Abstract
Lissoclimides are cytotoxic compounds produced by shell-less molluscs through chemical secretions to deter predators. Chlorinated lissoclimides were identified as the active component of a marine extract from Pleurobranchus forskalii found during a high-throughput screening campaign to characterize new protein synthesis inhibitors. It was demonstrated that these compounds inhibit protein synthesis in vitro, in extracts prepared from mammalian and plant cells, as well as in vivo against mammalian cells. Our results suggest that they block translation elongation by inhibiting translocation, leading to an accumulation of ribosomes on mRNA. These data provide a rationale for the cytotoxic nature of this class of small molecule natural products.
INTRODUCTION
Small molecule ligands that inhibit the process of translation have provided exquisite insight into ribosome function and translation factor activity in both prokaryotes and eukaryotes (Pestka 1977; Vazquez 1979). Inhibitors targeting a specific step of protein synthesis have enabled dissection of the translation pathway by allowing the characterization of events leading to the assembly of active polysomes, trapping intermediates of the initiation and elongation cycles, as well as providing insight into the molecular functions of protein factors (Pestka 1977; Vazquez 1979). Recently, there has been renewed interest in identifying additional inhibitors of translation as a consequence of trying to interdict this process in disease. Specifically, the ribosome recruitment phase of translation initiation is usurped in many human cancers (for review, see Mamane et al. 2004). Hence, blocking signaling events that impinge on this process may have significant therapeutic potential (Wendel et al. 2004).
Several inhibitors of elongation have been previously tested as anti-cancer agents in preclinical animal models and clinical trials. The selectivity of these general inhibitors for cancer cells may stem from the fact that transformed cells have higher translation rates than do their nontransformed counterparts (Heys et al. 1991). Structure–activity relationship studies on two translation elongation inhibitors, sparsomycin and didemnin B, demonstrated equivalent rank order potency for inhibition of translation inhibition and for their anti-proliferative effects (Ottenheijm and van den Broek 1988; van den Broek et al. 1989; Ahuja et al. 2000), indicating that the anti-tumor activity of these compounds is a direct consequence of protein synthesis inhibition. Currently, phase II clinical trials are in progress to assess the efficacy of homoharringtonine (an inhibitor of translation elongation), with imatinib (a tyrosine kinase inhibitor that selectively targets the ABL, PDGFR, and KIT kinases) in chronic myeloid leukemia (CML) or as salvage therapy in patients with refractory acute promyelocytic leukemia (see http://clinicaltrials.gov/ct).
We have undertaken a research effort to identify novel inhibitors of eukaryotic translation based on the ability of small molecules to block translation in a Krebs-2 in vitro translation extract programed with a bicistronic reporter (Novac et al. 2004). Extracts prepared from Krebs-2 ascites are well suited for these studies because they faithfully recapitulate the cap-dependent and poly(A)-dependent features of in vivo translation (Svitkin and Sonenberg 2004). This screening campaign has lead to the identification of novel elongation (Chan et al. 2004) and initiation (Bordeleau et al. 2005) inhibitors.
In the course of screening extracts prepared from marine organisms, we identified an extract active against both cap-dependent and IRES-mediated translation. Bioassay-guided fractionation of the Pleurobranchus forskalii extract led to identification of chlorolissoclimide and dichlorolissoclimide as the responsible inhibitory agents. Chlorolissoclimide and dichlorolissoclimide were first isolated in 1991 from a New Caledonian ascidian Lissoclinum voeltzkowi Michaelson, and in 2004, they were reisolated from P. forskalii collected near St. Rosa in the Philippines (Malochet-Grivois et al. 1991; Fu et al. 2004). The latter was the first report of the isolation of Lissoclinum metabolites from its predator P. forskalii. The mollusks are known to feed on ascidians. Chlorolissoclimide and dichlorolissoclimide are cytotoxic bicyclic diterpene alkaloids effective against a range of transformed cell lines (Malochet-Grivois et al. 1992; Roussakis et al. 1994; Uddin et al. 2001; Fu et al. 2004). They are part of a family of a larger class of natural molecules, referred to as labdanes, and for which no mode of action has yet been elucidated (Hanson 2005). Herein, it is demonstrated that these compounds are potent inhibitors of eukaryotic translation elongation. They cause accumulation of ribosomes on mRNA templates and interfere with the translocation step of elongation.
RESULTS AND DISCUSSION
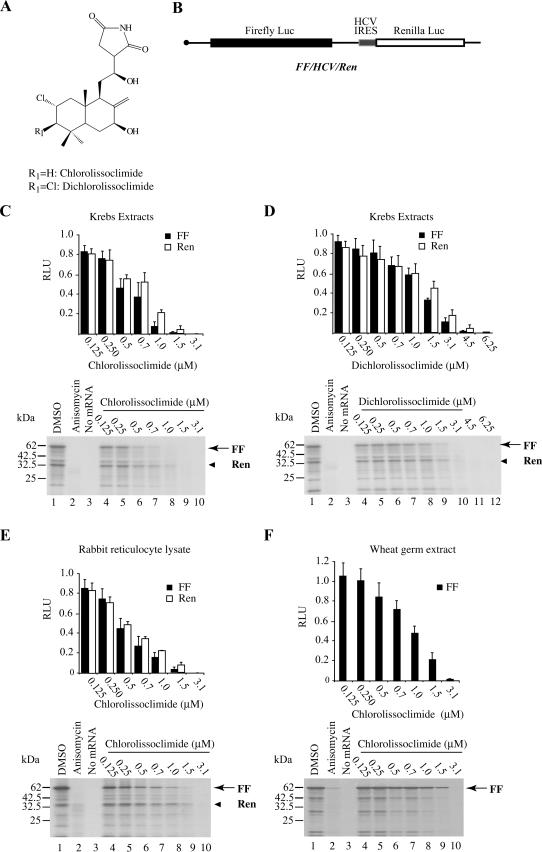
By utilizing a high-throughput screen in which the translational output of a bicistronic mRNA encoding firefly (FF) and renilla (Ren) luciferases (lucs) is monitored, we have screened a large collection of natural product extracts (Novac et al. 2004). During the course of this screen, we identified an inhibitory activity in extracts derived from the marine slug, P. forskalii. Bioassay-guided fractionation identified two active components, chlorolissoclimide and dichlorolissoclimide (Fig. 1A). When tested in Krebs-2 extracts programed with FF/HCV/Ren mRNA (Fig. 1B), both compounds exerted a dose-dependent inhibition of translation from the FF and Ren luc cistrons (Fig. 1C,D). Chlorolissoclimide appeared slightly more potent at inhibiting protein synthesis (IC50 = ∼0.7 μM) compared with dichlorolissoclimide (IC50 = ∼1.25 μM). Chlorolissoclimide was also active in translation extracts from rabbit reticulocyte lysates (Fig. 1E) and wheat germ extracts (Fig. 1F). Dichlorolissoclimide was also active in these extracts, although with an approximately twofold higher IC50 (data not shown). Neither chlorolissoclimide nor dichlorolissoclimide inhibited translations in Escherichia coli S30 extracts or exerted an effect on bacterial growth (data not shown).
FIGURE 1.
Chlorolissoclimides are new inhibitors of eukaryotic protein synthesis. (A) Schematic diagram illustrating the chemical structure of chlorolissoclimide and dichlorolissoclimide isolated from P. forskalii. (B) Schematic diagram of FF/HCV/Ren mRNA. (C) Translation mediated by cap-dependent and HCV-driven translation is sensitive to the presence of chlorolissoclimide in Krebs-2 cell extracts. Translations were performed in Krebs-2 cell extracts programed with 10 μg/mL FF/HCV/Ren mRNA and supplemented with 35S-methionine and the indicated final concentrations of chlorolissoclimide. (Top) The obtained luciferase activities were normalized to the activity obtained in the control translations (0.5% methanol) of the same mRNA species (which was set at one). Each data point represents the average of three translations, and the SEM is shown. (Bottom) The protein products were analyzed by SDS-PAGE, which were subsequently treated with EN3Hance, dried, and exposed to X-OMAT film (Kodak). The positions of migration of the firefly and renilla products are indicated by arrows and arrowheads, respectively. (D) Translation mediated by cap-dependent and HCV-driven translation is sensitive to the presence of dichlorolissoclimide in Krebs-2 cell extracts. (E) Inhibition of translations in rabbit reticulocyte lysate by chlorolissoclimide. Translation data and products were processed and analyzed as in C. (F) Chlorolissoclimide inhibits cap-dependent translation in wheat germ extracts. Translation data and products were processed and analyzed as in C.
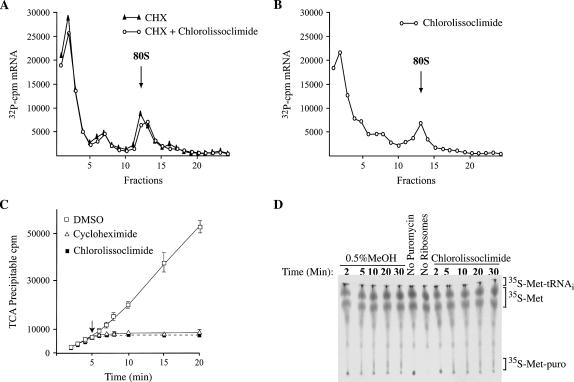
To identify the phase of translation at which chlorolissoclimide is exerting its inhibitory effect, we assessed the ability of this compound to inhibit initiation complex formation in the presence of cycloheximide (Fig. 2A). The failure to prevent 80S complex formation indicates that chlorolissoclimide is not blocking translation initiation. On its own, chlorolissoclimide is capable of trapping an 80S ribosome on an mRNA template, suggesting that the compound is inhibiting translation by blocking the elongation phase of translation (Fig. 2B). To assess if actively translating ribosomes are sensitive to chlorolissoclimide or if only newly initiated ribosomes were being targeted by this compound, it was added to a Krebs extracts 5 min after the start of translation (Fig. 2C). Immediate cessation of protein synthesis was observed with chlorolissoclimide and cycloheximide (Fig. 2C). These results indicate that chlorolissoclimide immediately blocks elongation and does not allow ribosomes to run-off the mRNA template, in which case a 2- to 3-min delay in the onset of inhibition would be expected (Chan et al. 2004).
FIGURE 2.
Chlorolissoclimide blocks elongation. (A) Chlorolissoclimide does not inhibit initiation complex formation. Rabbit reticulocyte lysate were preincubated with 0.6 mM cycloheximide (CHX) ± 50 μM chlorolissoclimide for 5 min at 30°C. 32P-radiolabeled CAT mRNA was then added to the reaction, and the incubation continued for an additional 10 min at 30°C. Translation complexes were resolved by centrifugation through 10%–30% glycerol gradients. Total counts recovered from each gradient and the percentage mRNA bound in 80S complexes were as follows: CAT mRNA/CHX, 17.3%; CAT mRNA/CHX + chlorolissoclimide, 18.4%. (B) Chlorolissoclimide can trap ribosomes on mRNA templates. Rabbit reticulocyte lysate were preincubated with 50 uM chlorolissoclimide for 5 min at 30°C. 32P-radiolabeled CAT mRNA was then supplemented to the reaction, and the incubation continued for an additional 10 min at 30°C. Translation complexes were resolved by centrifugation through 10%–30% glycerol gradients. The total counts recovered from the gradient and the percentage mRNA bound in 80S complexes was 17.8%. (C) Effect of adding chlorolissoclimide to an actively translating extract. Translations in Krebs extracts were initiated in the presence of 35S-methionine, and aliquots for TCA precipitation were removed at various time points. Five minutes after initiation of translation, compound (50 uM cycloheximide or 50 μM chlorolissoclimide) or vehicle (0.5% methanol) was added to the reaction (represented by a downward arrow), and the translation was continued for another 15 min with aliquots for TCA precipitation being removed at the indicated times. Each data point is the average of two TCA precipitations (performed in duplicate). (D) Chlorolissoclimide does not inhibit peptidyl transferase activity. Formation of 35S-methionine-puromycin product by the 80S ribosome was performed over the course of 30 min and detected by thin layer chromatography in the presence of vehicle (0.5% methanol) or 50 μM chlorolissoclimide. The position of the reaction products (35S-methionine-tRNA, free 35S-methionine and 35S-methionyl-puromycin) is indicated to the right.
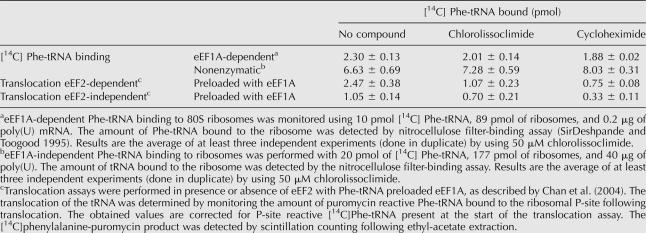
We characterized the events during elongation that might be affected by chlorolissoclimide by analyzing various steps of the process. We assessed the ability of these compounds to interfere with eEF1A-dependent aminoacyl-tRNA delivery, peptidyl transferase, and eEF2-dependent translocation by using salt-washed ribosomes isolated from rabbit reticulocytes lysate. Chlorolissoclimide did not inhibit eEF1A-dependent or nonenzymatic loading of aminoacyl-tRNA onto ribosomes (Table 1). Also, the compound had no effect on the peptidyl transferase reaction since the amounts of 35S-Met-puromycin dipeptide produced in the absence or presence of chlorolissoclimide were similar (Fig. 2D). However, inhibition of eEF2 dependent and eEF2-independent translocation was observed by chlorolissoclimide, similar to what was noted with cycloheximide (Table 1).
TABLE 1.
Effect of chlorolissoclimide on Phe-tRNA recruitment and ribosome translocation
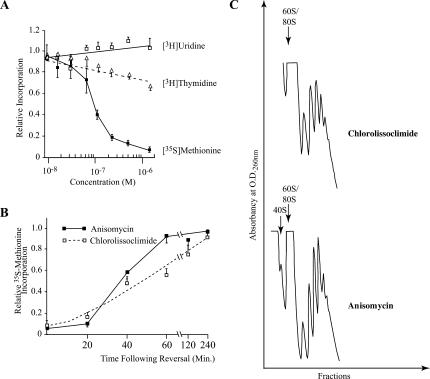
To determine whether chlorolissoclimide could inhibit cellular protein synthesis in vivo, and to establish the selectivity of this compound, we exposed HeLa cells to increasing concentrations of chlorolissoclimide and monitored protein, RNA, and DNA synthesis (Fig. 3A). In this experiment, protein synthesis was dramatically inhibited, whereas DNA synthesis was only slightly affected (30% inhibition) (Fig. 3A). RNA transcription was not affected under our experimental conditions (Fig. 3A). The observed protein synthesis inhibition was reversible, since following washout of compound, it took ∼4 h for cells to recover from the inhibitory effects of the compound (Fig. 3B). This contrasts to the 1-h recovery period required for cells exposed to anisomycin (Fig. 3B). Treatment of cells with chlorolissoclimide did not lead to a loss of polysomes but rather was consistent with a block in elongation in which ribosomes were stalled on the mRNA template (Fig. 3C). This mechanism of action differs from some other inhibitors of elongation, such as phyllanthoside and nagilactone C, in which inhibit of elongation results in a disruption of polysomes (Chan et al. 2004). As expected, anisomycin blocked elongation and trapped ribosomes in polysomes (Grollman 1967).
FIGURE 3.
Effect of chlorolissoclimide on protein, DNA and RNA synthesis in vivo. (A) The rate of macromolecular synthesis (TCA precipitable cpm for the indicated labeling period) obtained in the presence of increasing amounts of chlorolissoclimide was normalized to the rate obtained in the presence of vehicle (0.5% methanol) and is plotted. The results are shown for three experiments, and the SEM is presented. The rate of 35S-methionine incorporation obtained with the control reaction was 21,782 cpm/μg of protein/15-min incubation. The rate of 3H-thymidine and 3H-uridine incorporation obtained with the control reaction was 4920 cpm/μg of protein/15 min and 5560 cpm/μg of protein/20 min, respectively. (B) Inhibition of protein synthesis by chlorolissoclimide is reversible in vivo. HeLa cells were incubated for 1 h with 6.25 μM chlorolissoclimide after which time cells were washed and fresh medium added. Fifteen minutes before harvesting, cells were incubated with 35S-methionine and incorporation into protein determined by TCA precipitation. The rate of 35S-methionine incorporation obtained in the control reaction was 39,725 cpm/μg of protein/15-min incubation. Results are expressed relative to the incorporation in the presence of 0.5% methanol and represent the average of two experiments (each performed in duplicate). (C) Chlorolissoclimide does not disrupt cellular polysomes. HeLa cells were incubated for 1 h with 50 uM of chlorolissoclimide, after which the cells were washed twice with PBS, harvested with a rubber policeman, and collected by brief centrifugation. The cell pellet was resuspended in lysis buffer (5 mM Tris-HCl at pH 7.5, 2.5 mM MgCl2, 1.5 mM KCl, 0.5% Triton X-100, 0.5% sodium deoxycholate, 2 mM DTT), vortexed, and centrifuged for 2 min at 14,000g. The supernatants were loaded onto 10%–50% sucrose gradients prepared in 20 mM Hepes (pH 7.6), 100 mM KCl, and 5 mM MgCl2 and centrifuged in an SW40 at 35,000 rpm for 2 h. Gradients were analyzed by piercing the tube with a Brandel tube piercer, passing 60% sucrose through the bottom of the tube, and monitoring the absorbance of the material eluting from the tube using an ISCO UA-6 UV detector.
The results presented in this study explain the cytotoxicity associated with chlorolissoclimides (Malochet-Grivois et al. 1992; Roussakis et al. 1994; Uddin et al. 2001; Fu et al. 2004). We note a degree of structural similarity between lissoclimides and cycloheximide, a specific inhibitor of eukaryotic translation that interferes with ribosomal E-site function and blocks translocation (Obrig et al. 1971; Pestova and Hellen 2003). Similar to cycloheximide, lissoclimides block elongation and do not allow release of ribosomes from polysomes. Chlorolissoclimides are members of a larger family of diterpenoids, called labdanes, that also show significant activity against many eukaryotic organisms (Hanson 2005). For instance, norditerpenes aldehydes isolated from the shrub Vitex trifolia showed potent activity against Trypanosoma cruzi, the causative agent of American trypanosomiasis (Kiuchi et al. 2004). Also a furanolabdane extracted from the plant Potamogeton lucens is a potent algicide (Waridel et al. 2004), while a similar compound isolated from Hypoestes purpurea is cytotoxic to carcinoma cell lines (Shen et al. 2004). Although direct evaluation of these compounds is required to determine if they also inhibit protein synthesis, this mode of action may be a common defense approach mechanism utilized by a wide range of organisms to ward off predators.
MATERIALS AND METHODS
Materials and general methods
Restriction endonucleases and T7 RNA polymerase were purchased from New England Biolabs. 5-3H-cytidine triphosphate (20.5 Ci/mmol), 35S-methionine (>1000 Ci/mmol), 5-3H-uridine (22 Ci/mmol), and 6-3H-thymidine (10 Ci/mmol) were obtained from Perkin Elmer Life Sciences. Preparation of plasmid DNA, restriction enzyme digestions, agarose gel electrophoresis of DNA and RNA, and SDS-PAGE analysis were carried out by using standard methods (Sambrook and Russell 2001). Chlorolissoclimide and dichlorolissoclimide were purified from 25 mg P. forskalii extract. The extract was subjected to reverse-phase HPLC (Phenomenex, Prodigy ODS 5 μ, 250 × 10 mm) by using a gradient elution system from 100% water to 100% acetonitrile. Purification of the active fractions was performed by using normal-phase HPLC (Phenomenex Luna silica (2) 5 μ, 250 × 4.6 mm, λ 254 nm) using a gradient system from 100% hexane to 100% dichloromethane. A mixture of chlorolissoclimide and dichlorolissoclimide was obtained, and the compounds were separated by using reverse-phase HPLC (Phenomenex Luna C18 5 μ, 150 × 4.6 mm) using a water/methanol gradient system. Chlorolissoclimide was eluted first followed by dichlorolissoclimide at 88% MeOH/H2O.
The identity of the purified compounds was established by 1H-NMR measurement and mass spectrometry and by comparison with the literature (Malochet-Grivois et al. 1991; Fu et al. 2004). The HRESIMS of dichlorolissoclimide revealed the presence of two chlorine atoms from the isotope cluster at 440.1424 and 442.1389 representing [M + Na]+ (C20H28 35Cl2NO4Na). The 1H-NMR spectrum of chlorolissoclimide showed that it has the same skeleton as does dichlorolissoclimide, with the only difference being replacement of an H atom by a Cl atom at C-3 (Fig. 1A). HRESIMS of chlorolissoclimide confirmed the presence of only one chlorine atom from the isotope cluster at 406.1809 and 408.1775 representing [M + Na]+ (C20H29 35ClNO4Na).
In vivo metabolic labeling studies
For metabolic labeling studies, HeLa cells were incubated in the presence of compound or vehicle alone for the indicated amounts of time. 35S-methionine (150–225 uCi/mL), 3H-uridine (24 uCi/mL), or 3H-thymidine (48 uCi/mL; Perkin Elmer Life Sciences) was added to cells. For protein labeling, 35S-methionine was added in methionine-free media supplemented with 10% dialyzed FCS. For 3H-uridine and 3H-thymidine labeling, the isotopes were added in DMEM supplemented with 10% dialyzed FCS. Cells were washed once with phosphate-buffered saline, trypsinized, and harvested by centrifugation. The supernatant was removed by aspiration and the cell pellet was lysed in RIPA buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS). For determination of 35S-methionine incorporation, the lysate was spotted onto Whatman 3 MM paper (that had been preblocked with 50× amino acids mix) (Invitrogen), dried, and placed in cold 10% TCA for 20 min. Filters were transferred to 5% TCA, boiled for 15 min, washed once with 5% TCA and once with 95% ethanol, and dried. Radioactivity was determined by scintillation counting. The obtained counts were normalized to protein concentration in each sample, which had been determined by using a modified Lowry assay (DC Protein Assay; Bio-Rad).
For detection of radiolabeled nucleic acid, 5% TCA was added to the lysate supplemented with 9 μg of yeast tRNA. This was incubated for 20 min at 4°C, after which the solution was filtered through a GF/C glass fiber filter (Whatman; preblocked with 0.1 M sodium pyrophosphate in 5% TCA, washed twice with 5 mL of 1% TCA, and rinsed with 95% ethanol. After drying, radioactivity was determined by scintillation counting. The obtained counts were normalized to the protein concentration in each sample (determined using a modified Lowry assay; DC Protein Assay; Bio-Rad).
In vitro translations
In vitro translation assays were performed with a bicistronic mRNA reporter, FF/HCV/Ren, in which the first cistron encodes the FF luc protein and the second cistron encodes the Ren luc protein (Fig. 1B). Expression of the second cistron is driven by HCV IRES sequences. In vitro transcriptions were performed by using BamHI linearized templates. Translations were performed in Krebs extracts as previously reported (Novac et al. 2004). Translations in rabbit reticulocyte lysate, wheat germ extracts, and E. coli S30 extracts were performed as recommended by the manufacturer (Promega Corp). FF and Ren luc activity (RLU) were measured on a Berthold Lumat LB 9507 luminometer.
Ribosome binding assays
Ribosome binding assays were performed by incubating 32P-labeled mRNA in 25 μL of rabbit reticulocyte lysate (Promega) for 10 min at 30°C in the presence of 50 μM chlorolissoclimide with or without 0.6 mM cycloheximide. The final potassium acetate concentration was adjusted to 150 mM. Initiation complexes formed were analyzed by sedimentation through 10%–30% glycerol gradients. Centrifugation was for 3.5 h at 39,000 rpm at 4°C in an SW40 rotor. Fractions of 500 μL were collected, and radioactivity was determined by Cherenkov counting in a liquid scintillation counter. The plasmid pSP6/CAT used to generate mRNA for binding experiments has been previously described (Svitkin et al. 2001).
Met-puromycin dipeptide assays
Met-puromycin dipeptide assays were performed essentially as previously described (Lorsch and Herschlag 1999). Briefly, reactions included 60 nM ribosomes, 2 nM 35S-methionyl-tRNA, 400 μM puromycin, and 1 μM model mRNA, in a buffer containing 500 μM GTP, 1.75 mM magnesium acetate, and other components as previously described (Lorsch and Herschlag 1999). 80S complexes were preformed prior to addition of puromycin and inhibitor. Reactions proceeded for 30 min at 26°C, with aliquots removed periodically and quenched in 3M sodium acetate (pH 5.1). Quenched samples were resolved by cation-exchange TLC and 35S-Met-puromycin product quantitated. The observed rate constant at several concentrations of inhibitor was measured and normalized to a positive control reaction performed in the absence of inhibitor.
eEF1A- and eEF2-dependent assays
Phe-tRNAPhe was prepared as previously described (Odom et al. 1990), except that yeast S100 was used as a source of tRNA synthetase. The tRNA was further purified by passage through a Sephadex G-50 column. Both enzymatic and nonenzymatic aminoacyl-tRNA binding experiments were performed following a published procedure (SirDeshpande and Toogood 1995). Reaction mixtures containing HEPES buffer (20 mM HEPES-KOH at pH 7.5, 10 mM MgCl2, 100 mM KCl, 1.0 mM DTT), 89 pmol of 80S ribosome and inhibitor incubated for 10 min at 37°C; 0.15 mM GMPPNP, 0.2 μg of poly(U), 2.3 μg of eEF1A, and 10 pmol of [14C]Phe-tRNAPhe was then added and incubated for another 30 min at 37°C. To determine the amount of ribosome bound [14C]Phe-tRNAPhe, two aliquots (10%) of the assay mixture were removed, diluted with 0.8 mL of HEPES buffer, and filtered through a Millipore Type HA nitrocellulose filter. The filter was rinsed with the same buffer (5 × 2 mL), dried, and counted.
To measure nonenzymatic binding of [14C]Phe-tRNAPhe to ribosome, 177 pmol of 80S ribosome was preincubated with inhibitor in the presence of Tris-HCl buffer (50 mM Tris-HCl at pH 7.5, 60 mM KCl) and 20 mM MgCl2 at 37°C for 10 min; poly(U) (40 μg) and [14C]Phe-tRNAPhe were incubated at 37°C for 30 min. Two aliquots constituting 10% of the reaction mixture were removed and diluted into 0.8 mL of Tris-HCl buffer containing 20 mM MgCl2 and filtered through a Type HA nitrocellulose filter. The filter was rinsed with the same buffer (2 × 5 mL), dried, and counted to measure the amount of ribosome bound [14C]Phe-tRNAPhe.
eEF1A-dependent [14C]Phe-tRNAPhe binding to 89 pmol of ribosome was performed as described above. The amount of P-site bound [14C]Phe-tRNAPhe was determined for an identical sample by using the puromycin reaction (Wurmbach and Nierhaus 1979). To determine the inhibition of translocation, inhibitors were added into the rest of the 80% reaction mixture and incubated for 10 min at 37°C. Then, eEF2 (0.13 μg), HEPES buffer to compensate with the reaction volume, and 0.5 mM puromycin were added followed by 1 mM GTP to initiate translocation. Following incubation for an additional 30 min at 37°C, two aliquots (10%) of reaction mixture were quenched in 30 μL of 1 mM sodium bicarbonate (pH 5.1), and the amount of P-site bound [14C]Phe-tRNAPhe was determined by extraction of [14C]phenylalanine-puromycin into ethyl acetate (1 mL) and corrected for the amount of tRNA present in the P-site before translocation. The amount of radioactivity was measured by scintillation counting of 900 μL aliquots of the organic layer.
ACKNOWLEDGMENTS
We thank the NIH/NCI Developmental Therapeutics Program for their generous supply of P. forskalii extract. F.R. held a Canderel fellowship from the McGill Cancer Center and M.D. was supported by an Egyptian Government Predoctoral Fellowship during the course of this work. This work was supported in part by an NCIC grant (no. 014313) to J.P., an NIH grant (GM26796) to W.C.M, and an NIH/NIAID no. 1RO1AI36596 to M.T.H. J.P. is a Canadian Institute of Health Research (CIHR) Senior Investigator.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2346806.
REFERENCES
- Ahuja D., Geiger A., Ramanjulu J.M., Vera M.D., SirDeshpande B., Pfizenmayer A., Abazeed M., Krosky D.J., Beidler D., Joullie M.M., et al. Inhibition of protein synthesis by didemnins: Cell potency and SAR. J. Med. Chem. 2000;43:4212–4218. doi: 10.1021/jm000168v. [DOI] [PubMed] [Google Scholar]
- Bordeleau M.E., Matthews J., Wojnar J.M., Lindqvist L., Novac O., Jankowsky E., Sonenberg N., Northcote P., Teesdale-Spittle P., Pelletier J. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc. Natl. Acad. Sci. 2005;102:10460–10465. doi: 10.1073/pnas.0504249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Khan S.N., Harvey I., Merrick W., Pelletier J. Eukaryotic protein synthesis inhibitors identified by comparison of cytotoxicity profiles. RNA. 2004;10:528–543. doi: 10.1261/rna.5200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Palomar A.J., Hong E.P., Schmitz F.J., Valeriote F.A. Cytotoxic lissoclimide-type diterpenes from the molluscs Pleurobranchus albiguttatus and Pleurobranchus forskalii . J. Nat. Prod. 2004;67:1415–1418. doi: 10.1021/np0499620. [DOI] [PubMed] [Google Scholar]
- Grollman A.P. Inhibitors of protein biosynthesis, II: Mode of action of anisomycin. J. Biol. Chem. 1967;242:3226–3233. [PubMed] [Google Scholar]
- Hanson J.R. Diterpenoids. Nat. Prod. Rep. 2005;22:594–602. doi: 10.1039/b501834j. [DOI] [PubMed] [Google Scholar]
- Heys S.D., Park K.G., McNurlan M.A., Calder A.G., Buchan V., Blessing K., Eremin O., Garlick P.J. Measurement of tumour protein synthesis in vivo in human colorectal and breast cancer and its variability in separate biopsies from the same tumour. Clin. Sci. (Lond.) 1991;80:587–593. doi: 10.1042/cs0800587. [DOI] [PubMed] [Google Scholar]
- Kiuchi F., Matsuo K., Ito M., Qui T.K., Honda G. New norditerpenoids with trypanocidal activity from Vitex trifolia . Chem. Pharm. Bull. (Tokyo) 2004;52:1492–1494. doi: 10.1248/cpb.52.1492. [DOI] [PubMed] [Google Scholar]
- Lorsch J.R., Herschlag D. Kinetic dissection of fundamental processes of eukaryotic translation initiation in vitro. EMBO J. 1999;18:6705–6717. doi: 10.1093/emboj/18.23.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malochet-Grivois C., Cotelle P., Biard J.F., Henichart J.P., Debitus C., Roussakis C., Verbist J.F. Dichlorolissoclimide, a new cytotoxic labdane derivative from Lissoclinum voeltzkowi Michaelson (Urochordata) Tetrahedon Lett. 1991;32:6701–6702. [Google Scholar]
- Malochet-Grivois C., Roussakis C., Robillard N., Biard J.F., Riou D., Debitus C., Verbist J.F. Effects in vitro of two marine substances, chlorolissoclimide and dichlorolissoclimide, on a non–small-cell bronchopulmonary carcinoma line (NSCLC-N6) Anticancer Drug Des. 1992;7:493–502. [PubMed] [Google Scholar]
- Mamane Y., Petroulakis E., Rong L., Yoshida K., Ler L.W., Sonenberg N. eIF4E—from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- Novac O., Guenier A.S., Pelletier J. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 2004;32:902–915. doi: 10.1093/nar/gkh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrig T.G., Culp W.J., McKeehan W.L., Hardesty B. The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J. Biol. Chem. 1971;246:174–181. [PubMed] [Google Scholar]
- Odom O.W., Picking W.D., Hardesty B. Movement of tRNA but not the nascent peptide during peptide bond formation on ribosomes. Biochemistry. 1990;29:10734–10744. doi: 10.1021/bi00500a004. [DOI] [PubMed] [Google Scholar]
- Ottenheijm H.C., van den Broek L.A. The development of sparsomycin as an anti-tumour drug. Anticancer Drug Des. 1988;2:333–337. [PubMed] [Google Scholar]
- Pestka S. Inhibitors of protein synthesis. Academic Press; New York: 1977. [Google Scholar]
- Pestova T.V., Hellen C.U. Translation elongation after assembly of ribosomes on the cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes & Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussakis C., Charrier J., Riou D., Biard J.F., Malochet C., Meflah K., Verbist J.F. Chemotherapeutic inhibition of erb-B2 oncogene expression on a non–small-cell cancer line (NSCLC-N6) by marine substances. Anticancer Drug Des. 1994;9:119–128. [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Shen C.-C., Ni C.-L., Huang Y.-L., Huang R.-L., Chen C.-C. Furanolabdane diterpenes from Hypoestes purpurea . J. Nat. Prod. 2004;67:1947–1949. doi: 10.1021/np0497402. [DOI] [PubMed] [Google Scholar]
- SirDeshpande B.V., Toogood P.L. Mechanism of protein synthesis inhibition by didemnin B in vitro. Biochemistry. 1995;34:9177–9184. doi: 10.1021/bi00028a030. [DOI] [PubMed] [Google Scholar]
- Svitkin Y.V., Sonenberg N. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol. Biol. 2004;257:155–170. doi: 10.1385/1-59259-750-5:155. [DOI] [PubMed] [Google Scholar]
- Svitkin Y.V., Pause A., Haghighat A., Pyronnet S., Witherell G., Belsham G.J., Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M.J., Kokubo S., Ueda K., Suenaga K., Uemura D. Haterumaimides F-I, four new cytotoxic diterpene alkaloids from an ascidian Lissoclinum species. J. Nat. Prod. 2001;64:1169–1173. doi: 10.1021/np010066n. [DOI] [PubMed] [Google Scholar]
- van den Broek L.A., Lazaro E., Zylicz Z., Fennis P.J., Missler F.A., Lelieveld P., Garzotto M., Wagener D.J., Ballesta J.P., Ottenheijm H.C. Lipophilic analogues of sparsomycin as strong inhibitors of protein synthesis and tumor growth: A structure-activity relationship study. J. Med. Chem. 1989;32:2002–2015. doi: 10.1021/jm00128a051. [DOI] [PubMed] [Google Scholar]
- Vazquez D. Inhibitors of protein biosynthesis. Mol. Biol. Biochem. Biophys. 1979;30:1–312. doi: 10.1007/978-3-642-81309-2. [DOI] [PubMed] [Google Scholar]
- Waridel P., Wolfender J.-L., Lachavanne J.-B., Hostettmann K. ent-Labdane glycosides from the aquatic plant Potamogeton lucens and analytical evaluation of the lipophilic extract constituents of various Potamogeton species. Phytochemistry. 2004;65:945–954. doi: 10.1016/j.phytochem.2004.01.018. [DOI] [PubMed] [Google Scholar]
- Wendel H.-G., de Stanchina E., Fridman J.S., Malina A., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- Wurmbach P., Nierhaus K.H. Codon-anticodon interaction at the ribosomal P (peptidyl-tRNA)site. Proc. Natl. Acad. Sci. 1979;76:2143–2147. doi: 10.1073/pnas.76.5.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]