Abstract
The scanning model of RNA translation proposes that highly stable secondary structures within mRNAs can inhibit translation, while structures of lower thermal stability also affect translation if close enough to the 5′ methyl G cap. However, only fragmentary information is available about the dependence of translation efficiency in live mammalian cells on the thermodynamic stability, location, and GC content of RNA structures in the 5′-untranslated region. We devised a two-color fluorescence assay for translation efficiency in single live cells and compared a wide range of hairpins with predicted thermal stabilities ranging from −10 to −50 kcal/mol and 5′ G cap-to-hairpin distances of 1–46 bases. Translation efficiency decreased abruptly as hairpin stabilities increased from ΔG = −25 to −35 kcal/mol. Shifting a hairpin as little as nine bases relative to the 5′ cap could modulate translation more than 50-fold. Increasing GC content diminished translation efficiency when predicted thermal stability and cap-to-hairpin distances were held constant. We additionally found naturally occurring 5′-untranslated regions affected translation differently in live cells compared with translation in in vitro lysates. Our study will assist scientists in designing experiments that deliberately modulate mammalian translation with designed 5′ UTRs.
Keywords: translation, hairpin, structure, RNA, fluorescence, thermal stability
INTRODUCTION
RNA not only serves as templates for protein synthesis, but also catalyzes biochemical reactions and forms supramolecular complexes (Gesteland 1999; Sonenberg 2000; Brantl 2002). RNA has also been found to participate in gene regulation in prokaryotes and eukaryotes. In prokaryotes, riboswitches regulate bacterial metabolism by transcriptionally or translationally controlling genes involved in vitamin, amino acid, and purine metabolism (Nudler and Mironov 2004). Furthermore, small noncoding RNAs can regulate gene expression in bacteria (Brantl 2002). In eukaryotes, siRNAs and micro RNAs can regulate expression of genes through chromosomal silencing and post-transcription repression mechanisms (Westhof and Filipowicz 2005).
Translation initiation is a discrete step in eukaryotic gene regulation (McCarthy 1998; Sonenberg 2000; Gebauer and Hentze 2004). Secondary structural features of the mRNA 5′-untranslated region (UTR) are important to this regulation by affecting ribosomal recruitment and positioning at a favorable initiation codon (Gingras et al. 1999). The major rate-limiting step of translation initiation is determined by the binding of the 43S pre-initiation complex (composed of the 40S ribosomal complex, initiation factors eIF3, eIF1, eIF1A, eIF5, and eIF2-GTP-met-tRNA) to mRNA via the eIF4 initiation factor complex (eIF4E, eIF4A, eIF4G, and eIF4B) (Sonenberg and Dever 2003; Gebauer and Hentze 2004). The 5′ methyl G cap is initially recognized by initiation factor eIF4E. Under favorable translation conditions, eIF4G serves as a scaffold bridging the ribosome to the mRNA cap by binding eIF4E, eIF4A, and eIF3. Initiation factor eIF4A exhibits RNA helicase activity and is thought to assist the eIF4F complex in unwinding mRNA secondary structure, creating a binding site for the 43S initiation complex (Pestova et al. 2001; Takyar et al. 2005). The 43S complex then scans along the mRNA until it reaches a favorable initiation codon (Kozak 1991b).
Ribosomal scanning through structural barriers is known to be an important regulatory step, since several RNA structures located in the 5′ UTRs of mammalian mRNA transcripts were shown to affect translation efficiency (Kozak 1991b; Gebauer and Hentze 2004). Kozak first found that 5′ UTR noncoding regions of the α- and β-globin mRNAs impact their translation efficiency (Kozak 1994). Several groups demonstrated that the 5′ UTR of the ferritin receptor mRNA and the 3′ UTR of the ferritin mRNA each contain iron regulatory elements (IRE) which recruit IRE binding proteins to control cellular iron metabolism (Hentze et al. 1987; Rouault et al. 1988; Goossen and Hentze 1992; Eisenstein 2000). Werstuck and Green (1998) further showed that they could control translation by inserting small RNA aptamer motifs into the 5′ UTR of mRNAs to repress protein production upon ligand addition. Jiang and Lucy (2001) demonstrated the significance of regulation by 5′ UTRs when they showed that translation of the bovine growth hormone receptor can vary over an 80-fold range depending on the 5′ UTR splice variant.
Despite continuing efforts to investigate the role of RNA structure in mRNA translation, gaps remain in our understanding of how predicted ΔG and 5′-UTR cap-to-structure distance in cellular mRNAs control translation efficiency in live cells. Most of our current knowledge comes from Kozak's studies demonstrating the scanning model of mRNA translation (Kozak 1992). Specifically, various hairpins were inserted into the 5′ UTR of reporter constructs to demonstrate that such mRNA hairpins affected translation efficiency. These initial studies in Cos 7 cells found that hairpins with predicted thermal stability of −30 kcal/mol had no effect on translation, while hairpins of −50 kcal/mol reduced translation by 85%–95% (Kozak 1986). Kozak further showed that stem—loop cap-to-hairpin distance plays a role in translation efficiency (Kozak 1989). In these in vitro studies, structures of −30 kcal/mol reduced translation initiation when situated 12 bases from the cap; however, when cap-to-hairpin distance was increased to 52 bases, there was no translation inhibition (Kozak 1989). Taken together, Kozak found that translation efficiency is sensitive to structures proximal to the G cap and can efficiently scan through RNA structures further downstream if thermal stability is low to moderate. However, it remains unclear whether hairpin distance from the cap affects translation in live cells, whether the dependence on hairpin stability is graded or abrupt, and whether GC content has an effect independent of overall stability.
The current study expands upon the above classic work to investigate in more detail how RNA structures in the 5′ UTR of mRNA affect translation efficiency in live mammalian cells. We created a reporter vector to allow ratiometric analysis of translation efficiency in intact cells and thoroughly characterize the translation efficiency of a dynamic range of predicted hairpin thermal stabilities and distances from the 5′ G cap. In some cases, we found that a shift of only nine bases affects translation efficiency more than 50-fold. In addition, we found that stem GC content affects translation efficiency independent of predicted thermal stability and 5′ cap distance. Finally, we found that our live cell studies of translation efficiency of α- and β-globin 5′ UTRs are opposite of the in vitro lysate trends observed by Kozak. These differences show that live cells experiments to characterize 5′-UTR translation efficiency are critical for understanding how cis-acting mRNA elements regulate translation in intact cells.
RESULTS
Translation reporter system
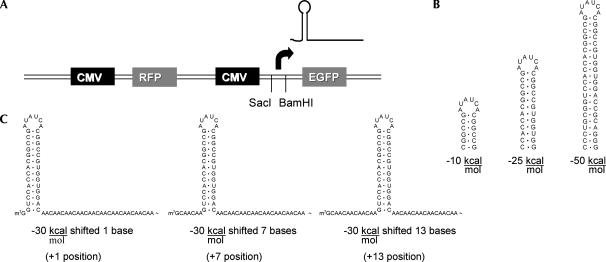
To investigate the effects that 5′-UTR structure had on mRNA transition, we constructed a translation reporter vector allowing facile manipulation of 5′-UTR sequences upstream of a GFP reporter protein. We cloned original enzyme sites encompassing a putative transcriptional start site. Using 5′-RACE assays, we determined RNA transcripts initiated primarily at base 837 of pcDNA3. This site was reverified with additional hairpin constructs using RNase protection and 5′-RACE procedures. We normalized for experimental variability by adding an RFP protein on the same vector. Transfection of this vector (GR1) into Cos 7 cells resulted in green and red emissions. A general map of vector GR1 is shown in Figure 1A.
FIGURE 1.
Placement of hairpin inserts in two color fluorescence vector. (A) Map showing placement of RFP and EGFP with respective CMV promoters. SacI and BamHI restriction sites encompass transcriptional start site for EGFP. The arrow indicates the relative location of the transcription start site. (B) Sample folding of the −10, −25, and −50 kcal/mol hairpin structures. A total of seven hairpins were utilized including −10, −20, −25, −30, −35, −40, and −50. (C) Example of the −30 kcal/mol hairpin placed at positions +1, +7, and +13. An insert length of 70 nucleotides was maintained with CAA repeats. Different hairpins were placed at positions +1, +4, +7, +10, +13, +16, +31, and +46. A complete listing of hairpins inserted into vectors GR1 and GR2 is shown in Supplemental Figure S1. Thermal stability predictions and hairpin structures were drawn with RNAstructure 3.7 (Mathews et al. 1999).
We next cloned a variety of RNA structures testing for thermal stability, cap-to-structure distance, and GC content. Although we could conceive of hundreds of structures to test, we started with hairpins due to their structural simplicity. We designed seven hairpin structures with predicted stabilities between −10 and −50 kcal/mol (Fig. 1B). Each hairpin was placed at distances between 1 and 16 bases from the cap, subsequently described as positions +1, +4, +7, +10, +13, and +16 (Fig. 1C). We tested effects of extended distance with hairpins −30 kcal/mol and greater, by repositioning cloning sites 30 bases downstream, positions +31 and +46. In addition, we tested GC content effects by designing four hairpins that maintained a predicted thermal stability near −30 kcal/mol and a +4 position. Insert lengths were maintained using CAA motifs as spacers, since these motifs are presumed to have no secondary structure (Hanson et al. 2003). We finally cloned control constructs composed entirely of CAA motifs for normalization to “structure-less RNAs”.
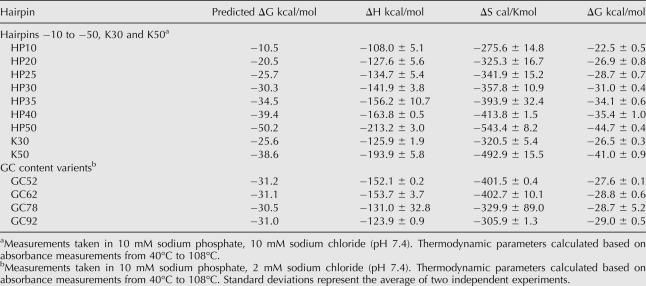
Predicted hairpin thermal stabilities (Mathews et al. 1999) were experimentally verified by measuring melting curves of transcribed RNAs. Empirical values showed the same trends as predicted, however, with a narrower range of ΔG values (Table 1). Supplemental Figure S1 (at http://www.tsienlab.ucsd.edu) summarizes all insert sequences used in this study.
TABLE 1.
ΔG Verification
Hairpin thermal stability and 5′-UTR distance influence translation efficiency
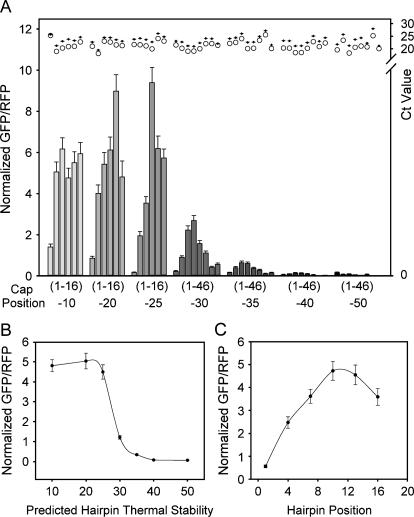
We evaluated how the hairpin library constructs affected translation efficiency by measuring the ratios of GFP to RFP with fluorescence microscopy. Figure 2A shows experimental translational efficiencies as a function of predicted thermal stability for all seven hairpin sets (−10 to −50 kcal/mol) with positions +1 to +46, a total of 50 constructs. Both thermal stability and cap-to-hairpin distance parameters affected translational efficiency, especially when the predicted hairpin stability was between −25 and −35 kcal/mol. Within this narrow −10 kcal/mol range, the dependence of translation on hairpin position was especially pronounced when hairpin structures were placed between +1 and +13. For example, in the −25 kcal/mol hairpin set, compared with a hairpin placed at +1, hairpins at +4 or +10 gave 11- or 50-fold higher translational efficiency. In addition, we observed a 9.6-fold difference between the +1 and +7 positions in the −30 kcal/mol hairpin set. Hairpins with predicted stabilities below −25 kcal/mol showed a slight distance dependence. Only when hairpins were placed at +1 did we observe inhibition effects. Translation efficiency was consistently inhibited with hairpins above −35 kcal/mol. In addition, distance effects were minimized. We did not observe an enhanced efficiency at +31 and +46. In fact, hairpins placed at extended distances translated less efficiently compared with structures at +16. This result differs from previous in vitro studies showing that a hairpin placed at +52 was beneficial for translation efficiency (Kozak 1989).
FIGURE 2.
Distance and thermal stability affect translation efficiency. (A) Each hairpin set is shown with increasing thermal stabilities (−10 to −50 kcal/mol) from left to right. Bars labeled (1–16) refer to hairpins placed at positions +1, +4, +7, +10, +13, and +16 from left to right for the −10, −20, and −25 kcal/mol hairpins. Bars labeled (1–46) refer to hairpins placed at positions +1, +4, +7, +10, +13, +16, +31, and +46 from left to right for the −30, −35, −40, and −50 kcal/mol hairpins. All values were normalized to their respective control CAA value. Error bars represent the standard error of the mean for 50 fields. Illustrated at top is quantitative RT–PCR data for each construct. Values represent the average of two Ct values for GFP (O) and RFP (+). (B) Average thermal stability effect on translation efficiency. Positions +1 to +16 for hairpins with predicted thermal stabilities between −10 and −25 kcal/mol and positions +1 to +46 for hairpins with predicted thermal stabilities between −30 and −50 kcal/mol were averaged. (C) Average distance effect on translation efficiency. Each position for hairpins with predicted thermal stabilities between −10 and −35 kcal/mol was averaged. Error bars are the standard error of the mean for B and C.
Quantitative RT–PCR cycle number results for GFP and RFP are displayed above each sample. There were no apparent trends in RNA stability providing verification that differences in constructs were due to translational efficiency. GFP to RFP transcript ratios remained consistent between samples, showing that the observed variations in translation could not be explained by variations in transcript abundance (Fig. 2A, top margin). This result also ruled out involvement of RNA silencing, since all but the −50 kcal/mol hairpins were below the 21-bp threshold needed to activate RNA silencing (Westhof and Filipowicz 2005), and translation was already minimal at −40 kcal/mol. We additionally verified that the observed trends were not due to the reporter protein itself, its 3′-UTR sequence, or nuclear export artifacts with RG1 vectors, by swapping the GFP reporter for an RFP and incorporating an IRES-driven GFP on the same transcript (Supplemental Fig. S2 and Protocol S5). We conclude that hairpin thermal stability and hairpin position dramatically and directly affect translation efficiency.
Figure 2B shows the results of increasing hairpin thermal stability by averaging all hairpin positions for each set. Between the stabilities of −10 and −25 kcal/mol, translation efficiencies remained consistent. As stabilities increased from −25 to −40 kcal/mol, we observed a dramatic inhibition of translation, a 40-fold drop over a narrow −15 kcal/mol range. Inhibition effects stabilized with a further increase to a thermal stability of −50 kcal/mol. These data suggest a narrow thermodynamic range, where RNA thermal stability dramatically affects translation efficiency. Figure 2C depicts the effect of hairpin position averaged over hairpin stabilities up to −35 kcal/mol. Translation efficiency increases linearly with hairpin position to +10, then drops slightly at greater distances.
GC content affects translation efficiency
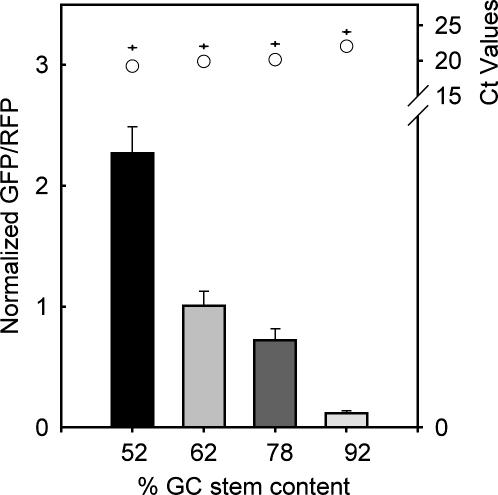
In addition to thermal stability and distance effects, we found translation efficiency was dependent upon RNA-hairpin GC stem content. We designed these hairpin constructs with different degrees of GC content while maintaining a position at +4 and thermal stability of −30 kcal/mol, empirically verified with absorbance measurements (Table 1). Although the insert length was maintained with CAA repeats, there was a 4-bp difference in hairpins with 52% and 92% GC content. As shown in Figure 3, as GC stem content was increased, translation efficiency decreased over 18-fold between stems with 52% and 92% GC content. Quantitative RT PCR (Fig. 3, top margin) and RG construct controls (Supplemental Figure S2 and Protocol S5) verified that observed trends were not due to the RNA abundance, the reporter protein itself, 3′-UTR sequence, or nuclear export artifacts. We conclude that RNA stem GC content affects translation efficiency.
FIGURE 3.
GC stem content affects translation efficiency in Cos7 cells. The percent stem GC content increases from left to right. Error bars represent the standard error of the mean for 50 fields. All values were normalized to their respective control CAA value. Illustrated at top is quantitative PCR data for each construct. Values represent the average of two Ct values for GFP (O) and RFP (+).
FACS analysis of hairpin effects on translation efficiency
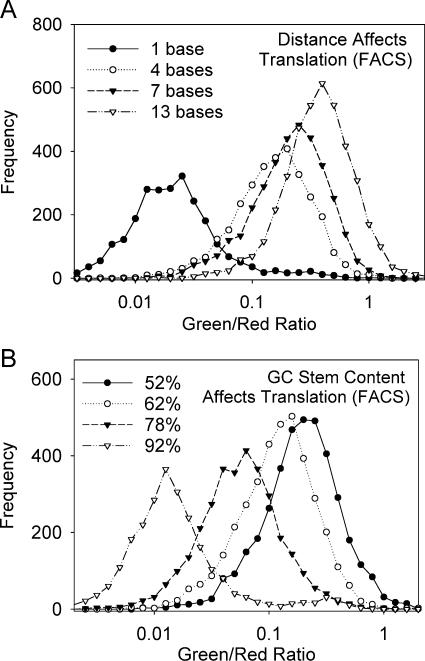
To examine distance and GC content differences at the single cell level with good statistics, we performed FACS analysis of the −25 kcal/mol hairpin set at positions +1 to +16 (Fig. 4A). Data curves for hairpins at +10 and +16 were not included for simplicity; however, they show similar curves at +7 and +13 positions, respectively. In addition, we evaluated population studies for hairpin constructs with GC contents between 52% and 92% (Fig. 4B). In both cases, FACS analysis confirmed microscopy data, since the mean translation efficiency increased as cap-to-hairpin distance was extended and GC stem content percentage was reduced. The populations remained unimodal in each case, verifying that differences were due to changes in the entire population as opposed to different quantities of finite values in a multimodal population.
FIGURE 4.
FACS analysis compares translation efficiencies in a population. (A) Cap-to-hairpin distance affects translation efficiency in a population. Data curves for positions +10 and +16 were not included, however, show similar curves as +7 and +13, respectively. (B) Stem GC content affects translation efficiency in a population.
Trends were similar with various cell types
To verify these trends were not cell-type specific, we compared translational efficiencies in primary mouse embryonic fibroblasts (MEFs) and Chinese hamster ovary cells (CHO). In both cases, we observed similar trends as Cos 7 cells with respect to thermal stability, hairpin position, and GC content (Supplemental Figs. S3 and S4).
We did, however, observe differences with each cell type analyzed. CHO cells (Supplemental Fig. S3) had a lower range of average ratios with respect to control constructs, values up to 2.5. Average thermal stability trends were similar to Cos7 cells, however, the −10 kcal/mol appeared to translate noticeably more relative to the −20 kcal/mol hairpin than with Cos7 cells. Average distance trends were similar to Cos7 cells to a +10 position. Ratio averages slightly dipped at +13, then increased again at +16. GC stem efficiencies differed only with the 62% and 78% stem content translating at similar levels.
MEF cells (Supplemental Fig. S4) had slightly different trends than Cos7 and CHO cell lines. RNA structure appeared to have minimal effects on thermal stability, since average levels between the −10 and −50 kcal/mol set only differed eightfold. The −10 kcal/mol hairpin set translated noticeably less than the −20 kcal/mol set. Furthermore, we observed distance effects up to the −50 kcal/mol hairpin (opposed to −35 kcal/mol with Cos7 and CHO cells lines). In addition, translation appeared to reach a peak at +13, with a local minimum at +10. Despite these small differences, the general effects of RNA hairpin thermal stability, cap-to-hairpin distance, and GC content on translation appear to be similar across three mammalian cell lines.
Translation efficiency with natural 5′ UTRs
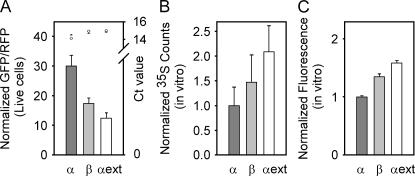
We next wanted to determine how the translation efficiencies of our designed hairpins compared with natural 5′ UTRs. Full-length 5′ UTRs were cloned by adding a restriction site as the second and third codons of GFP (vector GR3). We initially tested α- and β-globin 5′ UTRs previously evaluated in vitro; both predicted to have a hairpin in their 5′ UTR (Kozak 1994). For further comparison, we tested an optimized globin 5′ UTR named α ext (Kozak 1994). We found the α-globin 5′ UTR had the greatest translation efficiency followed by the β globin, then α ext, in ratios of 2.4:1.4:1, respectively (Fig. 5). This was a surprisingly opposite trend to that reported in vitro (Kozak 1994). Quantitative RT–PCR controls confirmed that trends were not due to mRNA stability. Instead, we found the α-globin mRNA was slightly less abundant in cells than the β-globin and α-ext mRNAs, so the translation efficiency of the α-globin 5′ UTR must be yet higher than the raw ratios alone. Efficiency values for the globin 5′ UTRs were much greater than the designed hairpin set. This is most likely due to weaker hairpins with predicted thermal stabilities of −8.4, −8.3, and −2 kcal/mol shifted 5, 7, and 34 bases from the cap for α globin, β globin, and α ext, respectively.
FIGURE 5.
Translation through natural 5′ UTRs differs in live cells vs. in vitro lysates. (A) Translation in live cells through α, β, or α ext globin 5′ UTRs was measured by fluorescence microscopy. Error bars represent the standard error of the mean for 40 fields. All values were normalized to their respective control CAA value. Illustrated at top is quantitative PCR data for each live cell construct. Values represent the average of two Ct values for GFP (O) and RFP (+). In vitro translation efficiency through α, β, or α ext globin was detected by either (B) radioactive 35S exposed to a PhosphorImaging screen, or (C) fluorescence intensity. Error bars represent the standard deviation of three independent experiments. Values were normalized to α globin.
Our present results differ from Kozak (1994) in translated protein (GFP and RFP vs. preCAT), assay method (ratiometric fluorescence measurements vs. 35S incorporation), and environment (in vitro translation vs. intact mammalian cells). To decide which factors mattered, we first measured translation in an in vitro system similar to Kozak's. We designed construct AgeI_Qc3c with an AgeI site just upstream of the first ATG of GFP. We then cloned a T7–5′ UTR with BamHI and AgeI sites (Suplemental Fig. S1). By 35S incorporation, we found ratios for the α , β, and α ext globins of 1:1.5:2 (Fig. 5B), almost identical to those reported previously (Kozak 1994). We also measured the fluorescence intensities of in vitro-translated fluorescent proteins (Fig. 5C). We found the same trends as with 35S, but less difference between α , β , and α ext globin 5′ UTRs with ratios of 1:1.4:1.6, respectively. Presumably, a fluorescence signal reflects properly translated and folded GFP, while the 35S experiment measures the incorporation of radioactive methionine into the protein regardless of secondary structure. Nevertheless, all methods agree on the rank order, α < β < α in vitro, in contrast to α > β > α ext in live cells.
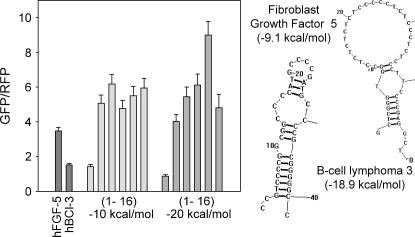
We additionally tested other natural 5′ UTRs similar in length, structure, and thermal stability to our hairpin sets. Figure 6 shows translation efficiencies of two natural mRNA transcripts adjacent to the −10 and −20 kcal/mol hairpin sets. The transcript coding for human fibroblast growth factor 5 (FGF-5) was predicted to form a stem–loop structure with a predicted stability of −9.1 kcal/mol and a +2 position. As predicted from the hairpin sets, FGF-5′s translation efficiency was between that of the −10 kcal/mol hairpin from positions +1 to +4. The transcript coding for human B-cell lymphoma 3 (BCL-3) was predicted to form a hairpin structure with an internal bulge, a predicted stability of −18.9 kcal/mol at position +3. As with FGF-5, BCL-3′s translation efficiency correlated with the designed hairpins sets with an efficiency level between that of the −20 kcal/mol hairpin from positions +1 to +4. We conclude that our designed hairpin sets should help predict translation efficiencies of natural 5′ UTRs with reasonably similar hairpin structures.
FIGURE 6.
Translation efficiency with natural 5′ UTRs. Translation efficiencies of fibroblast growth factor 5 (FGF-5) and B cell lymphoma 3 (BCL-3) are compared with translation efficiencies from the −10 and −20 kcal/mol hairpin sets (Fig. 2). Bars labeled (1–16) refer to hairpins placed at positions +1, +4, +7, +10, +13, and +16 from left to right for the −10 and −20 kcal/mol hairpins. Error bars represent the standard error of the mean for 20 fields. Shown to the right are predicted structures drawn with RNAstructure 3.7 (Mathews et al. 1999) for FGF-5 and BCL-3.
DISCUSSION
The goal of this study was to investigate the effects of hairpin distance, thermal stability, and GC content on mRNA translation efficiency in live cells and determine the correlation to natural hairpin structures. To our knowledge, this is the first study that systematically examines a wide range of both hairpin thermal stabilities and hairpin positions. In addition, this is the first report to show that GC content affects protein-translation efficiency independent of hairpin thermal stability and hairpin position.
In regard to hairpin thermal stability, we find that in live cells, increasing predicted mRNA thermal stability leads to a decrease in translation, which agrees with previous studies (Kozak 1986). We find the steepest falloff in translation efficiency occurs when hairpin-predicted stabilities increase from ΔG = −25 to −35 kcal/mol. This range differs slightly from previously reported values in live cells, which saw that a −30 kcal/mol hairpin translated efficiently, while a −50 kcal/mol hairpin inhibited translation (Kozak 1986). These predicted thermal stabilities, however, were based upon an algorithm published in the early 1970s (Tinoco et al. 1973). Current algorithms calculate that these −30 and −50 kcal/mol hairpins actually have thermal stabilities of −25.6 and −38.6 kcal/mol, respectively, showing Kozak's values to be consistent with our results (Mathews et al. 1999). Our experimentally determined Tms verify these predicted values as well (Table 1).
Our study also examines the significance of hairpin position on mRNA translation in live cells. Similar to in vitro studies, we confirm in live cells that mRNA structure is inhibitory when proximal to the 5′-mRNA cap (Kozak 1989). Previous in vitro studies found a +12 position inhibitory, while extending distances to +52 released this inhibition (Kozak 1989). In agreement with these findings, we find in live cells that RNA structures with positions +4 or less are inhibitory, perhaps perturbing mRNA recognition and binding of preinitiation complex initiation factors. In contrast to Kozak's studies, we find positions +7 to +13 bases actually enhance translation, provided that hairpin stabilities lie within the −25 to −40 kcal/mol range. This distance may promote favorable initiation complex formation to assist in ribosome binding and melting of mRNA structure. In further contrast to previous studies, we find that hairpins at +31 and +46 did not relieve hairpin inhibition, with exception of the weaker −30 kcal/mol hairpin at +31. In summary, we find that in live cells, mRNA cap-to-hairpin distances have the greatest control over translation when situated within the first 16 bases of the mRNA transcript.
An interesting finding was when translation efficiencies reached a basal minimum as hairpins approached predicted thermal stabilities of −50 kcal/mol, even when hairpins were situated at +1. We were surprised to find that any translation occurred under such inhibitory conditions. We postulate that under extremely inhibitory conditions, ribosomal shunting over or skipping over hairpin barriers may occur.
Although it is well known that GC content can affect translation initiation (Kozak 1991a), the discovery that stem GC content had a dramatic effect on mammalian translation efficiency independent of predicted thermal stability and hairpin position was quite unexpected. We observed an 18-fold difference between stems with 52% and 92% stem GC content. We later discovered a similar effect occurred in yeast with hairpins placed in the vicinity of the translation start codon (Vega Laso et al. 1993). It appears local stability per base has an effect on translation efficiency, since compact, thermal stable hairpins composed of GC bonds are harder to melt than hairpins composed of weaker AU bonds. With this information in mind, predicting the efficiency of translation becomes more complex. Perhaps hairpins with predicted thermal stabilities stronger than −50 kcal/mol may still be efficiently translated as long as stem GC content is relatively low. Consequently, less stable structures with high stem GC content may inhibit translation more than stable structures with low GC content. The simplest explanation is that the scanning ribosome does not melt the entire hairpin in a concerted event, but rather tries to bulldoze through the obstacle in a progressive, relatively local unzippering. High GC content confers high stability per base pair, perhaps exceeding the force (free energy per unit distance) available to the translation machinery.
Many natural mRNAs coding for regulatory proteins use 5′-UTR composition as a mechanism for gene regulation. As a result, many of these transcripts have structured 5′ UTRs with high GC content (between 70% and 90%) (Kozak 1991a). These transcripts code for transcription factors and DNA-binding proteins such as human NF-κB, mouse jun-D, and human GATA-1; ligand receptors such as human n-acetylcholine, human β2 adrenergic, and human insulin; and transcripts involved in signal transduction and growth control such as human bcl-3, human TGF-β1, and human PDGF (Kozak 1991a). Hershey and Miyamoto hypothesized that a cell can regulate gene expression by having different initiation potentials, or the ability to overcome structural barriers (Sonenberg 2000). When the initiation potential is low, highly structured mRNAs are poorly translated; however, when the potential is high, these mRNAs are allowed to translate efficiently (Sonenberg 2000). They believe the availability of initiation factor eIF4E can determine whether the initiation potential is high or low and is regulated by mechanisms including eIF2 phosphorylation and inhibitory actions of 4E-BP on eIF4E (Sonenberg 2000). As a result, cancer cells shift the initiation potential by overexpressing initiation factors such as eIF4E, allowing efficient translation through oncogenic transcripts including a cyclin D-1, c-Myc, FGF-2, and VEGF (de Benedetti and Harris 1999; Sonenberg 2000).
Finally, we compared translation efficiencies in live cells and in vitro lysates with α, β, and α ext globin 5′ UTRs (Kozak 1994). Interestingly, we found the opposite trends in translation efficiencies in live cells to those found in vitro. The α-globin mRNA translated the best in live cells and the worst in the in vitro lysates. We postulate these differences may be due to protein factors found in live cells not present in the lysates, or because coupled transcription—translation in live cells may follow different rules from translation divorced from transcription in vitro.
A comparable amount of in vitro and in vivo studies in yeast have also been dedicated to studying the effects of RNA structure, position, and thermal stability on mRNA translation (Vega Laso et al. 1993; Koloteva et al. 1997; McCarthy 1998). Consistent with these studies, we find RNA structures with stable secondary structures generally inhibit RNA translation with little regard for 5′-UTR position. Furthermore, we also find that less stable RNA structures can inhibit RNA translation when placed in a favorable position. However, in yeast, this position effect is dependent upon the RNA structure's proximity to the translation start site, not the 5′ G cap. Potentially higher eukaryotes are more sensitive to the initial interaction between the mRNA cap and the ribosome, whereas lower eukaryotes are more sensitive to ribosomal scanning (Vega Laso et al. 1993).
A study by Mattick estimates that 98% of the human genome is made up of nonprotein-coding RNA sequence. He hypothesizes that higher eukaryotes use this excess of noncoding RNA to regulate cellular processes (Mattick 2003, 2004). Our findings may lend support to Mattick's theory showing the effects of RNA structure on protein translation. In 1994, Kozak postulated that structure in 5′ UTR of mRNAs could act as regulatory sequences to enhance translation by allowing proteins to bind and shift structure downstream (Kozak 1994). Functioning in a similar capacity, noncoding RNAs could also bind these structures and release thermodynamic constraints, allowing for translation enhancement. This underscores the significant effect that a noncoding sequence of mRNA could have on gene expression.
In addition to controlling natural processes in the cell, these mechanisms we have used to modulate mRNA translation will also be useful in the design of artificial systems for controlling transgene expression in eukaryotic cells. In bacteria, a method has recently been described to control gene expression using small noncoding RNAs to modulate secondary structure at the Shine-Dalgarno site (Isaacs et al. 2004; Davidson and Ellington 2005). Bayer and Smolke (2005) recently modulated trans-binding RNAs to control reporter protein expression in yeast.
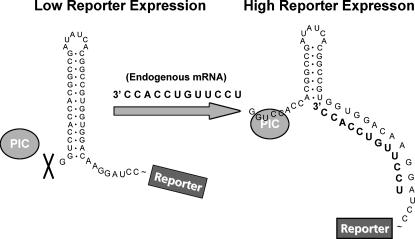
Our results suggest that a similar strategy of controlling gene expression through modulation of RNA secondary structure in cis may also be possible in mammalian cells. Figure 7 depicts a potential design in which a hairpin with moderate thermal stability (between −25 and −35 kcal/mol) is placed near the cap in the 5′ UTR of a reporter transcript. Upon strand hybridization, cap proximal structures would be shifted downstream, releasing cap proximal bases, allowing translation initiation to occur. Our studies indicate that as much as a 50-fold difference exists between −25 kcal/mol hairpin structures placed 1 base and 10 bases from the cap. In theory, these differences could be harnessed with this system. Such a strategy could be useful, for example, in rendering expression of a reporter gene or therapeutic transgene, dependent on the presence of specific endogenous mRNAs or possibly organic compounds.
FIGURE 7.
General strategy for mRNA modulation. In the low reporter expression state, an RNA structural blockade inhibits the ribosomal pre-initiation complex (PIC) from binding. As a result, the ribosome never reaches the translation start site for the reporter protein. Upon binding RNA (presumably endogenous mRNA), the structural blockade is altered, potentially by shifting hybridized RNA structures downframe. This shift leads to PIC binding, allowing the ribosome to reach the translation start-site translating reporter protein.
MATERIALS AND METHODS
Materials
Enzymes T4 PNK, T4 DNA ligase, and T7 RNA polymerase were purchased from New England Biolabs; Pfu Turbo and Lipofectamine 2000 were from Invitrogen; Taq DNA Polymerase was from Promega; Fugene 6 was from Roche; DNA Oligos were from Allele Biotechnology and Integrated DNA Technology; DNA and RNA isolation kits were from QIAGEN; 5′RACE reagents, RNA Surescript, and Rabbit Reticulate Lysates were from Ambion.
Vector construction
Vector Qc3c was created from an EGFP-containing PCDNA3 vector to contain unique restriction sites on both sides of the putative transcriptional start site. A Quickchange with primers A and B removed the SacI site downstream from the SP6 promoter, while a Quickchange with primers C and D removed SacI and BamHI sites in the cloning region. An original SacI site remained 10 bases upstream of the putative transcriptional start site. A BamHI site was next introduced nine bases downstream of the putative transcriptional start site by PCR with primers E and F.
A: 5′-CACCTAAATGCTAGACCTCGCTGATCAGCCT
B: 5′-AGGCTGATCAGCGAGGTCTAGCATTTAGGTG
C: 5′-GCTTGGTACCGAGCTGCGATCCGAATTCGCCACC
D: 5′-GGTGGCGAATTCGGATCGCAGCTCGGTACCAAGC
E: 5′-TGGGTTCTCTAGTTAGC
F: 5′-GGATCCACTGGCTTATCGAAATT
Vector GR1 was created by cloning a dimeric red fluorescent protein (RFP) derivative (Campbell et al. 2002) or mCherry (Shaner et al. 2004) into Qc3c, thus creating a plasmid expressing both green and red proteins, each driven by separate CMV promoters. To maintain original SacI and BamHI sites, an intermediate vector Qc4c was cloned removing SacI and BamHI sites from Qc3c with cassette insertion of annealed oligos G and H. RFP or mCherry was amplified with primers I and J and cloned into vector Qc4c using EcoRI and XbaI sites. Finally, the entire gene encompassing CMV, coding sequence, and BGH poly(A) tail was amplified with primers K and L and inserted into vector Qc3c with MfeI sites.
G: 5′-GTCTGGTAACTAGAGAACCCAC
H: 5′-GATCGTGGGTTCTCTAGTTAGCCAGACAGCT
I: 5′-CGGAATTCGCCACCATGGTGAGCAAGGGCGA
J: 5′-GGCTCTAGATTACTTGTACAGCTCGTC
K: 5′-GTCGTAGAACACGTAGTGCGCGAGCAAAAT
L: 5′-GGCACTCAATTGGCCGAAATCCCCAAAAT
Vector GR2 was created by inserting annealed oligos M and N into vector GR1, shifting original SacI and BamHI restriction sites 30 bases downstream.
M: 5′-GTCTGGCTAACTAGGCGCAACAACAACAACAACAACAACAAGAGCTCCAACAACCCTGCGGTCCACCCTACGGCCTGG
N: 5′-GATCCCAGGCCGTAGGGTGGACCGCAGGGTTGTTGGAGCTCTTGTTGTTGTTGTTGTTGTTGTTGCGCCTAGTTAGCCAGACAGCT
Vector GR3 was created by PCR amplification of an original AgeI restriction site after the first ATG of the GFP-coding frame with primers O and P. This fragment was then inserted into vector Qc3c with BamHI and XbaI restriction sites. RFP was added in using MfeI restriction sites as described above.
O: 5′-GATCCGAATTCGCCACCATGACCGTTGTGAGCAAGGGCGAGGAGC
P: 5′-CATTTAGGTGACACTA
Determining the transcription start site
Construct HP1 was created by ligating annealed oligos Q and R into the GR1 vector. A total of 4 μg of HP1 plasmid was transfected with Fugene 6 into a 70% confluent 10-cm dish of Cos7 cells for 24 h. RNA was then isolated using RNAeasy kit, and a rapid amplification of cDNA ends (RACE) procedure adapted from Ambion identified 5′-RNA ends. Isolated RNA was initially treated with Calf Intestinal Phosphatase (CIP) and Tobacco Acid Pyrophosphatase (TAP) to remove 5′ phosphates from degraded nucleic acids and 5′ G caps from full-length RNAs, respectively. The 5′-RACE adaptor S was next ligated to full-length mRNA transcripts with T4 RNA ligase. M-MLV Reverse Transcriptase next produced cDNAs. Nested PCR with outer primers T and U and inner primers V and W amplified cDNA products with added PstI and XbaI restriction sites. Finally, amplified products were cloned into a puc18 vector and sequenced.
Q: 5′-CTCTGGCTAACTAGGCCGGTTCGGTGGCCGAGCTTCGGCCACCTATCGGCCAG
R: 5′-GATCCTGGCCGATAGGTGGCCGAAGCTCGGCCACCGAACCGGCCTAGTTAGCCAGAGAGCT
S: 5′-GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA
T: 5′-CCGGAATTCGATGTTGCCGTCCTCCTTGA
U: 5′-GCTGATGGCGATGAATGAACACTG
V: 5′-GAACTGCAGGAACACTGCGTTTGCTGGCTTTGATG
W: 5′-CGCTCTAGAGTTGCCGTCGTCCTTGAAGAAGAT
Hairpin library construction
Each set of oligos was gel purified, phosphorylated with T4 PNK, and annealed before ligating into CIPed GR1, GR2, or GR3 vector digested with SacI and BamHI or GR3 digested with SacI and AgeI. Oligos ligated into vectors had the general sequence of oligos IA and IB for GR1, IC and ID for GR2, and IE and IF for GR3. A complete listing of inserts is shown in Supplemental Figure S1.
IA: 5′-CTCTGGCTAACTAGGC—insert 55/70 bases—G
IB: 3′-TCGAGAGACCGATTGATCCG—insert 55/70 bases—CCTAG
IC: 5′ C—insert 63 bases—G
ID: 3′-TCGAG—insert 63 bases—CCTAG
IE: 5′-CTCTGGCTAACTAGGC—5′-UTR insert—ATGA
IF: 3′-TCGAGAGACCGATTGATCCG—5′-UTR insert—TACTGGCC
Tm measurements
RNA was transcribed with T7 RNA polymerase and double-stranded DNA oligos containing a T7 promoter site 5′ of the hairpin sequence. RNA products were PAGE purified and added to a concentration of 0.105 mg/mL in 10 mM Sodium Phosphate buffer (pH 7.4) with either 2 or 10 mM NaCl. Absorbance at 260 was measured on a Beckman DU640 Spectrophotometer from 25°C to 108°C. Thermodynamic parameters ΔH, ΔS, and ΔH were calculated using Meltwin 3.5.
Cell culture
Cos 7 cells were maintained in DMEM containing 4.5 g/L glucose supplemented with 10% FBS and 1% penicillin/streptomycin. One day prior to transfection, cells were seeded into 12-well plates or imaging dishes. The next day, cells were transfected with 750 ng of DNA and 4 μL of lipofectamine 2000 reagent. Twenty-four hours later, cells were analyzed by fluorescence microscopy or FACS analysis.
Fluorescence microscopy
Cells in imaging dishes were washed once with HBSS and left in 2 mLs of the same medium. Images were acquired using a Zeiss Axiovert with a 40X NA 1.3 objective. GFP was analyzed using 505 nm dichroic, 480/30 nm excitation, and 525/25 nm emission filters. RFP was analyzed using 560 nm dichroic, 540/55 nm excitation, 595/50 nm emission filters. Data was analyzed with Metafluor Software. Average green to red ratios were computed after subtracting a background region from the green and red images. Ratios for at least 20 fields were averaged and normalized to the “structureless” constructs HPO, HPO2, or HPO_GR2.
FACS analysis
Cells on each 12-well dish were washed once with DPBS and released with 100 μL of trypsin solution. Cells were further diluted with 400 μL of HBSS. FACS analysis was performed on a BD FACSVantage DiVA. GFP was excited with a 488-nm laser and emission collected using a 510/21-nm emission filter. MCherry was excited at 565 nm and analyzed with a 615/40-nm emission filter. A total of 10,000 events were collected for each construct. Events greater than two standard deviations from the average GFP and mCherry emission of background Cos7 cells were further analyzed. The GFP to mCherry ratio was computed after background subtraction. Each data point represents the number of events that occurred within ratio bins of 0.04. An optimized mCherry aided in FACS analysis with its enhanced quantum yield and red-shifted emission (Shaner et al. 2004). Its fluorescence intensity remained consistent in all populations.
Quantitative RT—PCR analysis
RNA from each 12-well dish was isolated using QIAGEN's RNAeasy kit. CDNA was synthesized using Ambion's M-MLV Reverse Transcriptase with random decamer primers. Primers GF and GR and Taqman primer GT detected GFP transcripts, while primers RF and RR and Taqman RT detected RFP transcripts. Real time PCR analysis was performed on an Applied Biosystems ABI Prism 7700 Sequence Detection System equipped with software version 1.6.3.
GF: 5′-AGCAAAGACCCCAACGAGAA
GR: 5′-TTACTTGTACAGCTCGTCCATGCCG
GT: 5′-CGCGATCACATGGTCCTGCTGG
RF: 5′-CCACCAGGCCCTGAAGCT
RR: 5′-GGCTTCTTGGCCATGTAGATG
RT: 5′-AAGGACGGCGGCCACTACCTGGT
In vitro transcription/translation
Double-stranded DNA oligos were ligated into AgeI-modified Qc3c vector with the general sequence of oligos IJ and IK. A total of 1 μg of vector linearized with BamHI and XbaI restriction sites was added to a Surescript Transcription kit reaction to yield 5′ G-capped mRNA transcripts. A total of 360 μg of PAGE-purified RNA was then added to each 25 μL of Rabbit Reticulate lysate reaction with or without 35S, and allowed to translate for 2 h at 30°C. Protein quantities were determined by fluorescence spectroscopy or incorporation of 35S. Fluorescence measurements were taken on a Spex Fluorolog, diluting the translation mix 1:60, and exciting GFP at 470 and reading emission from 480 to 560. Background fluorescence was subtracted from each sample. Samples with incorporated 35S were run on SDS-PAGE and exposed to a PhosphorImaging screen for 3 d. Counts were measured on a STORM 860.
IJ: 5′-GATCCAATACGACTCACTATAG—5′-UTR insert—ATGA
IK: 5′-GTTATGCTGAGTGATATC—5′-UTR insert—TACTGGCC
ACKNOWLEDGMENTS
We thank Drs. Stephen Adams, Ben Giepmans, Simpson Joseph, Michael Lin, Amy Pasquinelli, Yitzhak Tor, Jiwu Wang, Nicholas Webster, Mike Whitney, and Charles Zuker for their helpful thoughts and suggestions with the manuscript; Paul Steinbach for help with fluorescence imaging; Coyt Jackson, Brent Martin, and Qing Xiong for help with the FACS analysis; and Nick Greco for help with Tm measurements. This work was supported by HHMI, NIH GM72033, NIH 27177, and DOE DE-FG03-01ER63276 NS27177, and the UCSD Pharmacology Training Grant.
Footnotes
SUPPLEMENTAL MATERIAL Supplemental material can be found at http://www.tsienlab.ucsd.edu.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2309906.
REFERENCES
- Bayer T.S., Smolke C.D. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- Brantl S. Antisense-RNA regulation and RNA interference. Biochim. Biophys. Acta. 2002;1575:15–25. doi: 10.1016/s0167-4781(02)00280-4. [DOI] [PubMed] [Google Scholar]
- Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E.A., Ellington A.D. Engineering regulatory RNAs. Trends Biotechnol. 2005;23:109–112. doi: 10.1016/j.tibtech.2005.01.006. [DOI] [PubMed] [Google Scholar]
- de Benedetti A., Harris A.L. eIF4E expression in tumors: Its possible role in progression of malignancies. Int. J. Biochem. Cell Biol. 1999;31:59–72. doi: 10.1016/s1357-2725(98)00132-0. [DOI] [PubMed] [Google Scholar]
- Eisenstein R.S. Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu. Rev. Nutr. 2000;20:627–662. doi: 10.1146/annurev.nutr.20.1.627. [DOI] [PubMed] [Google Scholar]
- Gebauer F., Hentze M.W. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R.F., Cech T.R., Atkins J.F. The RNA world: The nature of modern RNA suggests a prebiotic RNA. Cold Spring Harbor Laboratory Press; New York: 1999. [Google Scholar]
- Gingras A.C., Raught B., Sonenberg N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Goossen B., Hentze M.W. Position is the critical determinant for function of iron-responsive elements as translational regulators. Mol. Cell. Biol. 1992;12:1959–1966. doi: 10.1128/mcb.12.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson S., Berthelot K., Fink B., McCarthy J.E., Suess B. Tetracycline-aptamer-mediated translational regulation in yeast. Mol. Microbiol. 2003;49:1627–1637. doi: 10.1046/j.1365-2958.2003.03656.x. [DOI] [PubMed] [Google Scholar]
- Hentze M.W., Caughman S.W., Rouault T.A., Barriocanal J.G., Dancis A., Harford J.B., Klausner R.D. Identification of the iron-responsive element for the translational regulation of human ferritin mRNA. Science. 1987;238:1570–1573. doi: 10.1126/science.3685996. [DOI] [PubMed] [Google Scholar]
- Isaacs F.J., Dwyer D.J., Ding C., Pervouchine D.D., Cantor C.R., Collins J.J. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- Jiang H., Lucy M.C. Variants of the 5′-untranslated region of the bovine growth hormone receptor mRNA: Isolation, expression and effects on translational efficiency. Gene. 2001;265:45–53. doi: 10.1016/s0378-1119(01)00356-0. [DOI] [PubMed] [Google Scholar]
- Koloteva N., Muller P.P., McCarthy J.E. The position dependence of translational regulation via RNA-RNA and RNA-protein interactions in the 5′-untranslated region of eukaryotic mRNA is a function of the thermodynamic competence of 40 S ribosomes in translational initiation. J. Biol. Chem. 1997;272:16531–16539. doi: 10.1074/jbc.272.26.16531. [DOI] [PubMed] [Google Scholar]
- Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of vertebrate mRNA sequences: Intimations of translational control. J. Cell Biol. 1991a;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991b;266:19867–19870. [PubMed] [Google Scholar]
- Kozak M. Regulation of translation in eukaryotic systems. Annu. Rev. Cell Biol. 1992;8:197–225. doi: 10.1146/annurev.cb.08.110192.001213. [DOI] [PubMed] [Google Scholar]
- Kozak M. Features in the 5′ non-coding sequences of rabbit α and β-globin mRNAs that affect translational efficiency. J. Mol. Biol. 1994;235:95–110. doi: 10.1016/s0022-2836(05)80019-1. [DOI] [PubMed] [Google Scholar]
- Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Mattick J.S. Challenging the dogma: The hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- Mattick J.S. RNA regulation: A new genetics? Nat. Rev. Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- McCarthy J.E. Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E., Mironov A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva V.G., Lomakin I.B., Pilipenko E.V., Shatsky I.N., Agol V.I., Hellen C.U. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault T.A., Hentze M.W., Caughman S.W., Harford J.B., Klausner R.D. Binding of a cytosolic protein to the iron-responsive element of human ferritin messenger RNA. Science. 1988;241:1207–1210. doi: 10.1126/science.3413484. [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Dever T.E. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Hershey J.W.B., Mathews M.B. Translational control of gene expression, Cold Spring Harbor Laboratory Press, Cold Spring Harbor; New York: 2000. [Google Scholar]
- Takyar S., Hickerson R.P., Noller H.F. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr., Borer P.N., Dengler B., Levin M.D., Uhlenbeck O.C., Crothers D.M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat. New Biol. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Vega Laso M.R., Zhu D., Sagliocco F., Brown A.J., Tuite M.F., McCarthy J.E. Inhibition of translational initiation in the yeast Saccharomyces cerevisiae as a function of the stability and position of hairpin structures in the mRNA leader. J. Biol. Chem. 1993;268:6453–6462. [PubMed] [Google Scholar]
- Werstuck G., Green M.R. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- Westhof E., Filipowicz W. From RNAi to epigenomes: How RNA rules the world. Chembiochem. 2005;6:441–443. doi: 10.1002/cbic.200400418. [DOI] [PubMed] [Google Scholar]