Abstract
The ribosome consists of two unequal subunits, which associate via numerous intersubunit contacts. Medium-resolution structural studies have led to grouping of the intersubunit contacts into 12 directly visualizable intersubunit bridges. Most of the intersubunit interactions involve RNA. We have used an RNA modification interference approach to determine Escherichia coli 16S rRNA positions that are essential for the association of functionally active 70S ribosomes. Modification of the N1 position of A702, A1418, and A1483 with DMS, and of the N3 position of U793, U1414, and U1495 with CMCT in 30S subunits strongly interferes with 70S ribosome formation. Five of these positions localize into previously recognized intersubunit bridges, namely, B2a (U1495), B2b (U793), B3 (A1483), B5 (A1418), and B7a (A702). The remaining position displaying interference, U1414, forms a base pair with G1486, which is a part of bridge B3. We contend that these five intersubunit bridges are essential for reassociation of the 70S ribosome, thus forming the functional core of the intersubunit contacts.
Keywords: 16S rRNA, 30S subunit, ribosome, intersubunit bridges, modification interference
INTRODUCTION
The ribosome is a two-subunit ribonucleoparticle with the function of decoding the genetic messages (in the small subunit), and catalyzing the formation of peptide bonds (in the large subunit). The reasons behind the two-subunit nature of the ribosome are not entirely clear but likely include the need for the accurate initiation of translation (Risuleo et al. 1976; Hartz et al. 1989) and the facilitation of translocation of the reaction products from the acceptor to the donor to the exit site (Pestka 1969; Frank and Agrawal 2000).
The presence of rRNA in intersubunit contacts was first detected by chemical footprinting (Chapman and Noller 1977; Herr and Noller 1979) and modification interference methods (Herr et al. 1979). The first unambiguously located intersubunit contact was placed by chemical cross-linking between the 23S rRNA helix 69 and the 16S rRNA helices 44 and 45 (Mitchell et al. 1992). More recent chemical footprinting studies determined the identities of several rRNA positions, which are protected by the formation of 70S ribosome (Merryman et al. 1999a,b). These are generally in good correspondence with the intersubunit bridges, derived from the crystallographic model of the Thermus thermophilus ribosome (Yusupov et al. 2001) and the Escherichia coli ribosome (Schuwirth et al. 2005) and with cryo-EM models of the E. coli (Gabashvili et al. 2000; Gao et al. 2003) and Saccharomyces cerevisiae (Spahn et al. 2001, 2004) ribosomes. Recently, Ghosh and Joseph (2005) identified 28 nonbridging oxygens in the backbone of 16S rRNA whose substitution with sulfur led to reduced ability of 70S ribosome formation by reconstituted phosphorothioate-containing 30S subunits. Surprisingly, only one of the positions interfering with 70S formation (C770) falls into an intersubunit bridge as defined by structural studies (Yusupov et al. 2001; Gao et al. 2003).
The structural studies define, in addition to RNA–RNA contacts, several protein–RNA and protein–protein interactions. The model of Yusupov et al. (2001) incorporates 12 intersubunit bridges, which translate into more than 30 individual interactions between the 30S and the 50S subunits. The bridges seem to be largely conserved between the three kingdoms of life (Spahn et al. 2001; Gao et al. 2003). The central intersubunit areas (i.e., those closer to the reaction center in the LSU and to the decoding center in the SSU) are occupied by RNA-only bridges, while protein-containing bridges are more peripheral. Centrally located bridges contribute >80% of the individual intersubunit contacts (Gao et al. 2003). Gao et al. (2003) and, more recently, Spahn et al. (2004) describe changes in intersubunit bridges in translocation upon EF-G•GTP binding, where central RNA–RNA contacts are preserved while more peripheral RNA–protein and protein–protein contacts are often broken or rearranged. While it is important to keep the subunits together during translocation, the 70S ribosome is dissociated during termination of protein synthesis by the concerted action of RRF and EF-G (Karimi et al. 1999; Hirokawa et al. 2005). Structural models of RRF bound to post-termination 70S ribosomes and 50S subunits suggest that RRF dissociates the 70S ribosome by specifically disrupting the central intersubunit bridges B2a and B3 (Gao et al. 2005; Wilson et al. 2005). In addition, dynamics of the intersubunit bridges could play a role in tRNA decoding by relaying information of a successful decoding event in the small subunit to the GTPase-associated center in the large subunit thus helping to orchestrate timely GTP hydrolysis on EF-Tu (Belanger et al. 2004). Recently Hennelly et al. (2005) used chemical modification combined with quench-flow technique to provide time-resolved details of 16S rRNA structural changes that occur as bridges are formed between the ribosomal subunits as they associate. Time-resolved data allowed them to construct a model wherein some regions of the rRNA bind initially and generate structural changes that allow the remaining contacts to be made for complete association of the two subunits. The first interactions between subunits are made by 16S rRNA positions A892, A908, A909 (900 region, helix 27), and A1408 (bridge B2a, helix 44). Then follows protection of the 16S rRNA bases A1418 (bridge B5, helix 44) and A1413 (helix 44) by association with the 50S subunit. At a much slower rate, subunit association also protects A702 (B7a, helix 23) and A1441 (helix 44).
Mutations in the 16S rRNA 900 tetraloop impair subunit association and translational fidelity, in agreement with the location of this tetraloop at the subunit interface and close to the decoding center (Belanger et al. 2004). Indeed, the 900 tetraloop bases A900 and A901 contribute to the intersubunit bridge B2c. Bridge B2c, which connects the body of the 30S subunit to the 50S subunit, is not displaced upon EF-G•GTP binding and could therefore contribute to the maintenance of the subunit association during the ratchet-like intersubunit movement (Gao et al. 2003; Valle et al. 2003; Spahn et al. 2004).
Maiväli and Remme (2004) identified by modification interference three adenosines in the E. coli 23S rRNA, N1-methylation of which strongly reduced the ability of 50S subunits to form 70S ribosomes. These adenosines, which are essential for subunit association in vitro, have been assigned to intersubunit bridges B2a (A1912 and A1918) and B4 (A715) (Yusupov et al. 2001; Gao et al. 2003). Here we extend the modification interference studies of 70S ribosome formation to cover the E. coli 16S rRNA. We report that modification of the N1 positions of A702, A1418, and A1483 or of the N3 positions of U793, U1414, and U1495 in E. coli 30S subunits strongly interferes with 70S ribosome formation. All interferences fall either into, or close to, designated intersubunit bridges.
RESULTS
Experimental system
The crux of the modification interference methodology is to assemble functional complexes using randomly modified macromolecules, and subsequently physically separate active and inactive subpopulations. Modifications at positions whose presence is tolerated in the inactive subpopulation and counterselected in the active subpopulations are termed as interfering with the functionality of the macromolecule in question.
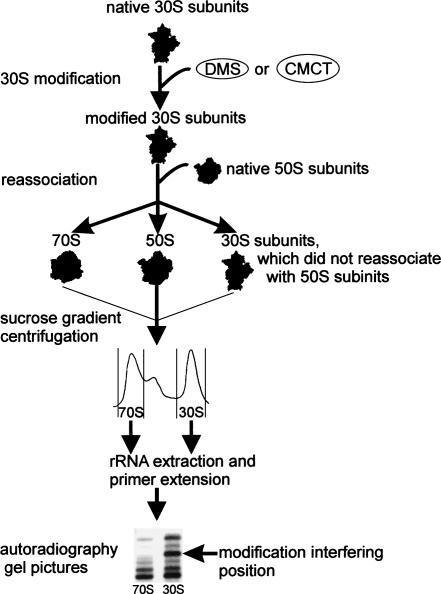
We chose to modify E. coli 30S ribosomal subunits with dimethyl sulfate (DMS; methylates N1 positions of adenosines, N7 positions of guanosines, and N3 positions of cytosines) or with 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate (CMCT; modifies N1 and N3 positions of uracils and N1 positions of guanosines). Modified 30S subunits were reassociated with unmodified 50S subunits to form 70S ribosomes. Resulting 70S and 30S populations were separated by sucrose gradient centrifugation, 16S rRNA was purified from the 30S and 70S gradient fractions, and 16S rRNA positions 1–1507 were scanned for DMS- or CMCT-specific reverse transcriptase stops (Fig. 1). Modifications, which were present in the 30S subunits but strongly reduced in the 70S ribosomes, were designated as interfering with 70S ribosome formation. 16S rRNA nucleotides that are accessible to DMS and CMCT modification in the 30S subunit are shown in Supplemental Figure S1. (Supplementary material can be found at http://www.ut.ee/~jremme/30Sarto.pdf; page 5, lines 17–19.)
FIGURE 1.
Scheme of the modification interference experiment. Native 30S subunits were modified with DMS or CMCT. Modified 30S subunits were reassociated with unmodified 50S subunits to form 70S ribosomes. Resulting 70S and 30S populations were separated by sucrose gradient centrifugation. Modified nucleotide positions were determined by primer extension analysis of 16S rRNA obtained from the 70S and 30S gradient fractions. Modifications, which were present in the 30S subunits but strongly reduced in the 70S ribosomes, were designated as interfering with 70S ribosome formation.
Optimization of the chemical modification procedure
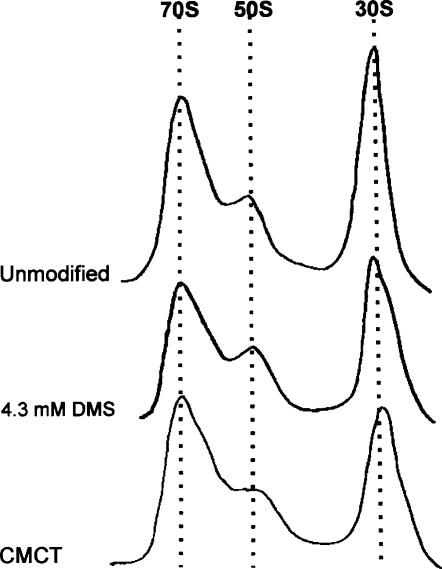
DMS is much more reactive toward RNA than CMCT. This increases the danger of inadvertent overmodification with DMS causing large-scale structural rearrangements in the structure of the 30S subunits. Therefore, while using standard conditions for CMCT modification resulting in no more than a few modifications per 16S rRNA molecule (Stern et al. 1988), we titrated DMS to ensure that minimal modification level still detectable by primer extension is used (Fig. 2). The chosen conditions (4 mM DMS) do not affect the sedimentation of the 30S subunits (Fig. 2). Therefore, modification of 30S subunits results in 30S populations, which are largely structurally homogeneous and able to form 70S ribosomes.
FIGURE 2.
Effect of chemical modification on the 30S subunits during ribosome subunit association. 30S subunits were modified with 4.3 mM DMS or 149 mM CMCT, and ribosomal particles were separated by sucrose gradient centrifugation. The positions of the 70S ribosomes and 50S and 30S subunits are indicated. All reassociation and selection experiments were done in 6 mM MgCl2.
If the chemical modification levels used resulted in nonspecific rupture of the 30S structure, the resulting ribosomes would very likely be inactive in toto. We have used a poly(U)-directed cell-free translation system to test the activity of 30S subunits previously subjected to various concentrations of DMS. This assay enables us to measure the net yield of translation in conditions in which translational yields are linearly dependent on ribosome concentrations. In no case did chemical modification reduce the level of poly-Phe synthesis by modified ribosomes (Table 1). This result shows that the vast majority of chemically modified 30S particles associated into functionally active 70S ribosomes. The fraction of 30S subunits, inactivated by DMS, cannot exceed the error margin of the poly(U) translation experiment, which we conservatively estimate to be <10%. Indeed, considering that treatment with 8 mM DMS does not noticeably reduce translational activity of the 30S subunits, it is highly probable that our working concentration of DMS (4 mM) does not cause large-scale structural perturbations of the 30S subunits. The expected minor fraction of functionally decapacitated particles would very likely be inactivated by specific single-hit mechanisms, rather than by combined action of several independent modification events. The presence of this minor inactivated subpopulation of 30S subunits can be successfully detected because of the very high sensitivity of the primer extension technique, since even a low proportion of modification in a given 16S rRNA position results in the emergence of a discrete stop in the primer extension reaction. It is usually possible to detect a reproducible band by autoradiography if 1%–3% of the reverse transcriptase stops at a given position (Maiväli et al. 2002).
TABLE 1.
Effect of DMS modification on the functional activity of 30S subunits during poly(U) translation
Determination of modified bases in 16S rRNA interfering with 70S reassociation
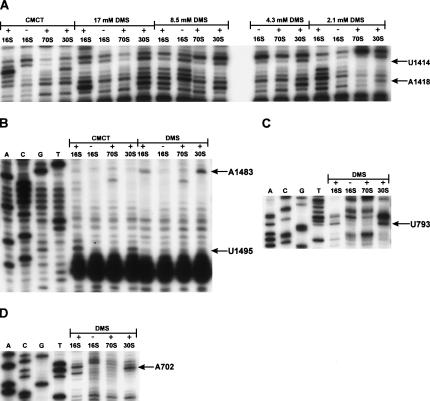
Scanning of 16S rRNA obtained from modified 30S subunits resulted in identification of 141 DMS-specific and 48 CMCT-specific primer extension stops (see Supplemental Fig. S1 at http://www.ut.ee/~jremme/30Sarto.pdf; page 5, lines 17–19). Of the 189 modifications seen in the 30S population, six were absent or considerably reduced in the 70S population (Fig. 3). 16S rRNA adenosines 702, 1418, and 1483 exhibited DMS-specific primer extension stops in the 30S fractions, while lacking stops in the 70S fractions (Fig. 3A,B,D). Therefore, methylation of each of these bases interfered with the ability of 30S subunits to reassociate with 50S subunits to form 70S ribosomes. Similarly, CMCT modification of 16S rRNA uridines 793, 1414, and 1495 interfered with 70S reassociation (Fig. 3A–C). The aforementioned six positions strongly interfered with 70S reassociation regardless if 30S or 50S subunits were limiting in the reassociation reaction (data not shown). All DMS-specific interferences were reproducibly observed when 4.3 mM, 8.5 mM, or 17 mM DMS was used in 30S modification (Fig. 3A,B,D; data not shown). DMS and CMCT also weakly modify N3 positions of cytosines and N1 positions of guanosines, respectively. The modification level of cytosines and guanosines in 16S rRNA was not sufficient to detect reproducible interferences (data not shown). We conclude that 16S rRNA bases at positions A702, U793, U1414, A1418, A1483, and U1495 are functionally important for the association of 70S ribosomes.
FIGURE 3.
Reverse transcriptase analysis of the positions of the DMS and CMCT modifications in 16S rRNA. (A) Positions U1414 and A1418, (B) positions U1483 and A1495, (C) position U793, and (D) position A702, whose modification interferes with 30S reassociation, are indicated by arrows. 16S, 30S, and 70S denote naked 16S rRNA or 16S rRNA extracted from the free 30S subunits or from the 70S ribosomes, respectively. The sequencing lanes are indicated by A, C, G, and T.
DISCUSSION
We have shown that modification of any of the given six bases in 16S rRNA (namely, A702, U793, U1414, A1418, A1483, or U1495) severely reduce the ability of modified 30S subunits to form 70S ribosomes in vitro. All modified bases interfering with formation of 70S ribosomes are located in or near the known intersubunit contact areas (Fig. 4; Gabashvili et al. 2000; Yusupov et al. 2001; Gao et al. 2003). It is possible that bases whose modification interferes with 70S ribosome formation make direct contacts with components of the 50S subunit. Alternatively, modification of the bases can disturb the local conformation of the 16S rRNA. In the 16S rRNA, there are 23 adenosines and 11 uracils, which are implicated in the intersubunit bridges (Yusupov et al. 2001; Gao et al. 2003) and/or are protected against chemical modification by the 50S subunit (Merryman et al. 1999b). Most of these bases are accessible to chemical modification (see Supplemental Fig. S1 at http://www.ut.ee/~jremme/30Sarto.pdf; page 5, lines 17–19). Thus, not all chemically modified positions that have been implicated in intersubunit contacts interfere with reassociation of 70S ribosomes.
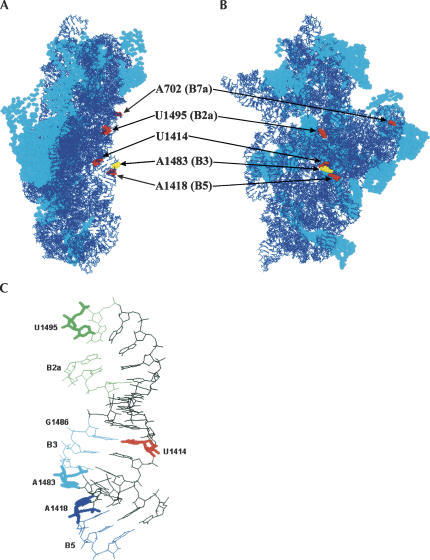
FIGURE 4.
Modeling of interfering positions into the crystal structure of T. thermophilus 30S subunits (Schluenzen et al. 2000; Wimberly et al. 2000) (PDB accession code 1FKA). (A) The left view is from the A-site side, and (B) the right view is from the subunit interface side. Arrows indicate interfering positions (E. coli numbering), and intersubunit bridges (B2a–B7a) are in the brackets. Interfering positions are highlighted in red and yellow spacefill. The rest of the rRNA is in blue wireframe, and r-proteins are in cyan spacefill (RasWin molecular graphics). Interfering position U793 is not resolved in the crystal structure of the T. thermophilus 30S subunit and therefore is not indicated in the figure. (C) Upper part of the 16S rRNA helix 44. Interfering positions (U1414, A1418, A1483, and U1495) are displayed in stick format. G1486 forms a base pair with U1414, which, in turn, is a part of bridge B3. Nucleotides, which form intersubunit bridges (B2a, B3, and B5), are indicated in different colors (B2a, green; B3, cyan; and B5, blue). The rest of the rRNA is in the black wireframe mode.
The interference data allow us to identify intersubunit contacts that are important for the reassociation of 70S ribosomes in the absence of tRNA. Of the six positions whose modification interferes with the reassociation of 70S ribosomes, five can be assigned to the five distinct intersubunit bridges (A702 to B7a, U793 to B2b, A1418 to B5, A1483 to B3, and U1495 to B2a) as defined by X-ray crystallography and cryo-EM (Fig. 4; Yusupov et al. 2001; Gao et al. 2003; Schuwirth et al. 2005). The remaining interfering base, U1414, pairs with G1486, a component of bridge B3 (Fig. 4C). In addition to the five bridges defined in the current work as functionally important, probing of modified 50S subunits under similar experimental conditions adds the bridge B4 and confirms the bridge B2a as necessary for 70S ribosome stability (Maiväli and Remme 2004). Moreover, point mutations A1912G, Ψ1917C, and A1919G of the helix–loop 69 of 23S rRNA have a severe effect on the translational activity both in vivo and in vitro, further emphasizing importance of the B2a bridge in ribosome function (Liiv et al. 2005).
We consider it very likely that each interference is caused by modification of a single 16S rRNA position because (1) a low level of modification is used, causing no more than a few modifications per molecule (see above), and (2) modification does not result in a significant reduction in the protein synthesis ability of the ribosomes (Table 1). Therefore, we suggest that no single intersubunit bridge by itself is sufficient to hold the 70S ribosome together. Disruption of any of the six bridges (B2a, B2b, B3, B4, B5, or B7a) leads to nonfunctional ribosomes, suggesting that combined stability of at least six distinct bridges is needed to ensure functional ribosomes. Although the bridges whose modification leads to 70S ribosome instability fall into both central (B2a, B2b, B3, and B5) and more peripheral (B7a and B4) regions of the intersubunit space, it is worth noting the important role of the 16S rRNA helix 44 in ribosomal subunit association. Of the four bridges associated with helix 44, namely, B2a, B3, B5, and B6, three (B2a, B3, and B5) are needed to keep the ribosome together in our assay. The functional role of B6 cannot be ascertained by our methodology because it lacks bases accessible to chemical modification in the 30S subunit (data not shown). Bridges B2a and B3 also seem to be crucial for stability of ribosomal subunit association in vivo, since subunit dissociation during termination of translation likely involves disruption of those two bridges by RRF (Gao et al. 2005; Wilson et al. 2005).
Methylation of A702 at the N1 position by DMS strongly interfered with 70S ribosome formation (Fig. 3D). A702 is a part of the loop of 16S rRNA helix 23, contributing to the intersubunit bridge B7a. More specifically, A702 appears to form an A-minor interaction with the minor groove of 23S rRNA helix 68 (Yusupov et al. 2001). Subunit association protects A702 from the action of DMS in the presence (Moazed and Noller 1989) and absence (Hennelly et al. 2005) of tRNA. This base is increasingly protected from DMS modification by the 50S subunit as subunit reassociation time was extended, implying that formation of bridge B7a is a late event in 70S ribosome formation (Hennelly et al. 2005). Our results indicate that bridge B7a contributes significantly to 70S association also in the absence of tRNA.
Methylation of the N1 positions of A1418 and A1483 of helix 44 strongly interferes with 70S reassociation (Fig. 3A,B). In the 30S subunit, A1418 and A1483 are not involved in intramolecular contacts. A1418 N-1 and A1483 N-6 lie close together (∼3 Å) on the same surface of helix 44 (Schluenzen et al. 2000; Wimberly et al. 2000; see also Fig. 4). According to Yusupov et al. (2001), A1418 participates in the intersubunit bridge B5 interacting with the minor groove of 23S rRNA helix 64. A1483 participates in bridge B3 interacting with 23S rRNA helix 71 at or around position 1947 (Yusupov et al. 2001). We suggest that both nucleobases are in direct contact with 23S rRNA. In a recent analysis, A1418 was found to be protected by the 50S subunit association in a time-dependent manner, being among the first nucleotides in 16S rRNA to become inaccessible to DMS in the 50-msec time scale (Hennelly et al. 2005). However, the conformation of bridges B3 and B5 does not appear to be significantly changed in the ratchet-like intersubunit movement of the ribosome effected by EF-G-GTP binding (Gao et al. 2003; Spahn et al. 2004).
We have found that CMCT-dependent modification of N3 position of the 16S rRNA nucleotide U1414 strongly interferes with 70S association (Fig. 3A). U1414 is too far from the 50S subunit to make an intersubunit contact. However, U1414 forms a base pair with G1486 in 16S rRNA (Schluenzen et al. 2000; Wimberly et al. 2000), which, in turn, is a part of bridge B3 (Fig. 4C; Yusupov et al. 2001). U1414 by itself is probably not interacting with 50S subunit but is maintaining the functional structure of the intersubunit bridge B3.
In a very recent publication Schuwirth et al. (2005) described an E. coli 70S ribosome structure at 3.5 Å resolution. N1 atoms of 16S rRNA nucleotides A702, A1418, and A1483 appear to be involved in hydrogen bonding with 23S rRNA. In contrast, N3 atoms of U793, U1414, and U1495 are not in hydrogen-bonding distance to 23S rRNA. It is possible that relatively bulky CMCT modification of uridines blocks subunit association through steric hindrances with nucleotides interacting with the 50S subunit. On the other hand, a possibility that the uridines are involved in a transient interaction with the 50S subunit cannot be excluded.
MATERIALS AND METHODS
Modification interference experiments
30S subunits were dissociated from tight-coupled 70S ribosomes by sucrose gradient centrifugation in 1 mM MgCl2 (Bommer et al. 1996). Modification of 30S subunits was conducted as in Stern et al. (1988). For this, 380 pmol of 30S and 2.4 μL of DMS stocks (16 μL of DMS—16 μL of ethanol, 2 μL of DMS—18 μL of ethanol, 2 μL of DMS—38 μL of ethanol, and 2 μL of DMS—78 μL ethanol) were used per 300 μL DMS reaction (17 mM, 8.5 mM, 4.3 mM, and 2.1 mM DMS, respectively) and 150 μL of CMCT stock (126 mg/mL) per 300-μL CMCT reaction (149 mM CMCT). Modification reactions commenced for 5 min at 37°C and were stopped by addition of 30 μL of 0.1% adenine on an ice bath. Stern et al. (1988) state that under conditions similar to our low DMS (17 mM, 8.5 mM, 4.3 mM, and 2.1 mM) or CMCT modification protocols, no more than a few modifications per 16S rRNA molecule occur. Ribosomes were further purified from modifying agents by Sephacryl S400 spin columns (Amersham Pharmacia), equilibrated in buffer M6 (6 mM MgCl2, 60 mM NH4Cl, 30 mM Tris-HCl at ph 7.5, 60 mM KCl, 5 mM 2-mercaptoethanol). 70S ribosomes were reassociated in buffer M6 by adding 160 pmol of unmodified 50S subunits to modified 30S subunits in 1 mL final volume. Reassociation was carried out for 30 min at 37°C. 70S ribosomes and 30S subunits were fractionated by 10%–20% sucrose gradient centrifugation in M6 (Beckmann rotor Sw 28, 20.4K rpm, 17 h), and rRNA was purified by silica binding as in Maiväli et al. (2002). Primer extension was done as in Stern et al. (1988). All 16S rRNA bases were scanned for modifications (except helix 45). The identity of interfering positions was ascertained by at least three independent replications of the modification-selection experiment. Band densitometric analysis and quantification of observed interferences at 6 mM MgCl2 revealed a 70%–95% reduction of band intensities in reassociated 70S ribosomes if compared with the 50S fractions (data not shown).
Poly(U)-dependent cell-free translation
70S ribosomes (10 pmol) were incubated in 50 μL of mixA [0.06 mg of poly(U), 20 mM HEPES at pH 8, 150 mM NH4OAc at pH 6, 2 mM spermidine, and 6 mM MgOAc] at 37°C for 15 min, followed by the addition of 50 μL of factor-containing mixB (20 mM HEPES at pH 8, 150 mM NH4OAc at pH 6, 2 mM spermidine, 6 mM MgOAc, 0.07 mg of bulk tRNA [Boehring Mannheim], 6 mM ATP, 4 mM GTP, 16 mM phosphoenolpyruvate [PEP], 16 μM Phe, 0.02 mM [14C]Phe [150 cpm/pmol; Amersham], 2 μM pyruvate kinase, 0.5 μg of Phe synthetase [PheRS], 4.2 μg of EF-Tu, 6.2 μg of EF-G, 0.2 μg of EF-Ts). After 30 min of incubation at 37°C, reactions were stopped by addition of 1 mL of 5% tricloroacetic acid (TCA) and heated for 20 min at 95°C. Precipitates were collected onto GF/A filters (Whatman) and counted for radioactivity.
ACKNOWLEDGMENTS
We thank Aivar Liiv and other colleagues in our laboratory for useful discussions and help. We are grateful to Aileen McLeod and Eva-Liis Loogväli for critical reading of the manuscript. The Howard Hughes Medical Institute International Research Grant No. 55000332 and Estonian Science Foundation Grant No. 5822 supported this work.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2275906.
REFERENCES
- Belanger F., Gagnon M.G., Steinberg S.V., Cunningham P.R., Braker-Gingras L. Study of the functional interaction of the 900 tetraloop of 16S ribosomal RNA with helix 24 within the bacterial ribosome. J. Mol. Biol. 2004;338:683–693. doi: 10.1016/j.jmb.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Bommer U., Burkhard N., Jünemann R., Spahn C.M.T., Triana-Alonso F.J., Nierhaus K.H. Ribosomes and polysomes. In: Graham J., Rickwoods D., editors. Subcellular fractionation. A practical approach. Oxford University Press; Oxford: 1996. pp. 271–301. [Google Scholar]
- Chapman N.M., Noller H.F. Protection of specific sites in 16S rRNA from chemical modification by association of 30S and 50S ribosomes. J. Mol. Biol. 1977;109:131–149. doi: 10.1016/s0022-2836(77)80049-1. [DOI] [PubMed] [Google Scholar]
- Frank J., Agrawal R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Gabashvili I.S., Agrawal R.K., Spahn C.M., Grassucci R.A., Svergun D.I., Frank J., Penczek P. Solution structure of the E. coli 70 S ribosome at 11.5 Å resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- Gao H., Sengupta J., Valle M., Korostelev A., Eswar N., Stagg S.M., Van Roey P., Agrawal R.K., Harvey S.C., Sali A., et al. Study of the structural dynamics of the E. coli 70 S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- Gao H., Zavialov A., Li W., Sengupta J., Valle M., Gursky R.P., Ehrenberg M., Frank J. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol. Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Joseph S. Nonbridging phosphate oxygens in 16S rRNA important for 30S subunit assembly and association with the 50S ribosomal subunit. RNA. 2005;11:657–667. doi: 10.1261/rna.7224305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D., McPheeters D.S., Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes & Dev. 1989;3:1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- Hennelly S.P., Antoun A., Ehrenberg M., Gualerzi C.O., Knight W., Lodmell J.S., Hill E.W. A time-resolved investigation of ribosomal subunit association. J. Mol. Biol. 2005;346:1243–1258. doi: 10.1016/j.jmb.2004.12.054. [DOI] [PubMed] [Google Scholar]
- Herr W., Noller H.F. Protection of specific sites in 23S rRNA and 5S rRNA from chemical modification by association of 30S and 50S ribosomes. J. Mol. Biol. 1979;130:421–432. doi: 10.1016/0022-2836(79)90432-7. [DOI] [PubMed] [Google Scholar]
- Herr W., Chapman N.M., Noller H.F. Mechanism of ribosomal subunit association: Discrimination of specific sites in 16S rRNA essential for association activity. J. Mol. Biol. 1979;130:433–449. doi: 10.1016/0022-2836(79)90433-9. [DOI] [PubMed] [Google Scholar]
- Hirokawa G., Nijman R.M., Raj V.M., Kaji H., Igarashi K., Kaji A. The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA. 2005;11:1317–1328. doi: 10.1261/rna.2520405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R., Pavlov M., Buchingham R., Ehrenberg M. Novel roles for classical factors at the interface between translation termination and termination. Mol. Cell. 1999;3:601–609. doi: 10.1016/s1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- Liiv A., Karitkina D., Maiväli Ü., Remme J. Analysis of the function of E. coli 23S rRNA helix-loop 69 by mutagenesis. BMC Mol. Biol. 2005;6:18. doi: 10.1186/1471-2199-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiväli Ü., Remme J. Definition of bases in 23S rRNA essential for ribosomal subunit association. RNA. 2004;10:600–604. doi: 10.1261/rna.5220504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiväli Ü., Pulk A., Loogväli E., Remme J. Accessibility of phosphates in domain I of 23S rRNA in the ribosomal 50S subunit as detected by Rp phosphorothioates. Biochim. Biophys. Acta. 2002;1579:1–7. doi: 10.1016/s0167-4781(02)00415-3. [DOI] [PubMed] [Google Scholar]
- Merryman C., Moazed D., Daubresse G., Noller H.F. Nucleotides in 23S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 1999a;285:107–113. doi: 10.1006/jmbi.1998.2243. [DOI] [PubMed] [Google Scholar]
- Merryman C., Moazed D., McWhirter J., Noller H.F. Nucleotides in 16S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 1999b;285:97–105. doi: 10.1006/jmbi.1998.2242. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Oswald M., Brimacombe R. Identification of intermolecular RNA cross-links at the subunit interface of the Escerichia coli ribosome. Biochemistry. 1992;31:3004–3011. doi: 10.1021/bi00126a023. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Pestka S. Translocation, aminoacryl-oligonucleotides, and antibiotic action. Cold Spring Harb. Symp. Quant. Biol. 1969;34:395–410. doi: 10.1101/sqb.1969.034.01.046. [DOI] [PubMed] [Google Scholar]
- Risuleo G., Gualerzi C., Pon C. Specificity and properties of the destabilization, induced by initiation factor IF-3, of ternary complexes of the 30S ribosomal subunit, aminoacyl-tRNA and polynucleotides. Eur. J. Biochem. 1976;67:603–613. doi: 10.1111/j.1432-1033.1976.tb10726.x. [DOI] [PubMed] [Google Scholar]
- Schluenzen F., Tociji A., Zarivach R., Harms J., Gluehmann M., Janell D., Bashan A., Bartels H., Agmon I., Franceschi F., et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H.D. Structures of the bacterial ribosome at 3.4 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Spahn C.M., Beckmann R., Eswar N., Penczek P.A., Sali A., Blobel G., Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae-RNA–ribosome and subunit–subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Spahn C.M., Gomez-Lorenzo M.G., Grassucci R.A., Jorgensen R., Andersen G.R., Beckmann R., Penczek P.A., Ballesta J.P., Frank J. Domain movement of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H.F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Wilson D.N., Schluenzen F., Harms J.M., Yoshida T., Ohkubo T., Albrecht R., Buerger J., Kobayashi Y., Fucini P. X-ray crystallography study on ribosome recycling: The mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 2005;24:251–260. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly B.T., Brodersen D.E., Clemons W.M., Morgan-Warren R.J., Carter A.P., Vonrhein C., Hartsch T., Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. Crystal structure procedure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]