Abstract
Recent studies have uncovered extensive presence and functions of small noncoding RNAs in gene regulation in eukaryotes. In particular, RNA interference (RNAi) has been the subject of significant investigations for its unique role in post-transcriptional gene regulation and utility as a tool for artificial gene knockdown. Here, we describe a novel strategy for post-transcriptional gene regulation in mammalian cells in which RNAi is specifically modulated through RNA aptamer–small molecule interaction. Incorporation of an RNA aptamer for theophylline in the loop region of a short hairpin RNA (shRNA) designed to silence fluorescent reporter genes led to dose-dependent inhibition of RNAi by theophylline. shRNA cleavage experiments using recombinant Dicer demonstrated that theophylline inhibited cleavage of an aptamer-fused shRNA by Dicer in vitro. Inhibition of siRNA production by theophylline was also observed in vivo. The results presented here provide the first evidence of specific RNA–small molecule interaction affecting RNAi, and a novel strategy to regulate mammalian gene expression by small molecules without engineered proteins.
Keywords: RNA interference (RNAi), aptamer, theophylline, Dicer
INTRODUCTION
Gene expression is regulated at multiple steps, ranging from transcription initiation to post-translational modification and/or degradation. Recently, small RNA-mediated gene-silencing phenomena in eukaryotes (Zamore and Haley 2005) have been extensively investigated due to their important regulatory roles in diverse organisms. The gene-silencing mechanism involves trans-acting small RNAs such as small interfering RNAs (siRNAs) and microRNAs (miRNAs) as essential regulatory RNA elements. siRNAs are key molecules in the RNA interference (RNAi) pathway, in which long double-stranded RNAs (dsRNAs) or short hairpin RNAs (shRNAs) are processed by the RNase III enzyme Dicer, and the resulting siRNAs of ~21 nt in length function as guides for specific mRNA degradation in the RNA-induced silencing complex (RISC) (for reviews, see Meister and Tuschl 2004; Mello and Conte 2004).
The discovery of RNAi added another layer of functionality of RNA molecules to the already versatile repertoire of biological functions. Furthermore, the universal nature of RNAi mechanism suggested its enormous potential as a tool to control gene expression in cultured cells and organisms. In fact, RNAi is now widely used as a simple and effective method to knock down gene expression in diverse organisms. Furthermore, therapeutic applications of siRNAs are being explored (for review, see Hannon and Rossi 2004).
In this report we describe a novel strategy for artificial gene regulation in mammalian cells using RNAi as a basal system. In this approach, RNAi is specifically modulated by theophylline thorough the binding to its aptamer. Aptamers are RNA or DNA oligonucleotides that can bind specific ligands with high affinity and specificity (Ellington and Szostak 1990; Tuerk and Gold 1990), and they have been used for artificial gene regulation in vivo (Werstuck and Green 1998; Harvey et al. 2002; Hanson et al. 2003; Suess et al. 2003; Buskirk et al. 2004; Desai and Gallivan 2004; Bayer and Smolke 2005). Our new approach to controlling gene expression in mammalian cells combines the versatility of siRNAs to target arbitrary genes and the powerful in vitro selection technology to develop RNA aptamers for a variety of molecules.
RESULTS AND DISCUSSION
RNAi induction with aptamer-fused shRNAs and its inhibition by theophylline
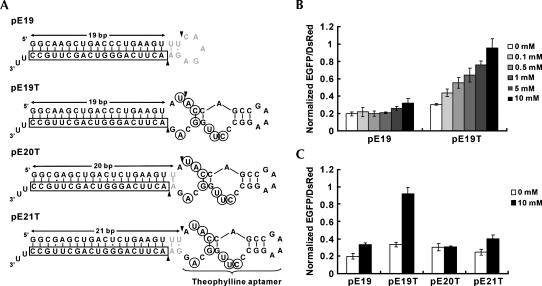
We constructed a human U6 promoter-driven vector that transcribes an shRNA in which the in vitro-selected RNA aptamer for theophylline (Jenison et al. 1994; Zimmermann et al. 1997) is fused to the loop region of the shRNA with a 19-bp double-stranded stem targeting the enhanced green fluorescent protein (EGFP) gene (pE19T) (Fig. 1A). When this construct was cotransfected into HEK293 cells with pEGFP-N1 and pDsRed1-N1 (EGFP and DsRed expression vector, respectively), EGFP silencing was inhibited in a dose-dependent manner to the theophylline concentration (Fig. 1B). DsRed fluorescence was used to normalize for transfection efficiency. The corresponding shRNA expression vector that lacks the aptamer (pE19) (Fig. 1A) retained its RNAi activity up to 1 mM theophylline with small increases in EGFP expression at 5 and 10 mM. The cells grew slightly slower at higher concentrations of theophylline (5 and 10 mM), which may account for the minor increase in EGFP expression, possibly due to nonspecific pleiotropic effects. Addition of up to 1 mM caffeine, which is structurally similar to theophylline but does not bind to the aptamer (Jenison et al. 1994), did not affect RNAi efficiency (data not shown), and higher concentrations of caffeine proved to be toxic to the cultured cells (data not shown).
FIGURE 1.
Design of aptamer-fused shRNAs and effect of theophylline on RNAi induced by the shRNAs. (A) Secondary structures of putative transcripts from pE19, pE19T, pE20T, and pE21T. Boxed nucleotides indicate the anti-sense strand targeting EGFP, gray nucleotides indicate the loop sequence from the pSilencer vector (Ambion) (Brummelkamp et al. 2002), circled nucleotides indicate bases that interact with theophylline (Zimmermann et al. 1997), and arrowheads indicate putative Dicer cleavage sites. RNA polymerase III transcripts are known to have heterogenous 3′ terminus with one to four U residues (Bogenhagen and Brown 1981; Miyagishi and Taira 2002). For simplicity, structures with two U residues are depicted. It should be noted that the pE19 transcript (E19 shRNA) was predicted to have a 21-bp stem region consisting of a 19-bp stem encoding an EGFP sequence and an additional 2-bp stem from the loop sequence recommended by the manufacturer. (B) Dose-dependent inhibition of RNAi with pE19T by theophylline. HEK293 cells were cotransfected with pEGFP-N1 and pDsRed1-N1, and pE19 (without aptamer) or pE19T (with aptamer) in a 96-well microplate. Fluorescence measurements were performed 48 h after transfection, and the ratios of EGFP and DsRed fluorescence intensity were calculated. The ratios were then normalized to those from cells transfected with an shRNA expression vector with a scrambled sequence that has no significant homology with the human genome. The data are averages of triplicate transfections and error bars represent standard deviations. (C) Effect of shRNA stem length on theophylline-dependent RNAi inhibition. HEK293 cells were cotransfected with pEGFP-N1, pDsRed1-N1, and either pE19, pE19T, pE20T, or pE21T in a 96-well microplate.
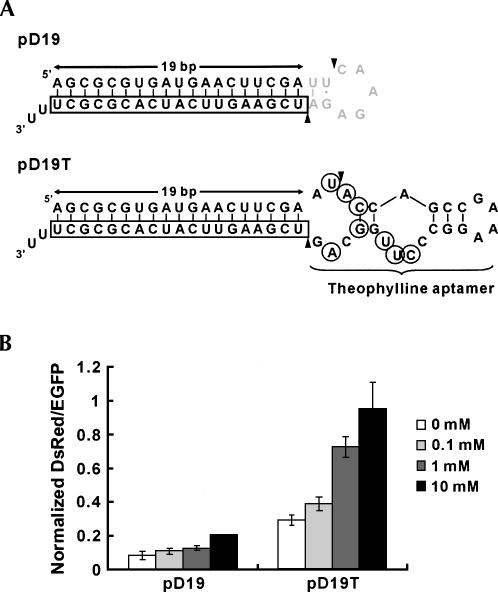
Similar RNAi inhibition by theophylline was obtained when DsRed gene was targeted instead of EGFP (Fig. 2). The results are consistent with the modular design of the aptamer-fused shRNAs comprised of a gene-targeting stem and a molecular-sensing aptamer that function independently.
FIGURE 2.
Design of an aptamer-fused shRNA targeting the DsRed1 gene and inhibition of RNAi by theophylline. (A) Secondary structures of putative transcripts from pD19 and pD19T. Boxed nucleotides indicate the anti-sense strand targeting DsRed, gray nucleotides indicate the loop sequence from the pSilencer vector, circled nucleotides indicate bases that interact with theophylline, and arrowheads indicate putative Dicer cleavage sites. Three U residues resulting in 2-nt 3′ overhang are depicted here for simplicity. Note that the putative transcript from pD19 has a 21-bp stem as well as that from pE19 (see the legend of Fig. 1A). (B) Dose-dependent inhibition of RNAi against the DsRed1 gene by theophylline. RNAi was induced for the DsRed1 gene with pD19 (without aptamer) or pD19T (with aptamer) in the presence or the absence of theophylline. The data shown are averages of normalized DsRed/EGFP values obtained from triplicate transfections with error bars representing standard deviations. Note that 10 mM theophylline pD19 data do not have an error bar because two of the transfection data did not reach the transfection efficiency standard as described in Materials and Methods (see the “Fluorescence measurements” section).
Two related constructs (pE20T and pE21T) (Fig. 1A) were examined to probe the operating mechanism of the system. pE20T and pE21T transcripts contain 20 and 21 double-stranded bp in the stem region, respectively. Both constructs retained gene-silencing activity but did not appreciably respond to theophylline (Fig. 1C). The expected Dicer cleavage sites of pE20T and pE21T transcripts lie outside of the theophylline-binding site within the aptamer (Fig. 1A), consistent with their lack of response to theophylline (Fig. 1C). These results suggested that theophylline binding to the aptamer of pE19T transcript interferes with Dicer cleavage due to the proximity of the binding pocket to the putative Dicer cleavage site (Fig. 1A).
Inhibition of Dicer-mediated shRNA cleavage by theophylline in vitro
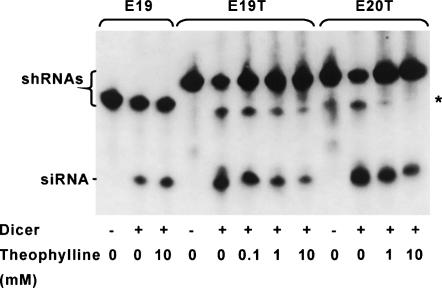
To test the possibility of Dicer-mediated cleavage of pE19T transcript being inhibited by theophylline, we prepared the predicted RNA transcripts of pE19 and pE19T (Fig. 1A) by in vitro transcription (E19 and E19T, respectively) and reacted them with recombinant Dicer in the absence and the presence of theophylline. As we expected, dose-dependent inhibition of Dicer-mediated siRNA production was observed for E19T but not for E19 (Fig. 3), supporting the proposed mechanism. The same reaction using E20T (Fig. 1A) also resulted in dose-dependent inhibition of Dicer cleavage by theophylline, but the degree of inhibition was smaller than that of E19T. These observations indicate that the proximity of Dicer cleavage site to the theophylline-binding site is important in modulating Dicer activity.
FIGURE 3.
Inhibition of Dicer-mediated shRNA cleavage by theophylline. In vitro transcribed E19, E19T, and E20T shRNAs were incubated with recombinant Dicer in the presence or the absence of theophylline. Reaction products were separated on a 15% denaturing polyacrylamide gel, and detected by Northern blotting using a 5′-biotinylated DNA oligonucleotide probe encoding an EGFP sense sequence. The asterisk indicates the position of partially cleaved shRNAs (putative). The sizes of shRNA precursors (49 nt for E19, 65 nt for E19T, 67 nt for E20T) and their diced products (siRNA, 21 nt) were verified using dsRNA Ladder (New England Biolabs) and SYBR Gold Nucleic Acid Gel Stain (Molecular Probes) (data not shown).
In addition to siRNA, longer cleavage products were detected in the Dicer cleavage reactions of E19T and E20T (indicated by an asterisk in Fig. 3). Judging from their sizes (between 30 and 50 nt; data not shown) and their detectability with the EGFP sense sequence probe, they are expected to be partial cleavage products between the 21st and 22nd nucleotides from the 5′ end of the shRNAs (a 44-nt fragment from E19T and a 46-nt fragment from E20T) (Fig. 1A). This observation suggests that insertion of theophylline aptamer decreased the efficiency of simultaneous cleavage of both strands by an intramolecular heterodimer of the two RNase III domains of Dicer (Zhang et al. 2004; Macrae et al. 2006), presumably due to the relatively large size of the aptamer and its unstructured nature in the absence of theophylline (Zimmermann et al. 1997). Addition of theophylline also resulted in reduction of the longer species, which was more evident with E20T than E19T (Fig. 3), suggesting that cleavage between A21 and U22 in E20T (cf. Fig. 1A) is more susceptible to theophylline binding than that between U21 and A22 in E19T (cf. Fig. 1A). These longer products were also observed in vivo (see below).
Siolas et al. (2005) recently reported that their shRNA with a 19-bp stem and a 4-bp loop was not cleaved by recombinant Dicer in vitro but was still able to induce RNAi in vivo. In contrast, our shRNA with a 19-bp stem (E19T) was clearly processed by Dicer in vitro in the absence of theophylline. The difference may be due to the distinct nature of the loop regions (length, structure, etc.) used in the two studies.
Inhibition of siRNA production by theophylline in vivo
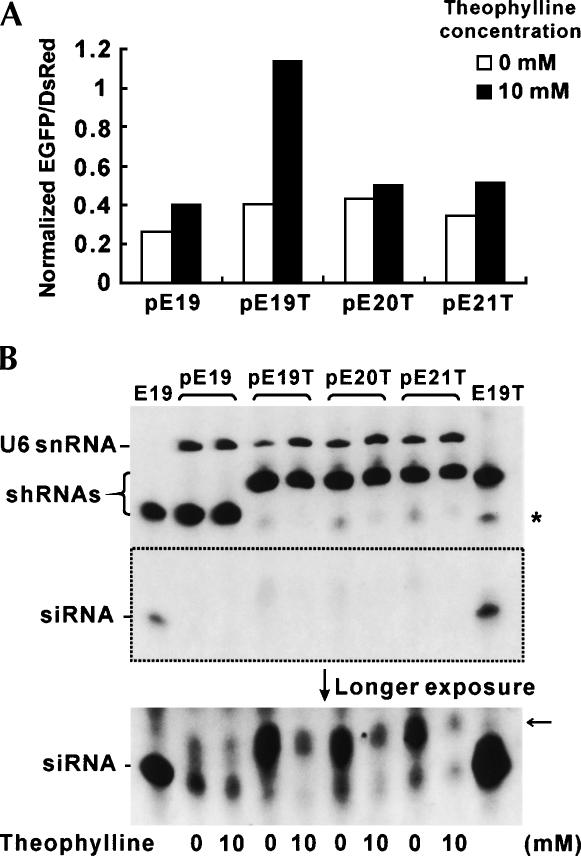
To investigate the effects of theophylline on siRNA accumulation in vivo, Northern analysis was performed using small RNAs isolated from HEK293 cells, which were cotransfected with pEGFP-N1, pDsRed1-N1, and one of the shRNA expression plasmids (pE19, pE19T, pE20T, or pE21T) and incubated in the presence or the absence of 10 mM theophylline. Strong bands corresponding to the unprocessed shRNAs were detected in all cell extracts regardless of theophylline addition (Fig. 4B). As evident from the bottom picture of Figure 4B, siRNA generation from E19T shRNA was almost completely inhibited by theophylline, whereas those from E20T and E21T were diminished but still detectable. No significant difference in the siRNA levels of pE19-transfected cells in response to theophylline was observed. These observations were consistent with the result of fluorescence measurement of the same samples (Fig. 4A) as well as the in vitro Dicer cleavage results (Fig. 3).
FIGURE 4.
Inhibition of shRNA cleavage by theophylline in vivo. HEK293 cells were cotransfected with pEGFP-N1 and pDsRed1-N1, and either pE19, pE19T, pE20T, or pE21T in 6-well plates. Small RNA species were isolated from the transfected cells after measuring EGFP/DsRed fluorescence. (A) Normalized fluorescence data of the transfected cells used to isolate small RNA species (cf. Fig. 1C). (B) Northern blotting of small RNAs from transfected cells. Expression of U6 snRNA was used as a loading control. In vitro-diced products of E19 and E19T in the presence of theophylline (cf. Fig. 3) were used as size markers. The asterisk indicates the position of partially cleaved shRNAs (putative), and the arrow indicates the position of unidentified cleavage products specifically observed in vivo (see text for details).
The putative partially cleaved RNA species observed in vitro were also detected in vivo (indicated by an asterisk in Fig. 4B). Additionally, RNA species of ~30 nt were also detected from the cell lysates transfected with the aptamer-fused shRNA expression vectors (indicated by an arrow in Fig. 4B). The intensity of these bands was also diminished in response to theophylline, suggesting that the cleavage site(s) responsible for these species is located within or near the theophylline-binding pocket. Although the exact nature of these by-products awaits further investigation, the similar levels of the RNA species detected in pE19T, pE20T, or pE21T-transfected cells in the presence of theophylline and the difference in observed RNAi among these cells suggest that these RNA species are not involved in the silencing of EGFP.
Taken together, theophylline clearly inhibits siRNA production from aptamer-fused shRNAs in vitro and in vivo, but the highest degree of inhibition is observed with E19T, in which one of the putative Dicer cleavage sites overlaps with the theophylline-binding site (Fig. 1A). Consequently, theophylline-induced RNAi inhibition in vivo was only observed in cells transfected with the aptamer-fused shRNA expression vectors with a 19-bp stem (pE19T and pD19T) (Figs. 1, 2).
Comparison with other RNAi regulation methods
Chiu et al. (2005) recently reported two ATP analogs that inhibit RNAi in mammalian cells discovered by screening of a chemical library. These molecules are likely to inhibit a cellular factor involved in the early RNAi pathway to induce general RNAi inhibition, whereas our system was designed to effect specific control of RNAi by a small molecule directly interacting with an shRNA. To our knowledge, our system is the first demonstration of sequence-specific RNAi inhibition by a small molecule at a defined mechanistic step.
Anti-miRNA oligonucleotides (AMOs) have been used to inhibit specific miRNAs and siRNAs in vivo (for review, see Weiler et al. 2005). AMOs are easy to design and can inhibit both endogenous miRNAs and synthetic siRNAs. However, the exact process in which AMOs exert their function in the RNAi pathway is not clear. Delivery and removal of AMOs are also more complicated than many small molecule drugs, which may be more advantageous for experiments requiring temporal control.
The most popular technique for achieving temporal control of RNAi in cultured cells is probably the use of inducible shRNA vectors based on the robust tetracycline or ecdysone-inducible systems using engineered transcription factors and promoters, or Cre-loxP recombination systems (for review, see Sandy et al. 2005). While these inducible systems offer various features needed for many applications, the aptamer-fused shRNA regulatory strategy operating at the Dicer cleavage step may be used in combination with the existing inducible shRNA expression systems to achieve more complex or improved regulation of RNAi in engineered cells. The ability to control RNAi at the Dicer cleavage step may result in a faster response compared with transcriptional control. Moreover, the modularity of the aptamer and the gene-targeting stem opens the possibility to regulate gene expression in response to endogenous metabolites or proteins for which RNA aptamers are selected in vitro. Reversibility of the RNA silencing effect will need to be demonstrated using stable transfectants of the aptamer-fused shRNA expression cassette, which will be the subject of future investigation.
pE19T and pD19T transcripts can be classified as RNA genetic switches that allow regulation of exogenous genes without exogenous protein factors. To our knowledge, only two such genetic switches that function in mammalian cells have been reported. Werstuck and Green (1998) inserted H33342 dye aptamers in the 5′-untranslated region (UTR) of a reporter transcript whose translation was negatively regulated in the presence of the cognate ligand. Yen et al. (2004) recently reported a system in which a ribozyme integrated in 5′ UTR of a reporter mRNA whose self-cleavage was inhibited by small molecule ribozyme inhibitors.
Although not directly demonstrated in this work, the modularity of our system provides a unique potential of regulating endogenous genes not possible with the previously described systems (Werstuck and Green 1998; Yen et al. 2004). RNA genetic switches have some advantages over those based on engineered protein transcription factors such as reduced risk of immunogenic complications and smaller genetic sizes that may be beneficial for in vivo applications. In light of the recent progress in natural and artificial RNA-based gene regulatory mechanisms in bacteria (Isaacs et al. 2004; Davidson and Ellington 2005) and yeast (Suess et al. 2003; Buskirk et al. 2004; Bayer and Smolke 2005), small molecule-regulated RNA genetic switches in mammalian cells should further advance our understanding of the roles of RNA in gene regulation as well as their applications.
MATERIALS AND METHODS
Plasmid construction
pEGFP-N1 and pDsRed1-N1 were obtained from Clontech. pSilencer 2.1-U6 hygro (Ambion) was used for constructing all RNAi vectors. pE19 (E19 stands for 19 bp of shRNA encoding an EGFP target sequence [Meister et al. 2004]), pE19T (T stands for theophylline aptamer inserted in the loop region of shRNA), pD19 (D stands for DsRed), and pD19T were produced by ligating phosphorylated and annealed oligonucleotides with pSilencer 2.1-U6 hygro. Oligonucleotides used were as follows (a pair of annealed oligonucleotides was indicated by “&”):
pE19: 5′-GATCCGGCAAGCTGACCCTGAAGT & 5′-TCTCTTGAAACTTCAGGGTCAGCTTGCCG, and 5′-TTCAAGAGAACTTCAGGGTCAGCTTGCCTTTTTTGGAAA & 5′-AGCTTTTCCAAAAAAGGCAAGCTGACCCTGAAGT;
pE19T: 5′-GATCCGGCAAGCTGACCCTGAAGT & 5′-GGTATACTTCAGGGTCAGCTTGCCG, and 5′-ATACCAGCCGAAAGGCCCTT & 5′-CTGCCAAGGGCCTTTCGGCT, and 5′- GGCAGACTTCAGGGTCAGCTTGCCTTTTTTGGAAA & 5′-AGCTTTTCCAAAAAAGGCAAGCTGACCCTGAAGT;
pD19: 5′-GATCCAGCGCGTGATGAACTTCGA & 5′-AGTTCATCACGCGCTG, and 5′-TTCAAGAGATCGAAG & 5′-TGAACTTCGATCTCTTGAATCGA, and 5′-TTCATCACGCGCTTTTTTGGA & 5′-AGCTTCCAAAAAAGCGCGTGA; and
pD19T: 5′-GATCCAGCGCGTGATGAACTTCGA & 5′-AGTTCATCACGCGCTG, and 5′-ATACCAGCCGAAAGGCCCTTGGCAGTCGAAG & 5′-TGAACTTCGACTGCCAAGGGCCTTTCGGCTGGTATTCGA, and 5′-TTCATCACGCGCTTTTTTGGA & 5′-AGCTTCCAAAAAAGCGCGTGA.
A target sequence of the DsRed1 gene (5′-AGCGCGTGATGAACTTCGA) was chosen according to the criteria established by Reynolds et al. (2004).
pE20T and pE21T were constructed from pE19B, which was engineered to contain two BbsI sites in the loop region of pE19. pE19B was produced by ligating phosphorylated and annealed oligonucleotides of the following sequences with pSilencer 2.1-U6 hygro: 5′-GATCCGGCGAGCTGACTCTGAAGTATGTCTTCTTTTTTCTCGAGA & 5′-GAGAAAAAAGAAGACATACTTCAGAGTCAGCTCGCCG, and 5′-CTTGAAGACTAACTTCAGGGTCAGCTTGCCTTTTTTGGA & 5′-AGCTTCCAAAAAAGGCAAGCTGACCCTGAAGTTAGTCTTCAAGTCTC. Phosphorylated and annealed oligonucleotides of the following sequences were ligated to the BbsI-digested pE19B to produce pE20T and pE21T. Oligonucleotides used to construct pE20T were 5′-AAGTTATACCAGCCGAAAGGCCCTTGGCAGA & 5′-AAGTTCTGCCAAGGGCCTTTCGGCTGGTATA. Oligonucleotides used to construct pE21T were 5′-AAGTTTATACCAGCCGAAAGGCCCTTGGCAGGA & 5′-AAGTTCCTGCCAAGGGCCTTTCGGCTGGTATAA.
Cell culture and transfection
HEK293 cells were maintained in a 5% CO2-humidified incubator at 37°C in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). One day before transfection, HEK293 cells were trypsinized and diluted 1:5 with fresh medium, and transferred to 96-well plates (130 μL/well). Cotransfection of pEGFP-N1, pDsRed1-N1, and an RNAi vector was carried out using PolyFect (QIAGEN) according to the manufacturer's instructions with modifications. Specifically, 10 ng of pEGFP-N1, 20 ng of pDsRed1-N1, and 100 ng of an RNAi vector were used with 2 μL of PolyFect reagent per well. Cells were incubated in a 5% CO2-humidified incubator at 37°C in the HEK293 medium supplemented with the indicated concentrations of theophylline or caffeine for 48 h before fluorescence measurements.
Fluorescence measurements
To quantitatively determine in vivo RNAi activity, dual fluorescence reporter systems using EGFP and DsRed (RFP) have been developed and used by many researchers (Chiu and Rana 2002, 2003; Chiu et al. 2004, 2005; Shah et al. 2005; Yang et al. 2005). Our assay procedure was established according to the previous reports with some modifications. EGFP and DsRed fluorescence were detected with Safire2 microplate reader (Tecan) according to the manufacturer's instructions. Before measuring fluorescence intensity, medium was removed from each well, and 150 μL of prewarmed (37°C) phosphate-buffered saline (PBS) was added and incubated at 37°C for 10 min in the instrument. Measurement conditions were as follows: measurement mode, fluorescence bottom well reading; excitation wavelength, 484 nm (for EGFP) or 558 nm (for DsRed); emission wavelength, 510 nm (for EGFP) or 583 nm (for DsRed); excitation bandwidth, 5 nm; emission bandwidth, 5 nm; gain, optimal; number of reads, five; temperature, 37°C. Data were processed as follows: First, fluorescence values of EGFP were divided by those of DsRed to obtain EGFP/DsRed (DsRed fluorescence was used to normalize for transfection efficiency), and the average values and standard deviations of EGFP/DsRed from triplicate samples were calculated. Next, the average values and the standard deviations were normalized to those from cells transfected with an shRNA expression vector that has a scrambled sequence with no significant homology with the human genome (Ambion, pSilencer 2.1-U6 hygro Negative Control, which expresses an shRNA with the following sequence: 5′-ACUACCGUUGUUAUAGGUGUUCAAGAGACACCUAUAACAACGGUAGUU-3′; double-stranded stem underlined) to obtain “Normalized EGFP/DsRed” values (for DsRed-targeting experiments, normalized DsRed/EGFP values were obtained). Fluorescence values at least 10 times higher than those from mock-transfected samples were used for calculations. We did not subtract autofluorescence values of mock-transfected samples from those of plasmid-transfected samples, because autofluorescence values from mock-transfected samples were usually <10% of those from plasmid-transfected samples, and the effect of the autofluorescence subtraction was negligible.
In vitro processing of shRNAs by recombinant Dicer
E19, E19T, and E20T shRNAs were prepared by in vitro transcription using AmpliScribe T7 High Yield Transcription Kit (Epicentre). Template DNAs were prepared by annealing and T4 DNA polymerase-mediated extension of synthetic oligonucleotides. Oligonucleotides used to generate an E19 template were 5′-GCGTAATACGACTCACTATAGGCAAGCTGACCCTGAAGTTTCAAGAGAAC & 5′-AAGGCAAGCTGACCCTGAAGTTCTCTTGAAAC (underline indicates T7 promoter sequence). Oligonucleotides used to generate an E19T template were 5′-GCGTAATACGACTCACTATAGGCAAGCTGACCCTGAAGTATACCAGCCGAAAG & 5′-AAGGCAAGCTGACCCTGAAGTCTGCCAAGGGCCTTTCGGCTGGTATAC (underline indicates T7 promoter sequence). Oligonucleotides used to generate an E20T template were 5′-GCGTAATACGACTCACTATAGGCGAGCTGACTCTGAAGTTATACCAGCCGAAAG & 5′-AAGGCAAGCTGACCCTGAAGTTCTGCCAAGGGCCTTTCGGCTGGTATAAC (underline indicates T7 promoter sequence). In vitro-transcribed shRNAs were purified with a 15% polyacrylamide/7 M urea denaturing gel prior to use.
shRNA (12 pmol) was mixed with 0.75 units of recombinant Dicer enzyme (Stratagene) in 10 μL of reaction mixture containing 27.5 mM Tris-HCl (pH 8.0), 225 mM NaCl, 2.5 mM MgCl2, and 0, 0.1, 1, or 10 mM theophylline, and incubated at 37°C for 18 h.
Isolation of small RNAs from transfected cells
A total of 150 ng of pEGFP-N1, 300 ng of pDsRed1-N1, and 1.5 μg of an RNAi vector were cotransfected into HEK293 cells using 20 μL of PolyFect reagent (QIAGEN) in 6-well plates. Cells were incubated in a 5% CO2-humidified incubator at 37°C in the HEK293 medium supplemented with 0 or 10 mM theophylline for 47 h. After measurement of fluorescence intensity as described above, small RNAs were isolated using mirVana miRNA Isolation Kit (Ambion) according to the manufacturer's instructions.
Northern blotting
A 0.5-μL aliquot of in vitro reaction products (0.6 pmol RNA; see above) or 1 μg of small RNAs from transfected cells (see above) was separated on a 15% polyacrylamide/7 M urea denaturing gel using NorthernMax Formaldehyde Load Dye (Ambion) and electroblotted to BrightStar-Plus Positively Charged Nylon Membrane (Ambion). After prehybridization in ULTRAhyb-Oligo Hybridization Buffer (Ambion) at 25°C for 30 min, blots were incubated with a 5′-biotin-labeled DNA oligonucleotide probe (100 ng/mL) for EGFP (5′-AAAAAGGCAAGCTGACCCTGAAGT; underline corresponds to the plus strand of shRNAs) (cf. Fig. 1A) in the same buffer at 25°C overnight. For small RNA analysis of transfected cells, a probe for U6 small nuclear (sn) RNA (5′-AAAATATGGAACGCTTCACGAATT) was also added (100 ng/mL). Blots were washed three times (once at 25°C for 5 min, once at 42°C for 5 min, once at 42°C for 15 min) using a low-stringency buffer (2×SSC, 0.1% SDS), and hybridized probes were detected using BrightStar BioDetect Kit (Ambion) according to the manufacturer's instructions. Chemiluminescence detection was performed using IVIS Imaging System (Xenogen).
ACKNOWLEDGMENTS
We thank Jim Trimmer for providing HEK293 cells, Kathryn Brayer and David Segal for electroblotting apparatus, and Chris Griesemer and Steve Rendig for chemiluminescence detection. The research was supported by the University of California, Davis, and by The Whitaker Foundation. V.B.T. was partially supported by an industry/campus supported fellowship under the Training Program in Biomolecular Technology (T32-GM08799) at the University of California, Davis.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2299306.
REFERENCES
- Bayer T.S., Smolke C.D. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D.F., Brown D.D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Buskirk A.R., Landrigan A., Liu D.R. Engineering a ligand-dependent RNA transcriptional activator. Chem. Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Chiu Y.L., Rana T.M. RNAi in human cells. Basic structural and functional features of small interfering RNA. Mol. Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- Chiu Y.L., Rana T.M. siRNA function in RNAi: A chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y.L., Ali A., Chu C.Y., Cao H., Rana T.M. Visualizing a correlation between siRNA localization, cellular uptake, and RNAi in living cells. Chem. Biol. 2004;11:1165–1175. doi: 10.1016/j.chembiol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Chiu Y.L., Dinesh C.U., Chu C.Y., Ali A., Brown K.M., Cao H., Rana T.M. Dissecting RNA-interference pathway with small molecules. Chem. Biol. 2005;12:643–648. doi: 10.1016/j.chembiol.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Davidson E.A., Ellington A.D. Engineering regulatory RNAs. Trends Biotechnol. 2005;23:109–112. doi: 10.1016/j.tibtech.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Desai S.K., Gallivan J.P. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Hannon G.J., Rossi J.J. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- Hanson S., Berthelot K., Fink B., McCarthy J.E., Suess B. Tetracycline-aptamer-mediated translational regulation in yeast. Mol. Microbiol. 2003;49:1627–1637. doi: 10.1046/j.1365-2958.2003.03656.x. [DOI] [PubMed] [Google Scholar]
- Harvey I., Garneau P., Pelletier J. Inhibition of translation by RNA-small molecule interactions. RNA. 2002;8:452–463. doi: 10.1017/s135583820202633x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs F.J., Dwyer D.J., Ding C., Pervouchine D.D., Cantor C.R., Collins J.J. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- Jenison R.D., Gill S.C., Pardi A., Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- Macrae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Meister G., Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Dorsett Y., Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C.C., Conte D., Jr. Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- Miyagishi M., Taira K. U6 promoter driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Sandy P., Ventura A., Jacks T. Mammalian RNAi: A practical guide. Biotechniques. 2005;39:215–224. doi: 10.2144/05392RV01. [DOI] [PubMed] [Google Scholar]
- Shah S., Rangarajan S., Friedman S.H. Light-activated RNA interference. Angew. Chem. Int. Ed. Engl. 2005;44:1328–1332. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]
- Siolas D., Lerner C., Burchard J., Ge W., Linsley P.S., Paddison P.J., Hannon G.J., Cleary M.A. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- Suess B., Hanson S., Berens C., Fink B., Schroeder R., Hillen W. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 2003;31:1853–1858. doi: 10.1093/nar/gkg285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Weiler J., Hunziker J., Hall J. Anti-miRNA oligonucleotides (AMOs): Ammunition to target miRNAs implicated in human disease? Gene Ther. 2005 doi: 10.1038/sj.gt.3302654. http://www.nature.com/gt/index.html [DOI] [PubMed] [Google Scholar]
- Werstuck G., Green M.R. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- Yang W., Wang Q., Howell K.L., Lee J.T., Cho D.S., Murray J.M., Nishikura K. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J. Biol. Chem. 2005;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen L., Svendsen J., Lee J.S., Gray J.T., Magnier M., Baba T., D'Amato R.J., Mulligan R.C. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Zimmermann G.R., Jenison R.D., Wick C.L., Simorre J.P., Pardi A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat. Struct. Biol. 1997;4:644–649. doi: 10.1038/nsb0897-644. [DOI] [PubMed] [Google Scholar]