Abstract
In organisms ranging from Arabidopsis to humans, Dicer requires dsRNA-binding proteins (dsRBPs) to carry out its roles in RNA interference (RNAi) and micro-RNA (miRNA) processing. In Caenorhabditis elegans, the dsRBP RDE-4 acts with Dicer during the initiation of RNAi, when long dsRNA is cleaved to small interfering RNAs (siRNAs). RDE-4 is not required in subsequent steps, and how RDE-4 distinguishes between long dsRNA and short siRNA is unclear. We report the first detailed analysis of RDE-4 binding, using purified recombinant RDE-4 and various truncated proteins. We find that, similar to other dsRBPs, RDE-4 is not sequence-specific. However, consistent with its in vivo roles, RDE-4 binds with higher affinity to long dsRNA. We also observe that RDE-4 is a homodimer in solution, and that the C-terminal domain of the protein is required for dimerization. Using extracts from wild-type and rde-4 mutant C. elegans, we show that the C-terminal dimerization domain is required for the production of siRNA. Our findings suggest a model for RDE-4 function during the initiation of RNAi.
Keywords: dsRNA, RNA interference, C. elegans, Dicer
INTRODUCTION
RNA interference (RNAi) is a conserved gene silencing mechanism that induces a sequence-specific degradation of mRNA in response to double-stranded RNA (dsRNA) (Tomari and Zamore 2005). The related micro-RNA (miRNA) pathway regulates expression of numerous genes, sometimes by mRNA degradation (Bagga et al. 2005), and in other cases by translational repression involving a mechanism that is as yet unclear (Murchison and Hannon 2004). Both pathways are mediated by small RNAs, termed small-interfering RNAs (siRNAs) or micro-RNAs (miRNAs), which are ∼22 nucleotides in length, and are products of dsRNA processing by the RNase III enzyme Dicer (Carmell and Hannon 2004). These small RNAs are incorporated into ribonucleoprotein complexes referred to as si- or mi-RISCs (RNA-induced silencing complexes) to carry out their effector functions (Tomari and Zamore 2005).
Dicer often requires accessory dsRNA-binding proteins (dsRBPs) to carry out its in vivo functions. For example, in Drosophila, which expresses two Dicer proteins, the dsRBP Loquacious associates with Dicer-1 for miRNA-mediated silencing (Forstemann et al. 2005; Saito et al. 2005), and another dsRBP, R2D2, forms a stable complex with Dicer-2 to initiate siRNA-mediated silencing (Liu et al. 2003; Pham and Sontheimer 2005). Arabidopsis contains four Dicer-like proteins of which three interact specifically with dsRBPs (Hiraguri et al. 2005). In addition, human Dicer requires the dsRBP TRBP (human immunodeficiency virus transactivating response RNA-binding protein) for both siRNA and miRNA-mediated gene silencing (Haase et al. 2005; Chendrimada et al. 2005).
Like the dsRBPs discussed above, the Caenorhabditis elegans dsRBP RDE-4 acts in concert with Dicer, in this case, to facilitate the production of siRNAs. The protein is required for RNAi in C. elegans, but is dispensable for miRNA-mediated silencing as evidenced by the lack of developmental defects in rde-4 mutant worms (Tabara et al. 1999). RDE-4 immunoprecipitates with DCR-1 (C. elegans Dicer) as well as with the proteins RDE-1, and DRH-1 (Tabara et al. 2002). In addition, RDE-4 immunoprecipitates with trigger dsRNA, but not siRNA (Tabara et al. 2002), and thus may differ from the Drosophila R2D2, which forms stable interactions with siRNA (Liu et al. 2003). C. elegans strains lacking RDE-4 show a marked decrease in the levels of siRNA derived from an injected dsRNA trigger, resulting in an RNAi defect that can be partially bypassed by the injection of synthetic siRNA (Parrish and Fire 2001). These observations show that RDE-4 facilitates the production of siRNA from a dsRNA trigger but is not essential for later steps in the pathway.
We set out to characterize the in vitro nucleic acid binding properties of RDE-4 in hopes that such studies would shed light on its in vivo functions. Two observations are interesting in this regard. First, we find that RDE-4 exists as a stable dimer, and that dimerization is required for RDE-4's role in production of siRNA. Second, we observe that RDE-4 binds long dsRNA with high affinity, and has low affinity for siRNA. The latter explains how a dsRBP that binds dsRNA of any sequence can act specifically in the first step of RNAi, and not affect later steps that also involve dsRNA, albeit the shorter siRNA. We speculate that cooperativity contributes to high-affinity binding. Taken together, our findings suggest a model for RDE-4 function during the initiation of RNAi.
RESULTS
RDE-4 has a higher affinity for longer dsRNA
Although the biochemical properties of RDE-4 have not been studied in detail, the protein was reported to bind to dsRNA during the initiation of RNAi (Tabara et al. 2002). Specifically, using worms grown on dsRNA-expressing bacteria, Tabara and colleagues were able to immunoprecipitate RDE-4 complexes containing the bacterially expressed dsRNA. Small RNAs were not observed in the complexes, suggesting that RDE-4 does not form stable complexes with the siRNA product, and preferentially interacts with longer dsRNAs (Tabara et al. 2002).
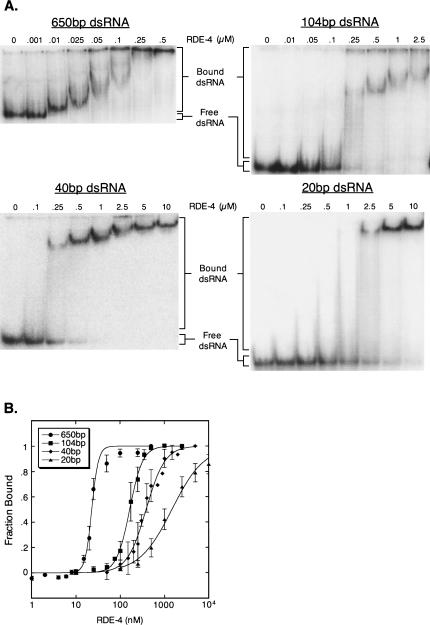
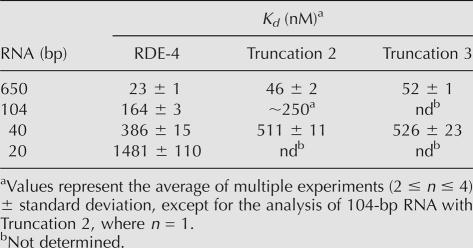
To characterize the nucleic acid binding properties of RDE-4 directly, we performed gel mobility shift assays with purified recombinant RDE-4, expressed in Saccharomyces cerevisiae (Fig. 1), and dsRNA substrates of different lengths. 32P-labeled dsRNA was incubated with increasing concentrations of RDE-4 and subsequently run on native polyacrylamide gels, enabling the visualization of protein/RNA complexes (Carey 1991). We tested the ability of RDE-4 to bind 650, 104, 40, and 20 base-pair (bp) dsRNA. RDE-4 readily formed stable complexes with all four substrates (Fig. 2A). However, the measured affinities were much higher with longer dsRNAs, with a Kd of ∼23 nM for the 650-bp dsRNA and a Kd of ∼1.5 μM for the 20-bp dsRNA (Fig. 2B; Table 1). The 20-bp RNA tested contained two nucleotide 3′ overhangs, mimicking an siRNA. In other experiments we analyzed binding to 40-bp RNA with and without overhangs. No differences were observed with the different termini (data not shown). Interestingly, distinct mobility shift intermediates were absent in all assays using longer dsRNA (Fig. 2A), which is suggestive of cooperative binding (see Discussion).
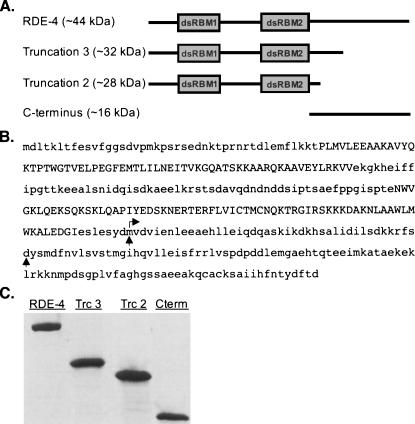
FIGURE 1.
RDE-4 protein constructs. (A) Schematic diagram of RDE-4 protein constructs with double-stranded RNA binding motifs (dsRBMs) shown as boxes and the molecular weights indicated. (B) Amino acid sequence of RDE-4. Vertical arrows indicate the last amino acid before the engineered stop codon of truncations 2 and 3. Horizontal arrow indicates start position for the C terminus protein. Truncation 2 contains eight additional nonnative amino acids (GMPPNNCS) at its C terminus (see Materials and Methods). (C) SYPRO Red-stained SDS-PAGE gel of purified RDE-4, Truncations 2 and 3 (Trc2 & Trc3) and C terminus (Cterm).
FIGURE 2.
Gel mobility shift analyses of RDE-4 and dsRNA substrates of different lengths. (A) Increasing concentrations of RDE-4 were added to 32P-labeled 650-bp dsRNA (top left), 104-bp dsRNA (top right), 40-bp dsRNA (bottom left), and 20-bp dsRNA (bottom right). Complex formation was analyzed by native gel electrophoresis. Bands corresponding to bound and free dsRNA are labeled. Previous studies note that cooperative ligands bind to nucleic acid lattices in clusters, and as the binding density increases, the average size of the bound clusters increases (Kowalczykowski et al. 1986). The gradual decrease in mobility for the 650-bp complex is likely due to additional protein binding events, possibly in “clusters,” but future studies will be required to confirm this. (B) RNA binding isotherms for RDE-4. Radioactivity in gels as in A were quantified to determine fraction bound = [dsRNA]bound/[dsRNA]total (see Materials and Methods). All RNA of slower mobility than dsRNAfree was considered as bound. Data points with error bars (standard deviation) represent average values (2 ≤ n ≤ 4) and were fit using the Hill formalism, where fraction bound = 1/(1 + (Kdn/[P]n)). Resulting Hill constants for the 650-bp, 104-bp, 40-bp, and 20-bp RNAs were 4.98 ± 1.0, 3.53 ± 0.1, 2.04 ± 0.2, and 1.18 ± 0.1, respectively, but it is important to note that these values may derive from statistical considerations rather than intrinsic cooperativity (see Discussion).
TABLE 1.
Summary of binding affinities
RDE-4 binds ssRNA independent of dsRNA binding
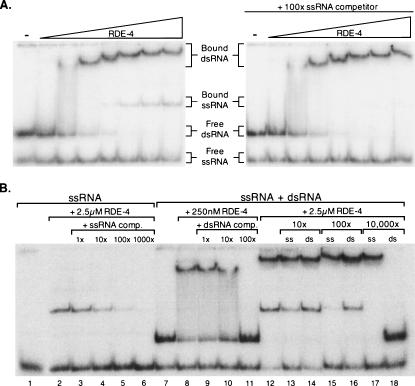
Occasionally, some of the purified 32P-labeled dsRNA contained a small amount of labeled ssRNA, either from copurification or from disassociation of the dsRNA in solution. When these RNAs were used in gel mobility shift assays, at high protein concentrations, we observed a second complex that migrated faster than the dsRNA/protein complex (Fig. 3A, left panel). The appearance of this faster-migrating complex was concomitant with the disappearance of the free ssRNA, suggesting the shift was due to RDE-4 binding to ssRNA. To verify the shift was due to ssRNA binding, we performed competition experiments with excess unlabeled ssRNA. The addition of 100-fold excess unlabeled 40-nt ssRNA successfully competed with the faster migrating shift while leaving the dsRNA shift unaffected (Fig. 3A, right panel). While all RNAs are likely to have some structure, the fact that the ssRNA did not diminish the amount of the dsRNA–RDE4 complex indicates the 40 mer ssRNA is largely single-stranded. We also assayed ssRNA binding directly (Fig. 3B), and measured a Kd of ∼2.5 μM for RDE-4 binding to ssRNA (Fig. 5B; data not shown).
FIGURE 3.
Competition assays. (A) Mobility shift assay showing that RDE-4 binds to dsRNA and ssRNA. Increasing concentrations of RDE-4, as specified in Figure 2A, were added to the preparation of 32P-labeled 40-bp dsRNA, in the absence (left panel) or presence (right panel) of competitor ssRNA (sequence matching one strand of the 40-bp dsRNA). The panel on the right verifies the second complex is RDE-4 binding to ssRNA, since it was competed by 100× unlabeled ssRNA without affecting dsRNA binding (compare complexes in left and right panels). (B) Competition assays at differing protein and competitor concentrations. RNAs are either purified 32P-labeled 40-nt ssRNA (lanes 1–6) or the mix of 32P-labeled dsRNA and 32P-labeled ssRNA used in part A (lanes 7–18). Competitor RNAs are 104 nt in length, either single stranded or double stranded, and are different in sequence to that of the labeled RNAs.
FIGURE 5.
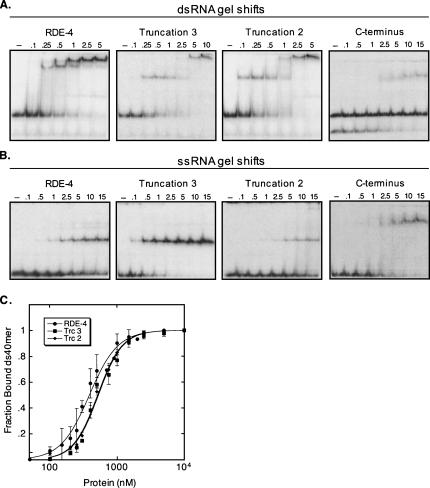
Gel mobility shift analyses comparing binding affinities of RDE-4, Truncation 3, Truncation 2, and the C terminus. (A) 32P-labeled 40-bp dsRNA was incubated with increasing amounts of protein (μM), as indicated above each gel. The 40-bp dsRNA preparations contained minimal amounts of contaminating ssRNA, except for that used with the C terminus protein, and in this case the observed complex is with ssRNA. (B) 32P-labeled 40-nt ssRNA was incubated with increasing amounts of protein (μM), as indicated above each gel. (C) Forty base-pair dsRNA binding isotherms for RDE-4, Truncation 3 and Truncation 2. The C terminus protein did not bind dsRNA and was not plotted. Data points with error bars (standard deviation) represent average values (2 ≤ n ≤ 3) and were fit using the Hill formalism as in Figure 2B. Resulting Hill coefficients for RDE-4, Truncation 2 and Truncation 3 were 2.04 ± 0.2, 2.39 ± 0.1 and 2.39 ± 0.2, respectively.
To test whether ssRNA and dsRNA binding was sequence-specific, we performed additional competition assays using unlabeled 104-nt ssRNA or dsRNA competitor RNA. Binding was visualized with 32P-labeled 40-nt ssRNA or a 32P-labeled ssRNA/dsRNA mix, both of which differed in sequence compared to the unlabeled competitor RNAs. The ssRNA shifts were successfully competed by the addition of the unlabeled 104-nt ssRNA (Fig. 3B, lanes 3–6). Likewise, the dsRNA shift observed at 250 nM RDE-4 was effectively competed by the addition of unlabeled 104-bp dsRNA (Fig. 3B, lanes 9–11), indicating that RDE-4 binds both ssRNA and dsRNA independent of sequence. Similarly, when dsRNA substrates of differing sequences were labeled with 32P and assayed directly by gel shift, similar affinities were measured (data not shown).
Competition of ssRNA binding using the 32P-labeled ssRNA/dsRNA mix was similar to that observed when 32P-labeled ssRNA was assayed alone (Fig. 3B, cf. lanes 13,15 and 4,5), suggesting the presence of dsRNA does not affect the relative affinity for ssRNA. Additionally, 10,000-fold excess competitor ssRNA left the dsRNA shift intact, while completely abolishing the ssRNA shift (Fig. 3B, lane 17), and 100-fold excess competitor dsRNA left the ssRNA shift relatively intact (Fig. 3B, lane 16). Taken together, these data suggest that the ssRNA and dsRNA binding sites of RDE-4 are mutually exclusive. The small amount of ssRNA competition seen at 100× dsRNA, and the complete competition at 10,000× dsRNA (Fig. 3B, lanes 16,18), were most likely due to contaminating ssRNA in our 104-bp dsRNA preparation, like that observed for our 40-bp dsRNA (Fig. 3A,B).
The C terminus of RDE-4 is required for dimerization
The final step of our purification protocol for RDE-4 was a Sephacryl S-200 gel filtration column. The protein consistently eluted earlier than predicted, at an MW of ∼80 kDa based on our standard calibration curve (data not shown). The calculated MW of the purified, recombinant RDE-4 is ∼43 kDa (Fig. 1A), suggesting RDE-4 formed a stable dimer in solution. When Truncations 2 and 3 were purified to homogeneity, they eluted as monomers from the gel filtration column, with observed MWs of ∼24 kDa, and ∼33 kDa, respectively (data not shown). The purified C terminus eluted as a monomer from the gel filtration column with an observed MW of ∼15 kDa, indicating that when expressed as a separate polypeptide, this sequence cannot dimerize (data not shown).
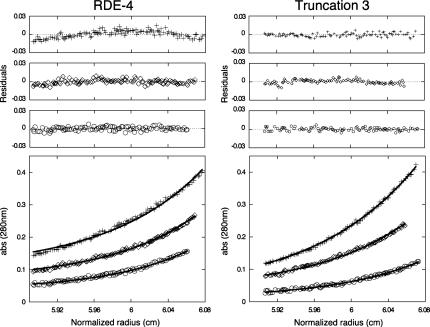
In order to confirm the oligomeric states of our recombinant proteins, we conducted sedimentation equilibrium experiments, which allow accurate molecular weight determination of macromolecules independent of their shape (Laue 1995). For RDE-4, Truncation 2 and Truncation 3, multiple data sets were obtained using different speeds and protein concentrations. Figure 4 shows characteristic experimental data, fits, and residuals for RDE-4 and Truncation 3. For the RDE-4 data, the best global fit using nonlinear regression analysis (Johnson et al. 1981) was to a single-species molecule with a molecular weight of 86,280 Da ± 3400 (Fig. 4, left panel). The MWobs/MWcalc = 1.99, confirming that RDE-4 formed a stable dimer in solution. Truncation 3 and Truncation 2 data fit best to single-species molecules with molecular weights of 35,863 Da ± 3500 (Fig. 4, right panel) and 29, 572 Da ± 1750 (data not shown), respectively. The MWobs/MWcalc for both Truncations 2 and 3 were ∼1.1, confirming the monomeric state of these proteins. The residuals for the fits were small and randomly distributed indicating good overall fits (Fig. 4). The calculated monomeric MWs for RDE-4 and Truncation 2 were confirmed by mass spectrometry, and showed 43.4 kDa and 28.4 kDa, respectively (data not shown). These results indicate that the 100 C-terminal amino acids of RDE-4 are required for dimerization.
FIGURE 4.
Sedimentation equilibrium data for RDE-4 and Truncation 3. The lower panels show data for three concentrations of RDE-4 (+ = 15 μM, ⋄ = 10 μM, ◯ = 5 μM) and Truncation 3 (+ = 20 μM, ⋄ = 10 μM, ◯ = 5 μM). RDE-4 data fit to a single-species of MW 86,280 Da ± 3400. The MWobs/MWcalc = 1.99, indicating RDE-4 formed a stable dimer in solution. Truncation 3 data fit to a single-species of MW 35,863 Da ± 3500 and Truncation 2 data fit to a single-species of MW 29,572 Da ± 1750 (data not shown) indicating both truncations are monomeric. The upper panels show the corresponding residuals for each fit.
To test if dimerization was required for RNA binding, the truncated protein constructs were assayed utilizing gel mobility shift assays. As shown in Figure 5A and C, Truncations 2 and 3 retained near wild-type dsRNA binding abilities, with apparent Kds of ∼0.5 μM for the 40-bp dsRNA. Furthermore, like full-length RDE-4, these two truncated proteins exhibited higher affinity for longer dsRNAs when tested with the 650-bp dsRNA (data not shown and Table 1). As expected, the C terminus construct was unable to bind dsRNA (Fig. 5A, right panel). In contrast, all three truncations retained the ability to bind ssRNA (Fig. 5B). However, Truncation 3 exhibited a higher affinity for ssRNA compared to RDE-4 and the C terminus protein, and binding by Truncation 2 was significantly impaired (Fig. 5B). Taken together, these data suggest that dimerization is not required for RNA binding, nor is it responsible for the increased affinity for long dsRNA.
RDE-4 C terminus is required for reconstitution of in vitro Dicer activity
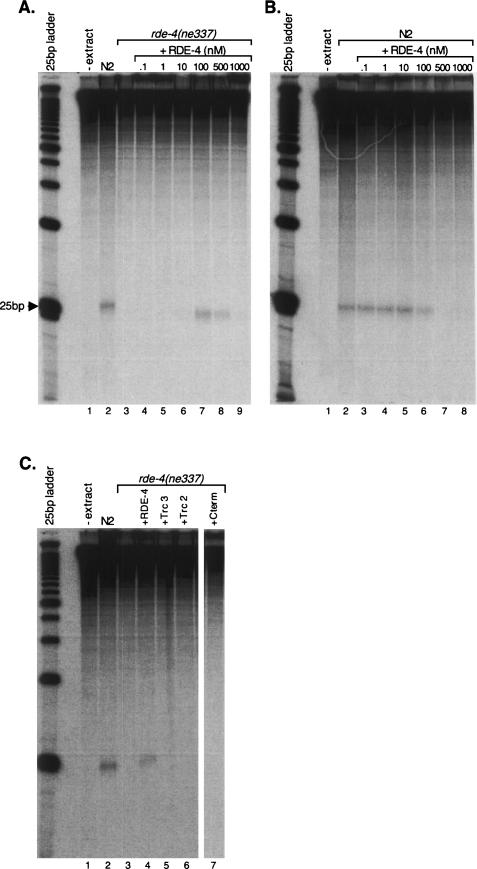
The production of siRNAs after injection of 32P-labeled dsRNA is greatly reduced in RDE-4 deficient worms compared to wild-type animals, suggesting that Dicer cannot process dsRNA without RDE-4 (Parrish and Fire 2001). So far, attempts to purify C. elegans Dicer in an active, recombinant form have been unsuccessful. However, when dsRNA is incubated in extracts of wild-type C. elegans, it is cleaved to siRNAs via endogenous Dicer (Ketting et al. 2001). Using a similar assay, we tested whether our purified, recombinant RDE-4 proteins could reconstitute Dicer activity in RDE-4-deficient C. elegans embryo extracts. We incubated 32P-internally labeled 650-bp dsRNA with protein extracts derived from C. elegans embryos. Wild-type (N2) extracts were competent for dsRNA processing, producing detectible siRNAs (Fig. 6A, lane 2), while extracts made from rde-4(ne337) embryos were deficient in siRNA production (Fig. 6A, lane 3). Importantly, the activity in extracts prepared from rde-4(ne337) embryos was restored by the addition of recombinant RDE-4 (Fig. 6A, lanes 4–9). The reconstitution was more effective when the dsRNA substrate was prebound by RDE-4 prior to adding worm protein extracts (data not shown, see Materials and Methods). Interestingly, the activity peaked around 100 nM RDE-4 (Fig. 6A, lane 7), and higher concentrations inhibited Dicer activity (Fig. 6A, lanes 8–9). This inhibition was also seen when RDE-4 was added to wild-type N2 extracts, at concentrations higher than 10 nM RDE-4 (Fig. 6B, lanes 3–8). The observed inhibition suggests that excess recombinant RDE-4 titrates a limiting factor.
FIGURE 6.
Reconstitution of siRNA production. Reactions were 30-min preincubations of a 650-bp 32P-internally labeled dsRNA ± recombinant RDE-4 proteins, followed by 1-h incubations with either wild-type (N2) or rde-4(ne337) embryo extracts (40-μg total protein). dsRNA cleavage products were analyzed by denaturing gel electrophoresis. (A) The autoradiogram shows products of reactions incubated without (-extract) or with extracts prepared from N2 and rde-4(ne337) embryos. The production of siRNA was reconstituted in the mutant extracts by adding recombinant RDE-4 at the concentrations indicated. Reconstituted Dicer activity is maximal at +100 nM RDE-4 and is inhibited by higher concentrations of RDE-4. The far left lane shows markers prepared from a 32P-end-labeled 25-bp DNA ladder. (B) As in A but with the addition of recombinant RDE-4 to N2 extracts. (C) As in A but showing reconstitution assays using full-length and truncated RDE-4; protein was added to a final concentration of 100 nM.
We also tested the ability of our truncated protein constructs to reconstitute Dicer activity. As shown in Figure 6C, none of the truncations were able to reconstitute Dicer activity over a range of protein concentrations (Fig. 6C, lanes 5–7; data not shown). Since Truncations 2 and 3 retained near wild-type dsRNA binding affinity (Fig. 5A,C), the lack of reconstitution with these two constructs was not due to defective substrate recognition. In support of this, the addition of increasing concentrations of Truncation 2 inhibited Dicer activity in N2 extracts (data not shown), possibly due to substrate competition with the endogenous RDE-4. Alternatively, as mentioned above, this inhibition could be due to titration of a limiting protein factor. These data indicate that the C terminus of RDE-4, which is required for dimerization, is also required for successful reconstitution of siRNA production, in a manner that is independent of substrate recognition.
DISCUSSION
RNA binding affinity of RDE-4
Our in vitro binding studies indicate that RDE-4 preferentially binds long dsRNA over short siRNA, and this is consistent with previously reported in vivo studies suggesting that RDE-4 acts early in RNAi and is not essential downstream of siRNA production (Parrish and Fire 2001; Tabara et al. 2002). RDE-4 may thus function differently from R2D2, its putative homolog in Drosophila, which is required for siRNA-mediated silencing by Dicer-2. R2D2 is not required for dsRNA processing by Dicer-2 in vitro, but instead is required for stabilization of the product siRNA (Liu et al. 2003). R2D2 has been implicated as a sensor of the thermodynamic asymmetry of the siRNA, helping determine which strand associates with RISC by binding to the siRNA end with the most double-stranded character (Tomari et al. 2004). Additionally, dsRNA-binding mutants of R2D2 had no adverse effect on processing of long dsRNA by Dicer-2, but downstream RISC activity was greatly impaired (Liu et al. 2003). Although the biochemical studies reported for the C. elegans and Drosophila systems are not directly comparable, the above observations suggest the in vivo roles of R2D2 do not require preferential binding to longer dsRNA.
Unexpectedly, RDE-4 also bound single-strand RNA (ssRNA), albeit with low affinity (Figs. 3, 5B). While this may point to additional roles of RDE-4 in RNAi, it also seems possible that the observed ssRNA binding is nonspecific, and future studies will be required to determine this.
Is RNA binding by RDE-4 cooperative?
Our studies suggest that RDE-4 binds dsRNA cooperatively. In this scenario, the intrinsic affinity of RDE-4 to a single binding site would be relatively weak, consistent with the low affinity of RDE-4 for the 20-bp RNA (Fig. 2B; Table 1). Higher affinity would occur with longer dsRNA due, at least in part, to interactions between RDE-4 molecules bound to adjacent sites. The idea that RDE-4 is cooperative is bolstered by the striking sigmoidicity we observe in the binding isotherms, which show steep transitions from dsRNAfree to dsRNAbound for all substrates except the 20bp dsRNA (Fig. 2B). Furthermore, the gel-shift analyses with RDE-4 described in this study are very different from our previous studies of another dsRNA-binding protein, 4F (Bass et al. 1994). In the latter study, increasing the length of dsRNA used for the gel-shift analysis resulted in an increase in the number of discrete gel shifts, corresponding to successive binding events on a single dsRNA. In contrast, we do not observe intermediate gel shifts in our studies of RDE-4, and we speculate this “all-or-none” behavior is due to cooperativity.
Describing the binding parameters of a protein that binds without sequence-specificity is complicated by the presence of overlapping binding sites. For example, a protein with an RNA interaction site size of 20 bp has 21 equivalent binding sites on a 40-bp RNA (length of RNA − binding site size + 1; McGhee and von Hippel 1974). The apparent affinity of such an interaction is the product of this statistical factor (21 in this case) and the intrinsic binding constant (Draper and von Hippel 1978). This relationship predicts that the apparent affinity will be a linear function of the lattice length (Draper and von Hippel 1978), and the binding constants measured for RDE-4 do follow a linear trend (Table 1). While it is possible that the higher affinity we observe for RDE-4 binding to long dsRNA is entirely due to the increased number of binding sites in the longer dsRNA, we speculate that the protein also has an intrinsic cooperativity. As mentioned, this idea is supported by the lack of discrete binding intermediates when RDE-4 binding is assayed by gel shift. However, the lack of discrete intermediates makes it difficult to measure binding events subsequent to the first, thus precluding an accurate determination of cooperativity.
In a classic paper, McGhee and von Hippel detailed an approach for analyzing binding of a sequence-independent binding protein to an infinite-length lattice, and described equations for determining the cooperativity of such a system as quantified by the parameter, ω (McGhee and von Hippel 1974). Additionally, Epstein developed a mathematical treatment to more accurately describe cooperative binding interactions with finite-length lattices (Epstein 1978). This latter treatment takes into account statistical probabilities of ligand arrangement on the lattice and potential “end effects” attributed to loss of cooperative interactions due to the greater ratio of “ends” to “middles” of smaller lattices (Kowalczykowski et al. 1986). Both the McGhee–von Hippel and Epstein models are applicable to our system, but both require knowledge of the fractional saturation of the RNA lattice, and this information cannot be obtained from our gel-shift analyses. That is, our assay monitors the loss of free dsRNA as protein/RNA is retarded in the gel but does not provide information in regard to the number of proteins bound to a particular RNA. In future studies we hope to employ alternative assay methods to definitively resolve the issue of whether RDE-4 has intrinsic cooperativity.
The role of the RDE-4 C terminus
Our gel filtration and sedimentation equilibrium data demonstrate that the C terminus of RDE-4 is required for dimerization. RDE-4 formed a stable dimer in solution, but truncation constructs lacking as few as 100 C-terminal amino acids were stable monomers (Fig. 4). However, the monomeric forms of RDE-4 were competent for dsRNA and ssRNA binding, with only slight differences in Kds compared to RDE-4 (Fig. 5; Table 1). The slight reduction in dsRNA affinity observed with Truncations 2 and 3 (∼twofold) suggests that the preformed dimers of RDE-4 stabilize the interaction with dsRNA (Fig. 5C; Table 1), perhaps by engaging dsRNA with all four dsRBMs present in the RDE-4 homodimers. The truncated proteins also exhibited higher affinities for longer dsRNAs (Table 1), indicating that dimerization is not required for high-affinity binding.
But if dimerization is not needed for dsRNA binding, why is it required for siRNA production, as indicated by the reconstitution experiments (Fig. 6)? One possibility is that RDE-4 dimerization facilitates formation of an active Dicer complex, either by bringing Dicer to the dsRNA, or by mediating interactions between Dicer molecules themselves. Existing literature provides several interesting precedents. For example, the dsRBP RNA-dependent protein kinase (PKR) requires dimerization for function and is activated by another dsRBP, PACT (Patel and Sen 1998). The N-terminal dsRBMs of PACT bind to PKR, while its C-terminal dimerization domain is essential for PKR activation (Peters et al. 2001; Huang et al. 2002), presumably because it facilitates PKR dimerization (Hitti et al. 2004). Another precedent comes from studies of the human RNase III enzyme Drosha, which processes primary miRNA. Drosha exists as a larger complex with its dsRBP partner, DGCR8, suggesting the formation of a heterotetramer consisting of two Drosha and two DGCR8 molecules (Han et al. 2004). Thus, one interpretation of our reconstitution data is that C. elegans Dicer acts as a dimer, and that dimerization is mediated by the homodimerization of RDE-4.
In addition to mediating binding to dsRNA, dsRBMs can function as protein–protein interaction domains (Tian et al. 2004). For example, a dsRBM of the putative RDE-4 homolog in Arabidopsis thaliana, HYL1, mediates interactions with itself as well as with the dsRBM of the Dicer protein, DCL1, in vivo (Hiraguri et al. 2005). Similarly, the third dsRBM of TRBP interacts with human Dicer in vivo (Haase et al. 2005). Thus, it is possible that interactions between RDE-4 and Dicer are mediated by the dsRBMs contained in both proteins. If RDE-4 dsRBMs interact with Dicer, we predict the full-length RDE-4 dimer, as well as the monomeric truncation constructs, which lack the C terminus but contain both dsRBMs, should bind to Dicer. In this light, the inability of the monomeric C-terminal deletion constructs to reconstitute extract activity would result from the inability to dimerize, rather than faulty substrate or Dicer interactions.
A model for RDE-4 function in the initiation of RNAi
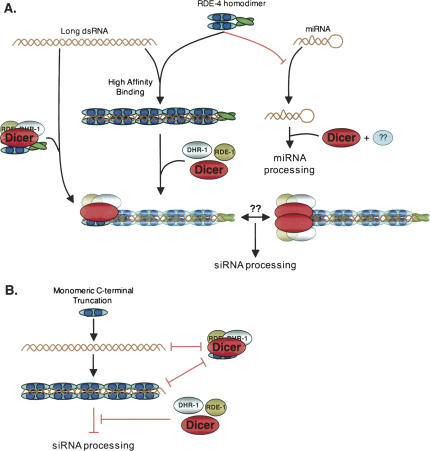
Our data suggest that smaller regions of dsRNA that occur naturally in a cell are not stably bound by RDE-4, preventing aberrant targeting by the RNAi machinery. Interestingly, Dicer processes miRNAs into their mature form from small stem–loop precursors, ∼22 bp in length, in an RDE-4-independent process (Bagga et al. 2005). Perhaps the low affinity of RDE-4 for short dsRNAs provides a simple explanation for its exclusion in miRNA processing (Fig. 7A, “miRNA” branch).
FIGURE 7.
Model of RDE-4 function during the initiation of RNAi. The C-terminal dimerization domain of RDE-4 is represented by a green oval and the dsRBMs by dark blue circles. (A) RDE-4 binds long dsRNA with high affinity. The left branch illustrates the siRNA production pathway, with RDE-4 shown in two complexes, one with dsRNA and one without. It is unknown if the Dicer/RDE-1/DRH-1 complex is preformed in the absence of RDE-4, so these proteins are shown separately in the center of the figure, before recruitment to the RDE-4-bound dsRNA. According to our model, RDE-4 dimerization is important for the assembly of active RDE-4/Dicer complexes via one of two proposed scenarios, proper Dicer recruitment to dsRNA, or facilitating Dicer dimerization. For simplicity, only one Dicer complex is illustrated for each scenario. The right branch depicts the miRNA production pathway. RDE-4 is potentially excluded from miRNA maturation due to its low affinity for short RNA duplexes. A dsRBP partner of Dicer during miRNA processing has yet to be reported for C. elegans, and its potential existence is represented by the blue oval with question marks. (B) C-terminal truncation proteins that cannot dimerize are able to bind dsRNA but are unable to form an active Dicer complex. See Discussion for details.
In contrast, based on our findings, long dsRNA is preferentially bound by preformed RDE-4 dimers potentially due to the higher affinity garnered by cooperative interactions (Fig. 7A, “long dsRNA” branch). The high-affinity binding of long dsRNA would effectively sequester “foreign RNA” and direct it specifically into the RNAi pathway. The model agrees with the observation that longer dsRNA is more potent in inducing RNAi in C. elegans (Fire et al. 1998; Parrish et al. 2000). As shown, the dsRNA/RDE-4 RNPs would then recruit the Dicer/RDE-1/DRH-1 complex via interactions between the dsRBMs of RDE-4 and Dicer. Previous work indicates that the RDE-4/Dicer/RDE-1/DRH-1 complex forms independent of dsRNA binding (Tabara et al. 2002) (Fig. 7A, far left branch). However, our finding that prebinding dsRNA with RDE-4 increases the efficacy of Dicer reconstitution, suggests that, at least sometimes, RDE-4 binds dsRNA first and then recruits the Dicer complex.
If two distinct RDE-4-containing complexes exist (dsRNA/RDE-4 and RDE-4/Dicer/RDE-1/DRH-1), the inhibition observed in extracts upon addition of high levels of recombinant RDE-4 could result from nonproductive binding to either complex, or because saturation of both complexes with RDE-4 precludes formation of the final active complex. According to our model, the C-terminal dimerization domain is essential for bringing the components of both complexes together to form the final active complex (dsRNA/RDE-4/Dicer/RDE-1/DRH-1). Thus, as we observe, truncated proteins that lack the C-terminal dimerization domain cannot reconstitute siRNA production. However, the truncated proteins can bind dsRNA, and possibly the Dicer/RDE-1/DRH-1 complex as well, thus explaining why they also inhibit siRNA production when added to extracts (Fig. 7B). After processing by Dicer, RDE-4 releases the siRNA product due to its low affinity for short duplexes, possibly handing it off to RDE-1, which is required downstream of siRNA production (Parrish and Fire 2001). The finding that RDE-4 binds ssRNA is intriguing, but its potential role in the RNAi pathway, if any, awaits further experimentation.
Our model presumes that the C terminus of RDE-4 is required for the assembly of a functional Dicer complex either by facilitating Dicer dimerization or its binding to dsRNA. Studies of recombinant human Dicer and Drosha suggest that both enzymes function through intramolecular dimerization of their two RNase III domains, arguing against homodimerization of either Dicer or Drosha in humans (Han et al. 2004; Zhang et al. 2004). However, the observation of a higher molecular weight complex of a size consistent with a Drosha/DGCR8 heterotetramer suggests that dimerization of RNase III enzymes might be critical for their in vivo functions (Han et al. 2004). Alternatively, dimerization of RDE-4 might be required for the proper recruitment of Dicer to dsRNA. In this scenario, one RDE-4 molecule of the homodimer binds RNA while the other binds Dicer, thereby forming a competent RNA-processing complex (Fig. 7A). Determining the stoichiometry of the C. elegans Dicer/RDE-4 complex will help in distinguishing between these two models.
MATERIALS AND METHODS
Construction, expression, and purification of RDE-4 protein constructs
The RDE-4 expression vector was constructed by PCR amplification of the RDE-4 open reading frame (ORF) from C. elegans cDNA. Forward and reverse primers including 5′ BamHI and a 3′ XhoI sites, respectively, were used to allow for ligation into the expression vector YEpTOP2PGAL1 (Giaever et al. 1988). The forward primer also included sequence for the 10-Histidine affinity tag and TEV protease recognition sequence. The following DNA oligonucleotides were used for amplification of RDE-4: RDE4BAMH1, GGGGGGGGATCCGTAACCATGTCACACCATCACCATCACCATCACCATCACCATGATTACGATATCCCAACGACCGAAAACCTGTATTTTCAGGGCGATTTAACCAAACTAACGTTTGAA; RDE4XHO1, GGGGGGCTCGAGTCAATCCGTGAAATCATAGGTGTT. Truncation 2 was constructed from a full-length RDE-4 PCR product that contained the third intron of the RDE-4 genomic sequence. This intron contained an in-frame stop codon, resulting in a truncated ORF with eight nonnative amino acids, GMPPNNCS, at the C terminus. Truncation 3 was PCR amplified from the RDE-4/YEpTOP2PGAL1 construct using the following primers: Trunc3BAMH1, GGGGGGGGATCCGTAACCATGTCACACCAT; Trunc3XHO1, GGGGGGCTCGAGTCAGTCTGAAAATCTTTTCTTGTC. The C terminus construct was PCR amplified from the RDE-4/YEpTOP2GAL1 construct using the following primers: CDom.pKM.Nde1, GGGGGGCATATGGTTGATGTGATTGAAAATTTG; CDom.pKM.BamH1, GGGGGGGGATCCTCAATCCGTGAAATCATAGGTGTT. The resulting PCR product was digested with restriction enzymes (Nde1 and BamH1) to allow ligation into the Escherichia coli expression vector pKM263 (Melcher 2000) which contains a Nco1 to Nde1 mutation.
RDE-4, Truncation 3, and Truncation 2 were expressed in the S. cerevisiae strain BCY123. Electrocompetent yeast cells were transformed with the expression vectors using an ECM 600 Electro Cell Manipulator (BTX Inc.), and plated on Complete Minimal–URA (CM-URA) + 2% dextrose plates. Ten milliliters of CM-URA + 2% dextrose media was inoculated with a single transformant and grown overnight at 30°C. Overnight cultures were diluted 1:100 in CM-URA + 3% glycerol + 2% lactate media and grown for ∼24 h at 30°C. Protein expression was induced by adding 3% galactose when cultures reached an O.D. of 1.0. Induction was for 6 h. Yeast cells were pelleted by centrifugation, washed once with ice-cold ddH2O, and stored at −80°C.
RDE-4, Truncation 3 and Truncation 2 were purified essentially as described (Macbeth et al. 2004), with the following modifications. For nickel column purification, buffer A (30 mM Tris pH 8.0, 1 mM 2-mercaptoethanol, 10% glycerol) contained 500 mM NaCl, and 30 mM imidazole. The column was washed in three successive steps with Buffer A containing 30 mM imidazole and NaCl concentrations of 500 mM, 250 mM, and 100 mM, respectively. Typical yields were 1–5 mg of pure protein per L of yeast culture.
The C terminus protein construct was expressed and purified as previously described (Macbeth et al. 2004).
RNA preparation
RNA (650 bp) was transcribed with T7 RNA polymerase from a template encoding sequence from the 10th exon of the C. elegans gene, fem-1. Fem-1 transcripts were labeled internally with α-32P ATP as previously described (Bass and Weintraub 1987), and purified on an 8% denaturing gel. Equimolar sense and antisense 650-nt ssRNAs were mixed in 10 mM Tris pH 7.5 and 40 mM KCl, boiled for 2 min, and allowed to anneal at room temperature for 1 h. Duplex RNA was purified on a 6% native gel. RNAs (104 bp) were transcribed from two separate (sense and antisense) T7 transcription templates to allow for complete base-pairing of the +1 G added by T7 RNA polymerase, resulting in the following sequence (sense): GGCAAUGAAAGACGGUGAGCUGGUGAUAUGGGAUAGUGUUCACCCUUGUUACACCGUUUUCCAUGAGCAAACUGAAACGUUUUCAUCGCUCUGGAGUGAAUACC. The 40-bp transcription templates were generated by annealing the following DNA oligos (written 5′ to 3′ with T7 promoter underlined): sense, TAATACGACTCACTATAGGGAAGCTCAGAATATTGCACAAGTAGAGCTTCTCGATCC and GGATCGAGAAGCTCTACTTGTGCAATATTCTGAGCTTCCCTATAGTGAGTCGTATTA. Antisense: TAATACGACTCACTATAGGATCGAGAAGCTCTACTTGTGCAATATTCTGAGCTTCCC and GGGAAGCTCAGAATATTGCACAAGTAGAGCTTCTCGATCCTATAGTGAGTCGTATTA. Transcribed 104 and 40 nt ssRNAs were dephosphorylated with calf intestinal phosphatase (New England Biolabs) followed by 5′ end-labeling with γ-32P (Amersham) using polynucleotide kinase (New England Biolabs), and then purified from 12% denaturing gels. Duplex purification was as for the 650-bp RNA. RNA (20 bp) was generated by annealing the following chemically synthesized ssRNAs: UGAGGUAGUAGGUUGUAUAGUU and CUAUACAACCUACUACCUCACC, resulting in a 20-bp duplex containing 2-nt 3′ overhangs. The ssRNAs were labeled and purified as described above.
Gel mobility shift assays
Gel-shift assays were performed as described (Bass et al. 1994), except for the following modifications. Twenty-microliter reactions containing 50 pM-labeled dsRNA and varying protein concentrations were incubated for 30 min at 4°C in a Mobility Shift Buffer containing a final concentration of 30 mM Tris pH 8.0, 150 mM NaCl, 1 mM 2-mercaptoethanol, and 10% glycerol. Gel shifts (650 bp) were assayed on 5% native gels; 104-, 40-, and 20-bp gel shifts were assayed on 8% native gels. All gels were electrophoresed at 4°C. Fraction bound was calculated as previously described (Bass et al. 1994). Briefly, radioactivity corresponding to dsRNAfree and dsRNAtotal (total radioactivity in entire lane) was quantified using a Molecular Dynamics PhosphorImager. All RNAs of slower mobility than dsRNAfree were considered as bound. The fraction bound = 1 − ([dsRNA]free/[dsRNA]total). Dissociation constants (Kds) and Hill coefficients were calculated as previously described (Henriet et al. 2005). All calculations assumed 100% active protein.
Competition experiments were performed as above, except constant concentrations of RDE-4 were included in each reaction as noted in Figure 3. Unlabeled competitor RNAs along with the labeled RNA were added to each reaction at indicated concentrations and allowed to equilibrate for 30 min. Gels were electrophoresed and analyzed as above.
Equilibrium sedimentation
Equilibrium sedimentation assays were performed as described (Andrews et al. 2005), except for the following modifications. The buffer used for all samples contained 30 mM Tris pH 8.0, 150 mM NaCl, and 1 mM 2-mercaptoethanol. RDE-4 samples (5, 10, and 15 μM) were centrifuged at 12,000 and 16,000 rpm. Truncation 2 and 3 samples (5, 10, and 20 μM) were centrifuged at 16,000 and 18,000 rpm. Cells were scanned radially with data resulting from five absorbance readings taken at 0.001-cm intervals. The partial specific volumes used for RDE-4, Truncation 3 and Truncation 2 were 0.7267, 0.7288, and 0.7262, respectively.
In vitro Dicer activity and reconstitution assays
C. elegans N2 and rde-4(ne337) strains were maintained as described (Brenner 1974; Sulston and Brenner 1974). Extracts were prepared from embryos isolated from 1L cultures of gravid N2 and rde-4(ne337) worms. Embryos were resuspended in an equal volume of lysis buffer (30 mM HEPES pH 7.4, 100 mM KOAc, 2 mM MgOAc, 50% glycerol, and 5 mM DTT), and lysed by 15 strokes with a dounce tissue grinder. The protein extract was cleared by centrifugation at 20,000g for 10 min. The supernatant was aliquoted and frozen at −80°C.
Dicer activity assays consisted of 50-μL reactions containing 40-μg protein extract and buffer final concentrations of 30 mM HEPES pH 7.4, 100 mM KOAc, 2 mM MgOAc, 1 mM DTT, 2 mM ATP, 10% glycerol, and 1 U/μL rRNasin (Promega). Reactions were incubated with 10 fmol of 32P-internally labeled 650-bp dsRNA for 1 h at 20°C. Reactions were then organic extracted with 1:1 phenol/CHCl3 and RNA products were ethanol precipitated after the addition of 20 μg of glycogen (Roche) as a carrier. The RNA was run on 12% denaturing gels at 40 W for ∼2 h, which were then fixed, dried, and visualized using a Storm 860 PhosphorImager (Molecular Dynamics). For Dicer reconstitution and inhibition reactions, RDE-4 protein was diluted in the reaction buffer described above and preincubated with RNA substrate for 30 min at 20°C, prior to the addition of embryo extracts. The reactions were then processed as described above.
ACKNOWLEDGMENTS
We thank members of the Bass lab for helpful discussions, former Bass lab member Dr. Scott Knight for developing the in vitro Dicer activity assay used in these studies and Dr. Lisa Joss for her assistance with sedimentation equilibrium experiments. We also thank Dr. Andrew Fire for critically reading this manuscript and Dr. Wes Sundquist for helpful discussions. This work was supported by funds to B.L.B. from the National Institutes of Health (GM067106) and the National Institutes of Health training grant (GM08537) to G.S.P. B.L.B. is a Howard Hughes Medical Institute investigator.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2338706.
REFERENCES
- Andrews D., Butler J.S., Al-Bassam J., Joss L., Winn-Stapley D.A., Casjens S., Cingolani G. Bacteriophage P22 tail accessory factor GP26 is a long triple-stranded coiled-coil. J. Biol. Chem. 2005;280:5929–5933. doi: 10.1074/jbc.C400513200. [DOI] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bass B.L., Weintraub H. A developmentally regulated activity that unwinds RNA duplexes. Cell. 1987;48:607–613. doi: 10.1016/0092-8674(87)90239-x. [DOI] [PubMed] [Google Scholar]
- Bass B.L., Hurst S.R., Singer J.D. Binding properties of newly identified Xenopus proteins containing dsRNA-binding motifs. Curr. Biol. 1994;4:301–314. doi: 10.1016/s0960-9822(00)00069-5. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey J. Gel retardation. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- Carmell M.A., Hannon G.J. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D.E., von Hippel P.H. Nucleic acid binding properties of Escherichia coli ribosomal proteinS1. II. Co-operativity and specificity of binding site II. J. Mol. Biol. 1978;122:339–359. doi: 10.1016/0022-2836(78)90194-8. [DOI] [PubMed] [Google Scholar]
- Epstein I.R. Cooperative and non-cooperative binding of large ligands to a finite one-dimensional lattice. A model for ligand–oligonucleotide interactions. Biophys. Chem. 1978;8:327–339. doi: 10.1016/0301-4622(78)80015-5. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Forstemann K., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G.N., Snyder L., Wang J.C. DNA supercoiling in vivo. Biophys. Chem. 1988;29:7–15. doi: 10.1016/0301-4622(88)87020-0. [DOI] [PubMed] [Google Scholar]
- Haase A.D., Jaskiewicz L., Zhang H., Laine S., Sack R., Gatignol A., Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha–DGCR8 complex in primary microRNA processing. Genes & Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriet S., Richer D., Bernacchi S., Decroly E., Vigne R., Ehresmann B., Ehresmann C., Paillart J.C., Marquet R. Cooperative and specific binding of Vif to the 5′ region of HIV-1 genomic RNA. J. Mol. Biol. 2005;354:55–72. doi: 10.1016/j.jmb.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Hiraguri A., Itoh R., Kondo N., Nomura Y., Aizawa D., Murai Y., Koiwa H., Seki M., Shinozaki K., Fukuhara T. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana . Plant Mol. Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- Hitti E.G., Sallacz N.B., Schoft V.K., Jantsch M.F. Oligomerization activity of a double-stranded RNA-binding domain. FEBS Lett. 2004;574:25–30. doi: 10.1016/j.febslet.2004.07.080. [DOI] [PubMed] [Google Scholar]
- Huang X., Hutchins B., Patel R.C. The C-terminal, third conserved motif of the protein activator PACT plays an essential role in the activation of double-stranded-RNA-dependent protein kinase (PKR) Biochem. J. 2002;366:175–186. doi: 10.1042/BJ20020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.L., Correia J.J., Yphantis D.A., Halvorson H.R. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys. J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R.F., Fischer S.E., Bernstein E., Sijen T., Hannon G.J., Plasterk R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans . Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski S.C., Paul L.S., Lonberg N., Newport J.W., McSwiggen J.A., von Hippel P.H. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: Experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986;25:1226–1240. doi: 10.1021/bi00354a006. [DOI] [PubMed] [Google Scholar]
- Laue T.M. Sedimentation equilibrium as thermodynamic tool. Methods Enzymol. 1995;259:427–452. doi: 10.1016/0076-6879(95)59055-2. [DOI] [PubMed] [Google Scholar]
- Liu Q., Rand T.A., Kalidas S., Du F., Kim H.E., Smith D.P., Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Macbeth M.R., Lingam A.T., Bass B.L. Evidence for auto-inhibition by the N terminus of hADAR2 and activation by dsRNA binding. RNA. 2004;10:1563–1571. doi: 10.1261/rna.7920904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J.D., von Hippel P.H. Theoretical aspects of DNA–protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Melcher K. A modular set of prokaryotic and eukaryotic expression vectors. Anal. Biochem. 2000;277:109–120. doi: 10.1006/abio.1999.4383. [DOI] [PubMed] [Google Scholar]
- Murchison E.P., Hannon G.J.2004. miRNAs on the move: miRNA biogenesis and the RNAi machinery Curr. Opin. Cell Biol. 16223–229. [DOI] [PubMed] [Google Scholar]
- Parrish S., Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans . RNA. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- Parrish S., Fleenor J., Xu S., Mello C., Fire A. Functional anatomy of a dsRNA trigger: Differential requirement for the two trigger strands in RNA interference. Mol. Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Patel R.C., Sen G.C. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G.A., Hartmann R., Qin J., Sen G.C. Modular structure of PACT: Distinct domains for binding and activating PKR. Mol. Cell. Biol. 2001;21:1908–1920. doi: 10.1128/MCB.21.6.1908-1920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham J.W., Sontheimer E.J. Molecular requirements for RNA-induced silencing complex assembly in the Drosophila RNA interference pathway. J. Biol. Chem. 2005;280:39278–39283. doi: 10.1074/jbc.M509202200. [DOI] [PubMed] [Google Scholar]
- Saito K., Ishizuka A., Siomi H., Siomi M.C. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J.E., Brenner S. The DNA of Caenorhabditis elegans . Genetics. 1974;77:95–104. doi: 10.1093/genetics/77.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H., Sarkissian M., Kelly W.G., Fleenor J., Grishok A., Timmons L., Fire A., Mello C.C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans . Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tabara H., Yigit E., Siomi H., Mello C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans . Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Tian B., Bevilacqua P.C., Diegelman-Parente A., Mathews M.B. The double-stranded-RNA-binding motif: Interference and much more. Nat. Rev. Mol. Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Zamore P.D. Perspective: Machines for RNAi. Genes & Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Matranga C., Haley B., Martinez N., Zamore P.D. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]