for the 40S subunit, but did increase Bmax to 0.8 (Table 2A). The increasing value of Bmax suggests that the different conformational states of the 43S complex we observe in the absence of mRNA are influenced by the structure of the tRNA and may involve distinct positions of ternary complex on the 40S subunit. Because eIF1A is the ortholog of bacterial IF1 (Kyrpides and Woese 1998), which binds over the A-site of the small ribosomal subunit (Carter et al. 2001), it seems unlikely that one of the binding modes consists of interaction of tRNAi with the A-site. Instead, the different modes may be distinct positions of tRNAi (as part of the ternary complex) within or near the P-site.
40S subunit binding by mutant initiators in the presence of mRNA
We also investigated the ability of ternary complexes containing mutant tRNAs to bind to the 40S subunit in the presence of a model mRNA containing an AUG codon. The binding of ternary complex containing WT Met-tRNAi to the 40S ribosomal subunit is thermodynamically coupled by at least 1000-fold to the binding of an AUG codon-containing mRNA (Maag et al. 2005). That is, binding of ternary complex increases the affinity of the 40S subunit for mRNA by at least 1000-fold and vice-versa. As the ternary complex is thought to bind to 40S subunits before mRNA in vivo, it is unlikely that this coupling is required for 43S formation. Instead, the thermodynamic coupling reflects changes in interactions within the complex that take place upon AUG recognition, and thus may be a readout of events involved in the response to start codon identification.
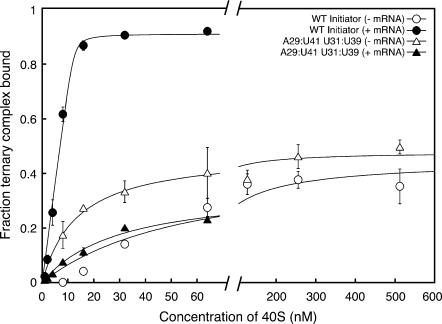
Thermodynamic coupling with mRNA binding was abrogated when the initiator-specific G:C pairs in the anticodon stem of tRNAi were replaced with their elongator counterparts (A29:U41 and U31:U39; Table 2B; Fig. 4). These data indicate that the G29:C41 and G31:C39 base pairs in the anticodon stem of tRNAi are required for thermodynamic coupling between the binding of ternary complex and mRNA to the 40S subunit. The fact that mutating these positions increases the affinity of the ternary complex for the 40S subunit in the absence of mRNA but dramatically decreases it in the presence of mRNA (Table 2A,B) suggests the possibility that the ternary complex or the Met-tRNAi within it binds in different modes on the 40S subunit in the absence of mRNA and following AUG codon recognition; in this case, mutations stabilizing the binding mode in the absence of mRNA would have a negative effect on the affinity in the presence of mRNA.
FIGURE 4.
Conversion of the anticodon stem G:C base pairs to their elongator identities eliminates the thermodynamic coupling between binding of eIF2•GTP•Met-tRNAi ternary complex, and mRNA to the 40S subunit. Points are the averages of at least two independent experiments. Errors are average deviations. Curve fits were as described in the methods section. All reactions contained eIFs 1 and 1A. Ternary complexes were formed using sufficient eIF2 to drive binding of all tRNAs tested (see Materials and Methods). No binding was detected in the absence of eIF2 (data not shown).
We also tested whether eIF3 would specifically suppress this defect, possibly by stabilizing a conformation of the complex destabilized by the anticodon stem mutations. Addition of a stoichiometric concentration of eIF3 (with respect to the highest 40S subunit concentration used, 250 nM) led to a sevenfold increase in the affinity of ternary complex containing the A29:U41 U31:U39 mutant Met-tRNAi (Kd of 4 nM; Table 2B). This effect is, however, similar to the two- to fivefold effect that yeast eIF3 has on binding of WT ternary complex to 40S subunits in the absence of mRNA in vitro (Maag et al. 2005) (we are unable to assess the thermodynamic effect of eIF3 on WT ternary complex binding in the presence of mRNA because it is too tight to accurately measure even in the absence of eIF3; Kds ≤ 1 nM with or without eIF3) and on ternary complex recruitment in yeast in vivo (Jivotovskaya et al. 2006). Thus it does not appear that eIF3 specifically suppresses the effect of the mutation of both anticodon stem G:C pairs, although, as with WT Met-tRNAi, it does enhance the affinity of the ternary complex for the 40S subunit.
Although mutant tRNAis with elongator identities in the anticodon stem were defective in responding to the presence of mRNA, the replacement of the initiator T-loop identities A54 and A60 with the elongator sequences U54 and C60 reduced the negative effect of these anticodon stem mutations. In the case of the U31:U39 mutant tRNAi, this effect was at least 20-fold, reducing the apparent Kd from 20 nM to ≤ 1 nM (Table 2B). On their own, the U54 and C60 elongator T-loop identities in the initiator tRNA had no detectable deleterious effects. These data suggest that the T-loop identity elements can influence the function of the conserved bases in the anticodon stem of the initiator tRNA. In contrast to their positive effect in the presence of mRNA, the U54 and C60 mutations had a negative effect when combined with the individual anticodon stem G:C pair mutations (A29:U41 and U31:U39) in the absence of mRNA (Table 2A), again suggesting that Met-tRNAi binds in a different mode in the 43S complex before and after AUG recognition.
Increasing initiator identity content in elongator tRNAs improves 40S subunit binding in the absence of mRNA, although no initiator identities are required for coupled binding with mRNA
40S subunit binding by mutant elongator tRNAs in the absence of mRNA
We also investigated the affinities of ternary complexes containing WT and mutant elongator tRNAs for the 40S subunit by measuring their binding in the absence of mRNA (Table 3A; Fig. 3, right). No binding of ternary complex containing WT Met-tRNAe could be detected up to a concentration of 512 nM 40S subunits. Based on the lowest detectable signal, we estimate the Kd for the 40S subunit and ternary complex with WT Met-tRNAe in the absence of mRNA to be ≥ 17 μM. Replacing the anticodon stem pair U31:U39 with the initiator sequence (G31:C39) or the acceptor stem G1:C72 base pair with the initiator A:U pair allowed detectable binding of ternary complex to the 40S subunit (Table 3A), although the endpoints were extremely low (< 0.1; in cases with Bmax < 0.1, it is not clear that changes in apparent Kds are significant, because of the very low signal). Combining A1:U72 with G31:C39 increased Bmax to 0.2 and gave an apparent Kd the same as that with WT Met-tRNAi (40 nM; Table 3A). Having both initiator identity G:C pairs (G29:C41 and G31:C39) in the anticodon stem increased Bmax to 0.8 and gave an apparent Kd of 210 nM, and combination of these two G:C pairs with the acceptor stem A1:U72 pair allowed ternary complex binding to 40S subunits with a Kd essentially indistinguishable from WT Met-tRNAi and increased Bmax twofold over the WT initiator (apparent Kd = 110 nM, Bmax = 0.9; Table 3A). These data support the conclusion that the initiator-specific A1:U72 base pair in the acceptor stem of tRNAi and the G:C pairs in the anticodon stem play roles in proper interaction of the ternary complex with the 40S ribosomal subunit.
TABLE 3.
40S subunit binding by ternary complexes containing mutant Met-tRNAes
40S subunit binding by mutant elongator tRNAs in the presence of mRNA
In contrast to the varied performances of the mutant initiator tRNAs, every elongator tRNA allowed for thermodynamic coupling between binding of ternary complex and mRNA to the 40S subunit, although with differing endpoints (Table 3B). In general, those tRNAe mutants that contained at least one of the initiator-specific G:C base pairs in their anticodon stems gave endpoints in excess of 0.8, while the other mutants gave endpoints of 0.6. It is interesting to note that replacing the initiator-specific anticodon stem G:C pairs in tRNAi with the elongator sequence (A29:U41, U31:U39) eliminated coupled binding of ternary complex and mRNA to the 40S subunit (Table 2), while the WT elongator tRNA, which also lacks these two G:C pairs, demonstrates significant coupling (≥1700-fold; Table 3). This suggests that other elements in the elongator tRNA, apparently not present in the initiator tRNA, allow coupled binding with mRNA even when the G:C base pairs in the anticodon stem are not present. It is possible that it is inherently easier to achieve coupled binding with mRNA for tRNAe because it binds to the ribosome in a different manner than the initiator tRNA.
Mutant initiator tRNAs show few defects in the formation of 80S initiation complexes
We next investigated the ability of 43S•mRNA complexes containing various mutant initiator tRNAs to proceed on to the next step in translation initiation, joining with a 60S subunit to form an 80S initiation complex. Following 43S•mRNA complex formation, eIF5, eIF5B, and 60S subunits were added and the formation of 80S complexes was monitored by a native gel assay which allows visualization of free Met-tRNA, unjoined 40S•mRNA•Met-tRNA complexes, and complete 80S initiation complexes. An increase in free Met-tRNA relative to what is observed with WT tRNAi suggests that the mutant tRNA is less stably bound in the 40S•mRNA•Met-tRNA or 80S initiation complex or that there is a decrease in subunit joining efficiency allowing Met-tRNA to dissociate from the 40S•mRNA•Met-tRNA complex. An increase in the observed level of 40S•mRNA•Met-tRNA complex and a decrease in the level of 80S complex relative to that with WT tRNAi suggests that the mutant tRNAi decreases the efficiency of subunit joining.
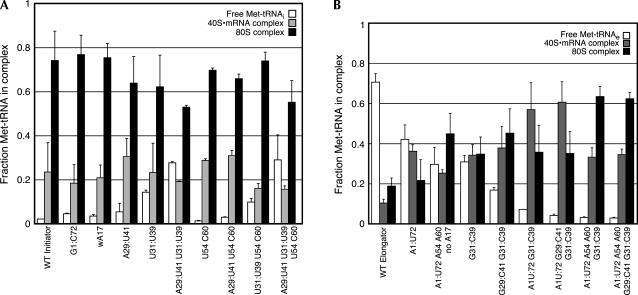
Although all the mutant initiator tRNAs examined were able to form 80S complexes well, alteration of the conserved G:C base pairs in the anticodon stem (29:41, 31:39) resulted in modest deficiencies in complex stability or subunit joining as indicated by the elevated fractions of free Met-tRNA released from the ribosome during the assay with these mutants (Fig. 5A). Nevertheless, it is clear that Met-tRNAs that displayed serious defects in their formation of 43S•mRNA complexes could be effectively driven into 80S complexes, due perhaps to a pulling of the equilibrium of 43S•mRNA complex formation or to the presence of eIF5 and/or eIF5B.
FIGURE 5.
Mutant initiator tRNAs show few defects in 80S complex formation (A), while the initiator T-loop identities are stimulatory for 80S complex formation with mutant elongator tRNAs (B). Data are the averages of at least two independent experiments and error bars indicate average deviations. In all cases, sufficient 40S subunits were used to drive 43S•mRNA complex formation (see Materials and Methods).
The A54 and A60 initiator T-loop identities promote 80S initiation complex formation by mutant elongator tRNAs
To determine whether the mutant elongator tRNAs were forming competent 43S•mRNA complexes, we examined their abilities to participate in 80S complex formation. The WT elongator tRNA and the A1:U72 acceptor stem mutant were defective in 80S complex formation, most likely because these tRNAs were less stably bound to the ribosome during or after the formation of 80S complexes (note the increased fractions of free Met-tRNA and decreased 80S complex in these lanes in Fig. 5B). The increased levels of 40S•mRNA•Met-tRNA complex and decreased levels of free Met-tRNA with the A1:U72 mutant relative to WT tRNAe, however, are further evidence that the A1:U72 base pair stabilizes the tRNA on the 40S subunit. Addition of the G31:C39 anticodon stem base pair to WT tRNAe alone or in combination with the G29:C41 base pair slightly enhanced stable 80S complex formation, at least in part by increasing the stability of the Met-tRNAe in the ribosomal complexes. The combination of the A1:U72 acceptor stem base pair with either the G31:C39 alone or both the G29:C41 and G31:C39 anticodon stem base pairs increased the stabilities of the 40S•mRNA•Met-tRNAe complexes relative to the tRNAs bearing these anticodon stem base pairs in isolation, although it did not further increase the extent of 80S complex formation. These data are consistent with the A1:U72 base pair's influencing the stability of Met-tRNA binding to the 40S subunit, as indicated by its effects on 43S complex formation.
Addition of the A54 and A60 T-loop identity elements to the A1:U72 mutant tRNAe along with deletion of A17 in the D-loop increased the level of stable 80S complex formation (Fig. 5B). When the A54 and A60 bases were combined with the acceptor stem A1:U72 base pair and the G:C base pairs in the anticodon stem (G29:C41 and G31:C39), 80S complex formation was nearly as efficient as with the WT initiator tRNA. Thus both the initiator-specific G:C pairs in the anticodon stem and the A54 and A60 elements in the T-loop influence the efficiency of 80S initiation complex formation.
Given these data one might have expected to observe more of a defect in the formation of 80S complexes by mutant initiator tRNAs bearing the elongator T-loop identities U54 and C60, since the initiator T-loop identities apparently were required in mutant elongator tRNAs for WT tRNAi levels of 80S complex formation. The lack of an effect of changing A54 and A60 in tRNAi to U54 and C60 suggests that other elements within the initiator sequence must suppress the effect of their absence. Alternatively, the A54 and A60 initiator identities may have induced structural changes in the elongator tRNA that are not necessarily functionally relevant but lead to enhanced 80S complex formation simply by reducing the off-rate of the tRNA from the 40S•mRNA•Met-tRNA complex following irreversible GTP hydrolysis by eIF2.
The elongator T-loop identities stimulate methionyl-puromycin synthesis with mutant initiator tRNAs
As a test of the competence of the 80S initiation complexes formed with various mutant initiator and elongator tRNAs, we employed the methionyl-puromycin assay, which mimics the step immediately following the conclusion of translation initiation, formation of the first peptide bond between the initiator tRNA in the ribosomal P-site, and the first elongator tRNA to be delivered to the ribosomal A-site. Although Met-Puromycin synthesis activity in our system depends on the full complement of core factors and other components (Algire et al. 2002) and, thus, appears to reflect the formation of competent 80S initiation complexes, it should be noted that the rate constant for Met-Puromycin synthesis from the assembled complexes is quite low relative to estimated rate constants for peptide bond formation during elongation in vivo (Palmiter 1975). The low rate of dipeptide formation in this assay may be because the minimal substrate puromycin does not bind in the ribosomal A-site in an optimal manner for reaction with the P-site tRNA (Youngman et al. 2004). Nevertheless, effects of alterations of the tRNA structure on the rate of Met-Puromycin formation are likely to reflect changes in the interactions and interplay between the tRNA and the other components of the initiation machinery.
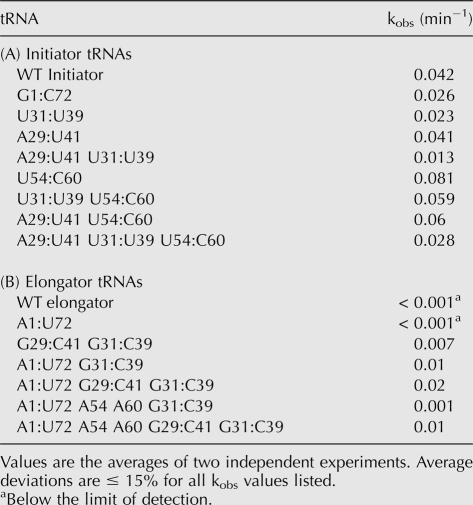
The observed rate constant for Met-Puromycin synthesis with 80S initiation complexes containing the G1:C72 mutant Met-tRNAi was slightly reduced compared with that of the WT initiator (0.026 vs. 0.042 min−1; Table 4A; note, average deviations for the kobs values are ≤ 15% for all tRNAs listed). This alteration did not significantly affect 80S complex formation efficiency, suggesting that the A1:U72 base pair in the initiator tRNA may influence reactivity of the tRNA in the initiation complex. Disruption of the G:C base pairs in the anticodon stem also decreased Met-Puromycin synthesis activity, twofold for the U31:U39 mutant tRNAi and threefold for the A29:U41 U31:U39 tRNAi, although this effect could be at least partially due to the reduction in 80S complex formation and/or stability (Fig. 5A).
TABLE 4.
Observed rate constants (kobs) for methionyl-puromycin synthesis
The U54 and C60 elongator T-loop identities, when placed into the initiator tRNA body, led to a doubling of the observed rate constant for methionyl-puromycin synthesis in comparison with that for the WT initiator (0.081 vs. 0.042 min−1). The elongator T-loop identities also improved the observed rate constants obtained with mutant initiator tRNAs bearing elongator identities in their anticodon stems (approximately twofold; Table 4; Fig. 6A). These changes did not induce any apparent increases in 80S complex formation or stability (Fig. 5B), suggesting that they increase the reactivity of the Met-tRNA within the complex.
FIGURE 6.
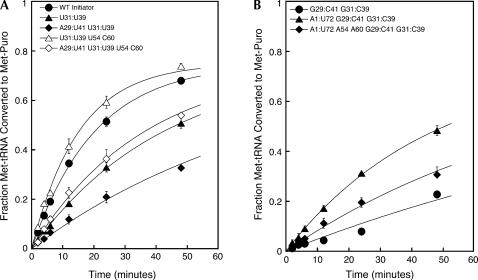
(A) Mutation of the anticodon stem G:C base pairs in mutant initiator tRNAs diminishes their abilities to participate in methionyl-puromycin synthesis, although their defects can be rescued by elongator T-loop identities. (B) The A1:U72 base pair is stimulatory for methionyl-puromycin synthesis by mutant elongator tRNAs, but the initiator T-loop identities are inhibitory. Points are the averages of at least two independent experiments. Errors are average deviations. In all cases, sufficient 40S subunits were used to drive 43S•mRNA complex formation (see Materials and Methods).
The acceptor stem A1:U72 and anticodon stem G:C base pairs are important for Met-tRNA reactivity in 80S initiation complexes
Met-Puromycin synthesis was not detected with the WT or A1:U72 elongator tRNAs, at least in part due to their deficiencies in forming 80S initiation complexes. Mutant tRNAe with the A1:U72 acceptor stem base pair and the single G31:C39 anticodon stem base pair resulted in an observed rate constant for Met-Puromycin formation of 0.01 min−1, at least 10-fold higher than with the A1:U72 mutant alone, but fourfold lower than with the WT tRNAi (Table 4B; Fig. 6B, diamonds). When the tRNAe had the A1:U72 pair and both anticodon stem G:C pairs (G29:C41, G31:C39) the observed rate constant for Met-Puromycin formation was doubled, to within twofold of the value with WT tRNAi (Table 4B; Fig. 6B, triangles). Note that although addition of the second G:C pair in this context increased the rate of Met-Puromycin synthesis, the same change had no detectable effect on 80S complex formation (Fig. 5B, cf. A1:U72 G31:C39 and A1:U72 G29:C41 G31:C39 tRNAs). These data suggest that the G:C base pairs in the anticodon stem have effects on the reactivity of the Met-tRNA in the 80S complex. The G29:C41 G31:C39 mutant tRNAe yielded an observed rate constant threefold lower than that for the A1:U72 G29:C41 G31:C39 tRNAe (0.007 vs. 0.02 min−1; Table 4B; Fig. 6B, solid circles). Because addition of the A1:U72 acceptor stem base pair to the tRNAe with both anticodon stem G:C pairs does not increase the level of 80S initiation complex formation (Fig. 5B), these data support the proposal that the A1:U72 base pair also affects the reactivity of the Met-tRNA in the 80S initiation complex.
Oddly, the addition of the initiator T-loop identities A54 and A60 to mutant elongator tRNAs with both the acceptor stem A1:U72 base pair and one or both of the anticodon stem G:C base pairs diminished their performance in methionyl-puromycin synthesis even though they enhanced 80S complex formation (Table 4B; Figs. 5B, 6B). The observed rate constant for Met-Puromycin synthesis was reduced 10-fold for the A1:U72 A54 A60 G31:C39 tRNAe (0.001 vs. 0.01 min−1), but only twofold for the A1:U72 A54 A60 G29:C41 G31:C39 tRNAe (0.01 vs 0.02 min−1; Table 4B; Fig. 6B, solid diamonds), in both cases relative to the same tRNAs with the elongator T-loop identities (U54 and C60). These data suggest that the initiator T-loop identity elements actually suppress dipeptide bond formation activity in the initial 80S complex. The difference in the effect of U54 and C60 depending on whether one or both of the anticodon stem G:C pairs were present in the tRNA also further supports the proposal that the T-loop and anticodon stem are functionally coupled.
DISCUSSION
The A1:U72 base pair influences multiple steps of initiation
Multiple studies, both in vivo and in vitro, have shown that the A1:U72 base pair in the acceptor stem of the initiator tRNA is a critical binding determinant for eIF2 (von Pawel-Rammingen et al. 1992; Astrom et al. 1993; Farruggio et al. 1996; Drabkin and RajBhandary 1998; Kapp and Lorsch 2004a). The results of our study indicate that, in addition to its key role in eIF2•GTP•Met-tRNAi ternary complex formation, the A1:U72 base pair influences later steps in the pathway. The largest effect of the A1:U72 pair (approximately 10-fold) was on the binding of the ternary complex to the 40S subunit (Tables 2A, 3A). A smaller effect (twofold) was also seen on the ability of the Met-tRNA in the 80S complex to participate in peptide bond formation (Table 4A,B).
The defects associated with mutation of the A1:U72 base pair in several steps of initiation may be a reflection of its importance in eIF2 recognition. Since a critical role of the A1:U72 base pair is to orient the methionine moiety into its pocket on eIF2, resulting in a 10-fold stabilization of Met-tRNA binding (Kapp and Lorsch 2004a), it is possible that G1:C72-containing Met-tRNAs bind to eIF2 aberrantly and are consequently inappropriately presented to the 40S subunit, resulting in decreased stability of the complex. Misplacement of Met-tRNAs in the 40S subunit after their release from eIF2 could also result in the defects observed downstream from 43S complex formation.
The initiator-specific G:C base pairs in the anticodon stem of the initiator tRNA are required for thermodynamic coupling between binding of ternary complex and mRNA to the 40S subunit
Other work from our laboratory has demonstrated that there is ≥1000-fold thermodynamic coupling between binding of ternary complex and mRNA to the 40S subunit, and that this coupling depends on the presence of an AUG codon in the mRNA (Maag et al. 2005). This coupling is unlikely to be important for ternary complex recruitment to the 40S subunit because recruitment is thought to precede mRNA binding. Instead, it reflects changes in interactions occurring within the complex upon start codon recognition that are likely involved in mediating the response to AUG identification. The results presented here indicate that this coupling is dependent upon the presence of the initiator-specific G:C base pairs in the anticodon stem of tRNAi.
S1 nuclease digestion studies (Wrede et al. 1979; Seong and RajBhandary 1987b) as well as crystallographic studies (Schevitz et al. 1979; Woo et al. 1980; Basavappa and Sigler 1991) have suggested that the three consecutive G:C base pairs in the anticodon stem of initiator tRNA impart a unique conformation to the anticodon loop, one that has been described as “skewed” and may be required for proper codon:anticodon pairing between mRNA and the initiator tRNA. The progressive mutation of the individual G:C base pairs in the anticodon stem of the Escherichia coli initiator tRNA to their corresponding elongator identities converted its S1 nuclease digestion pattern to that of an elongator tRNA (Seong and RajBhandary 1987a), and such mutations have produced defects in the functioning of mutant initiator tRNAs in vivo (Mandal et al. 1996). The G31:C39 base pair lies at the bottom of the anticodon stem and closes the anticodon loop, perhaps explaining why its replacement by U31:U39 is slightly more deleterious for the recognition of mRNA than the conversion of the G29:C41 base pair near the middle of the anticodon stem to A29:U41. Several mitochondrial tRNAs lack the G29:C41 base pair, but to our knowledge all known initiator tRNAs contain the equivalent of the G31:C39 base pair (Marck and Grosjean 2002). Taken together with our results, these data suggest that the anticodon stem G:C base pairs in the initiator tRNA communicate with the anticodon loop, allowing it to interact properly with the mRNA in the P-site or allowing the tRNA to signal to eIF2 and/or the 40S subunit that it has formed base pairs with the AUG codon.
Crystal structures of 70S and 30S ribosomal complexes have indicated that 16S rRNA bases G1338 and A1339 interact with the initiator tRNA anticodon stem G:C pairs 29:41 and 30:40 (Carter et al. 2000; Yusupov et al. 2001). In bacteria, IF3 is crucial for discriminating between initiator and elongator tRNAs, a role that requires the initiator-specific G:C pairs in the anticodon stem (Hartz et al. 1990). Chemical foot-printing and cryo-EM experiments indicated that IF3’s position on the 30S subunit is too far from these tRNA base pairs for a direct interaction (McCutcheon et al. 1999; Dallas and Noller 2001). Based on these results, it was proposed that IF3 may stabilize a conformation of the 30S subunit that allows G1338 and A1339 in the rRNA to interact with the 29:41 and 30:40 G:C pairs in the initiator tRNA, which would at least partially explain how IF3 helps discriminate between initiator and elongator tRNAs (Dallas and Noller 2001). Recent site-directed mutagenesis studies of the 16S rRNA support this proposal (Lancaster and Noller 2005). Although IF3 and eIF1 do not appear to be homologous proteins, they are both involved in maintaining the fidelity of start codon selection and they bind to similar sites on the small ribosomal subunit (Lomakin et al. 2003). It was recently shown that eIF1 and IF3 can function in one another's place in bacterial and mammalian in vitro translation initiation systems, respectively, in several ways, including discriminating against non-AUG codons and initiator tRNAs with mutations in the conserved G:C pairs in the anticodon stem (Lomakin et al. 2006) (interestingly, for the latter activity in the mammalian system IF3 was actually more effective than eIF1). These data suggest that eIF1 is involved in promoting recognition of the initiator-specific G:C pairs in the anticodon stem, and part of the effect we have observed of mutating these positions may be due to disruption of the eIF1-induced inspection of these base pairs by the 18S rRNA equivalents of G1338 and A1339.
Studies examining mutant E. coli initiator tRNAs have indicated the importance of the conserved anticodon stem G:C base pairs in initiation in vivo (Mandal et al. 1996) and replacing these base pairs in human initiator tRNA with the elongator sequences reduced the efficiency of translation in eukaryotic cell lysate systems (Drabkin et al. 1993). In yeast, however, the expression of initiator tRNAs bearing elongator identities at base pairs 29:41 and 31:39 in the anticodon stem as the sole copies of initiator tRNAs produced only slight growth defects (von Pawel-Rammingen et al. 1992). In experiments in which mutant elongator tRNAs were expressed in yeast strains lacking initiator tRNA genes, however, addition of the anticodon stem G:C base pairs was found to be critical for allowing the tRNAs to function in initiation (Astrom et al. 1993). In our in vitro system, it is clear that the anticodon stem G:C base pairs are required in initiator tRNAs for coupling between the binding of ternary complex and mRNA to the 40S subunit (Table 2A,B; Fig. 4) and in elongator tRNAs to enable them to bind to the 40S subunit in the absence of mRNA (Table 3A) and with maximal endpoints in the presence of mRNA (Table 3B). It is possible that the thermodynamic coupling between mRNA and ternary complex is part of the mechanism of signaling within the 43S complex that the AUG codon has been found, and that disrupting it in the context of an initiator tRNA body produces only subtle defects in yeast growing under optimal conditions, perhaps due to partial suppression by redundant interactions mediated by other components of the complete initiation complex in vivo (e.g., eIF4F). It will be interesting to determine whether tRNAis lacking the initiator-specific G:C base pairs produce initiation codon selection fidelity defects in vivo in yeast (Sui- phenotypes).
Conformational coupling of tRNA domains in the yeast initiator tRNA is suggested by the effects of mutations of the T-loop identities
Previous studies of the roles of the A54 and A60 T-loop identities in the initiator tRNA indicated that these bases functioned as anti-elongator elements, preventing the initiator tRNA from participating in the elongation phase of translation (Wagner et al. 1989; von Pawel-Rammingen et al. 1992; Astrom et al. 1993; Drabkin et al. 1993, 1998). Our data suggest that these bases may also play a subtler role in the proper functioning of the tRNA during translation initiation.
Replacement of A54 and A60 with the corresponding elongator tRNA bases (U54 and C60) did not have any detectable negative effects on initiator tRNA function in our assays. Addition of A54 and A60 to mutant elongator tRNAs, however, significantly increased the efficiency of stable 80S initiation complex formation (Fig. 5B), suggesting that they influence the subunit joining step or the stability of the resulting complex. Oddly, however, these mutant elongator tRNAs performed worse in Met-Puromycin synthesis than did the corresponding tRNAs without the initiator T-loop identities (Table 4B). In contrast, replacing the T-loop identities in initiator tRNAs with the elongator sequences (U54 and C60) actually increased Met-Puromycin synthesis activity from the resulting 80S initiation complexes (Table 4A). These data suggest that the initiator T-loop bases A54 and A60 may be involved in altering the binding mode or conformation of the tRNA such that it is not fully reactive for the formation of the first peptide bond in the 80S initiation complex. It is possible that to reduce the chance of spurious hydrolysis of the ester bond linking the methionine moiety to the initiator tRNA, the 80S initiation complex starts in a low-reactivity state and is only fully activated when EF1A delivers the first aminoacyl tRNA to the A-site.
The negative effects of mutations of the conserved G:C base pairs in the anticodon stem of initiator tRNAs on the thermodynamic coupling between binding of mRNA and ternary complex to the 40S subunit and on Met-Puromycin synthesis were ameliorated by the addition of the elongator T-loop identities U54 and C60 (Tables 2, 4). This was surprising given that there are no direct contacts between the anticodon stem or loop and the T-loop in the yeast initiator tRNA (Basavappa and Sigler 1991). Several previous studies have indicated that the cognate codon:anticodon pairing between elongator tRNAs and mRNA in the A-site induces conformational changes in the tRNA resulting in the loosening of the association between the D- and T-loops (Schwarz et al. 1976; Farber and Cantor 1980; Bertram et al. 1983; Moras et al. 1986; Rodnina et al. 1994; Dabrowski et al. 1995; Cochella and Green 2005). In addition, a deformation of the anticodon stem, described as a “kinking,” about a hinge region formed from the junction of the D-stem and anticodon stem has been observed in tRNAs paired with their cognate codons in both the ribosomal A- and P-sites in cryo-EM and crystal structures of the bacterial 70S ribosome as well as in cryo-EM reconstructions of the yeast 80S ribosome (Spahn et al. 2001; Yusupov et al. 2001; Valle et al. 2002, 2003). It, thus, seems possible that the effects of replacing the initiator T-loop identities with the elongator identities are due to their lowering the energy barrier for the alteration of the D-loop–T-loop connections during mRNA decoding; the A54 and A60 initiator T-loop identities allow a stronger hydrogen bonding network between the D- and T-loops of the initiator tRNA compared with that of elongator tRNAs (Basavappa and Sigler 1991). Thus, a mutation affecting either the geometry of the tRNA about the hinge region or the conformation of the anticodon stem leading to difficulty in mRNA recognition by the tRNA could be counteracted by lowering the energy barrier for the deformation of the D- and T-loop interactions, since mRNA recognition might be coupled to an anticodon stem deformation/tRNA accommodation event. It will be interesting to examine the effects of mutations of both the T-loop and the anticodon stem on the fidelity of AUG codon selection during initiation, as it has previously been demonstrated that mutation of identities within the D-stem, which may affect the interaction of the D- and T-loops or the position of the anticodon stem relative to the rest of the tRNA, alters the decoding specificity of tRNAs during elongation (Hirsh 1971; Smith and Yarus 1989a,b; Schultz and Yarus 1994a,b; Cochella and Green 2005).
MATERIALS AND METHODS
Protein and ribosome purification
eIF1, eIF1A, eIF2, eIF5, and eIF5B were purified as previously described (Maag et al. 2005). 40S subunits and salt-washed 80S ribosomes were prepared essentially as described (Algire et al. 2002, 2005).
RNA preparation
Templates encoding mutant initiator tRNAs for run-off transcription by T7 RNA polymerase were based on a previously generated initiator methionine tRNA construct within the vector pUC19 (Invitrogen) (Kapp and Lorsch 2004a). Because there is a site for the restriction enzyme BstN1 within the 3′ acceptor stem of the yeast elongator methionine tRNA, a Fok1 restriction site was introduced downstream from the CCA end, such that the Fok1 enzyme would cut immediately 5′ to the T encoding the 3′ terminal adenosine of the tRNA. Mutations of tRNA identity elements were introduced via PCR by the quick change method (Stratagene). The resulting plasmid constructs were subjected to large-scale purification from E. coli and linearized with either BstN1 or Fok1 for run-off transcription by T7 RNA polymerase as described (Kapp and Lorsch 2004a). All constructs were verified by complete sequencing of the tRNA genes.
tRNAs were purified on 12% denaturing gels followed by HPLC first using a semi-preparative monomeric C18 column (238TP54, Vydac) with Solvent A = 20 mM Tris-Acetate pH 6.0, 20 mM Mg(OAc)2, and 400 mM NaCl and Solvent B = the same buffer with 60% methanol and then via a semi-preparative ion exchange column (VHP552, Vydac) with Solvent A = 25 mM KBr, 30% acetonitrile and Solvent B = 1 M KBr, 30% acetonitrile.
To make 35S-Met-tRNA for binding assays, aminoacylation of tRNA was conducted in 75 mM Tris-Cl pH 7.6, 10 mM Mg(OAc)2, and 1 mM DTT using 1–4 μg tRNA, 2 mM ATP, 2 μg glycogen, 8.7 pmol 35S-methionine (NEN), and 5 μL semi-purified GST-tagged methionine aminoacyl-tRNA synthetase from an S. cerevisiae strain kindly provided by Franco Fasiolo (Institut de Biologie Moléculaire et Cellulaire, Strasbourg) in a volume of 20 μL. Reactions were processed as previously described (Kapp and Lorsch 2004a).
Although initiator tRNAs with mutations in their anticodon stems and the U54C60 mutant initiator, in particular, displayed defects in aminoacylation (data not shown), all Met-tRNAs were chargeable. We have shown previously that the methionylated initiator tRNA obtained from S. cerevisiae bulk tRNA performs the same as a purified methionylated T7 transcript of the initiator tRNA in ternary complex formation, 43S•mRNA complex formation, and methionyl-puromycin synthesis (Kapp and Lorsch 2004a).
tRNA affinity measurements
Measurement of eIF2–GTP–Met-tRNA Kds
Filter binding assays measuring ternary complex formation by Met-tRNAs were performed as previously described (Kapp and Lorsch 2004a).
40S subunit binding and 80S complex formation assays
40S subunit binding assays were conducted in 1X Recon Buffer (30 mM Hepes pH 7.4, 100 mM KOAc pH 7.5, 3 mM Mg(OAc)2, and 2 mM DTT) using 200 nM eIF2, 200 nM eIF1 and eIF1A (in the presence of mRNA), or 1.6 μM eIF1 and eIF1A (in the absence of mRNA), 500 μM GDPNP•Mg+2, 1 nM 35S-Met-tRNA, and 1 μM model mRNA when required (Algire et al. 2002). Ternary complexes were preformed by the addition of eIF2 to Met-tRNA and GDPNP with incubation for 10 min at 26°C and then added to 40S subunits, eIF1, eIF1A, and mRNA, when required. Incubation was for a further 10 min in the presence of mRNA or 20 min in the absence of mRNA before reactions were mixed with sucrose-loading dye and loaded onto 4% native gels already running at 20 W. The binding of mutant tRNAs to 40S ribosomal subunits was dependent on the presence of eIF2 (data not shown).
To form 80S initiation complexes 43S•mRNA complexes were formed as indicated above, with the following modifications: 150 μM GTP•Mg+2 was used along with 400 nM each eIF1 and eIF1A, 200 nM 40S subunits and 1 μM mRNA. After the 10-min incubation following the addition of the ternary complex to the 40S subunits and factors, 200 nM eIF5, 1 μM eIF5B, 200 nM 60S subunits, and 1.5 mM GDPNP-Mg+2 to prevent the recycling of eIF2•GDP were added and incubated for 20 min prior to the addition of loading dye and electrophoresis. Incubation times as short as 2 min or as long as 40 min did not produce significantly different results in these experiments (data not shown).
Binding curves for Met-tRNA were either fit to the equation (fraction bound) = Bmax[S]/(Kd + [S]), where Bmax is the maximum fraction bound at infinite [S] or, for experiments in which binding is tight (titrations), to the equation:
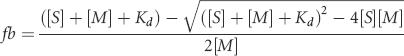 |
using the program Kaleidagraph. In both cases, S is the ligand whose concentration is varied.
Methionyl-puromycin assay
Reactions were performed in 1X Recon Buffer with 200 nM eIF2, 2 mM DTT, 500 μM GTP•Mg+2, 1 nM 35S-Met-tRNA, 1 μM eIF1, 1 μM eIF1A, 200 nM eIF5, 25 nM eIF5B, 400 nM salt-washed 80S ribosomes, 2 μM model mRNA, and 3% DMSO. 80S complexes were formed as described above and then puromycin was added to 1 mM. Reaction products were resolved via cation exchange TLC plates (Polygram Ionex-25 SA-Na, Machery-Nagel) as described (Lorsch and Herschlag 1999; Algire et al. 2002) and developed in 1 M ammonium acetate pH 5.3, 25% acetonitrile. The dried TLC plates were exposed to phosphorimager screens (Molecular Dynamics) overnight for analysis. All curves were fit according to the equation (fraction Met-Puro formed) = A0 × (1−exp−kt) using the program Kaleidagraph, where A0 is the maximum fraction Met-Puro formed at infinite time.
ACKNOWLEDGMENTS
We thank Luisa Cochella and members of our lab for critical reading of the manuscript. We also thank Luisa Cochella and Rachel Green for discussions and our reviewers for insightful comments. This work was funded by NIH grant GM62128 to J.R.L.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2263906.
REFERENCES
- Algire M.A., Maag D., Savio P., Acker M.G., Tarun S.Z., Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L., et al. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002;8:382–397. doi: 10.1017/s1355838202029527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algire M.A., Maag D., Lorsch J.R. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Astrom S.U., von Pawel-Rammingen U., Bystrom A.S. The yeast initiator tRNAMet can act as an elongator tRNAMet in vivo. J. Mol. Biol. 1993;233:43–58. doi: 10.1006/jmbi.1993.1483. [DOI] [PubMed] [Google Scholar]
- Astrom S.U., Nordlund M.E., Erickson F.L., Hannig E.M., Bystrom A.S. Genetic interactions between a null allele of the RIT1 gene encoding an initiator tRNA-specific modification enzyme and genes encoding translation factors in Saccharomyces cerevisiae . Mol. Gen. Genet. 1999;261:967–976. doi: 10.1007/s004380051045. [DOI] [PubMed] [Google Scholar]
- Basavappa R., Sigler P.B. The 3 Å crystal structure of yeast initiator tRNA: Functional implications in initiator/elongator discrimination. EMBO J. 1991;10:3105–3111. doi: 10.1002/j.1460-2075.1991.tb07864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Goringer U., Wagner R. Structural investigation of Phe-tRNAPhe from E. coli bound to the ribosomal A-site. Nucleic Acids Res. 1983;11:575–589. doi: 10.1093/nar/11.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P., Clemons W.M., Brodersen D.E., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Carter A.P., Clemons W.M., Jr., Brodersen D.E., Brodersen D.E., Morgan-Warren R.J., Hartsch T., Wimberly B.T., Ramakrishnan V.V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- Chakrabarti A., Maitra U. Function of eukaryotic initiation factor 5 in the formation of an 80S ribosomal polypeptide chain initiation complex. J. Biol. Chem. 1991;21:14039–14045. [PubMed] [Google Scholar]
- Cochella L., Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowski M., Spahn C.M., Nierhaus K.H. Interaction of tRNAs with the ribosome at the A and P sites. EMBO J. 1995;14:4872–4882. doi: 10.1002/j.1460-2075.1995.tb00168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas A., Noller H.F. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- Desgres J., Keith G., Kuo K.C., Gehrke C.W. Presence of phosphorylated O-ribosyl-adenosine in T-psi-stem of yeast methionine initiator tRNA. Nucleic Acids Res. 1989;17:865–882. doi: 10.1093/nar/17.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabkin H.J., RajBhandary U.L. Initiation of protein synthesis in mammalian cells with codons other than AUG and amino acids other than methionine. Mol. Cell. Biol. 1998;18:5140–5147. doi: 10.1128/mcb.18.9.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabkin H.J., Helk B., RajBhandary U.L. The role of nucleotides conserved in eukaryotic initiator methionine tRNAs in initiation of protein synthesis. J. Biol. Chem. 1993;268:25221–25228. [PubMed] [Google Scholar]
- Drabkin H.J., Estrella M., RajBhandary U.L. Initiator-elongator discrimination in vertebrate tRNAs for protein synthesis. Mol. Cell. Biol. 1998;18:1459–1466. doi: 10.1128/mcb.18.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber N., Cantor C.R. Comparison of the structures of free and ribosome-bound tRNAPhe by using slow tritium exchange. Proc. Natl. Acad. Sci. 1980;77:5135–5139. doi: 10.1073/pnas.77.9.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farruggio D., Chaudhuri J., Maitra U., RajBhandary U.L. The A1 x U72 base pair conserved in eukaryotic initiator tRNAs is important specifically for binding to the eukaryotic translation initiation factor eIF2. Mol. Cell. Biol. 1996;16:4248–4256. doi: 10.1128/mcb.16.8.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster C., Chakraburtty K., Sprinzl M. Discrimination between initiation and elongation of protein biosynthesis in yeast: Identity assured by a nucleotide modification in the initiator tRNA. Nucleic Acids Res. 1993;21:5679–5683. doi: 10.1093/nar/21.24.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar N.J., Kinzy T.G., Scherer B.J., Humbelin M., Hershey J.W., Merrick W.C. Translation initiation factor eIF-2. Cloning and expression of the human cDNA encoding the gamma-subunit. J. Biol. Chem. 1994;269:3415–3422. [PubMed] [Google Scholar]
- Guillon J.M., Heiss S., Soutourina J., Mechulam Y., Laalami S., Grunberg-Manago M., Blanquet S. Interplay of methionine tRNAs with translation elongation factor Tu and translation initiation factor 2 in Escherichia coli . J. Biol. Chem. 1996;271:22321–22325. doi: 10.1074/jbc.271.37.22321. [DOI] [PubMed] [Google Scholar]
- Hartz D., Binkley J., Hollingsworth T., Gold L. Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes & Dev. 1990;4:1790–1800. doi: 10.1101/gad.4.10.1790. [DOI] [PubMed] [Google Scholar]
- Hirsh D. Tryptophan transfer RNA as the UGA suppressor. J. Mol. Biol. 1971;58:439–458. doi: 10.1016/0022-2836(71)90362-7. [DOI] [PubMed] [Google Scholar]
- Huang H., Yoon H., Hannig E.M., Donahue T.F. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes & Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jivotovskaya A.V., Valasek L., Hinnebusch A.G., Nielsen K.H. eIF3 and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol. Cell. Biol. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp L.D., Lorsch J.R. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J. Mol. Biol. 2004a;335:923–936. doi: 10.1016/j.jmb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Kapp L.D., Lorsch J.R. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004b;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Kiesewetter S., Ott G., Sprinzl M. The role of modified purine 64 in initiator/elongator discrimination of tRNAi Met from yeast and wheat germ. Nucleic Acids Res. 1990;18:4677–4682. doi: 10.1093/nar/18.16.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides N.C., Woese C.R. Universally conserved translation initiation factors. Proc. Natl. Acad. Sci. 1998;95:224–228. doi: 10.1073/pnas.95.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster L., Noller H.F. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Lomakin I.B., Kolupaeva V.G., Marintchev A., Wagner G., Pestova T.V. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes & Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin I.B., Shirokikh N.E., Yusupov M.M., Hellen C.U., Pestova T.V. The fidelity of translation initiation: Reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch J.R., Herschlag D. Kinetic dissection of fundamental processes of eukaryotic translation initiation in vitro. EMBO J. 1999;18:6705–6717. doi: 10.1093/emboj/18.23.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D., Fekete C.A., Gryczynski Z., Lorsch J.R. A conformational change in the eukaryotic translation pre-initiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Maag D., Algire M.A., Lorsch J.R. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J. Mol. Biol. 2006;356:724–737. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- Mandal N., Mangroo D., Dalluge J.J., McCloskey J.A., RajBhandary U.L. Role of the three consecutive G:C base pairs conserved in the anticodon stem of initiator tRNAs in initiation of protein synthesis in Escherichia coli . RNA. 1996;2:473–482. [PMC free article] [PubMed] [Google Scholar]
- Marck C., Grosjean H. tRNomics. Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C., Stortchevoi A., Kohrer C., Varshney U., RajBhandary U.L. Initiator tRNA and its role in initiation of protein synthesis. Cold Spring Harb. Symp. Quant. Biol. 2001;66:195–206. doi: 10.1101/sqb.2001.66.195. [DOI] [PubMed] [Google Scholar]
- McCutcheon J.P., Agrawal R.K., Philips S.M., Grassucci R.A., Gerchman S.E., Clemons W.M., Jr., Ramakrishnan V., Frank J. Location of translational initiation factor IF3 on the small ribosomal subunit. Proc. Natl. Acad. Sci. 1999;96:4301–4306. doi: 10.1073/pnas.96.8.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D., Dock A.C., Dumas P., Westhof E., Romby P., Ebel J.P., Giege R. Anticodon-anticodon interaction induces conformational changes in tRNA: Yeast tRNAAsp, a model for tRNA-mRNA recognition. Proc. Natl. Acad. Sci. 1986;83:932–936. doi: 10.1073/pnas.83.4.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter R.D. Quantitation of parameters that determine the rate of ovalbumin synthesis. Cell. 1975;4:189–197. doi: 10.1016/0092-8674(75)90167-1. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Hellen C.U. Preparation and activity of synthetic unmodified mammalian tRNAi Met in initiation of translation in vitro. RNA. 2001;7:1496–1505. doi: 10.1017/s135583820101038x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Borukhov S.I., Hellen C.U.T. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Lomakin I.B., Lee J.H., Choi S.K., Dever T.E., Hellen C.U. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- RajBhandary U.L. Initiator transfer RNAs. J. Bacteriol. 1994;176:547–552. doi: 10.1128/jb.176.3.547-552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina M.V., Fricke R., Wintermeyer W. Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry. 1994;33:12267–12275. doi: 10.1021/bi00206a033. [DOI] [PubMed] [Google Scholar]
- Schevitz R.W., Podjarny A.D., Krishnamachari N., Hughes J.J., Sigler P.B., Sussman J.L. Crystal structure of a eukaryotic initiator tRNA. Nature. 1979;278:188–190. doi: 10.1038/278188a0. [DOI] [PubMed] [Google Scholar]
- Schmitt E., Blanquet S., Mechulam Y. The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors. EMBO J. 2002;21:1821–1832. doi: 10.1093/emboj/21.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D.W., Yarus M. tRNA structure and ribosomal function. I. tRNA nucleotide 27-43 mutations enhance first position wobble. J. Mol. Biol. 1994a;235:1381–1394. doi: 10.1006/jmbi.1994.1095. [DOI] [PubMed] [Google Scholar]
- Schultz D.W., Yarus M. tRNA structure and ribosomal function. II. Interaction between anticodon helix and other tRNA mutations. J. Mol. Biol. 1994b;235:1395–1405. doi: 10.1006/jmbi.1994.1096. [DOI] [PubMed] [Google Scholar]
- Schwarz U., Menzel H.M., Gassen H.G. Codon-dependent rearrangement of the three-dimensional structure of phenylalanine tRNA, exposing the T-Y-C-G sequence for binding to the 50S ribosomal subunit. Biochemistry. 1976;15:2484–2490. doi: 10.1021/bi00656a035. [DOI] [PubMed] [Google Scholar]
- Seong B.L., RajBhandary U.L. Escherichia coli formylmethionine tRNA: Mutations in GGGCCC sequence conserved in anticodon stem of initiator tRNAs affect initiation of protein synthesis and conformation of anticodon loop. Proc. Natl. Acad. Sci. 1987a;84:334–338. doi: 10.1073/pnas.84.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong B.L., RajBhandary U.L. Mutants of Escherichia coli formylmethionine tRNA: A single base change enables initiator tRNA to act as an elongator in vitro. Proc. Natl. Acad. Sci. 1987b;84:8859–8863. doi: 10.1073/pnas.84.24.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek M., RajBhandary U.L. The primary structure of yeast initiator transfer ribonucleic acid. Biochem. Biophys. Res. Commun. 1972;49:508–515. doi: 10.1016/0006-291x(72)90440-8. [DOI] [PubMed] [Google Scholar]
- Smith D., Yarus M. Transfer RNA structure and coding specificity. I. Evidence that a D-arm mutation reduces tRNA dissociation from the ribosome. J. Mol. Biol. 1989a;206:489–501. doi: 10.1016/0022-2836(89)90496-8. [DOI] [PubMed] [Google Scholar]
- Smith D., Yarus M. Transfer RNA structure and coding specificity. II. A D-arm tertiary interaction that restricts coding range. J. Mol. Biol. 1989b;206:503–511. doi: 10.1016/0022-2836(89)90497-x. [DOI] [PubMed] [Google Scholar]
- Spahn C.M., Beckmann R., Eswar N., Penczek P.A., Sali A., Blobel G., Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae–tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Vassilenko K.S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M., Sengupta J., Swami N.K., Grassucci R.A., Burkhardt N., Nierhaus K.H., Agrawal R.K., Frank J. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- von Pawel-Rammingen U., Astrom S., Bystrom A.S. Mutational analysis of conserved positions potentially important for initiator tRNA function in Saccharomyces cerevisiae . Mol. Cell. Biol. 1992;12:1432–1442. doi: 10.1128/mcb.12.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T., Rundquist C., Gross M., Sigler P.B. Structural features that underlie the use of bacterial Met-rRNAf Met primarily as an elongator in eukaryotic protein synthesis. J. Biol. Chem. 1989;264:18506–18511. [PubMed] [Google Scholar]
- Woo N.H., Roe B.A., Rich A. Three-dimensional structure of Escherichia coli initiator tRNAf Met . Nature. 1980;286:346–351. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]
- Wrede P., Woo N.H., Rich A. Initiator tRNAs have a unique anticodon loop conformation. Proc. Natl. Acad. Sci. 1979;76:3289–3293. doi: 10.1073/pnas.76.7.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman E.M., Brunelle J.L., Kochaniak A.B., Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]