Abstract
In plant pararetroviruses, pregenomic RNA serves both as a template for replication through reverse transcription and a polysictronic mRNA. This RNA has a complex leader sequence preceding the first large ORF. The leader contains multiple short ORFs and strong secondary structure, both inhibiting ribosome scanning. Translation on this RNA is initiated by shunting, in which scanning ribosomes bypass a large portion of the leader with the inhibitory secondary structure and short ORFs. In Cauliflower mosaic virus (CaMV), the ribosome shunting mechanism involves translation of the 5′-proximal short ORF terminating in front of the secondary structure that appears to force ribosomes to take off and resume scanning at a landing site downstream of the structure. Using two plant protoplast systems and shunt-competent wheat-germ extracts, we demonstrate that in Rice tungro bacilliform virus (RTBV) shunting also depends on the first short ORF followed by strong secondary structure. Swapping of the conserved shunt elements between CaMV and RTBV revealed the importance of nucleotide composition of the landing sequence for efficient shunting. The results suggest that the mechanism of ribosome shunting is evolutionary conserved in plant pararetroviruses.
Keywords: ribosome shunting, translation initiation, sORF, pararetrovirus, Rice tungro bacilliform virus
INTRODUCTION
Ribosome shunting is a mode of translation initiation in eukaryotes that combines features of 5′-end-dependent scanning and internal initiation (for review, see Ryabova et al. 2002). This unusual initiation process has been first discovered in Cauliflower mosaic virus (CaMV)(Fütterer et al. 1990, 1993) and then in Rice tungro bacilliform virus (RTBV) (Fütterer et al. 1996), both belonging to plant pararetroviruses. Similar phenomena have later been described for several animal viruses, including Sendai paramyxovirus (Latorre et al. 1998; de Breyne et al. 2003), human type C adenovirus (Yueh and Schneider 1996, 2000; Xi et al. 2004), human papillomavirus (Remm et al. 1999), and duck hepatitis B pararetrovirus (Sen et al. 2004), and for some cellular mRNAs (Yueh and Schneider 2000; Rogers et al. 2004). Shunting operates in plant, animal, and yeast translational systems, both in vivo and in vitro (Schmidt-Puchta et al. 1997; Hemmings-Mieszczak et al. 2000; Ryabova and Hohn 2000; Pooggin et al. 2001), thus suggesting its evolutionary conservation.
At the molecular level, a mechanism of shunting has been investigated most extensively for CaMV (Fütterer et al. 1990, 1993; Dominguez et al. 1998; Hemmings-Mieszczak et al. 1998; 2000, 2001; Ryabova and Hohn 2000; Ryabova et al. 2000). A current model of the CaMV shunting states that 40S ribosomal subunits with a complement of initiation factors (hereafter, scanning ribosomes) engage the CaMV pregenomic 35S RNA at the capped 5′-end, scan for a short distance of 60 nt to encounter a first AUG, the start codon of the 5′-proximal short ORF (sORF A), and a majority of the scanning ribosomes assemble at this AUG into complete 80S ribosomes to initiate translation. After the short translation event and peptide release, the 80S ribosomes disassemble at the stop codon of sORF A (a take-off site) and a fraction of the released 40S subunits (hereafter, shunting ribosomes) shunt over an ~480-nt downstream sequence that forms strong secondary structure and contains several sORFs. Downstream of this structure, shunting ribosomes resume scanning for a distance of 45 nt and reinitiate translation at the AUG start codon of the first large viral ORF (ORF VII). Consistent with this “reinitiation” model, the viral transactivator/viroplasmin (TAV) that serves as a reinitiation factor to promote translation of several consecutive large ORFs on the polycistronic 35S RNA (Bonneville et al. 1989; Park et al. 2001) can also increase the efficiency of shunting (Fütterer et al. 1993; Pooggin et al. 2000, 2001).
Other cases, in which shunting mechanisms have been investigated in sufficient detail (Yueh and Schneider 2000; de Breyne et al. 2003), appear to differ from the CaMV case in that they do not involve translation of an sORF. However, a common theme might be pausing of shunting ribosomes at a take-off site. In CaMV, such a pausing might be caused by strong secondary structure six nucleotides downstream of the first sORF and lack of certain initiation factors required for efficient unwinding of the structure following the translation event at sORF A. In adenovirus late RNAs and hsp70 mRNA, pausing might be caused by interaction of scanning ribosomes with the leader regions complementary to 18S ribosomal RNA, which are required for efficient shunting in those cases (Yueh and Schneider 2000). In Sendai virus, pausing might occur during initiation at an upstream non-AUG start codon of the C′ gene (de Breyne et al. 2003).
In this study, we investigated the mechanism of ribosome shunting in RTBV that belongs to a distinct genus of plant pararetroviruses, infecting monocot rice plants (Hull 1999, 2002). In contrast to CaMV, RTBV does not encode any homolog of TAV and polycistronic translation on its pgRNA occurs by leaky scanning (Fütterer et al. 1997; for review, see Pooggin et al. 2002). In both cases, however, translation of the first viral ORF downstream of the long structured leader is initiated by ribosome shunting (Fütterer et al. 1993, 1996). Here we demonstrate that shunting in RTBV depends on the 5′-proximal sORF (sORF 1) followed by a stable secondary structure element, a configuration conserved in the whole family of plant pararetroviruses (Pooggin et al. 1999).
RESULTS AND DISCUSSION
The 5′-proximal sORF is required for shunt-mediated translation in RTBV
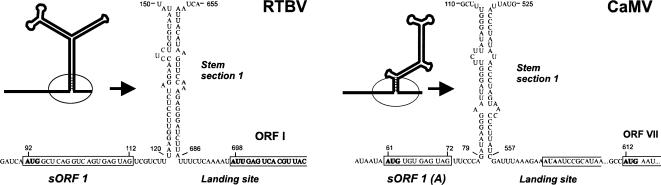
In RTBV, the shunt landing site at the 3′-end of the pgRNA leader has been precisely defined (Fütterer et al. 1996). It includes a non-AUG ORF I start codon (AUU), from which translation is drastically increased upon replacement by AUG, while the same strong AUG introduced 12 nts upstream of the AUU is not recognized at all. The take-off site has been only roughly mapped to a large 5′-proximal region of the leader between positions 89 and 470 by introducing at those positions the Kozak stem structure that blocks scanning (Fütterer et al. 1996). This RTBV leader contains 12 sORFs, and formation of a predicted stem–loop secondary structure would bring the first sORF into close spatial proximity to the landing site (Pooggin et al. 1999; Fig. 1). This strikingly resembles the shunt configuration in the CaMV leader, in which the first sORF A terminates a few bases upstream of “Stem section 1” (Fig. 1), suggesting that the corresponding elements in RTBV are also required for shunting. To investigate a role of the first ORF in RTBV shunting, we introduced two critical point mutations in the full-length RTBV leader. The first mutation knocked out the sORF 1 start codon (AUG to uaG), thus preventing any translation event in front of the stem. The second mutation eliminated the sORF stop codon (TAG to TAc), thus extending the first sORF by an additional seven codons. In CaMV, similar mutations abolish shunt-mediated translation and infectivity of the virus (Pooggin et al. 2001). Wild-type and mutant versions of the RTBV and CaMV leaders were introduced in a plasmid construct between a CaMV 35S promoter and a chloramphenicol acetyl transferase (CAT) reporter ORF followed by a CaMV terminator, and the effect of these mutations on shunt-mediated expression of CAT was tested in two protoplast systems derived from cell suspensions of Oryza sativa (rice; a host plant for RTBV) and Orychophragmus violaceus (a host plant for CaMV).
FIGURE 1.
The conserved shunt configuration in the pgRNA leaders of RTBV and CaMV. The large stem–loop structures of the leaders predicted by GCG MFold for RTBV (left) and CaMV (right) and experimentally verified for CaMV (Hemmings-Mieszczak et al. 1997) are schematically drawn with thick lines. The 5′- and 3′-sequences flanking the main structure are shown in open conformation since they do not form any extensive structures. The stable structural element at the stem base (stem section 1) and adjacent regions are enlarged and their sequences shown. The nucleotide numbering is from the pgRNA 5′-end (cap-site). The 5′-proximal short ORF (sORF 1) is boxed. The AUG start codons and the non-AUG initiating codons in the shunt landing site are in bold.
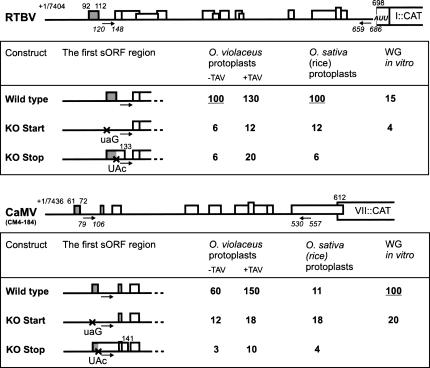
The mutations of RTBV sORF 1 reduced CAT expression eight- and 17-fold in rice and O. violaceus protoplasts, respectively (Fig. 2). Similar strong negative effects of the CaMV sORF A start and stop codon mutations were observed in O. violaceus protoplasts, thus confirming our previous results (Pooggin et al. 2000; Fig. 2). These dramatic negative effects indicate that a translational event at the first sORF terminating at a proper position is essential for ribosome shunting on the RTBV leader.
FIGURE 2.
The 5′-proximal sORF is required for ribosome shunting in RTBV. The RTBV and CaMV leader sequences preceding the first long ORF (ORF VII or ORF I) fused to the CAT reporter are depicted as thick lines; the sORFs are indicated by boxes, with internal start codons indicated by vertical lines. The genome position of the pgRNA cap-site is given. The numbering within the leader is from the pgRNA 5′-end (+1). Arrows under the leader define the complementary sequences that form stem section 1. Relative values of CAT expression downstream of the wild-type and mutated versions (“KO start”and “KO stop”) of the RTBV (top panel) and CaMV (bottom panel) leaders in the three translation systems are given. Expression from the wild-type RTBV construct in O. violaceus and O. sativa protoplasts or from the CaMV wild-type construct in the wheat-germ (WG) in vitro system is set to 100%. In O. violaceus protoplasts, relative expression levels in the absence (−TAV) and the presence (+TAV) of the CaMV reinitiation factor TAV are shown.
In rice protoplasts, CAT expression downstream from the wild-type CaMV leader was inefficient, constituting only 11% of that driven by the RTBV leader, and the mutation in the sORF A start codon slightly increased CAT expression (Fig. 2B). This suggests that the CaMV shunt does not function properly in this particular system, and the observed inefficient translation is possibly achieved by a scanning-dependent mechanism. Mutation in the sORF A stop codon reduced expression about threefold, suggesting that sORF A is nevertheless translated in rice cells and that scanning-dependent initiation at the CAT start codon is influenced by this 5′-proximal translational event.
Previously, the CaMV leader was shown to inhibit gene expression much stronger in nonhost than in host plant protoplast systems (Fütterer et al. 1989). The poor expression downstream from the CaMV leader observed here in yet another nonhost system is consistent with those results. It should be noted, however, that some cryptic events (aberrant transcription initiation, RNA splicing, or degradation) that target the CaMV leader sequence in rice and other nonhost cells may account for inefficient expression observed by us.
The CaMV transactivator protein TAV is a reinitiation factor that directs polycistronic translation on the viral pregenomic (pg) RNA (Bonneville et al. 1989; Park et al. 2001). TAV can also enhance CaMV shunting (Pooggin et al. 2000), which appears to be a special case of reinitiation following translation of sORF A (Ryabova and Hohn 2000). We tested the effect of TAV on RTBV shunting by coexpressing TAV protein from a separate plasmid under the 35S promoter. In O. violaceus protoplasts, CAT expression downstream of the RTBV leader was only slightly increased by TAV (Fig. 2), while as expected, it had a strong effect on expression directed by the CaMV leader (Fig. 2). Since RTBV does not encode any homolog of TAV and its polycistronic translation occurs by leaky scanning (Fütterer et al. 1997) rather than reinitiation, it appears that the RTBV shunt has evolved as independent of any viral reinitiation factor and to direct a more efficient basal translation. The latter is indeed evident in both protoplast systems (Fig. 2).
In rice protoplasts, TAV had no influence on CAT expression downstream of either the CaMV or the RTBV leader (data not shown), suggesting that it may not function in the monocot plant cells. However, we cannot exclude that TAV was not expressed to sufficient levels in this particular system.
To investigate whether the RTBV shunt can also operate in vitro, as shown for CaMV (Dominguez et al. 1998; Ryabova and Hohn 2000; Ryabova et al. 2000), the capped CAT transcripts containing the RTBV and CaMV leader sequences or their mutant versions were produced in vitro using T7 polymerase and then translated in the shunt-competent wheat-germ extracts. As with the plant protoplast system, accumulation of CAT protein was measured by CAT ELISA. Translation downstream from the RTBV leader was about seven times less efficient than that directed by the CaMV leader. But in both cases, mutation of the first sORF start codon resulted in a drastic decrease in translation (Fig. 2). This suggests that the sORF-mediated RTBV shunt also functions in wheat-germ extracts, albeit much less efficiently than the CaMV shunt (see below).
A minimal shunt configuration in RTBV
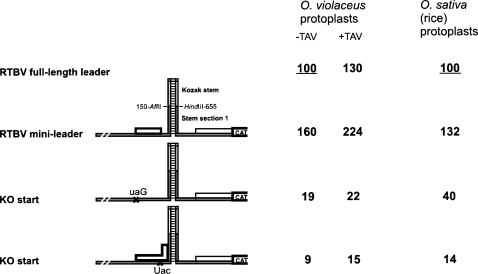
To investigate whether shunting mechanisms of RTBV and CaMV are similar, we chose the O. violaceus protoplast system that supports efficient translation downstream of both CaMV and RTBV leaders. Previously, we have designed a minimal shunt-competent construct based on the CaMV leader (Pooggin et al. 2000), in which a 415-nt central region, normally bypassed by shunting ribosomes, was replaced with a 40-nt Kozak stem sequence that blocks scanning (Kozak 1989). This minimal shunt construct has retained all of the characteristics of the full-length CaMV leader-directed translation initiation (Pooggin et al. 2000), including the requirement for a proper translational event at sORF A terminating a few nucleotides in front of the stem section 1; the primary sequence of the latter was not critical and could be dramatically altered provided that a secondary structure was maintained by compensatory mutations on its ascending and descending arms (Dominguez et al. 1998; Pooggin et al. 2000). To investigate whether a similar structural configuration, which has been recognized in RTBV (Pooggin et al. 1999; Fig. 1), is also sufficient to drive shunting, a 504-nt portion of the RTBV leader, involved in formation of secondary structure above the RTBV stem section 1, was replaced by the Kozak stem sequence (Fig. 3). Basal expression from the resulting construct was 1.6 times higher than from the full-length RTBV leader construct and transcativated by TAV to a comparable degree (1.4-fold). Moreover, mutation of the sORF1 start or stop codon nearly abolished downstream expression (Fig. 3), thus strikingly resembling the results obtained for the minimal CaMV shunt construct (Pooggin et al. 2000). Furthermore, a chimeric RTBV-CaMV construct comprising the RTBV 5′-sequences (sORF followed by ascending arm of the structure) and the CaMV 3′-sequences (descending arm of the structure followed by landing sequence) did not support any significant expression (Fig. 4). Obviously, in this latter construct, secondary structure cannot be formed at a critical distance of several nucleotides downstream of the sORF stop codon, the juxtaposition absolutely required for shunting (Hemmings-Mieszczak et al. 2000; Pooggin et al. 2000).
FIGURE 3.
The RTBV mini-leader that supports ribosome shunting. The mini-leader construct primary (sORF 1, ORF I::CAT fusion) and secondary (stem section 1 extended by the Kozak stem–loop) structures are depicted schematically. The RTBV nucleotide positions between which the 40-nt Kozak stem sequence was inserted (flanked with AflII and HindIII sites) are indicated. RTBV ORF I starting with an AUU codon (in-frame with the CAT ORF) is drawn by a thin line. In constructs “KO start and “KO stop,”mutation of sORF1 (shown by cross) eliminates this sORF or extends it into the ascending arm of the stem (as depicted), respectively. Relative levels of CAT expression in the two plant protoplast systems are given. Expression from the full-length RTBV leader construct in the absence of the CaMV transactivator TAV (−TAV) is set to 100%.
FIGURE 4.
Swapping of the conserved shunt element between CaMV and RTBV. Primary and secondary structure elements of the RTBV and CaMV mini-leaders and their chimeric derivatives are depicted schematically. The viral sequences from CaMV are represented by filled thick lines, while those from the RTBV by nonfilled thick lines. The 5′-proximal sORF is shown as filled (CaMV) or nonfilled (RTBV) box. RTBV ORF I starting with an AUU codon (in-frame with the CAT ORF) and the corresponding non-AUG short ORF of CaMV (overlapping ORF VII::CAT), both originating from the shunt landing site (see Fig. 1), are drawn by a thin line. The unique restriction sites of the CaMV mini-leader construct used for swapping of the shunt elements are indicated. Relative levels of CAT expression in the three translation systems are given. Expression from the full-length RTBV leader construct in O. violaceus and O. sativa protoplasts or from the CaMV mini-leader construct in the wheat-germ (WG) in vitro system is set to 100%.
To further compare the shunting mechanisms of the two viruses, we swapped the three shunt elements—sORF, stem structure, and landing site—individually and in all possible combinations between RTBV and CaMV minimal shunt constructs. In O. violaceus protoplasts, each of the three elements of the CaMV shunt configuration could be functionally substituted by the corresponding RTBV sequences (Fig. 4). Also, replacements of those elements pair-by-pair or all of them together did not cause any dramatic effect on shunt-mediated expression (Fig. 4). Notably, in the presence of the CaMV stem, the chimeric constructs showed slightly lower basal expression than those containing the RTBV stem (Fig. 2). A pairwise comparison indicates that the RTBV stem always mediates about twofold more efficient shunting than the CaMV stem (CaMV “Wild type” vs. “Stem,” “Land” vs. “Stem + Land,” “sORF” vs. “sORF + Stem”). Thus, this secondary structure element alone accounts for higher basal efficiency of RTBV shunting, which is evident for both minishunt and full-length constructs.
In the presence of TAV, expression from all of the swapped constructs was transactivated 1.4- to 2.3-fold. Notably, the lower basal expression levels, especially for those constructs containing the CaMV stem, correlated with the higher degrees of transactivation (two- to 2.3-fold), and vice versa.
Requirements for efficient shunting in rice and wheat-germ systems
In rice protoplasts, the RTBV mini-leader construct expressed CAT as efficiently as the full-length RTBV leader, which also depended on the proper translation of sORF 1 (Fig. 3), whereas CAT expression driven by the mini-CaMV construct was as low as that by the full-length CaMV leader (data not shown). The latter confirms that CaMV shunting does not operate properly in rice cells. In contrast, reciprocal results were obtained in the in vitro wheat-germ system, both full-length and mini leaders of CaMV directed efficient translation, whereas the corresponding RTBV constructs were much less efficient (Fig. 4).
In rice protoplasts, swapping of the shunt elements revealed that expression mediated by the CaMV mini-leader could be significantly improved in the presence of the RTBV landing sequence (threefold), while the RTBV sORF or stem structure alone had only a slight effect or no effect at all. In combinations, the RTBV elements had more pronounced positive effects. The three RTBV elements together, being flanked with the remaining CaMV sequences, further improved expression up to 32% of that driven by the RTBV mini-leader (Fig. 3, “sORF + Stem + Land”). It has been shown that the RTBV leader region preceding sORF 1 contains enhancer elements acting both at the transcriptional and post-transcriptional levels (He et al. 2002). A corresponding sequence from the CaMV leader may not possess an enhancer active in rice protoplasts, which would explain the threefold difference in the expression levels of the latter constructs. Alternatively, a short CaMV sequence following the RTBV landing site may have a negative effect. Taken together, the CaMV leader-containing constructs are poorly expressed in rice protoplasts, not only due to inefficient shunting, but also because of additional factors that remain to be identified.
In the wheat-germ system, swapping of the shunt elements revealed that the RTBV landing sequence is a main inhibitory element. Indeed, none of the chimeras containing this sequence showed efficient translation and its individual replacement by the CaMV landing sequence increased translation 10- to 150-fold in any context (Fig. 4). On the other hand, the RTBV stem or sORF 1 individually improved shunt-mediated translation significantly. All of the three RTBV elements together, being flanked by the CaMV sequences, supported only 10% of the CaMV shunt-mediated translation (Fig. 4, “sORF + Stem + Land”), apparently due to the presence of the inhibitory landing site. Notably, mutation of the sORF start or stop codon in the latter construct totally abolished translation (Fig. 5B, KO start and KO stop), further indicating that inefficient translation directed by RTBV sequences in wheat-germ extracts is achieved by ribosome shunting following translation of RTBV sORF 1.
FIGURE 5.
A 25-nt poly(A) can function as the shunt landing sequence. (A) For the CaMV mini-leader and its derivatives, the sequence in the shunt landing site just downstream of stem section 1 is shown. Relative levels of CAT expression (measured by CAT ELISA) in O. violaceus protoplasts and wheat-germ (WG) extracts are given. Expression from the CaMV mini-leader construct is set to 100%. (B) Translation of capped CAT mRNAs carrying the CaMV mini-leader or its derivatives in WG extracts supplied with [35S]Met.
We next investigated why the RTBV landing sequence failed to function efficiently in the wheat-germ system. In both CaMV and RTBV, sequences downstream of the stem section 1 are very AU-rich (72% and 76%, respectively) (Fig. 5), which ensures low propensity to form secondary structure. However, a U-to-A ratio in RTBV is two times higher than in CaMV, which may account for the observed difference in translation. Indeed, introduction of a 25-nt poly(U) sequence in place of the landing site in the context of the CaMV mini-leader nearly abolished shunt-mediated translation, thus resembling the effect of the RTBV landing sequence (Fig. 5A,B). In contrast, a 25-nt poly(A) sequence introduced at the same location could support 50% of shunt-mediated translation directed by the CaMV landing sequence. Poly(C) or poly(G) sequences failed to support shunting (Fig. 5A,B). Thus, A-richness is a major parameter of an optimal shunt landing sequence in the wheat-germ system.
Testing the four poly(N)-replacement constructs in O. violaceus protoplasts revealed that only the poly(A) sequence could support relatively efficient shunting (37% of the CaMV wild-type sequence), which was also responsive to TAV (about threefold), whereas poly(U), poly(C), or poly(G) failed to functionally replace the shunt landing sequence (Fig. 5A). Taken together, nucleotide composition of the shunt landing site appears to determine efficiency of ribosome shunting in different translational systems and possibly under different conditions.
Conclusions
The mechanism of ribosome shunting in RTBV resembles that in CaMV. In both cases, translation of the 5′-proximal sORF terminating a few nucleotides upstream of the base of a stem–loop structure is absolutely required for efficient shunting, bringing ribosomes to the landing site located just downstream of the structure. The structural configuration of an sORF followed by a secondary structure element is recognized in the pgRNA leader sequences of most plant pararetroviruses sequenced so far (Pooggin et al. 1999; Geering et al. 2005), with the notable exception of Cestrum yellow leaf curl virus, where an sORF in front of the pgRNA leader stem–loop structure is missing (Stavolone et al. 2003), and we therefore predict that the sORF-mediated shunting mechanism is conserved in this viral family. Swapping of the shunt elements, individually and in combination, between CaMV and RTBV revealed that these elements are functionally equivalent in dicot plant cells, even though their primary nucleotide sequences differ considerably. However, in the monocot systems (rice protoplasts and wheat-germ extracts), the shunting mechanism shows some preference for certain cis-acting features. In fact, the landing site sequence of the RTBV leader failed to function in wheat-germ extracts, while it operated well in the two protoplast systems. In rice protoplasts, the CaMV shunt elements together, or in certain combinations with complementing RTBV elements, did not support efficient translation (Fig. 4). In general, the RTBV shunt turned out to direct higher basal translation than the CaMV shunt, as evident from both protoplast systems, and to be less responsive to the CaMV reinitiation factor TAV. This correlates with a difference in the strategies of polycistronic translation from viral pgRNA that relies on leaky scanning in RTBV versus the viral factor-mediated reinitiation in CaMV. It is worth mentioning that the observed differences in shunting efficiency may account for tissue specificity of viral replication, with CaMV infecting most cell types, with RTBV being phloem-limited (Sta Cruz et al. 1993).
MATERIALS AND METHODS
Plasmid constructs
The full-length RTBV leader construct (“Wild type”) (Fig. 2), in which the RTBV pgRNA sequence from the cap-site to position +730 is inserted between the CaMV 35S promoter and the CAT reporter ORF followed by a CaMV terminator, has been described previously as CIC-21A.ATG (Fütterer et al. 1996). Note that this construct and its derivatives tested here contain the strong in-frame AUG start codon downstream from the authentic RTBV ORF I AUU start codon, and ∼90% of the shunting ribosomes, which have scanned through the AUU, initiate at this AUG codon (Fütterer et al. 1996). In the latter construct, point mutations of the RTBV sORF 1 start (ATG to taG) or stop (TAG to Tac) codons were introduced by PCR ligation, yielding constructs “KO start” and “KO stop,” respectively. The corresponding constructs with wild-type and mutated versions of the full-length CaMV leader fused to the CAT reporter under the control of the CaMV 35S promoter and terminator have been described by Pooggin et al. (2000).
The RTBV mini-leader and its derivatives “KO start” and “KO stop” (Fig. 3) were constructed in two steps, based on the CaMV mini-leader construct (Fig. 4) that has been described earlier as pKSXAHA (Pooggin et al. 2000). First, a fragment of the full-length RTBV leader construct from position +655 of the RTBV leader to a unique EcoRI site of CAT, was amplified by PCR and introduced between HindIII and EcoRI sites of pKSXAHA in place of the corresponding CaMV leader-CAT region. Second, in the resulting construct, the CaMV sequence between EcoRV and AflII sites, containing part of the CaMV promoter followed by the CaMV leader sequence up to position +110, was replaced by PCR-amplified segments of the full-length RTBV leader constructs “Wild type,” “KO start,” or “KO stop,” which comprised part of the CaMV promoter (starting from EcoRV) and the RTBV leader sequence up to position +150. The chimeric “RTBV-CaMV” mini-leader was constructed by introducing the “Wild-type” segment between EcoRV and AflII of pKSXAHA.
Swapping of individual shunt elements and their combinations were done in the context of the CaMV mini-leader construct (pKSXAHA) (Pooggin et al. 2000) by making use of four unique restriction sites (see Fig. 4): XhoI (upstream of sORF A), AflII (at the junction between the ascending arm of stem section 1 and the Kozak stem), HindIII (at the junction between the Kozak stem and the descending arm of stem section 1), and AatII (just downstream of the landing sequence). The following chimeric oligonucleotides were ligated between corresponding sites (underlined):
(a) aagcttgtatttaccctatataccctagtaaccccttatctttctcaaaatattgagtcacgttacgacgtc, yielding construct “Land”;
(b) ctcgagatcaatggctcaggtcagtgagtagtcgtcttgataagggaattagggttcttatagggtttcgcttaag, yielding “sORF”;
(a) + (b), yielding construct “sORF + Land”;
(c) ctcgataataatgtgtgagtagttcccataaggttcctctaggaacctctgtgtaatatcttaag and (d) aagcttactaattacataagttccgaagaggaatcttagatttaaagaaataatccgcataagacgtc, yielding construct “Stem”;
(e) ctcgagatcaatggctcaggtcagtgagtagtcgtctttaaggttcctctaggaacctctgtgtaatatcttaag and (d), yielding construct “sORF+Stem”;
(c) and (f) aagcttactaattacataagttccgaagaggaatcttatttctcaaaatattgagtcacgttacgacgtc, yielding construct “Stem + Land”; and
(e) and (f), yielding construct “sORF + Stem + Land.”
Also, the landing sequence of the CaMV mini-leader construct was replaced with four 25-nt homopolymers by cloning the chimeric oligonucleotides consisting of the descending arm of the CaMV stem section 1 followed by poly(N) (shown in Fig. 5A) between HindIII and AatII sites, yielding “Poly(A),” “Poly(U),” “Poly(C),” and “Poly(G).”
For the in vitro translation experiments, the T7 promoter was introduced just upstream of the RTBV full-length and mini-leaders and their variants with the sORF1 mutations by replacing the 35S promoter sequence between EcoRV and ClaI sites with the following sequence: gatatctccactgtaatacgactcactatagggatcgat (the sites underlined). The T7 constructs with the full-length CaMV leader and its mutant version “KO start” have been described earlier as Lm-CAT and LaTAG-CAT, respectively (Dominguez et al. 1998). The CaMV mini-leader and its mutant and chimeric versions constructed above (Figs. 4, 5) were introduced downstream of T7 promoter of Lm-CAT (Dominguez et al. 1998) in place of the full-length CaMV leader by taking advantage of two unique sites of both types of constructs, i.e., XhoI, located between the 5′-untranslated sequence (S1) and sORF A, and SphI, located 117 bp downstream from the CAT ORF stop codon.
Transient expression in plant protoplasts
Protoplasts form suspension cultures of O. violaceus and O. sativa were prepared and transfected with plasmid DNA by electroporation and polyethylene glycol methods, respectively, as described previously (Fütterer et al. 1996; Pooggin et al. 2000). Routinely, 2 × 106 (O. violaceus) or 0.6 × 106 (O. sativa) protoplasts were transfected with 10 μg of CAT-expressing plasmid and 2 μg of β-glucuronidase (GUS)-expressing plasmid (pMONOGUS) (Bonneville et al. 1989). The latter served as an internal control of transfection efficiency. For transactivation, 5 μg of TAV-expressing plasmid (pHELP7) (Bonneville et al. 1989) was also added. Following incubation for 19–24 h at 27°C in the dark, protoplasts were harvested and protein extracts prepared and assayed for CAT and GUS accumulation, as described previously (Pooggin et al. 2000). Relative GUS activities were taken for normalization of CAT expression levels. For each construct, the values given are the means of at least three experiments in independent batches of protoplasts. Deviations from the mean values did not exceed 20%.
In vitro transcription and translation
The in vitro experiments were performed essentially as described earlier (Dominguez et al. 1998) with a few modifications. Briefly, the T7-promoter plasmids were linearized by SphI and transcribed in the presence of the cap analog 7mGpppG (in sixfold molar excess over GTP) by incubation with T7 RNA polymerase (Biofinex) according to the protocol of Gurevich (1996). The integrity of the synthesized transcripts was evaluated on a 6% denaturing polyacrylamide gel. Equimolar amounts of capped transcripts (0.5 pmol) were translated for 1 h at 27°C in a wheat-germ extract prepared according to Roberts and Paterson (1973), in the absence or the presence of [35S]Met. Accumulation of CAT protein in the “cold” translation mixture was measured in duplicate by CAT ELISA (Roche) as recommended by the manufacturer. One-tenth (2.5 μL) of the “hot” translation mixture was resolved on a high cross-linking Tricine SDS–polyacrylamide gel (Schagger and von Jagow 1987). The gel was fixed in the presence of 10% glycerol, dried at 50°C overnight, and exposed to X-ray film (Fuji). For each construct, in vitro translation was performed at least three times with freshly prepared capped RNA, yielding similar results.
ACKNOWLEDGMENTS
We thank Matthias Müller and Sandra Corsten for expert technical assistance. We are grateful to Prof. Christian Körner and Prof. Thomas Boller for providing the lab space and infrastructure in the Institute of Botany. This work was supported in part by a FEBS long-term fellowship to M.M.P. in 1999–2001, and by the Novartis Research Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2285806.
Abbreviations: CaMV, Cauliflower mosaic virus; CAT, chloramphenicol acetyl transferase; GUS, β-glucuronidase; pgRNA, pregemomic RNA; KO, knock out; RTBV, Rice tungro bacilliform virus; sORF, short open reading frame; TAV, transactivator/viroplasmin; WG, wheat germ.
REFERENCES
- Bonneville J.M., Sanfaçon H., Fütterer J., Hohn T. Posttranscriptional trans-activation in cauliflower mosaic virus. Cell. 1989;59:1135–1143. doi: 10.1016/0092-8674(89)90769-1. [DOI] [PubMed] [Google Scholar]
- de Breyne S., Simonet V., Pelet T., Curran J. Identification of a cis-acting element required for shunt-mediated translational initiation of the Sendai virus Y proteins. Nucleic Acids Res. 2003;31:608–618. doi: 10.1093/nar/gkg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D.I., Ryabova L.A., Pooggin M.M., Schmidt-Puchta W., Fütterer J., Hohn T. Ribosome shunting in cauliflower mosaic virus. Identification of an essential and sufficient structural element. J. Biol. Chem. 1998;273:3669–3678. doi: 10.1074/jbc.273.6.3669. [DOI] [PubMed] [Google Scholar]
- Fütterer J., Gordon K., Pfeiffer P., Sanfaçon H., Pisan B., Bonneville J.-M., Hohn T. Differential inhibition of downstream gene expression by the cauliflower mosaic virus 35S RNA leader. Virus Genes. 1989;3:45–55. doi: 10.1007/BF00301986. [DOI] [PubMed] [Google Scholar]
- Fütterer J., Gordon K., Sanfaçon H., Bonneville J.M., Hohn T. Positive and negative control of translation by the leader sequence of cauliflower mosaic virus pregenomic 35S RNA. EMBO J. 1990;9:1697–1707. doi: 10.1002/j.1460-2075.1990.tb08293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J., Kiss-Laszlo Z., Hohn T. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell. 1993;73:789–802. doi: 10.1016/0092-8674(93)90257-q. [DOI] [PubMed] [Google Scholar]
- Fütterer J., Potrykus I., Bao Y., Li L., Burns T.M., Hull R., Hohn T. Position-dependent ATT initiation during plant pararetrovirus rice tungro bacilliform virus translation. J. Virol. 1996;70:2999–3010. doi: 10.1128/jvi.70.5.2999-3010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J., Rothnie H.M., Hohn T., Potrykus I. Rice tungro bacilliform virus open reading frames II and III are translated from polycistronic pregenomic RNA by leaky scanning. J. Virol. 1997;71:7984–7989. doi: 10.1128/jvi.71.10.7984-7989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geering A.D., Pooggin M.M., Olszewski N.E., Lockhart B.E., Thomas J.E. Characterisation of Banana streak Mysore virus and evidence that its DNA is integrated in the B genome of cultivated Musa. Arch. Virol. 2005;150:787–796. doi: 10.1007/s00705-004-0471-z. [DOI] [PubMed] [Google Scholar]
- Gurevich V.V. Use of bacteriophage RNA polymerase in RNA synthesis. Methods Enzymol. 1996;275:382–397. doi: 10.1016/s0076-6879(96)75023-1. [DOI] [PubMed] [Google Scholar]
- He X., Fütterer J., Hohn T. Contribution of downstream promoter elements to transcriptional regulation of the rice tungro bacilliform virus promoter. Nucleic Acids Res. 2002;30:497–506. doi: 10.1093/nar/30.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings-Mieszczak M., Hohn T. A stable hairpin preceded by a short open reading frame promotes nonlinear ribosome migration on a synthetic mRNA leader. RNA. 1999;5:1149–1157. doi: 10.1017/s1355838299990325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings-Mieszczak M., Steger G., Hohn T. Alternative structures of the cauliflower mosaic virus 35S RNA leader: Implications for viral expression and replication. J. Mol. Biol. 1997;267:1075–1088. doi: 10.1006/jmbi.1997.0929. [DOI] [PubMed] [Google Scholar]
- Hemmings-Mieszczak M., Steger G., Hohn T. Regulation of CaMV 35S RNA translation is mediated by a stable hairpin in the leader. RNA. 1998;4:101–111. [PMC free article] [PubMed] [Google Scholar]
- Hemmings-Mieszczak M., Hohn T., Preiss T. Termination and peptide release at the upstream open reading frame are required for downstream translation on synthetic shunt-competent mRNA leaders. Mol. Cell. Biol. 2000;20:6212–6223. doi: 10.1128/mcb.20.17.6212-6223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull R. Plant papraretroviruses—rice tungro bacillifom virus. In: Granoff A., Webster R.G., editors. Encyclopedia of virology. 2d ed. Academic Press; San Diego, CA: 1999. pp. 1292–1296. [Google Scholar]
- Hull R. Matthews' plant virology. 4th ed. Academic Press; San Diego, CA: 2002. [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre P., Kolakofsky D., Curran J. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol. Cell. Biol. 1998;18:5021–5031. doi: 10.1128/mcb.18.9.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.S., Himmelbach A., Browning K.S., Hohn T., Ryabova L.A. A plant viral “reinitiation” factor interacts with the host translational machinery. Cell. 2001;106:723–733. doi: 10.1016/s0092-8674(01)00487-1. [DOI] [PubMed] [Google Scholar]
- Pooggin M.M., Hohn T., Fütterer J. Forced evolution reveals the importance of short open reading frame A and secondary structure in the cauliflower mosaic virus 35S RNA leader. J. Virol. 1998;72:4157–4169. doi: 10.1128/jvi.72.5.4157-4169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooggin M.M., Fütterer J., Skryabin K.G., Hohn T. A short open reading frame terminating in front of a stable hairpin is the conserved feature in pregenomic RNA leaders of plant pararetroviruses. J. Gen. Virol. 1999;80:2217–2228. doi: 10.1099/0022-1317-80-8-2217. [DOI] [PubMed] [Google Scholar]
- Pooggin M.M., Hohn T., Fütterer J. Role of a short open reading frame in ribosome shunt on the cauliflower mosaic virus RNA leader. J. Biol. Chem. 2000;275:17288–17296. doi: 10.1074/jbc.M001143200. [DOI] [PubMed] [Google Scholar]
- Pooggin M.M., Fütterer J., Skryabin K.G., Hohn T. Ribosome shunt is essential for infectivity of cauliflower mosaic virus. Proc. Natl. Acad. Sci. 2001;98:886–891. doi: 10.1073/pnas.98.3.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooggin M.M., Ryabova L.A., Hohn T. Translation strategies in members of the family Caulimoviridae. In: Khan J.A., Dijkstra J., editors. Plant viruses as molecular pathogens. Haworth Press Inc; New York: 2002. pp. 317–338. [Google Scholar]
- Remm M., Remm A., Ustav M. Human papillomavirus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J. Virol. 1999;73:3062–3070. doi: 10.1128/jvi.73.4.3062-3070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B.E., Paterson B.M. Efficient translation of tobacco mosaic virus RNA and rabbit globin 9S RNA in a cell-free system from commercial wheat germ. Proc. Natl. Acad. Sci. 1973;70:2330–2334. doi: 10.1073/pnas.70.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G.W., Jr., Edelman G.M., Mauro V.P. Differential utilization of upstream AUGs in the β-secretase mRNA suggests that a shunting mechanism regulates translation. Proc. Natl. Acad. Sci. 2004;101:2794–2799. doi: 10.1073/pnas.0308576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabova L.A., Hohn T. Ribosome shunting in the cauliflower mosaic virus 35S RNA leader is a special case of reinitiation of translation functioning in plant and animal systems. Genes & Dev. 2000;14:817–829. [PMC free article] [PubMed] [Google Scholar]
- Ryabova L.A., Pooggin M.M., Dominguez D.I., Hohn T. Continuous and discontinuous ribosome scanning on the cauliflower mosaic virus 35 S RNA leader is controlled by short open reading frames. J. Biol. Chem. 2000;275:37278–37284. doi: 10.1074/jbc.M004909200. [DOI] [PubMed] [Google Scholar]
- Ryabova L.A., Pooggin M.M., Hohn T. Viral strategies of translation initiation: Ribosomal shunt and reinitiation. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:1–39. doi: 10.1016/S0079-6603(02)72066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N., Cao F., Tavis J.E. Translation of duck hepatitis B virus reverse transcriptase by ribosomal shunting. J. Virol. 2004;78:11751–11757. doi: 10.1128/JVI.78.21.11751-11757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schmidt-Puchta W., Dominguez D., Lewetag D., Hohn T. Plant ribosome shunting in vitro. Nucleic Acids Res. 1997;25:2854–2860. doi: 10.1093/nar/25.14.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sta Cruz F.C., Koganezawa H., Hibino H. Comparative cytology of rice tungro viruses in selected rice cultivars. J. Phytopathol. 1993;138:274–282. [Google Scholar]
- Stavolone L., Ragozzino A., Hohn T. Characterization of Cestrum yellow leaf curling virus: A new member of the family Caulimoviridae. J. Gen. Virol. 2003;84:3459–3464. doi: 10.1099/vir.0.19405-0. [DOI] [PubMed] [Google Scholar]
- Xi Q., Cuesta R., Schneider R.J. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes & Dev. 2004;18:1997–2009. doi: 10.1101/gad.1212504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yueh A., Schneider R.J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes & Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- Yueh A., Schneider R.J. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes & Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]