Abstract
Objective:
Mouse Twisted gastrulation gene (Twsg1) expression is found throughout embryonic development, including substantial levels in the first branchial arch that gives rise to the submandibular salivary gland (SMG). We addressed the proposition that normal Twsg1 expression is critical to normal SMG ontogenesis.
Design
Utilizing C57BL/6 embryos that were Twsg1−/− homozygotes, as well as wild type and heterozygote littermates, we investigated SMG development from gestational day 13 to newborn.
Results
Twsg1 protein is immunodetected in epithelia throughout SMG development. Twsg1−/− embryos display widely variable craniofacial phenotypes that range from normal to severe holoprosencephaly/agnathia with no mandibular arch or stomodeum. The SMG phenotypes are correlated with the external craniofacial phenotype, ranging from normal to agenesis/aplasia.
Conclusions
It is evident that normal Twsg1 expression is critical for normal mouse SMG ontogenesis. Twsg1 loss of function is ultimately epistatic to the epigenome under normal physiologic conditions, but not always so. The reduced penetrance and variable expressivity seen in the SMGs of Twsg1−/− embryos is a challenging enigma.
Keywords: Twisted gastrulation gene, Mouse, Salivary glands, Development, Embryonic
Abbreviations: BMP, bone morphogenetic protein; BSA, bovine serum albumin; FGF8, fibroblast growth factor 8; Pitx1, paired-like homeodomain transcription factor 1; Shh, sonic hedgehog; SMG, submandibular salivary gland; Tsg, twisted gastrulation; Twsg1, twisted gastrulation gene
Introduction
Mutation of the twisted gastrulation gene in Drosophila (tsg) is associated with dysmorphic embryonic salivary glands, specifically changes in shape and position.1 tsg encodes a secreted, cysteine-rich, protein which is expressed in early embryogenesis and regulates the activity of decapentaplegic (dpp), an ortholog of the vertebrate bone morphogenetic proteins BMP2 and BMP4.2,3
The mammalian (mouse and human) twisted gastrulation gene (Twsg1) is highly conserved relative to the Drosophila tsg gene, including all 24 cysteine residues and a hydrophobic signal sequence.2 In contrast to Drosophila tsg expression, mammalian Twsg1 expression is found throughout embryonic development, including substantial levels in the first branchial arch that gives rise to salivary glands.2 Though newborn Twsg1 mutants display background-dependent craniofacial defects,4 the salivary gland phenotype has heretofore not been investigated, neither at birth nor during embryonic development. Further, there is a conserved interaction with BMPs, which may be agonistic, or antagonistic in a context-dependent manner.4–10 This interaction appears to mediate the expression of two key molecules that regulate craniofacial development, sonic hedgehog (Shh) and fibroblast growth factor 8 (FGF8).4,9,11,12
Mouse submandibular salivary gland (SMG) organogenesis is initiated with a thickening of the oral epithelium of the mandibular arch around E11 and its subsequent development is best conceptualised in stages.13,14 During the Initial Bud stage, the thickening epithelium grows down into the arch mesenchyme adjacent to the tongue to form the primordial SMG. With continued epithelial proliferation and downgrowth, the SMG primordium becomes a solid, elongated epithelial stalk terminating in a bulb. In the Pseudoglandular stage, the primordium branches by repeated furcation in the distal ends of successive buds to produce a bush-like structure comprised of a network of epithelial buds. These branches and buds hollow out by epithelial cell apoptosis during the Canalicular and Terminal Bud stages to form the ductal system and presumptive acini.
This progressive differentiation is consequent to the functional integration of parallel and broadly related signalling pathways.14,15 Of particular note here, BMP2, BMP4, FGF8, and Shh are immunolocalised to SMG epithelia throughout organogenesis, Early Initial Bud to Late Terminal Bud. Shh and FGF8 appear essential to SMG development.16,17 In Shh null mice, the SMG fails to progress beyond the Early Pseudoglandular stage.16 Fgf8 hypomorphs have hypoplastic SMGs, whereas conditional null SMGs exhibit ontogenic arrest at the Initial Bud stage, followed by involution and absence by E18.5.17
Given the Twsg1 regulation of BMP function and the BMP mediation of Shh and FGF8 protein expression, it is reasonable to propose that, as with Drosophila tsg, mutant Twsg1 would be associated with dysmorphic salivary glands, that is, normal Twsg1 expression is critical to normal SMG ontogenesis. To address this question, we investigated SMG development in inbred C57BL/6 embryos who were Twsg1−/− (null) homozygotes, as well as wildtype and heterozygote littermates.
Materials and methods
The generation of Twsg1-deficient mice has previously been described.4 Briefly, inbred mice carrying a heterozygous conditional allele of Twsg1 (Twsg1neo/+) in a mixed background (129Sv/Ev, C57BL/6) were bred to Bactin-Cre mice to remove exon 3, which encodes part of the N-terminal domain of Twsg1 (aa 74—162) that has the BMP-binding activity. Mice heterozygous for deletion of exon 3 are called Twsg1+/−. Heterozygous mice were backcrossed to C57BL/6 background for nine generations and then intercrossed to generate homozygotes (Twsg1−/−).
Pregnant females were euthanized by CO2 narcosis and cervical dislocation on days 12.5 (E12.5) through 18.5 (E18.5) of gestation. Twsg1−/− newborns with craniofacial abnormalities are stillborn. The embryos were dissected in cold phosphate-buffered saline (PBS). Yolk sacs were collected for genotyping. Genomic DNA was extracted by standard methods18 and amplified by PCR as previously described.4 Heads were fixed in 4% paraformaldehyde, embedded in paraplast, and serial coronal sections were stained with haematoxylin and eosin. A minimum of 12 heads per gestational age were analysed. Twsg1 genotype and external craniofacial phenotype were correlated to SMG phenotype.
Immunolocalisation to detect Twsg1 protein was done using commercially available monoclonal anti-mouse TSG antibody (R&D Systems). Wildtype E12.5—E18.5 SMGs were fixed in cold 10% neutral buffered formalin for 4 h and processed as described above. The sections were deparaffinised, rehydrated, placed in antigen retrieval solution (10 mM citrate acid, pH 6), microwaved for 5 min, and rinsed in water. The blocking was performed in TNBTT (50 mM Tris—Cl pH 7.5; 150 mM NaCl; 0.1% BSA; 0.1% Triton X-100), first with the addition of 2% bovine serum albumin (BSA, Sigma) at room temperature for 1 h, then with 5% goat serum (MP Biomedicals) at room temperature for 30 min. The slides were rinsed with TNBTT and incubated with anti-mouse TSG antibody at 1:20 dilution at 4 °C overnight. Following several washes with PBS and TNBTT, the slides were incubated with goat anti-rat antibody Alexa Fluor 488 at 1:50 dilution (Molecular Probes) for 2 h at room temperature. The slides were washed with PBS and covered using permafluor mountant medium (Thermo Electron Corporation).
In situ hybridisation was performed on paraffin embedded tissue sections as described.19 Paired-like homeodomain transcription factor 1 (Pitx1) cDNA probe was a 512-bp fragment amplified from I.M.A.G.E. clone ID4192818 using primers: Pitx1For: 5′-cgccgctgtctaccaagagc-3′ and Pitx1Rev: 5′-caaaaccaacctggaggcgg-3′. The probe was subcloned into pCRII-TOPO vector (Invitrogen). The plasmid was linearized with BamHI and transcribed with T7 polymerase (Roche).
Results
Twsg1 protein is immunodetected throughout the development of SMG, from Early Initial Bud to Late Terminal Bud stages (Fig. 1). The protein is localized exclusively to SMG epithelia, not mesenchyme. Twsg1 protein is primarily seen in epithelial cell membranes; this is consistent with it being a secreted protein.
Figure 1.

Twsg1 protein localization during embryonic SMG development. (A) Initial Bud stage: the SMG consists of a solid, elongated epithelial stalk terminating in a bulb. (B) Negative control section. (C) Pseudoglandular stage: the SMG appears as a bush-like structure composed of a network of epithelial branches and end-buds, the presumptive ducts and terminal buds. At this stage, no lumina are seen in either the ducts or terminal buds. (D) Terminal Bud stage SMG: the number of branches and buds has markedly increased and the presumptive ducts and terminal buds exhibit distinct lumina lined by cuboidal epithelial cells. In all stages, Twsg1 protein is immunodetected in the epithelia and not in the mesenchyme. Bar: A and B—30 μm; C and D—50 μm.
Embryonic Twsg1−/− mice displayed craniofacial phenotypes that range from normal or minimally affected (microphthalmia) (Fig. 2B) to simple aganthia with a hypoplastic mandibular arch, low set ears and microstomia (Fig. 2C) to severe holoprosencephaly/agnathia with no mandibular arch or grossly evident stomodeum (Fig. 2D).
Figure 2.

Twsg1−/− embryos display widely variable craniofacial phenotypes at E13.5. Normal craniofacial phenotypes are seen in Twsg1+/− (A). Twsg1−/− embryo display craniofacial phenotypes that range from normal mandibular arch development (with or without associated defects such as microphthalmia (B) to simple agnathia with a hypoplastic mandibular arch (arrow), low set ears and microstomia (C) to severe holoprosencephaly/agnathia with no mandibular arch (arrowhead) or grossly evident stomodeum (D). e—eye. Bar, 200 mm.
Twsg1 genotype and external craniofacial phenotype was correlated to SMG phenotype. The widely variable phenotypic expression noted previously in the craniofacies of Twsg1−/− embryonic (Fig. 2) and newborn4 was also found in submandibular glands. Twsg1−/− embryos with normal mandibular arches (Fig. 2B) exhibit age-appropriate Canalicular stage development in E15 SMGs (Fig. 3C), identical to that seen in their normal Twsg1+/+ and Twsg1+/− littermates (Fig. 3A and B). Twsg1−/− embryos with a primitive mandibular arch (Fig. 2C), at E15 exhibit no evidence of several arch-derived structures (Meckel’s cartilage, bone, or tongue) and SMGs that are abnormally positioned and ontogenically arrested at the Early Initial Bud stage (Fig. 3D). At birth, such newborns display one of four SMG phenotypes (Fig. 4): normal, hypoplasia, dysplasia, or absence of the gland (aplasia). Finally, Twsg1−/− embryos with severe holoprosencephaly/agnathia (Fig. 2D) never exhibit SMG ontogeny, either early on (Fig. 3E and F) or at birth (not shown). This agenic phenotype occurs with or without a histologically demonstrable primitive stomodeum (Fig. 3E and F), and verified by Pitx1 expression (Fig. 3G and H). Pitx1 is a marker of oral epithelium and its derivatives, including SMG.20
Figure 3.
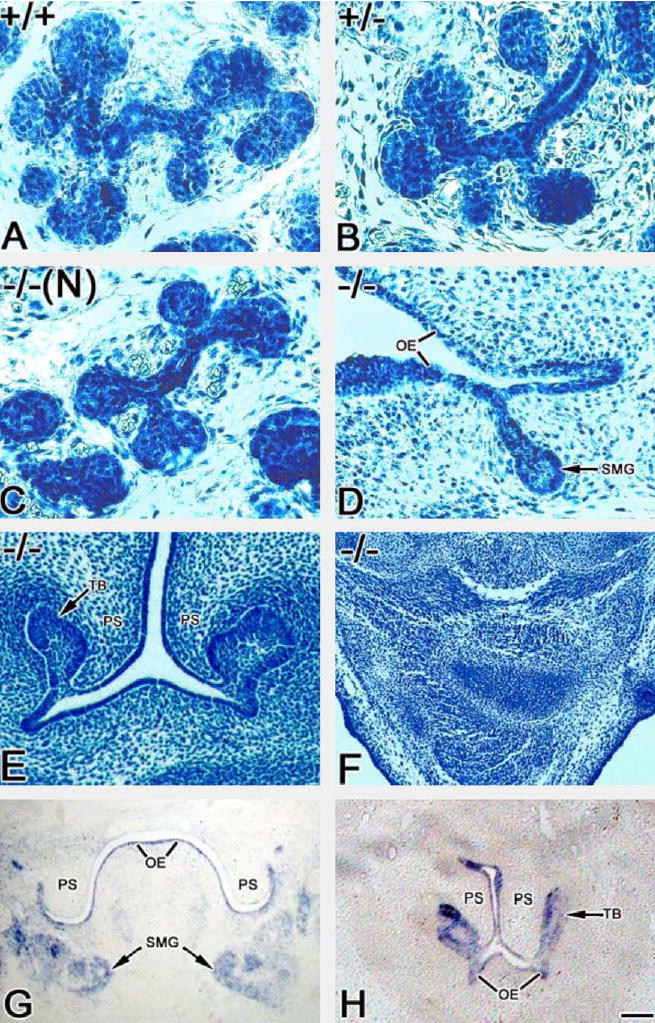
Twsg1−/− mutant SMG phenotypes. Age-appropriate Canalicular stage SMGs are seen in E15.5 Twsg1+/+ (A), Twsg1+/− (B) and craniofacially-normal or minimally affected Twsg1−/− (C) embryos. The Twsg1−/− SMG (D) found in mutants with hypoplastic mandibular arches is abnormally positioned and arrested at the Early Initial Bud stage. Twsg1−/− embryos with severe holoprosencephaly/agnathia (E and F) never exhibit SMG ontogeny. This agnathic phenotype occurs with (E) or without (F) a primitive stomodeum. Note the presence of maxillary tooth buds (TB) in severely abnormal mutants with a primitive stomodeum (E). (G and H) Pitx1 expression. Pitx1 is expressed in the oral epithelium and SMG of E13.5 wildtype embryos (G). SMG agenesis is seen in the presence of a rudimentary oral cavity in Twsg1−/− embryos at E13.5 −/− (N): craniofacially-normal Twsg1−/− mice; OE—oral epithelium; PS—palatal shelves. Bar, 50 μm.
Figure 4.

Newborn SMG phenotypes. (A) Late Terminal Bud stage SMGs are seen in newborn Twsg1+/+ mice. (B) Hypoplastic SMGs are seen in agnathic Twsg1−/− mice, characterized by fewer epithelial ducts and terminal buds. (C) Severely abnormal SMG consisting of very few epithelial branches and disorganized mesenchyme are seen in Twsg1−/− mice with an extremely small mandibular arch. Bar, 50 μm.
Discussion
It is clear that normal Twsg1 expression is critical to normal mouse SMG ontogenesis. The reduced penetrance and variable expressivity seen in the SMGs of Twsg1−/− embryos is a challenging enigma.
Salivary gland epithelial branching is a multi-factorial trait, largely dependent upon a series of interrelated genetic circuits through which morphogenesis is realized (i.e., its epigenotype).14,15 The functional epigenome changes with progressive differentiation (time and space). As we see presently, Twsg1 loss of function is ultimately epistatic to the epigenome under normal physiologic conditions (i.e., no other gene mutations nor untoward environments), but not always so (Fig. 2). Since epistasis of declining Twsg1 function is a nonlinear emergent property of the complete functional epigenotype, it cannot escape the effects of pathway noise.21–23
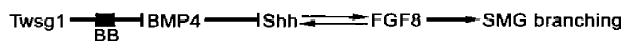
The underlying reactions of molecular pathways, typically involve small numbers of macromolecules (e.g. transcription factor and signal transduction factors) which are distributed in a Poissonian fashion and inevitably exhibit stochastic fluctuations that generate phenotypic diversity.24,25 Generally, two types of noise may be distinguished: intrinsic noise attributable to stochastic events during gene expression, and extrinsic noise due to cellular heterogeneity (size, shape, cycle stage, etc.) or to stochastic events in upstream signal transduction; extrinsic noise dominates intrinsic noise by a factor of 5–50 to 1.26 It is, thus, reasonable to expect that stochastic fluctuations in any one or a number of other parallel molecular SMG branching pathways [e.g. EGF and IGF14,15,27] would result in a more normal SMG phenotype even in the presence of the usually epistatic Twsg1 loss of function, what Schrodinger24 refers to as “the statistical mechanism which produces order from disorder.” This redundancy in signal integration, as well as cell—cell and extracelluar interactions, has been well-modelled.28,29 Although the system of genetic networks is robust, it moves toward criticality (between stable and chaotic).
The variable expressivity notwithstanding, it seems apparent that the abnormal phenotype seen in Twsg1−/− embryos (Fig. 2) is associated with an upregulation of BMP signalling and a concomitant downregulation of Shh signalling in oral epithelia.4 It has been consistently demonstrated that increased BMP signalling results in decreased Shh and FGF8 protein expression.11,12 Downregulation of Shh signalling in embryonic SMGs also results in a very significant decline in FGF8 expression and in SMG morphogenesis.16 Some have argued that induction of FGF8 protein expression does not depend on Shh signalling, but that its maintenance depends on Shh signaling.12 This is supported by our studies of SMG ontogenesis: with Fgf8 loss of function, SMG development is only partially rescued by enhanced Shh expression17; with Shh loss of function, SMG development is fully rescued by enhanced FGF8 expression.17 All this suggests an intriguing Twsg1-mediated epistatic molecular pathway in which Shh and FGF8 signalling reciprocally maintain each other: where BB = “Black Box” (Twsg1 genotype + genomic background + epigenotype + contingent environment). Investigating Twsg1 and Bmp4 conditional mutants in which Twsg1 and Bmp4 function has been completely ablated in its expression domain in the first branchial arch ectoderm from the time of arch formation, would greatly enhance our understanding of this epistatic pathway.
Acknowledgments
The authors wish to thank Yan-Min Zhou, Pablo Bringas Jr., and Michael Jarcho for technical assistance. This work was supported by NIH (NIDCR) RO1 DE014535 (TJ/MM) and NIH (NICHD) K08-HD043138 (AP), and NIH (NIDCR) T32 DE07288 (ADP).
References
- 1.Lammel U, Saumweber H. X-linked loci of Drosophila melanogaster causing defects in the morphology of the embryonic salivary glands. Dev Genes Evol. 2000;210:525–35. doi: 10.1007/s004270000096. [DOI] [PubMed] [Google Scholar]
- 2.Graf D, Timmons P, Hitchins M, Episkopou V, Moore G, Ito T, et al. Evolutionary conservation, developmental expression, and genomic mapping of mammalian Twisted gastrulation. Mam Gen. 2001;12:554–60. doi: 10.1007/s0033501-0005-x. [DOI] [PubMed] [Google Scholar]
- 3.Ross JJ, Shimmi O, Vilmos P, Petryk A, Kim H, Gaudenz K, et al. Twisted gastrulation is conserved extracellular BMP antagonist. Nature. 2001;410:479–83. doi: 10.1038/35068578. [DOI] [PubMed] [Google Scholar]
- 4.Petryk A, Anderson R, Jarcho M, Leaf I, Carlson C, Klingen-smith J, et al. The mammalian twisted gastrulation gene functions in foregut and craniofacial development. Dev Biol. 2004;267:374–86. doi: 10.1016/j.ydbio.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Oelgeschlager M, Larrain J, Geissert D, De Robertis EM. The evolutionary conserved BMP-binding protein Twisted gastrulation promotes BMP signaling. Nature. 2000;405:757–63. doi: 10.1038/35015500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu K, Srinivasan S, Shimmi O, Biehs B, Rashka KE. Processing of the Drosophila Sog protein creates a novel BMP inhibitory activity. Development. 2000;127:2143–54. doi: 10.1242/dev.127.10.2143. [DOI] [PubMed] [Google Scholar]
- 7.Scott IC, Blitz IL, Pappano WN, Maas SA, Cho KWY, Greenspan DS. Homologues of Twisted gastrulation are extracellular cofactors in antagonism of BMP signalling. Nature. 2001;410:475–8. doi: 10.1038/35068572. [DOI] [PubMed] [Google Scholar]
- 8.Nosaka T, Morita S, Kitamura H, Nakajima H, Shibata F, Morikawa Y, et al. Mammalian Twisted gastrulation is essential for skeleto-lymphogenesis. Mol Cell Biol. 2003;23:2969–80. doi: 10.1128/MCB.23.8.2969-2980.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zakin L, De Robertis E. Inactivation of mouse Twisted gastrulation reveals its role in promoting Bmp4 activity during forebrain development. Development. 2004;131:413–24. doi: 10.1242/dev.00946. [DOI] [PubMed] [Google Scholar]
- 10.Zakin L, Reversade B, Kuroda H, Lyons KM, De Robertis EM. Sirenomelia in BMP7 and Tsg compound mutant mice: requirement for BMP signaling in the development of ventral posterior mesoderm. Development. 2005;132:2489–99. doi: 10.1242/dev.01822. [DOI] [PubMed] [Google Scholar]
- 11.Anderson R, Lawrence A, Stottmann R, Bachiller D, Klingen-smith J. Chordin and noggin promote organizing centers of forebrain development in the mouse. Development. 2002;129:4975–87. doi: 10.1242/dev.129.21.4975. [DOI] [PubMed] [Google Scholar]
- 12.Ohkubo Y, Chiang C, Rubenstein J. Coordinate regulation and synergistic actions of BMP4, SHH and FGF8 in the rostral prosencephalon regulate morphogenesis of the telencephalic and optic vesicles. Neuroscience. 2002;111:1–17. doi: 10.1016/s0306-4522(01)00616-9. [DOI] [PubMed] [Google Scholar]
- 13.Melnick M, Jaskoll T. Mouse submandibular gland morphogenesis: a paradigm for embryonic signal processing. Crit Rev Oral Biol. 2000;11:199–215. doi: 10.1177/10454411000110020401. [DOI] [PubMed] [Google Scholar]
- 14.Jaskoll T, Melnick M. Embryonic salivary gland branching morphogenesis. In: Davies J, editor. Branching morphogenesis Georgetown, TX: Landes Bioscience; 2004.
- 15.Melnick M, Chen H, Zhou Y, Jaskoll T. The functional genomic response of developing embryonic submandibular glands to NF-κB inhibition. BMC Dev Biol. 2001;1:15. doi: 10.1186/1471-213X-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaskoll T, Leo T, Witcher D, Ormestad M, Astorga J, Bringas P, et al. Sonic hedgehog signaling plays an essential role during embryonic salivary gland epithelial branching morphogenesis. Dev Dyn. 2004;229:722–32. doi: 10.1002/dvdy.10472. [DOI] [PubMed] [Google Scholar]
- 17.Jaskoll T, Witcher D, Toreno L, Bringas P, Moon A, Melnick M. FGF8 dose-dependent regulation of embryonic submandibular salivary gland morphogenesis. Dev Biol. 2004;268:457–69. doi: 10.1016/j.ydbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the mouse embryo. A laboratory manual New York: Cold Spring Harbor Laboratory Press; 1994.
- 19.Person AD, Garriock RJ, Krieg PA, Runyan RB, Klewer SE. Frzb modulates Wnt-9a-mediated β-catenin signaling during avian atrioventricular cardiac cushion development. Dev Biol. 2004;278:35–48. doi: 10.1016/j.ydbio.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Lanctot C, Lamolet B, Drouin J. The bicoid-related homeo-protein Ptx1 defines the most anterior domain of the embryo and differentiates posterior from anterior lateral mesoderm. Development. 1997;124:2807–17. doi: 10.1242/dev.124.14.2807. [DOI] [PubMed] [Google Scholar]
- 21.Isaacs FJ, Blake WJ, Collins JJ. Signal processing in single cells. Science. 2005;307:1886–8. doi: 10.1126/science.1110797. [DOI] [PubMed] [Google Scholar]
- 22.Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science. 2005;307:1965–9. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- 23.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single cell level. Science. 2005;307:1962–5. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 24.Schrodinger E. What is life?Cambridge: Cambridge University Press; 1994.
- 25.Spudich J, Koshland D. Non-genetic individuality: chance in the single cell. Nature. 1976;262:467–71. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- 26.Raser J, O’Shea E. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–4. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlessinger J. Common and distinct elements in cellular signaling via EGF and FGF receptors. Science. 2004;306:1506–7. doi: 10.1126/science.1105396. [DOI] [PubMed] [Google Scholar]
- 28.Kauffman S, Peterson C, Samuelsson B, Troein C. Random Boolean network models and the yeast transcriptional network. Proc Natl Acad Sci USA. 2003;100:14796–9. doi: 10.1073/pnas.2036429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kauffman S, Peterson C, Samuelsson B, Troein C. Genetic networks with canalyzing Boolean rules are always stable. Proc Natl Acad Sci USA. 2004;101:17102–7. doi: 10.1073/pnas.0407783101. [DOI] [PMC free article] [PubMed] [Google Scholar]


