Abstract
Eukaryotic cells utilize a cycle of ribosome trafficking on the endoplasmic reticulum (ER) to partition mRNAs between the cytosol and ER compartments. In this process, ribosomes engaged in the synthesis of signal sequence-bearing proteins are trafficked to the endoplasmic reticulum via the signal-recognition particle pathway and are released from the ER upon translation termination. Though the processes governing ribosome trafficking to the ER are well understood, little is known regarding the complementary ribosome release process. In this study, Coxsackie B virus (CBV) infection was used to inactivate the initiation stage of protein synthesis, thereby limiting translation to the elongation and termination stages. Ribosome partitioning between the cytosol and ER compartments was examined to determine the role of termination in ribosome release from the ER. CBV infection resulted in efficient cleavage of eIF4G and PABP, coincident with polyribosome breakdown in the cytosol and ER compartments. Termination resulted in the continued association of ribosomes with the ER compartment, rather than the expected process of ribosome release. Analyses of ribosome/mRNA loading patterns in the cytosol and ER revealed that CBV infection was accompanied by a suppression of mRNA translation in the cytosol and the sustained, although reduced, translation in the ER compartment. Direct biosynthetic labeling experiments demonstrated that protein synthesis on the ER was enhanced relative to the cytosol following CBV infection. In total, these data demonstrate that ribosome and mRNA release from the ER is regulated independent of translation termination and identify the ER as a privileged site for protein synthesis.
Keywords: endoplasmic reticulum, picornavirus, protein synthesis, ribosome, termination
INTRODUCTION
In eukaryotic cells, mRNAs are partitioned between the cytosol and endoplasmic reticulum (ER) compartments. As described in current models, this ubiquitous mRNA partitioning process is driven by a cotranslational cycle of ribosome binding and release on the ER (Blobel and Dobberstein 1975a,b; Lingappa and Blobel 1980; Walter and Johnson 1994; Rapoport et al. 1996). In brief, all mRNAs initiate translation in the cytosol. Early in synthesis, those mRNA/ribosome/nascent polypeptide chain complexes (RNCs) engaged in the synthesis of secretory/membrane proteins are trafficked to the ER via the signal-recognition particle (SRP) pathway. At the ER, a process of cotranslational protein translocation ensues, with secretory proteins being translocated into the lumen of the ER and membrane proteins integrated into the ER membrane. At the termination of protein synthesis, the ribosome dissociates into its component 60S and 40S subunits and is released from the ER membrane to rejoin a common cytosolic pool of ribosomal subunits. In this model, the global partitioning of mRNAs to the ER is intrinsically linked to a dynamic, translation-coupled cycle of ribosome traffic on the ER (Blobel and Dobberstein 1975a,b; Lingappa and Blobel 1980; Walter and Johnson 1994; Rapoport et al. 1996).
Extensive in vitro studies have unequivocally demonstrated that the SRP pathway mediates the cotranslational trafficking of RNCs to the ER (Walter and Blobel 1981a,b; Walter et al. 1981; Gilmore et al. 1982a,b; Meyer et al. 1982). How and when ribosomes are released from the ER is, however, poorly understood. One experimental approach to this question utilized pharmacological inhibitors of the initiation stage of protein synthesis and ultrastructural analyses of ER-bound ribosome density to assess termination-coupled ribosomes release (Seiser and Nicchitta 2000). Though pharmacological inhibition of initiation yielded a dramatic loss of polyribosome structure, the predominant fate of post-termination, ER-bound ribosomes was continued ER association (Seiser and Nicchitta 2000; Potter and Nicchitta 2002). In a related series of studies, it was reported that ER membrane-bound ribosomes were capable of the de novo synthesis of both secretory and soluble proteins, results suggesting that that the ribosome cycle is not obligatory for mRNA partitioning to the ER (Potter and Nicchitta 2000; Potter et al. 2001; Nicchitta 2002). Consistent with this hypothesis, global studies of mRNA partitioning between the cytosol and ER compartments of yeast, fly, and mammalian cells have demonstrated that mRNAs encoding soluble proteins are broadly represented and, in a limited number of cases, highly enriched, on ER-bound ribosomes (Mechler and Rabbitts 1981; Mueckler and Pitot 1981; Kopczynski et al. 1998; Diehn et al. 2000; Lerner et al. 2003; Stephens et al. 2005).
An alternative experimental approach to analyze the ribosome cycle was recently reported in a study of protein synthesis during the unfolded protein response (UPR). The UPR is activated upon accumulation of malfolded proteins in the ER and broadly comprises two processes (Chapman et al. 1998; Harding et al. 2002; Schroder and Kaufman 2005). One, in mammalian cells, global protein synthesis is suppressed through activation of the PKR-like ER kinase, PERK, an ER-resident eukaryotic initiation factor (eIF) 2α kinase (Shi et al. 1998; Harding et al. 1999, 2000b). Phosphorylation of eIF2α results in inhibition of initiation of protein synthesis and polyribosome breakdown. Two, synthesis of the UPR-linked transcription factors Hac1p (yeast), or ATF4 and XBP1, is enhanced, yielding activation of a stress response transcriptional program (Chapman et al. 1998; Harding et al. 2000a, 2002; Yoshida et al. 2001; Schroder and Kaufman 2005). Similar to prior studies with pharmacological inhibitors of the initiation stage of protein synthesis, UPR activation was accompanied by polyribosome breakdown and the accumulation, rather than the expected release of ER-bound 80S ribosomes (Stephens et al. 2005). Surprisingly, following stress induction, the ER was reported to serve as the primary compartment for the synthesis of soluble and secretory proteins alike (Stephens et al. 2005). These investigators proposed that the continued post-termination association of ribosomes with the ER membrane allows this compartment to serve as a “privileged” site for protein synthesis during cell stress.
The observations discussed above identify new and unexpected roles for the ER compartment in the regulation of cellular protein synthesis. In particular, the demonstration that protein synthesis can be sustained on the ER compartment under conditions where protein synthesis in the cytosol is strongly suppressed suggests that protein synthesis in the two compartments is under separate regulatory control (Stephens et al. 2005). To further examine this proposal, we analyzed ribosome partitioning and compartment-restricted protein synthesis using an alternate physiological scenario, a picornavirus infection, to inhibit the initiation stage of protein synthesis. The picornavirus utilized, Coxsackie B3 virus (CBV), inactivates cap-dependent initiation through encoded viral proteases 2A and 3C. Protease 2A cleaves the eIF4G component of the eIF4F complex, and proteases 2A and 3C cleave poly(A)-binding protein (PABP) (Lamphear et al. 1993; Ventoso et al. 1998; Joachims et al. 1999; Kerekatte et al. 1999). The eIF4F complex is comprised of cap-binding protein eIF4E, RNA helicase eIF4A, and scaffold protein eIF4G (Hershey and Merrick 2000). Cleavage of eIF4G disables the eIF4F complex, thereby blocking cap-dependent initiation. In addition, cleavage of eIF4G and PABP would be expected to disrupt circularization of host-cell mRNAs and further disrupt translation of host-cell mRNAs (Gallie 1998; Sachs and Varani 2000). Translation of the viral RNA is unaffected by cleavage of eIF4G and PABP, as initiation on the CBV RNA occurs through a cap-independent mechanism. Like the closely related poliovirus RNA, CBV RNA is uncapped, but encodes an internal ribosome entry site (IRES) within its 5′ noncoding region (Pelletier and Sonenberg 1988). The IRES is thought to directly recruit the 43S preinitiation complex, allowing for IRES-dependent initiation (Belsham and Sonenberg 1996; Hellen and Sarnow 2001).
As expected, cap-dependent protein synthesis was profoundly suppressed following CBV infection, with suppression occurring coincident with proteolytic cleavage of eIF4G and PABP. Under these conditions, polyribosome breakdown occurred in both the cytosolic and ER membrane compartments, yielding a substantial accumulation of 80S monosomes. Similar to results obtained using pharmacological inhibitors or UPR activation, the majority of membrane-bound ribosomes retained their association with the ER following CBV-mediated suppression of host-cell protein synthesis. Of particular interest, the cytosol and ER compartments of CBV-infected cells could be clearly distinguished in their capacity to support protein synthesis, with the ER serving as the primary protein synthesis site. Based on these findings, we propose that ER-bound ribosomes provide a unique, privileged mRNA translation function during physiological scenarios in which the initiation reaction of protein synthesis is suppressed.
RESULTS
Picornavirus infection as a tool to study protein synthesis dynamics
The primary goal of this study was to determine whether and how ribosome exchange on the endoplasmic reticulum (ER) is coupled to protein synthesis. In current models, ribosome trafficking to the ER is initiated early in the synthesis of a signal sequence-bearing protein, and ribosome release from the ER is thought to accompany the ribosome disassembly events of termination (Blobel et al. 1979; Alberts et al. 2002). The picornavirus Coxsackie B3 provides a physiologically relevant means to examine this fundamental question—the CBV genome encodes proteases (2A, 3C) that target the eIF4G component of the eIF4F cap-binding complex and poly(A)-binding protein (PABP), to yield the inactivation of cap-dependent initiation and the loss of mRNA circularization (Gallie 1998; Kerekatte et al. 1999). Elongation and termination proceed under these conditions, to yield global polyribosome breakdown.
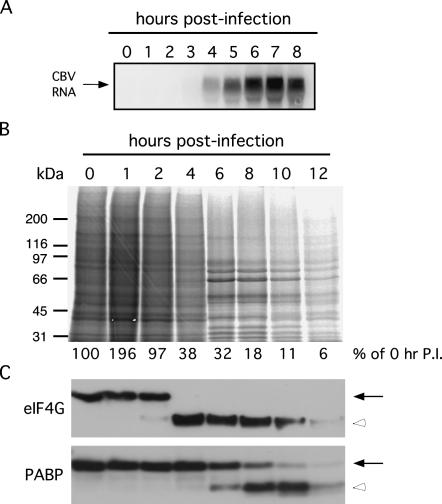
In the experiments depicted in Figure 1, the sequence of CBV infection, as assayed by viral genome replication, loss of cap-dependent translation, and proteolytic cleavage of eIF4G and PABP, is shown. Northern blot analysis of CBV RNA revealed the replication of the viral RNA at 4 h post-infection (PI), reaching peak levels by 6 h post-infection (Fig. 1A). Profiles of total protein synthesis in CBV-infected cells, obtained by [35S]methionine labeling, demonstrated that by 6 h post-infection, cellular protein synthesis levels had decreased significantly, to ∼30% of mock-infected control cells (Fig. 1B). As viral proteins account for the majority of proteins synthesized at that time point (for example, predominant bands seen at 6 and 8 h post-infection at ∼50, 66, 80, and 90 kDa), host-cell protein synthesis levels were likely to be decreased significantly more than the indicated 30%. Cleavage of eIF4G and PABP occurs following synthesis of CBV proteases 2A and 3C (Lamphear et al. 1993; Ventoso et al. 1998; Joachims et al. 1999; Kerekatte et al. 1999). Proteolysis of eIF4G was detectable by 2 h post-infection, and was complete by 4 h post-infection (Fig. 1C). Cleavage of PABP was delayed relative to eIF4G cleavage, with 25% of PABP cleaved by 6 h post-infection and 75% by 8 h post-infection (Fig. 1C). This timeline of eIF4G cleavage preceding both PABP cleavage and the substantial decrease in cellular protein synthesis has been noted in previous studies (Kerekatte et al. 1999). Cell death, as assessed by cell rounding and detachment, occurred within 10–12 h post-infection (data not shown). The results of the above experiments identified the 6-h post-infection time point as an appropriate time to examine ribosome cycling on the ER.
FIGURE 1.
Inhibition of cap-dependent translation in HeLa cells by CBV infection. (A) Total RNA was isolated from HeLa cells infected with CBV from 0 to 8 h. Replication of CBV RNA was analyzed by Northern blot. (B) Radiolabeled extracts from HeLa cells infected with CBV from 0 to 12 h were separated by SDS-PAGE. Total protein synthesis was examined by autoradiograph. (C) Extracts from samples shown in B were separated by SDS-PAGE and immunoblotted with antibodies directed against eIF4G and PABP to detect full-length (solid arrows) and cleavage products (open arrowheads) of the respective proteins.
CBV infection elicits polyribosome breakdown in the cytosol and ER compartments
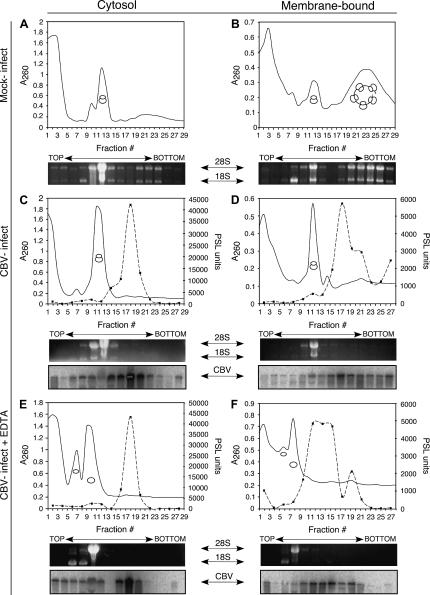
Viral protease-mediated cleavage of eIF4G and PABP is accompanied by polyribosome breakdown. To examine the effects of CBV infection on polyribosome structure in the cytosol and ER compartments, a previously validated detergent fractionation procedure was used (Potter and Nicchitta 2002; Lerner et al. 2003; Le Gall et al. 2004; Stephens et al. 2005). In these experiments, HeLa cells were either mock-infected or infected with CBV for 6 h and fractionated to release cytosolic contents containing the free ribosome fraction, followed by solubilization of ER membranes, to obtain the membrane-bound ribosome fraction. The ribosome composition of the two fractions was analyzed by velocity sedimentation on continuous sucrose gradients and is depicted in Figure 2. The ribosomal subunit composition of the gradient fractions was determined by denaturing RNA gel analysis of 18S and 28S rRNAs. The gradient A260nm (top) and rRNA (bottom) profiles of HeLa cytosol-derived polyribosomes from mock-infected cells showed distinct peaks representing 40S and 60S ribosomal subunits (distinguished by their component 18S rRNA in fraction 8 and 28S rRNA in fraction 10, respectively), 80S monosmes (fraction 12), and modest levels of polyribosomes (fractions 20–24) (Fig. 2A). This biochemical result was confirmed by electron microscopy, which detected an abundance of 80S monosomes and few polyribosomes in HeLa cytosol (data not shown). The HeLa ER membrane-bound ribosomes display a different profile. In mock-infected cells, membrane-bound ribosomes were largely in the form of polysomes (fractions 18–29), with the remainder of the bound ribosome population in the form of ribosomal subunits (40S subunit in fraction 8, 60S subunit in fraction 10), or monosomes (fraction 12) (Fig. 2B).
FIGURE 2.
CBV infection elicits breakdown of free and membrane-bound polysomes. HeLa cells were mock-infected (A,B) or infected with CBV (C–F) for 6 h and processed to yield cytosol and ER fractions. Following isolation, fractions were untreated (A–D) or treated with 15 mM EDTA (E,F) for 30 min on ice. Polyribosome profiles of cytosolic (free) (A,C,E) and ER (membrane-bound) (B,D,F) ribosomes were determined by velocity sedimentation of fractions on 15%–50% linear sucrose gradients. Gradient fractions were collected and analyzed by UV spectroscopy (solid line). The sedimentation of ribosomal subunits and ribosomes was determined through analysis of ribosomal RNA by agarose denaturing gels. For C–F, gels were transferred to nylon membranes and processed for Northern analysis using a probe directed against CBV RNA. Quantification of Northern blot signals was plotted as PhosphorImager (PSL) units (dashed line, solid squares).
Following infection with CBV, polysome structures were absent in both free and membrane-bound ribosome pools (Fig. 2C,D). The predominant monosome peak present in the free ribosome pool (fraction 12) increased 1.6-fold, whereas the membrane-bound ribosomes underwent a dramatic loss of polyribosomes and accumulation of 80S monosomes (fraction 12) (1.9-fold increase in monosome peak absorbance) following CBV infection. The identification of the primary accumulated ribosome species as 80S monosomes was confirmed by denaturing RNA gel analysis of the relevant gradient fractions.
To determine the translation state of the CBV RNA, Northern blot analysis was performed on cytosol and ER fractions of CBV-infected cells. In the cytosol fraction, CBV RNA was detectable throughout the gradient, though most heavily concentrated in the area of the gradient approximating the disome (two ribosome) region, at approximately fraction 18 (Fig. 2C). In contrast, gradient fractions derived from the membrane-bound fraction showed the CBV RNA more strongly represented in the faster sedimenting (polyribosome) region, at fractions 17–22 (Fig. 2D). As seen in the cytosol fraction, a strong peak of CBV RNA was also present in the disome region of the gradient (fraction 18), which represents CBV virions, known to sediment at 155S (Mapoles et al. 1985; Fig. 2D).
To determine whether the ∼160S CBV peak reflected ribosome-associated or virion-derived RNA, HeLa cells were infected with CBV for 6 h, fractionated into cytosol and ER fractions, and the fractions supplemented with 15 mM EDTA. Under these conditions, ribosomes dissociate into their component subunits, whereas CBV virions are unaffected. Samples were then centrifuged on continuous sucrose gradients as above, and polyribosome structure was assessed by UV spectrometry (Fig. 2E,F). EDTA treatment yielded the loss of polyribosome/monosome structure and the accumulation of 40S (in the cytosol, fractions 6–7; in the membrane-bound fraction 6) and 60S ribosomal subunits (in the cytosol, fractions 10; in the membrane-bound fraction 8). Northern blot analysis of the gradient fractions demonstrated that the high S value (e.g., polyribosome associated) CBV RNA fraction was lost (fractions 25–29), with an accumulation of CBV RNA at the lower-density fractions. EDTA treatment did not significantly alter the relative migration of the ∼155 S CBV component, indicating that it represented the virion form.
Interestingly, the EDTA-dependent loss of CBV RNA from the large polysome pool occurred in both free and membrane-bound populations of ribosomes. This suggests that the CBV RNA is translated in both compartments, even though this monocistronic viral RNA does not encode a signal sequence. Plus-strand RNA viruses, such as poliovirus and CBV, are known to alter host-cell membrane structures, creating double-lipid bilayer structures containing ER, trans-Golgi, and lysosomal markers in association with viral proteins (Schlegel et al. 1996; Suhy et al. 2000). While these membranous structures containing ER components are known to be important sites of viral replication (Egger et al. 2000), translation of picornavirus RNAs on ER-bound ribosomes has not been previously observed in vivo.
Membrane-bound ribosomes maintain their association with the ER following CBV infection
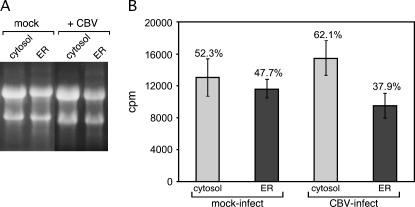
Coincident with CBV infection, there is a dramatic shift from abundant polyribosomes to predominantly monosomes in both the cytosol and ER compartments (Fig. 2A–D). Current models propose, however, that termination is accompanied by ribosome release from the ER (Blobel and Dobberstein 1975a; Mechler and Vassalli 1975; Lingappa and Blobel 1980). To further examine the consequences of CBV-mediated inactivation of cap-dependent initiation on ribosome exchange at the ER, the following experiment was performed: HeLa cell cultures were metabolically labeled by overnight culture in [3H]-uridine, and subsequently either mock-infected or infected with CBV. During the 6-h mock infection or CBV infection, medium containing radiolabeled nucleotide was replaced with medium containing excess unlabeled uridine to prevent metabolic labeling of viral RNAs. Cells were then fractionated to yield cytosol and ER, and total RNA was isolated from each fraction. The RNA was electrophoresed on denaturing RNA gels, and the region of the gel ranging from 28S to 18S RNA excised and counted by liquid scintillation to determine the distribution of ribosomes between the cytosol and ER compartments.
As depicted in Figure 3, in mock-infected HeLa cells ribosomes are distributed approximately equally between the cytosol and ER compartments (Fig. 3A,B). Interestingly, this distribution was only modestly altered following CBV infection (Fig. 3A,B). In mock-infected HeLa cells, [3H]-ribosomes were distributed almost equally between the membrane-bound and cytosolic compartments, with membrane-bound ribosomes representing ∼48% of total cellular ribosomes, and cytosolic ribosomes making up the remaining 52% (Fig. 3B). Following a 6-h CBV infection, ∼38% of total ribosomes remained bound to the ER (Fig. 3B). Thus, the majority (79%) of membrane-bound ribosomes maintain their attachment to the ER following CBV infection. Based upon these findings, we conclude that the termination of protein synthesis is not accompanied by a global release of membrane-bound ribosomes.
FIGURE 3.
Ribosomes maintain their association with the ER during CBV infection. HeLa cells were labeled overnight with [3H]5,6-uridine, then chased with 50 mM uridine during either a mock or a CBV infection. At 6 h post-infection, cells were processed to yield cytosol and ER fractions. Total RNA was purified from cell fractions and separated on denaturing agarose gels, an example of which is pictured in A. The [3H]5,6-uridine content of the 28S to 18S bands was determined by liquid scintillation spectrometry of gel slices. The results of four separate experiments are shown in B.
To determine whether the relative distribution of radiolabeled ribosomes in the cytosol and ER fractions of CBV-infected cells was a consequence of any CBV-dependent alterations in ribosome synthesis/degradation, cytosolic and membrane-bound [3H]-ribosomes were isolated from either mock-infected or CBV-infected cells. If ribosome degradation occurred at different rates in mock-infected versus CBV-infected cells, the total [3H]rRNA content of CBV-infected cells would be expected to be less than that of control cells. We observed that the total [3H]rRNA content of control and CBV-infected cells was not significantly different (data not shown). The differences seen in relative partitioning of ribosomes between the cytosol and ER compartments following CBV infection therefore reflect ribosome repartitioning rather than ribosome metabolism.
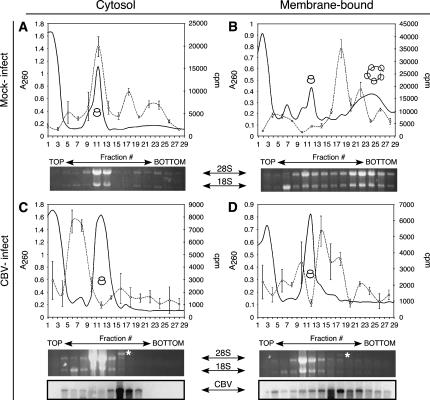
mRNAs are released from free ribosomes, yet retained on membrane-bound polysomes following CBV infection
The data presented above demonstrate that membrane-bound ribosomes remain in association with the ER following CBV-mediated inactivation of cap-dependent initiation; the fate of the mRNAs was unknown. To determine whether mRNA association with free and membrane-bound ribosomes was affected by CBV infection, poly(A+) RNA distribution was determined in mock- and CBV-infected cells. For these experiments, control and CBV-infected HeLa cells were processed to yield cytosol and ER fractions, and were subsequently separated by sucrose gradient velocity sedimentation. Poly(A+) RNA in each fraction was assessed by oligo d(T)-primed reverse-transcription in the presence of [32P]dCTP (Stephens et al. 2005). The results of these experiments are shown in Figure 4. In mock-infected cells, poly(A+) RNA were recovered in association with free or membrane-bound monosomes and polysomes (Fig. 4A,B). In the cytosol fraction (free), poly(A+) RNA was predominately recovered in the monosome fraction (fractions 9–13), whereas in the membrane-derived fraction, poly(A+) RNA was recovered predominately in the polyribosome fraction (fractions 17–25). Thus, in mock-infected cells, poly(A+) RNAs are primarily engaged in actively translating monosomes and polyribosomes in the cytosol and on the ER.
FIGURE 4.
Global mRNA translation is compartmentalized to the ER following CBV infection. HeLa cells were mock-infected (A,B) or infected with CBV (C,D) for 6 h and fractionated to yield cytosol and ER. Polyribosome profiles of cytosolic (free) (A,C) and ER (membrane-bound) (B,D) ribosomes were determined by velocity sedimentation on 15%–50% linear sucrose gradients. Gradient fractions were collected and analyzed by UV spectroscopy (solid line). Total RNA was isolated from individual gradient fractions. mRNA content was measured by quantitative incorporation of [32P]dCTP-radiolabeled nucleotide precursor into cDNA using an oligo (dT)-primed reverse transcription assay (dashed line, open circles). The sedimentation patterns of ribosomal subunits and ribosomes were determined through analysis of ribosomal RNA (indicated by 28S and 18S), and CBV virions (*) were determined by agarose denaturing gels. For C and D, RNA gels were transferred to nylon membranes and processed for Northern analysis using a probe directed against CBV RNA.
CBV infection dramatically altered the distribution of poly(A+) RNA in the cytosol fraction (Fig. 4C). In the cytosol of CBV-infected cells, the primary poly(A+) RNA peak was present in fraction migrating at <80S, indicating that following CBV infection, cytosolic poly(A+) RNAs are primarily present as RNPs (fractions 4–8) (Fig. 4C). A small peak of poly(A+) RNA was also seen in the disome/trisome region of the gradient (fraction 16); however, that peak aligns with the sedimentation of virion particles as determined by denaturing RNA gel and Northern blot analysis for the CBV RNA. In marked contrast to the poly(A+) RNA distribution in the cytosol fraction, the membrane-associated poly(A+) RNA was recovered in the small polysome region of the gradient (fractions 13–19). This peak is likely composed of both cellular mRNAs in addition to the CBV RNA, in the form of CBV virions (Mapoles et al. 1985; Fig. 4D). These data suggest that following CBV infection, the translation activity of the cytosol and ER compartments is differentially influenced; whereas cytosolic polyribosomes disassemble into component ribosome and mRNA fractions, membrane-bound polyribosomes are relatively resistant to breakdown and maintain their association with cellular mRNAs.
mRNAs are preferentially associated with membrane-bound polyribosomes following CBV infection
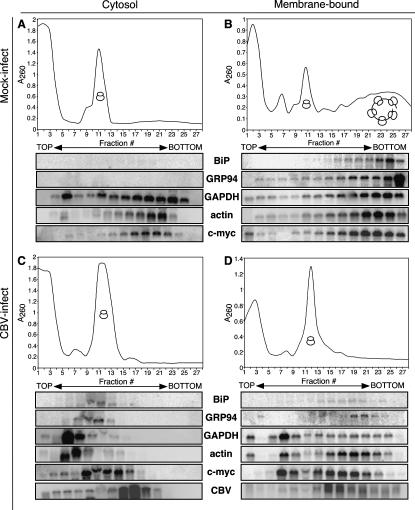
The data presented above describe a rather unexpected finding; CBV-mediated inactivation of cap-dependent translation dramatically suppressed translation in the cytosol compartment, whereas translation in the ER compartment was, on a relative basis, sustained. To further evaluate these findings, the ribosomal loading patterns of individual mRNAs were examined by Northern blot analyses. mRNAs encoding proteins residing in different subcellular locations or displaying alternative translational regulation characteristics were examined. GAPDH and actin served as control mRNAs, as both mRNAs encode soluble proteins and lack both encoded signal sequences and IRES elements. The soluble transcription factor c-myc was selected, as it encodes an IRES and lacks an encoded signal sequence (Nanbru et al. 1997; Stoneley et al. 1998). BiP and GRP94 mRNAs both encode ER chaperones; therefore, both mRNAs encode a signal sequence and each is thought to encode an IRES (Sarnow 1989; Qin and Sarnow 2004). For comparative purposes, the ribosomal loading pattern of CBV RNA was also examined. The results of these experiments are shown in Figure 5.
FIGURE 5.
mRNAs partition to free RNPs or membrane-bound polysomes in response to CBV infection. HeLa cells were mock-infected (A,B) or infected with CBV (C,D) for 6 h and processed to yield cytosol and ER fractions. Polyribosome profiles of cytosolic (free) (A,C) and ER (membrane-bound) (B,D) ribosomes were determined by velocity sedimentation on 15%–50% linear sucrose gradients. Gradient fractions were collected and analyzed by UV spectroscopy. Total RNA was isolated from individual gradient fractions and the precise sedimentation of ribosomal subunits and ribosomes was determined through analysis of ribosomal RNA by agarose denaturing gels. Gels were transferred to nylon membranes and processed for Northern analysis using probes directed against BiP, GRP94, GAPDH, actin, c-myc mRNAs, and CBV RNAs.
In the cytosol fraction of control (mock-infected) cells, mRNAs encoding BiP and GRP94 were not detected; mRNAs encoding GAPDH, actin, and c-myc were predominately present in the heavy polyribosome fractions (Fig. 5A). In the membrane-derived fraction (Fig. 5B), all assayed mRNAs were detected, and all were recovered in the heavy polyribosome fraction. The presence of mRNAs encoding soluble proteins on ER-bound polyribosomes has been extensively described in a number of past and recent studies and will not be further discussed here (Mechler and Rabbitts 1981; Mueckler and Pitot 1981; Kopczynski et al. 1998; Diehn et al. 2000; Lerner et al. 2003; Stephens et al. 2005). Following CBV infection, the ribosome loading patterns of the cytosol and ER-derived fractions differed markedly from one another and, in the case of the cytosol-derived fractions, from the mock-infected cells as well (Fig. 5, cf. A/B and C/D). In the cytosol fraction derived from CBV-infected cells, mRNAs encoding BiP, GRP94, GAPDH, and actin were now detected in the pre-80S/RNP portion of the gradients, suggesting that the ribosome/mRNA complexes had undergone substantial disassembly (Fig. 5C). The appearance in the cytosol fraction of mRNAs encoding BiP and GRP94 was unexpected, and may represent newly synthesized mRNAs that have not yet trafficked to the ER, mRNAs released from the ER as a consequence of CBV-mediated eIF4G/PABP cleavage, or an accumulation of preinitiation complexes. The origin of this mRNA fraction is under continuing investigation. In contrast to the cytosol fraction, mRNAs encoding BiP, GRP94, GAPDH, and actin were detected in the heavy polyribosome fractions of CBV-infected cells, and therefore likely to be undergoing active translation (Fig. 5D). These data are consistent with the global poly(A+) RNA analysis presented in Figure 4.
Both c-myc and CBV mRNAs contain IRES elements and therefore should remain competent for translation following loss of eIF4G/PABP integrity (Johannes and Sarnow 1998; Hellen and Sarnow 2001). Nonetheless, the ribosome loading patterns of c-myc and CBV mRNAs differed in the two compartments. In the cytosol fraction of CBV-infected cells (Fig. 5C), both mRNAs were predominately present in the monosome fractions. While all other host-cell mRNAs examined were in the pre-80S/RNP region of the gradient in the cytosol of CBV-infected cells, the c-myc mRNA alone was associated with 80S monosomes (Fig. 5C). In contrast, in the membrane-derived fraction of CBV-infected cells, c-myc and CBV mRNAs were present in heavy polysome fractions (Fig. 5D). Comparisons of the ribosome loading patterns in the membrane-derived fractions of mock- versus CBV-infected cells indicated that the efficiency of ribosome loading of c-myc was decreased following CBV infection in a manner similar to the other messages analyzed. Significantly, though c-myc and CBV RNAs encode IRES elements, only in the membrane-derived fraction were c-myc and CBV mRNAs detected in the heavy polyribosome fractions. These findings are consistent with the hypothesis that the cytosol and ER compartments are under differing regulatory control, and demonstrate that the observed differences in ribosome loading patterns in the cytosol and ER compartment are evident regardless of whether the mRNA encodes a signal sequence or an IRES element.
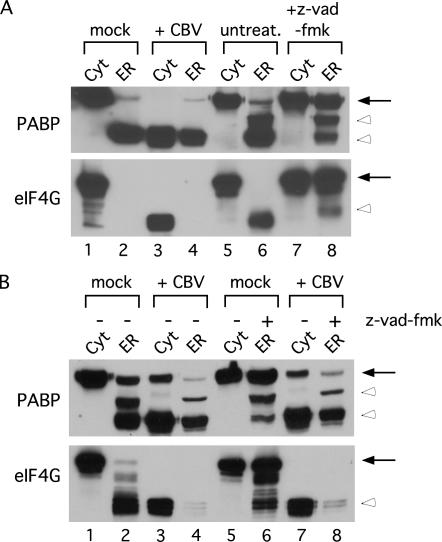
Cytosol- and ER-localized PABP and eIF4G are cleaved by viral proteases
At both the global [poly(A+)] and mRNA-specific level, CBV infection disrupted the polyribosome loading patterns of cytosolic mRNAs to a far greater degree than that of ER-associated mRNAs. To explain these differences, we postulated that viral protease–mediated cleavage of ER-localized eIF4G and PABP might be less efficient than that occurring in the cytosol. To test this prediction, we examined the structural integrity of eIF4G and PABP in the cytosol and ER of mock-infected and CBV-infected cells (Fig. 6). As shown, intact eIF4G and PABP were readily detected in the cytosol fraction of mock-infected cells; as expected, infection with CBV resulted in the proteolysis of eIF4G and PABP in the cytosol fraction (Fig. 6A, lane 3). Unexpectedly, however, intact eIF4G was not detected in the ER fraction and the majority of the PABP was recovered as a cleaved intermediate from both mock- and CBV-infected cells (Fig. 6A, lanes 2,4). eIF4G and PABP are cleaved not only in response to CBV infection, but also during apoptosis. eIF4G is known to be cleaved by caspase-3 during apoptosis, and PABP is also cleaved during apoptosis by an as yet unidentified protease (Marissen and Lloyd 1998; Marissen et al. 2004). We thus considered that the conditions used for CBV infection and/or the processing of the cells for polyribosome analyses were sufficient to elicit an early apoptotic response leading to preferential cleavage of the ER-associated eIF4G and PABP. To test this prediction, the integrity of cytosolic and ER-associated eIF4G and PABP was examined in untreated cultured cells. Again, as in mock-infected cells, cytosolic intact eIF4G and PABP were detected in the cytosol fraction, whereas ER-associated eIF4G and PABP were cleaved; therefore, this ER-specific cleavage event was not due to activation of a pro-apoptotic cascade (Fig. 6A, lanes 5,6). It appeared, then, that ER-localized eIF4G and PABP were being processed by a latent resident eIF4G/PABP protease whose activity was activated upon solubilization of the ER membrane. As eIF4G and PABP are proteolyzed during apoptosis, we attempted to block cleavage of eIF4G and PABP through treatment with pan-caspase inhibitor benzyloxycarbonyl-VAD-fluoromethyl ketone (z-vad-fmk) (Marissen and Lloyd 1998; Marissen et al. 2004). When cells were fractionated as above, with the inclusion of z-vad-fmk during solubilization of the ER, the stability of the ER-localized eIF4G and PABP was enhanced. These results demonstrate that a eIF4G/PABP-directed protease activity(s) is indeed present in the endoplasmic reticulum (Fig. 6A, cf. lanes 6 and 8). On the basis of these findings, we re-examined the state of ER-localized eIF4G and PABP in mock-infected and CBV-infected cells, including z-vad-fmk during solubilization of the ER. As shown, cytosolic eIF4G and PABP were intact in mock-infected cells and cleaved in CBV-infected cells (Fig. 6B, lanes 1,3,5,7). If z-vad-fmk was not included during ER solubilization in both mock-infected and CBV-infected cells, eIF4G and PABP appeared to be primarily in a cleaved state (Fig. 6B, lanes 2,4). When z-vad-fmk was included in the ER solubilization, eIF4G and PABP were primarily intact in mock-infected cells, yet still cleaved in CBV-infected cells (Fig. 6B, lanes 6,8). These results demonstrate that although there exists a latent membrane-associated eIF4G/PABP protease(s), ER-localized eIF4G and PABP are efficiently processed by viral proteases during CBV infection. Therefore, another as yet unidentified cellular mechanism(s) is responsible for the privileged translation status of the ER, as revealed during the cellular response to CBV infection.
FIGURE 6.
ER-localized eIF4G and PABP are susceptible to cleavage by viral proteases. (A) HeLa cells were mock-infected (lanes 1,2) or CBV-infected (lanes 3,4) for 6 h, or untreated (lanes 5–8) and fractionated to yield cytosol and ER fractions via digitonin extraction. The ER fraction in lane 8 was treated with 100 μM z-vad-fmk during solubilization. Protein was isolated from cell fractions, separated by SDS-PAGE, and immunoblotted with antibodies directed against eIF4G and PABP to detect full-length (solid arrows) and cleavage products (open arrowheads) of the respective proteins. (B) HeLa cells were mock-infected (lanes 1,2,5,6) or CBV-infected (lanes 3,4,7,8) for 6 h and fractionated to yield cytosol and ER fractions via digitonin extraction. ER fractions in lanes 6 and 8 were treated with 100 μM z-vad-fmk during solubilization. Protein was isolated from cell fractions, separated by SDS-PAGE, and immunoblotted with antibodies directed against eIF4G and PABP to detect full-length (solid arrows) and cleavage products (open arrowheads) of the respective proteins.
Protein synthesis is sustained on the ER during CBV infection
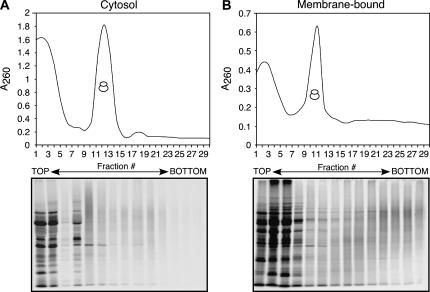
The data presented thus far suggest that following inactivation of cap-dependent translation, the ER serves as a favored site of mRNA translation. This hypothesis extends primarily from analyses of the ribosome loading patterns of cytosolic and ER mRNAs. And though the existence of ER polyribosomes in CBV-infected cells is consistent with this hypothesis, it remained uncertain whether the ER polyribosome fraction was translationally active. To determine whether the observed ribosome loading profiles were indeed representative of actively translating ribosomes, HeLa cells were infected with CBV for 5.5 h, methionine-starved for 30 min, pulsed with [35S]methionine/cysteine for 2 min, and subsequently treated with cycloheximide for 15 min. Cells were then fractionated to yield cytosol and ER and the two fractions separated by sucrose gradient velocity sedimentation. The protein component of the sucrose gradient fractions was recovered by TCA precipitation and [35S]methionine incorporation determined by PhosphorImager analysis of SDS–polyacrylamide gels.
For both cytosol and ER fractions, many radiolabeled proteins were detected in fractions at the top of the gradients (Fig. 7A,B). These represent proteins that had completed protein synthesis during the 2-min pulse. The polyribosome regions of the gradients from the cytosol and membrane fractions were, however, quite different. Whereas only a modest incorporation of radiolabeled methionine was apparent in cytosol-derived ribosome fractions, the polyribosomes derived from the membrane fraction displayed the established pattern of nascent chain radiolabeling, with a “smear” of radioactivity appearing in the higher molecular weight fractions, coincident with an increased degree of ribosomal mRNA loading and ongoing protein synthesis (Fig. 7, cf. A and B).
FIGURE 7.
Protein synthesis is compartmentalized to the ER during CBV infection. At 5.5 h post-infection, HeLa cells infected with CBV were methionine-starved for 30 min, then pulsed for 2 min with [35S]methionine/cysteine. To inhibit further incorporation of radiolabeled amino acid, cells were treated with 200 μM cycloheximide and subsequently fractionated to yield cytosol and ER. Polyribosome profiles of cytosolic (free) (A) and ER (membrane-bound) (B) ribosomes were determined by velocity sedimentation on 15%–50% linear sucrose gradients. Gradient fractions were collected and analyzed by UV spectroscopy. Total protein was isolated from individual gradient fractions, separated by SDS-PAGE, and analyzed by [35S]methionine incorporation determined by PhosphorImager analysis.
DISCUSSION
In this communication, we report two primary observations: One, the predominant fate of ER-bound ribosomes upon termination is continued membrane association; two, the ER serves as a privileged site of protein synthesis following the inactivation of cap-dependent translation, as occurs upon CBV infection. This latter conclusion extends from the finding that following proteolytic cleavage of eIF4G and PABP, mRNA translation was sustained in the ER compartment and markedly suppressed in the cytosol compartment. Interestingly, preferential mRNA translation on the ER was observed regardless of whether the mRNA encoded a signal sequence or an IRES element. These findings suggest that the protein synthesis machinery of the cytosol and ER compartments is under distinct regulatory control.
While the cellular processes governing the trafficking of ribosomes to the ER has been studied in great detail, the complementary ribosome release process is relatively uncharacterized. It has long been assumed that ribosome release from the ER membrane occurs coincident with the termination of protein synthesis. Recent findings, combined with results described in this report, indicate, however, that termination is not itself a necessary and sufficient signal for ribosome release. For example, following the pharmacological inhibition of initiation, ribosomes were found to reside in stable association with the ER membrane, as determined by both ultrastructural and biochemical analyses (Seiser and Nicchitta 2000; Potter and Nicchitta 2002). Similarly, we recently reported that physiological inactivation of eIF2α activity, via activation of the unfolded protein response (UPR), resulted in polyribosome breakdown and the continued association of ribosomes with the ER membrane (Stephens et al. 2005). In the present study, Coxsackievirus B was utilized as a physiological means to inactivate cap-dependent translation and thereby promote the polyribosome breakdown events thought to initiate ribosome dissociation from the ER. Here as well, polyribosome breakdown was not accompanied by the release of membrane-bound ribosomes to the cytosol. Rather, and consistent with previous studies, the predominant fate of membrane-bound ribosomes upon termination was continued association with the ER. Thus, regardless of whether the biochemical events of protein synthesis initiation are compromised by pharmacological inhibitors, phosphorylation of eIF2α, or CBV-mediated cleavage of eIF4G and PABP, the resulting physiological termination events are not functionally linked to the process of ribosome exchange on the ER. This conclusion is further strengthened by the recent findings that physiological inactivation of eIF2α is accompanied by the appearance of ER-bound 80S ribosomes lacking associated mRNAs (Stephens et al. 2005). In this context, it should also be noted that purified ribosomes bind to ER membranes with equilibrium binding constants in the low nanomolar range, and thus undergo spontaneous exchange only infrequently (time constants of tens of minutes) (Borgese et al. 1974; Kalies et al. 1994; Raden et al. 2000). In fact, when direct ribosome exchange was examined in vitro, it was found that large ribosomal subunits, through which binding to the ER membrane occurs, were refractory to exchange (Borgese et al. 1973).
The finding that ribosomes retain their association with the ER following physiological termination is significant because it suggests a more global role for the ER compartment in cellular protein synthesis than previously appreciated. In particular, we have previously demonstrated that post-termination, ER-bound ribosomes are capable of de novo translation regardless of whether or not the mRNA encodes a signal sequence (Potter and Nicchitta 2000). In principle, then, ER-bound ribosomes can be envisioned as participating in the translation of all cellular mRNAs, a suggestion consistent with global studies of mRNA partitioning between cytosolic and ER-bound polyribosomes (Mechler and Rabbitts 1981; Mueckler and Pitot 1981; Kopczynski et al. 1998; Diehn et al. 2000). What remains to be understood is how the relative partitioning of individual mRNAs between the two compartments is regulated. Presumably, such partitioning is influenced by localization information present in the mRNA and RNA-binding proteins functioning in the recognition of such information (Nicchitta 2002; Nicchitta et al. 2005).
A particularly surprising finding described in the present study concerns the status of the ER compartment as a preferred site for protein synthesis. In examining the ribosome loading profiles of cytosol and ER-derived ribosomes, as well as their biosynthetic activity, it was evident that protein synthesis on the ER was sustained, albeit at a substantially reduced rate, following CBV-mediated proteolytic inactivation of eIF4G and PABP. In contrast, protein synthesis in the cytosol was dramatically reduced upon CBV infection, with the majority of mRNAs being recovered in the pre-80S/RNP fractions. This difference was particularly evident in comparisons of c-myc mRNA translation in the cytosol and ER compartments. c-myc mRNA encodes an IRES element and can undergo translation when cap-dependent translation is inhibited (Nanbru et al. 1997; Stoneley et al. 1998). Following CBV infection, c-myc mRNA was assembled into 80S monosomes in the cytosol, but only in the ER compartment was c-myc mRNA efficiently assembled into polyribosomes. An earlier study using an attenuated poliovirus infection demonstrated the continued association of IRES-containing mRNAs (including c-myc) with polyribosomes during infection (Johannes and Sarnow 1998). Our results confirm findings from this report, and further reveal a compartmental restriction of mRNA translation to the ER. Interestingly, we recently reported that physiological inhibition of eIF2α function, via induction of the UPR, also results in a compartmentalization of protein synthesis to the ER (Stephens et al. 2005). These parallels are particularly striking in that when two distinct physiological scenarios yielding the inhibition of cap-dependent translation were examined, both scenarios revealed a very similar cellular response—protein synthesis on the ER was favored over that in the cytosol regardless of whether the mRNA encoded a soluble cytosolic protein or a secretory/membrane protein (Stephens et al. 2005; present study). These findings highlight an emerging view of the ER as a dynamic compartment functioning in the translation of the global population of mRNAs (Potter et al. 2001; Nicchitta 2002; Nicchitta et al. 2005). Interestingly, it has recently been proposed that the ER can undergo additional levels of functional specialization, with extrinsic cues and local interactions with other organelles contributing to a higher order spatial compartmentalization and organization of mRNA translation (Levine and Rabouille 2005).
Though evidence has accumulated supporting the view that protein synthesis in the cytosol and ER compartments is differentially regulated, the precise mechanism for how this occurs is not known. As noted above, protein synthesis on the ER is favored following physiological inactivation of eIF2α or proteolytic inactivation of eIF4G/PABP (Stephens et al. 2005; present study). For both scenarios, there are established examples of specific mRNA elements that support ongoing synthesis under conditions where global, cap-dependent protein synthesis is suppressed. In the case of eIF2α inactivation, the translation of mRNAs encoding multiple upstream open reading frames, i.e., the transcription factor ATF4, is enhanced (Lu et al. 2004; Vattem and Wek 2004). Similarly, mRNAs encoding IRES elements undergo translation under conditions where eIF4G and PABP have been proteolytically inactivated (Johannes and Sarnow 1998; Johannes et al. 1999). Although these mechanisms provide compelling explanations for why particular mRNAs can undergo continued translation following a reduction in eIF2α or eIF4G/PABP activity, they do not explain the observation that global translation on the ER is favored following the loss of eIF2α or eIF4G/PABP functionality. Regarding the latter, we can eliminate the possibility that the ER functions to “protect” eIF4G and PABP from proteolytic inactivation, and thus we suggest that the ER environment provides a regulatory environment that can be distinguished from the cytosol by its reduced dependence on eIF4G/PABP function (Fig. 6). One possible way that this could be achieved is through provision of alternative sources of eIF4G- and PABP-like functions, albeit in an ER-restricted manner. For example, export of newly transcribed mRNAs is known to occur coincident with a “remodeling” of the cap and tail-binding proteins, with the nuclear forms (CBP80/CBP20 and PABP2) being exchanged for the eIF4F complex and PABP1. Perhaps CBP80/CBP20 and PABP2 are utilized by ER-bound ribosome/mRNA complexes upon inactivation of eIF4GI/II and PABP to enable sustained, though reduced protein synthesis of host cells mRNAs on the ER (Ishigaki et al. 2001; Chiu et al. 2004).
An alternative possibility is that the fragments of eIF4G and PABP are sufficient for continued ER-localized translation, while unable to perform functions of their intact counterparts in the cytosol. Apoptosis is another physiological cell stress that elicits eIF4G and PABP cleavage. During apoptosis, eIF4G is cleaved by caspase-3 and PABP is cleaved by an as yet unidentified protease (Marissen and Lloyd 1998; Marissen et al. 2004). Interestingly, fragments of eIF4G are known to mediate both IRES-mediated translation of both transcription factor c-myc and pro-apoptotic mRNAs Apaf-1 and DAP5 and low levels of cap-dependent initiation (Ali et al. 2001; Nevins et al. 2003; Hundsdoerfer et al. 2005). It is possible that fragments of eIF4G and/or PABP derived by viral protease cleavage are not competent to elicit initiation events within the cytosol but are able to specifically stimulate ER-localized translation. Ongoing studies are under investigation to determine whether pharmacological or physiological induction of apoptotic pathways, which similarly result in cleavage of eIF4G and PABP, will also restrict mRNA translation to the ER.
At a more speculative level, the constraining of the complex reactions of protein synthesis to the two-dimensional plane of the ER membrane may offer a considerable kinetic advantage and thereby favor translation on the ER versus the cytosol compartments. In this vein, it is also worth noting that ER-bound polysomes have long been known to be highly structured, forming well-defined “rosettes” and spirals on the ER membrane (Fawcett 1981; Christensen et al. 1987). Perhaps the highly ordered structure of ER-bound polyribosomes enables alternative routes of initiation/ribosome recycling that display a reduced requirement for the protein machinery functioning in de novo initiation.
MATERIALS AND METHODS
Reagents
All tissue-culture reagents were obtained from GIBCO. Molecular biology reagents were acquired from Promega, New England Biolabs, and Invitrogen. Radioactive reagents were obtained from Amersham Pharmacia Biotech, Moravek, and ICN. Other reagents were from Sigma-Aldrich unless otherwise indicated.
Cell culture
HeLa human epithelial cells were cultured in DMEM supplemented with 10% fetal calf serum and100 U/mL penicillin and 100 U/mL streptomycin (“complete DMEM”). Cells were maintained at 37°C and 5% CO2 and were subcultured at 2–3 d intervals.
Viral infection
Infection of HeLa cells with Coxsackie B3 virus (CBV), kindly provided by Dr. Matthias Gromeier (Duke University Medical Center), was carried out as follows: HeLa cells were grown to ∼90% confluence and washed twice with serum-free DMEM. CBV was added to cells in a minimal volume of serum-free DMEM (multiplicity of infection = 20). Cells were incubated with virus at room temperature for 30 min, washed twice with serum-free DMEM, and subsequently incubated at 37°C and 5% CO2 in excess volume of fresh serum-free DMEM. Mock-infected cells underwent the above procedure, except that during viral binding discussed above, cells were exposed to a minimal volume of serum-free DMEM lacking virus.
Cell fractionation
HeLa cells were grown to 80%–90% confluence, and treated with 200 μM cycloheximide at 37°C for 15 min prior to fractionation. Cells were then placed on ice and washed with 4°C DPBS containing 200 μM cycloheximide. Cells were incubated with a cytosol buffer containing 110 mM KOAc, 25 mM K-HEPES (pH 7.5), 2.5 mM Mg(OAc)2, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, 200 μM cycloheximide, 50 U/mL RNase Out (Invitrogen), and the plasma membrane permeabilized by addition of 150 μg/mL digitonin (Calbiochem) (Lerner et al. 2003; Stephens et al. 2005). Cells were incubated in cytosol buffer on ice for 5 min, and the cytosol-buffer supernatant was collected and saved on ice. Cells were rinsed twice on ice with cytosol buffer lacking detergent to remove any remaining cytosolic components. Permeabilized cells were scraped into wash buffer lacking detergent and centrifuged at 500g for 5 min at 4°C. Cell pellets were resuspended in an ER-solubilization buffer containing 400 mM KOAc, 25 mM K-HEPES (pH 7.5), 15 mM Mg(OAc)2, 1 mM DTT, 1 mM PMSF, 200 μM cycloheximide, 50 U/mL RNase Out (Invitrogen), and either 20 mg/mL digitonin (Calbiochem) or 20 mg/mL n-dodecyl-β-D-maltoside (Calbiochem). For samples treated to inhibit caspase activity, 100 μM z-vad-fmk was included in the ER-solubilization buffer. This resuspended cell pellet was incubated on ice for 30 min, then centrifuged at 10,000g for 10 min at 4°C to remove nuclei, mitochondria, and incompletely solubilized cells. The supernatant was saved and used as a source of ER-bound polyribosomes. The cytosol-buffer supernatant was also centrifuged at 10,000g for 10 min at 4°C to remove contaminating cellular components. This supernatant was used as a source of cytosolic polyribosomes. All buffers described above were made with DEPC-treated solutions.
Polyribosome analysis
HeLa cells were mock-infected or infected with CBV for 6 h, then treated with 200 μM cycloheximide at 37°C for 15 min. Cells were fractionated as described above to yield cytosolic and membrane-bound polyribosome fractions. Fractions containing 2 × 107 cell equivalents of free or membrane-bound polyribosomes were loaded onto continuous 15%—50% sucrose gradients containing 400 mM KOAc, 25 mM K-HEPES (pH 7.5), 15 mM Mg(OAc)2, 200 μM cycloheximide, and 10 U/mL RNase Out (Invitrogen). For samples treated with EDTA, HeLa-purified cytosol and membrane-bound polyribosome fractions were brought to 15 mM EDTA and incubated on ice for 30 min. The EDTA-treated fractions were loaded onto continuous 15%–50% sucrose gradients containing 400 mM KOAc, 25 mM K-HEPES (pH 7.5), 15 mM EDTA, 200 μM cycloheximide, and 10 U/mL RNase Out (Invitrogen). Gradients were centrifuged for 3 h at 150,000g at 4°C in a SW41 rotor (Beckman). Gradient fractions (∼350–400-μL fractions) were manually collected and the A260 nm was determined. RNA was isolated from gradient fractions as described below. All buffers described above were made with DEPC-treated solutions.
Protein synthesis assays
To detect alterations in protein synthesis over the course of CBV infection, radioisotope-labeling experiments were performed as follows: For analysis of total protein synthesis over a time course of CBV infection, cells were mock-infected or infected with CBV, and, 40 min prior to each time point, washed three times with labeling medium (DMEM lacking serum and methionine supplemented with 1 mM sodium-pyruvate, 0.25 mM HEPES at pH 7.3, 2 mM L-glutamine, 100 U/mL penicillin and 100 U/mL streptomycin) and methionine “starved” in labeling medium for 30 min at 37°C. Isotopic labeling was initiated by addition of 150 μCi/mL [35S]methionine/cysteine, and cells incubated for 10 min at 37°C. Medium was removed, cells washed with cold 1xDPBS, and scraped into cold DPBS. Cells were collected by centrifugation at 2500g for 5 min at 4°C, and cell pellets resuspended in lysis buffer containing 50 mM Tris (pH 7.5), 5 mM EDTA, 150 mM NaCl, 1 mM PMSF, and 1% NP-40. Cell pellets were gently vortexed, incubated on ice for 10 min, and centrifuged at 10,000g to pellet nuclei and unbroken cells. Supernatants were saved and processed as described below. For analysis of total protein synthesis in association with the free and membrane-bound ribosomes, cells were treated similarly to above, except they were infected for 5.5 h, starved for 30 min, and pulse-labeled with 250 μCi/mL [35S]methionine/cysteine for 2 min. Cells were further treated with 200 μM cycloheximide, fractionated to yield cytosol and ER subcellular fractions, centrifuged on continuous sucrose gradients, and gradient fractions isolated for further analysis. Fractions were analyzed by UV spectrometry and further processed as described below.
Proteins from total, cytosol, or ER fractions, or sucrose gradient fractions, were precipitated by addition of cold trichloroacetic acid to 10%, incubation on ice for 30 min, and centrifugation at 20,000g for 15 min at 4°C. Pellets were washed once with 5% TCA and once with cold acetone. Final protein pellets were resuspended in sample buffer (50 mM Tris at pH 6.8, 2% SDS, 2% glycerol) and 1% β-mercaptoethanol or 1 mM DTT, heated at 65°C, and processed for SDS-PAGE. Gels were either fixed and dried for autoradiography or transferred to nitrocellulose for Western blot.
Immunoblotting
PABP antibody was a kind gift from Dr. Jack Keene (Duke University Medical Center), and eIF4GI antibody was provided by Dr. Robert Rhoads (Louisiana State University Health Sciences Center, Shreveport). Protein samples were separated by SDS-PAGE and transferred to nitrocellulose membranes using a semidry transfer apparatus (Bio-Rad) in transfer buffer containing 50 mM CAPS (pH 11.0), 20% methanol, and 0.075% SDS. Membranes were blocked in phosphate-buffered saline in 9% milk and 0.1% Tween 20 at 4°C overnight. Primary incubations were performed for 1 h at room temperature in phosphate-buffered saline containing 1% milk and 0.1% Tween 20, and secondary incubations were performed for 30 min at room temperature in the identical buffer using secondary antibodies at 1:2000 dilution. Immunoblots were visualized using ECL Western Blotting System (Amersham Pharmacia Biosciences).
RNA isolation
RNA from total, cytosol, or ER fractions was obtained using Trizol reagent (Invitrogen) as per the manufacturer's instructions. RNA was isolated from sucrose gradient fraction samples (∼350—400 μL) by addition of 400 μL guanidine thiocynate buffer (4 M guanidine thiocynate, 25 mM sodium citrate, 0.5% N-lauryl sarcosine, 5 mM EDTA, and 0.1M β-mercaptoethanol) and 250 μL of saturated phenol. Samples were rotated for 15 min at room temperature, supplemented with 200 μL chloroform, mixed vigorously, and centrifuged at 5000g for 10 min at 4°C. The aqueous layer was saved and RNA precipitated by addition of 1 vol of isopropanol and 1/10 vol of 3 M NaOAc (pH 5.2) at −20°C. RNA was pelleted at 20,000g for 15 min at 4°C, washed with 70% ethanol, and resuspended in DEPC-treated water.
Northern blot analysis
Northern analyses were conducted by standard protocols (Sambrook et al. 1989). For Northern analyses done from total cellular RNA, 10 μg of total HeLa cell RNA per sample was separated on 1% agarose gels containing 3% formaldehyde. For analysis of gradient samples by Northern blot, total RNA was purified from each gradient sample and separated on agarose gels as described above. Note that RNA levels vary among gradient samples due to different sedimentation rates for ribosomal subunits, monosomes, and polyribosomes. Gels were transferred in 5× SSC, 10 mM NaOH by downward capillary flow onto Hybond charged nylon (Amersham Pharmacia Biotech) for 2 h. Nylon membranes were cross-linked, then prehybridized in 1× Church buffer (7% SDS, 1 mM EDTA, and 0.5 M phosphate buffer) containing 100 μg/mL salmon sperm DNA at either 50° or 55°C for 1 h. Blots were hybridized in 1× Church buffer containing 100 μg/mL salmon sperm DNA and 50–100 μL of radiolabeled probe at 5°C above the prehybridization temperature overnight. Northern blots were washed twice in 0.1x Church buffer, 30 min per wash, at the prehybridization temperature. Hybridization products were detected by PhosphorImager analysis using a Fuji MacBAS 1000 PhosphorImager or GE Typhoon Trio. Blots were often stripped and reprobed twice to view three separate probes per blot; blots were stripped in boiling 0.5% SDS twice and checked for probe removal by Geiger counter before being reprobed.
Membranes were probed with a KpnI fragment of a GRP94 cDNA (975 bp), a BamHI/KpnI fragment of GAPDH cDNA (498 bp), an EcoRI/SalI fragment of c-myc cDNA (1.32 kb), a PstI/EcoRI fragment of BiP cDNA (1.5 kb), and a XbaI/SacII fragment of actin cDNA (500 bp). A 1-kb probe to the CBV RNA was generated through PCR of a Coxsackievirus B3 chimera (CBV-RICO) plasmid (Dobrikova et al. 2003) using forward primer 5′-GGACACTCCTTTCATTTCGCAGG-3′ and reverse primer 5′-CGTTGTGCACTGACATCTGGC-3′. All probes were internally labeled with [32P]dCTP using a random hexanucleotide primer kit (Boehringer-Mannheim or Roche). Probes were purified away from unincorporated [32P]dCTP using Nick columns (Amersham Pharmacia Biosciences) as per the manufacturer's instructions.
Ribosome quantitation
Briefly, HeLa cells were grown to ∼60%–70% confluence and ribosomes were radiolabeled with 5 μCi/mL (350 nM) [3H]5,6-uridine overnight at 37°C in 5% CO2. Complete DMEM containing uridine radiolabel was removed, cells were washed twice with complete DMEM, then chased for 2 h at 37°C in 5% CO2 with complete DMEM containing 50 μM uridine. This chase step was essential to prevent incorporation of radiolabeled nucleotide into the replicating viral RNAs. HeLa cells were then mock-infected or infected with CBV for 6 h and fractionated to yield purified cytosol and ER fractions by detergent extraction, all as described above. RNA was purified from each fraction in triplicate using Trizol reagent (Invitrogen) and resolved on 1% agarose, 3% formaldehyde gels. Bands corresponding to the 18S and 28S ribosomal RNAs were excised from the gel, melted at 95°C for 5 min, and measured by liquid scintillation spectrometry.
Reverse transcription assay
As described in Stephens et al. (2005), HeLa cells were mock-infected or infected with CBV for 6 h, fractionated to yield purified cytosol and ER fractions by detergent extraction, and subcellular fractions centrifuged on 15%–50% sucrose gradients, all as described above. RNA was isolated from each gradient fraction using guanidine thiocynate isolation described above, and resuspended in 5 μL of nuclease-free water. RNA samples were brought to a total volume of 11 μL with nuclease-free water and 100 ng of oligo-(dT)20 primer, and heated at 65°C for 10 min. Samples were cooled on ice for 2 min and supplemented with 8 μL of a reaction mix (4 μL of 5× first strand buffer [Invitrogen], 2 μL 0.1 M DTT, 1 μL dNTPs [10 mM dATP, dGTP, dTTP, and 8.33 mM dCTP], 10 U RNAse Out [Invitrogen], and 5 μCi [32P]dCTP [1.67 mM]). Samples were heated for 10 min at 42°C and 200 U of Superscript RNaseH- (Invitrogen) reverse transcriptase were added. Reactions were incubated for another 50 min at 42°C, and enzymatic activity stopped by incubation at 65°C for 15 min. Radiolabeled cDNA–RNA products were precipitated with cold 10% TCA and 10 μg of carrier yeast tRNA on ice for a minimum of 10 min. Samples were collected in triplicate by vacuum filtration onto GC/F glass fiber filters (Wheaton) presoaked in 10% TCA. Filters were washed once with cold 10% TCA and twice with cold 95% ethanol, then dried and counted by liquid scintillation spectrometry.
ACKNOWLEDGMENTS
We thank Matthias Gromeier (Duke University Medical Center) and members of his laboratory for providing Coxsackie B3 virus, CBV-RICO plasmid, primers used for synthesis of the CBV RNA Northern probe, pBabe-h-c-myc plasmid, and generous technical assistance. We thank Jack Keene (Duke University Medical Center) and Robert Rhoads (Louisiana State University Health Sciences Center) for anti-PABP and anti-eIF4G antibodies, respectively. We thank Shelton Bradrick for critical reading of the manuscript. We also thank members of the Nicchitta laboratory, especially Samuel Stephens, Brook Pyhtila, and Rebecca Dodd for stimulating and helpful discussions regarding the work described here. This work was supported by NIH grant DK-47897 to C.V.N.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2318906.
Abbreviations:CBV, Coxsackie B3 virus; eIF, eukaryotic initiation factor; ER, endoplasmic reticulum; IRES, internal ribosome-entry site; PABP, poly(A)-binding protein; SRP, signal recognition particle; UPR, unfolded protein response; z-vad-fmk, benzyloxycarbonyl-VAD-fluoromethyl ketone.
REFERENCES
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. Molecular biology of the cell. Garland Science; New York: 2002. [Google Scholar]
- Ali I.K., McKendrick L., Morley S.J., Jackson R.J. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 2001;20:4233–4242. doi: 10.1093/emboj/20.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham G.J., Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 1975a;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer to proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J. Cell Biol. 1975b;67:852–862. doi: 10.1083/jcb.67.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Walter P., Chang C.N., Goldman B.M., Erickson A.H., Lingappa V.R. Translocation of proteins across membranes: The signal hypothesis and beyond. Symp. Soc. Exp. Biol. 1979;33:9–36. [PubMed] [Google Scholar]
- Borgese D., Blobel G., Sabatini D.D. In vitro exchange of ribosomal subunits between free and membrane-bound ribosomes. J. Mol. Biol. 1973;74:415–438. doi: 10.1016/0022-2836(73)90037-5. [DOI] [PubMed] [Google Scholar]
- Borgese N., Mok W., Kreibich G., Sabatini D.D. Ribosomal-membrane interaction: In vitro binding of ribosomes to microsomal membranes. J. Mol. Biol. 1974;88:559–580. doi: 10.1016/0022-2836(74)90408-2. [DOI] [PubMed] [Google Scholar]
- Chapman R., Sidrauski C., Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu. Rev. Cell Dev. Biol. 1998;14:459–485. doi: 10.1146/annurev.cellbio.14.1.459. [DOI] [PubMed] [Google Scholar]
- Chiu S.Y., Lejeune F., Ranganathan A.C., Maquat L.E. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes & Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen A.K., Kahn L.E., Bourne C.M. Circular polysomes predominate on the rough endoplasmic reticulum of somatotropes and mammotropes in the rat anterior pituitary. Am. J. Anat. 1987;178:1–10. doi: 10.1002/aja.1001780102. [DOI] [PubMed] [Google Scholar]
- Diehn M., Eisen M.B., Botstein D., Brown P.O. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat. Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- Dobrikova E., Florez P., Bradrick S., Gromeier M. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′ nontranslated region. Proc. Natl. Acad. Sci. 2003;100:15125–15130. doi: 10.1073/pnas.2436464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger D., Teterina N., Ehrenfeld E., Bienz K. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 2000;74:6570–6580. doi: 10.1128/jvi.74.14.6570-6580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett D.W. The cell. W.B. Saunders Co; Philadelphia, PA: 1981. [Google Scholar]
- Gallie D.R. A tale of two termini: A functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- Gilmore R., Blobel G., Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 1982a;95:463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R., Walter P., Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J. Cell Biol. 1982b;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000a;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Bertolotti A., Zeng H., Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 2000b;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Harding H.P., Calfon M., Urano F., Novoa I., Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- Hellen C.U., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Hershey J.W.B., Merrick W.C. The pathway and mechanism of initiation of protein synthesis. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 33–88. [Google Scholar]
- Hundsdoerfer P., Thoma C., Hentze M.W. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl. Acad. Sci. 2005;102:13421–13426. doi: 10.1073/pnas.0506536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y., Li X., Serin G., Maquat L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- Joachims M., Van Breugel P.C., Lloyd R.E. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 1999;73:718–727. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G., Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G., Carter M.S., Eisen M.B., Brown P.O., Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalies K.U., Gorlich D., Rapoport T.A. Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J. Cell Biol. 1994;126:925–934. doi: 10.1083/jcb.126.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerekatte V., Keiper B.D., Badorff C., Cai A., Knowlton K.U., Rhoads R.E. Cleavage of Poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: Another mechanism for host protein synthesis shutoff? J. Virol. 1999;73:709–717. doi: 10.1128/jvi.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopczynski C.C., Noordermeer J.N., Serano T.L., Chen W.Y., Pendleton J.D., Lewis S., Goodman C.S., Rubin G.M. A high throughput screen to identify secreted and transmembrane proteins involved in Drosophila embryogenesis. Proc. Natl. Acad. Sci. 1998;95:9973–9978. doi: 10.1073/pnas.95.17.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear B.J., Yan R., Yang F., Waters D., Liebig H.D., Klump H., Kuechler E., Skern T., Rhoads R.E. Mapping the cleavage site in protein synthesis initiation factor eIF-4 γ of the 2A proteases from human Coxsackievirus and rhinovirus. J. Biol. Chem. 1993;268:19200–19203. [PubMed] [Google Scholar]
- Le Gall S., Neuhof A., Rapoport T. The endoplasmic reticulum membrane is permeable to small molecules. Mol. Biol. Cell. 2004;15:447–455. doi: 10.1091/mbc.E03-05-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner R.S., Seiser R.M., Zheng T., Lager P.J., Reedy M.C., Keene J.D., Nicchitta C.V. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T., Rabouille C. Endoplasmic reticulum: One continuous network compartmentalized by extrinsic cues. Curr. Opin. Cell Biol. 2005;17:362–368. doi: 10.1016/j.ceb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Lingappa V.R., Blobel G. Early events in the biosynthesis of secretory and membrane proteins: The signal hypothesis. Recent Prog. Horm. Res. 1980;36:451–475. doi: 10.1016/b978-0-12-571136-4.50018-8. [DOI] [PubMed] [Google Scholar]
- Lu P.D., Harding H.P., Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapoles J.E., Krah D.L., Crowell R.L. Purification of a HeLa cell receptor protein for group B coxsackieviruses. J. Virol. 1985;55:560–566. doi: 10.1128/jvi.55.3.560-566.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marissen W.E., Lloyd R.E. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 1998;18:7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marissen W.E., Triyoso D., Younan P., Lloyd R.E. Degradation of poly(A)-binding protein in apoptotic cells and linkage to translation regulation. Apoptosis. 2004;9:67–75. doi: 10.1023/B:APPT.0000012123.62856.20. [DOI] [PubMed] [Google Scholar]
- Mechler B., Rabbitts T.H. Membrane-bound ribosomes of myeloma cells. IV. mRNA complexity of free and membrane-bound polysomes. J. Cell Biol. 1981;88:29–36. doi: 10.1083/jcb.88.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechler B., Vassalli P. Membrane-bound ribosomes of myeloma cells. III. The role of the messenger RNA and the nascent polypeptide chain in the binding of ribosomes to membranes. J. Cell Biol. 1975;67:25–37. doi: 10.1083/jcb.67.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D.I., Krause E., Dobberstein B. Secretory protein translocation across membranes—The role of the “docking protein.”. Nature. 1982;297:647–650. doi: 10.1038/297647a0. [DOI] [PubMed] [Google Scholar]
- Mueckler M.M., Pitot H.C. Structure and function of rat liver polysome populations. I. Complexity, frequency distribution, and degree of uniqueness of free and membrane-bound polysomal polyadenylate-containing RNA populations. J. Cell Biol. 1981;90:495–506. doi: 10.1083/jcb.90.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbru C., Lafon I., Audigier S., Gensac M.C., Vagner S., Huez G., Prats A.C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J. Biol. Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- Nevins T.A., Harder Z.M., Korneluk R.G., Holcik M. Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J. Biol. Chem. 2003;278:3572–3579. doi: 10.1074/jbc.M206781200. [DOI] [PubMed] [Google Scholar]
- Nicchitta C.V. A platform for compartmentalized protein synthesis: Protein translation and translocation in the ER. Curr. Opin. Cell Biol. 2002;14:412–416. doi: 10.1016/s0955-0674(02)00353-8. [DOI] [PubMed] [Google Scholar]
- Nicchitta C.V., Lerner R.S., Stephens S.B., Dodd R.D., Pyhtila B. Pathways for compartmentalizing protein synthesis in eukaryotic cells: The template partitioning model. Biochem. Cell Biol. 2005;83:687–695. doi: 10.1139/o05-147. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Potter M.D., Nicchitta C.V. Regulation of ribosome detachment from the mammalian endoplasmic reticulum membrane. J. Biol. Chem. 2000;275:33828–33835. doi: 10.1074/jbc.M005294200. [DOI] [PubMed] [Google Scholar]
- Potter M.D., Nicchitta C.V. Endoplasmic reticulum-bound ribosomes reside in stable association with the translocon following termination of protein synthesis. J. Biol. Chem. 2002;277:23314–23320. doi: 10.1074/jbc.M202559200. [DOI] [PubMed] [Google Scholar]
- Potter M.D., Seiser R.M., Nicchitta C.V. Ribosome exchange revisited: A mechanism for translation-coupled ribosome detachment from the ER membrane. Trends Cell Biol. 2001;11:112–115. doi: 10.1016/s0962-8924(00)01905-x. [DOI] [PubMed] [Google Scholar]
- Qin X., Sarnow P. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem. 2004;279:13721–13728. doi: 10.1074/jbc.M312854200. [DOI] [PubMed] [Google Scholar]
- Raden D., Song W., Gilmore R. Role of the cytoplasmic segments of Sec61α in the ribosome-binding and translocation-promoting activities of the Sec61 complex. J. Cell Biol. 2000;150:53–64. doi: 10.1083/jcb.150.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport T.A., Jungnickel B., Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Sachs A.B., Varani G. Eukaryotic translation initiation: There are (at least) two sides to every story. Nat. Struct. Biol. 2000;7:356–361. doi: 10.1038/75120. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular cloning: A laboratory manual. 2d ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sarnow P. Translation of glucose-regulated protein 78/immunoglobulin heavy-chain binding protein mRNA is increased in poliovirus-infected cells at a time when cap-dependent translation of cellular mRNAs is inhibited. Proc. Natl. Acad. Sci. 1989;86:5795–5799. doi: 10.1073/pnas.86.15.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A., Giddings T.H., Jr., Ladinsky M.S., Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M., Kaufman R.J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Seiser R.M., Nicchitta C.V. The fate of membrane-bound ribosomes following the termination of protein synthesis. J. Biol. Chem. 2000;275:33820–33827. doi: 10.1074/jbc.M004462200. [DOI] [PubMed] [Google Scholar]
- Shi Y., Vattem K.M., Sood R., An J., Liang J., Stramm L., Wek R.C. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens S.B., Dodd R.D., Brewer J.W., Lager P.J., Keene J.D., Nicchitta C.V. Stable ribosome binding to the endoplasmic reticulum enables compartment-specific regulation of mRNA translation. Mol. Biol. Cell. 2005;16:5819–5831. doi: 10.1091/mbc.E05-07-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M., Paulin F.E., Le Quesne J.P., Chappell S.A., Willis A.E. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- Suhy D.A., Giddings T.H., Jr., Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: An autophagy-like origin for virus-induced vesicles. J. Virol. 2000;74:8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem K.M., Wek R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventoso I., MacMillan S.E., Hershey J.W., Carrasco L. Poliovirus 2A proteinase cleaves directly the eIF-4G subunit of eIF-4F complex. FEBS Lett. 1998;435:79–83. doi: 10.1016/s0014-5793(98)01027-8. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 1981a;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Blobel G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981b;91:551–556. doi: 10.1083/jcb.91.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Johnson A.E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Walter P., Ibrahimi I., Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]