Abstract
Loss of CD4+ T cells, the hallmark of HIV pathogenesis, was suggested to be partly due to apoptosis. We recently reported that IFN-α produced by HIV-1-activated plasmacytoid dendritic cells (pDCs) contributes to CD4+ T cell apoptosis by the TNF-related apoptosis-inducing ligand (TRAIL)/death receptor (DR)5 pathway. Here, we show that HIV-1-induced intracellular expression of IFN-α in pDCs is coupled to increased expression of IFN regulatory factor 7 and MyD88 by pDCs in vivo and in vitro. Expression of IFN-α was increased in lymphoid tonsillar tissue (LT) of patients with progressive (HIVprog) compared with nonprogressive (HIVNP) HIV-1 disease and to uninfected controls. LT from HIVprog exhibited higher TRAIL and DR5 mRNA levels than LT from HIVNP or controls. TRAIL mRNA levels in LT correlated with plasma viral load. We show that HIV-1 induces IFN-α and the TRAIL/DR5 apoptotic pathway in LT, suggesting a role for these cytokines in HIV-1 immunopathogenesis.
Keywords: pDC, IRF-7, MyD88, AT-2 HIV-1
Type I IFN (IFN-α/β) provides essential innate immunity against viruses (1) and inhibits HIV-1 replication in vitro (2, 3). Plasmacytoid dendritic cells (pDCs), located in blood and lymphoid tissue (1, 4), are specialized cells that produce up to 1,000-fold more IFN-α than other blood cell types in response to viral stimulation (5). The number of circulating pDCs was decreased in HIV-1 infection (6), and the lack of IFN-α production was suggested to be responsible for HIV-1 disease progression (7, 8). However, induction of type I IFN could be a double-edged sword and might exert pathogenic in addition to protective effects. Thus, high plasma titers of IFN-α are found during acute HIV-1 infection and reappear during late-stage disease as an indicator of poor clinical prognosis (9). Furthermore, the loss of uninfected T cells in vitro due to HIV-1 is induced by IFN-α and can be blocked by anti-IFN-α antibodies (10).
IFN-α could also contribute to HIV-1 pathogenesis by inducing expression of death molecules, such as TNF-related apoptosis-inducing ligand (TRAIL) (11). TRAIL may contribute to HIV-1 immunopathogenesis, because CD4+ and CD8+ T cells from HIV+ patients are more susceptible to TRAIL-induced apoptosis in vitro than T cells from healthy control (HIV−) donors (12, 13). TRAIL was shown to be involved in selective apoptosis of uninfected CD4+ T cells in an in vitro human model (14) and in HIV-1-infected (HIV+) human-peripheral-blood lymphocyte–nonobese-diabetic–severe combined immunodeficient mice (15). We recently reported that the TRAIL/death receptor (DR)5 pathway contributed to selective apoptosis of CD4+ T cells in vitro and that TRAIL levels and CD4+ T cells expressing DR5 were elevated in blood of untreated HIV+ patients (16). Isolated pDCs cultured with infectious or noninfectious HIV-1 produced large amounts of IFN-α, (17, 18) that induced TRAIL expression on primary CD4+ T cells (19). We hypothesize that IFN-producing cells, pDCs, and/or other dendritic cells (20) activated by HIV-1 migrate to lymphoid organs and contribute to HIV-1-induced CD4+ T cell depletion by producing IFN-α, which, in turn, regulates TRAIL expression in lymphoid tissue, where extensive T cell depletion occurs (21–23).
Results
Regulation of IFN-α Expression in Circulating pDCs of HIV+ Patients.
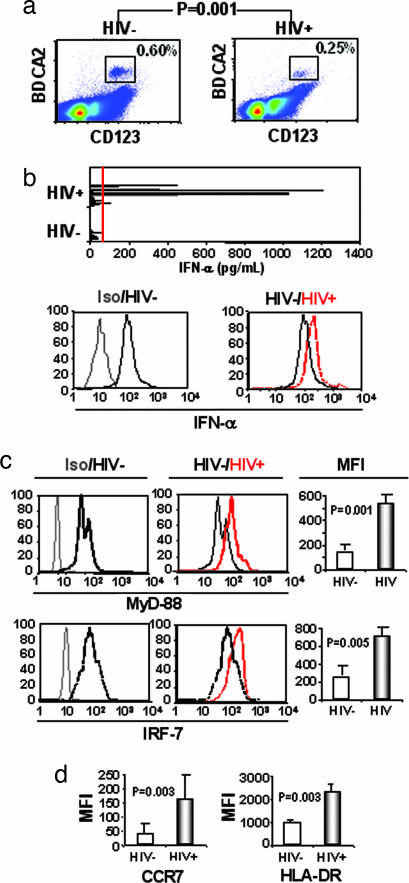
Twenty-eight untreated HIV+ patients were studied and compared with 22 HIV− donors for their circulating pDC numbers. We defined pDCs as CD4+ CD123+ CD11c− BDCA-2+ cells. The percentage of pDCs in the blood of HIV+ patients was decreased compared with HIV− controls (0.25% versus 0.60%, P = 0.001) (Fig. 1a). However, 7 of 28 HIV+ patients, but 0 of 22 HIV− controls, exhibited high serum IFN-α levels (Fig. 1b Upper). Furthermore, intracellular levels of IFN-α were higher in pDCs from 11 HIV+ patients than in pDCs from 10 HIV− controls (Fig. 1b Lower, representative examples shown). Three HIV+ patients exhibited IFN-α levels similar to HIV− controls (data not shown).
Fig. 1.
Characterization of circulating pDCs from HIV+ patients (HIV+) compared with HIV− donors (HIV−). (a) Percentage of pDCs in HIV+ and HIV− individuals. CD123+ BDCA-2+ CD11c− pDC percentages were determined in PBMCs from 22 HIV− and 28 HIV+ individuals. (b Upper) IFN-α in serum from HIV− and HIV+ individuals. Red line indicates limit of detection. Compare intracellular levels of IFN-α in pDCs from isotype to HIV− (Lower Left) and from HIV− to HIV+ (Lower Right). Samples representative of 11 patients (HIV+) and 10 controls (HIV−) are shown. (c) Intracellular expression of MyD88 and IRF7 in pDCs from HIV− and HIV+ individuals. (Left and Center) Comparison is shown between irrelevant isotype-matched antibody to MyD88 and IRF7 expression in uninfected donors. (Right) Compare IRF7 and MyD88 levels in pDCs from HIV− to HIV+ individuals. Mean fluorescence intensity (MFI) of controls and patients for MyD88 and IRF7. (d) Activation of pDC determined by extracellular expression of CCR7 and HLA-DR by pDCs from HIV− and HIV+ patients. P values were determined by using two-tailed Student’s t test.
IFN-α production by pDCs after influenza or herpes simplex virus infections depends on the IFN regulatory factor (IRF)7 and the adaptor molecule MyD88 (24, 25). Peripheral blood mononuclear cells (PBMCs) from HIV− and HIV+ individuals were tested for intracellular expression of MyD88 and IRF7 in pDCs. We found that pDCs, but not myeloid dendritic cells (data not shown), from HIV− donors’ PBMCs constitutively expressed MyD88 and IRF7. However, MyD88 and IRF7 expression were increased in HIV+ patients compared with HIV− controls (P = 0.001 and P = 0.0005, respectively) (Fig. 1c).
The culture of pDCs with HIV-1 up-regulated the chemokine receptor CCR7, which was suggested to induce pDC migration to T cell-rich areas in lymphoid tonsillar tissue (LT) (17). We found increased expression of CCR7 on pDCs from HIV+ patients compared with HIV− controls (P = 0.003) (Fig. 1d). The activation marker HLA-DR was also increased in the patients (P = 0.003) (Fig. 1d). These results demonstrate that HIV-1 induces activation of pDCs and suggests migration of pDCs to lymph nodes, which could account for the decrease of pDCs in patients’ blood.
HIV-1 Activation of IFN-α Expression by pDCs in Vitro.
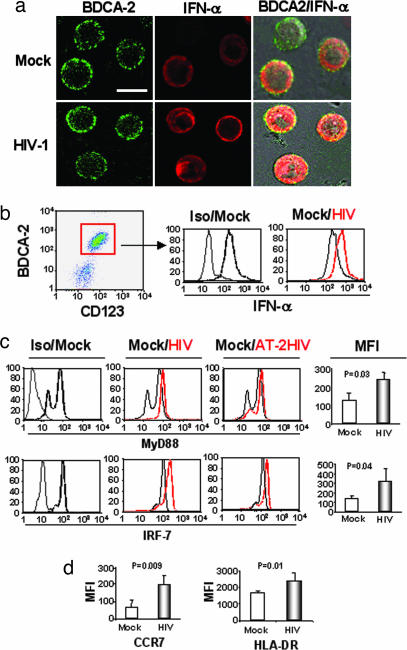
The above results show both functional and phenotypical activation of pDCs in HIV+ patients. We then tested whether in vitro stimulation of pDCs by HIV-1 would mimic our in vivo observations. Therefore, we enriched pDCs from healthy donors’ PBMCs and cultured them for 18 h with HIV-1. Because the vast majority of HIV-1 particles in patients are not infectious (26), we compared infectious and noninfectious [aldrithiol-2 (AT-2)] HIV-1 for stimulation of pDCs. Cells were stained for BDCA-2 and intracellular IFN-α and analyzed by confocal microscopy after culture with AT-2 HIV-1 or with the negative control microvesicles (27) (Fig. 2a, Mock). Most of the pDCs were positive for both BDCA-2 and IFN-α (Fig. 2a Upper). The number of IFN-α-positive pDCs and the intensity of IFN-α staining were increased by HIV-1 exposure (Fig. 2a Lower). Flow cytometry analysis confirmed that IFN-α expression was higher in pDCs exposed to either infectious or AT-2 HIV-1 than in pDCs cultured with microvesicles (Fig. 2b Right, Mock). Both infectious and noninfectious HIV-1 increased MyD88 and IRF7 expression in pDCs compared with microvesicles (Fig. 2c). IRF7 expression was partly regulated by IFN-α, because blocking IFN-α antibodies reduced HIV-1-mediated IRF7 overexpression by 50–60%, and recombinant IFN-α (1 μg/ml) increased IRF7 expression in pDCs (data not shown). HIV-1 also up-regulated in vitro expression of CCR7 and HLA-DR as reported (2, 17) (Fig. 2d). Taken together, these results are consistent not only with our previous demonstration that both infectious and noninfectious HIV-1 induce IFN-α production (19) but also with our in vivo data in Fig. 1.
Fig. 2.
HIV-1 activation of human pDCs from HIV− donors. (a) Confocal immunofluorescence images of BDCA-2 (green) and IFN-α (red) staining in pDCs cultured with AT-2 HIV-1 (Lower) and pDCs cultured with media alone (Upper). Combined BDCA-2/IFN-α images overlaid onto corresponding images are shown (Right). (Scale bar, 10 μm.) (b Left) FACS analysis of pDCs using BDCA-2 and CD123. (b Center) FACS profiles of IFN-α staining of CD123+ BDCA-2+ CD4+-gated cells cultured with microvesicles [Mock versus isotype (Iso)]. (b Right) pDCs cultured overnight with AT-2 HIV-1 compared with pDCs cultured with microvesicles (Mock). (c) Intracellular expression of IRF7 and MyD88 in pDCs cultured with infectious or AT-2 HIV-1. (Left) Compare isotype controls (Iso, light gray line) with MyD88 and IRF7 expression in pDCs cultured with microvesicles (Mock, heavy black line). (Center) Compare MyD88 and IRF7 expression in pDCs cultured with microvesicles (Mock, light black line) and infectious HIV-1 (HIV, heavy red line). (Right) Compare MyD88 and IRF-7 expression in pDCs cultured with microvesicles (Mock, light black line) and noninfectious HIV-1 (AT-2 HIV, heavy red line). (d) Extracellular expression of HLADR and CCR7. (c and d) Mean fluorescence intensity (MFI) for pDCs cultured with microvesicles (Mock), HIV-1, or AT-2 HIV-1 (combined data).
IFN-α Protein Production and TRAIL and Fas mRNA Expression in LT from HIV+ Patients.
If HIV-1-activated pDCs migrate to lymphoid tissue, we would expect an increased frequency of IFN-α-expressing cells in the lymphoid tissue of untreated HIV+ patients compared with HIV− controls. Therefore, we studied IFN-α expression by immunohistochemistry in LT sections from nine HIV− controls, from six HIV+ patients with progressive (HIVprog), and from five nonprogressive HIV-1 disease (HIVNP). Nonprogressor patients have maintained viral loads <5,000 copies per ml and CD4 counts >400 per mm3 without treatment (Table 1).
Table 1.
Clinical parameters for HIVNP patients
| Patient | M/F | Age | CD4 | VL | Treatment | CDC-stage | Infection duration, yr |
|---|---|---|---|---|---|---|---|
| L2 | F | 41 | 410 | 4,300 | None | A1 | 14 |
| L3 | M | 36 | 510 | 300 | None | A1 | 15 |
| L4 | M | 43 | 760 | <50 | None | A1 | 15 |
| L5 | F | 27 | 920 | <50 | None | A1 | 5 |
| L6 | F | 34 | 620 | 200 | None | A1 | 5 |
M/F, male/female; VL, viral load (copies of HIV-1 RNA/mL); CDC, Centers for Disease Control.
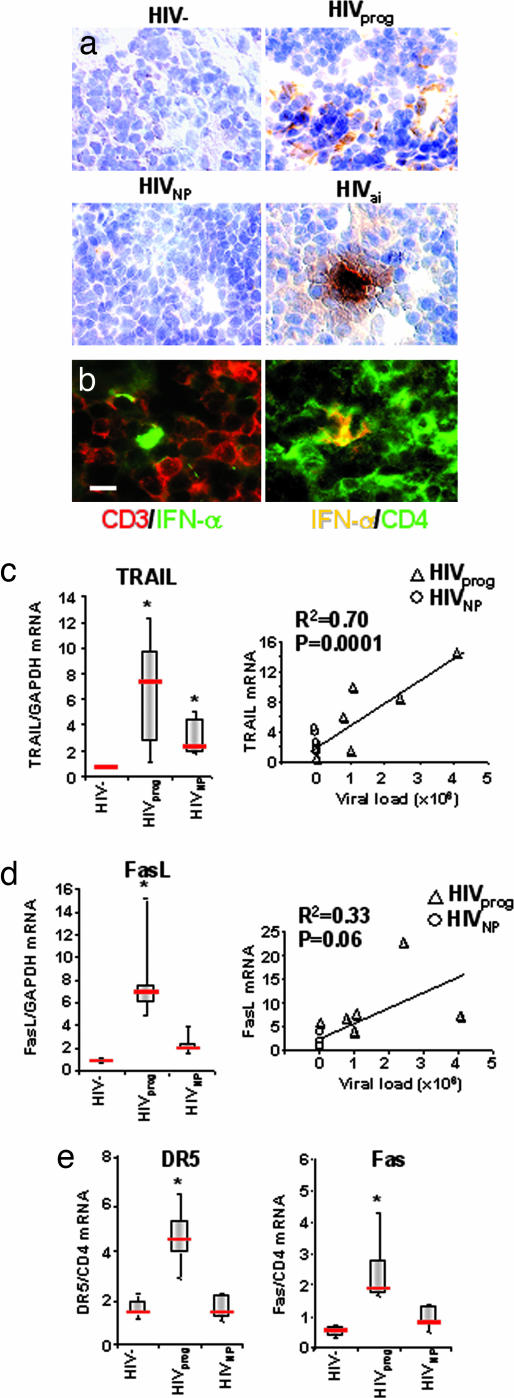
We also compared HIVprog and HIVNP samples with LT from six acutely infected HIV-1 patients (HIVai). HIVai patients served as positive controls because they have high viral load in their LT (28, 29) and high IFN-α plasma levels (9) and exhibit extensive CD4+ T cell depletion in gut-associated lymphoid tissue (21). The frequency of IFN-α-expressing cells in LT from controls was <1 per 104 cells (Fig. 3a Upper Left); from HIVprog was 5 ± 3 of 104 cells (Fig. 3a Upper Right); and from HIVNP was <1 of 104 cells (Fig. 3a Lower Left). Furthermore, LT from HIVai patients contained an even higher number of IFN-α-expressing cells (28 ± 19 of 10,000 cells) than from HIVprog (Fig. 3a Lower Right). To investigate whether the IFN-α expressed in patients’ LT would be found in the vicinity of CD4+ T cells, we tested LT T cell-rich regions for expression of IFN-α in conjunction with either CD3 or CD4 by immunofluorescence. IFN-α-expressing cells were localized in close proximity to both CD3- (Fig. 3b Left) and CD4- (Fig. 3b Right) expressing cells. These results indicate that expression of high levels of IFN-α occurred in the T cell-rich regions of LT from HIV+ patients. For the definition of T cell-rich areas, see Fig. 4, which is published as supporting information on the PNAS web site.
Fig. 3.
IFN-α production and apoptotic pathway analyses in human LT. (a) IFN-α expression (brown) in LT sections from HIV− (Upper Left), HIVprog (Upper Right), HIVNP (Lower Left), and an HIVai individual (Lower Right). Cell nuclei were counterstained with hematoxylin (blue). Magnification, ×380. (b) An LT section from HIVprog was stained with IFN-α (green) and CD3 (red) (Left) or IFN-α (red) and CD4 (green) (Right). Images show IFN-α expression in parafollicular T cell-rich areas. (Scale bar, 10 μm.) IFN-α-expressing cells were proximal to CD3+ and CD4+ cells in LT. (c Left) TRAIL mRNA expression in LT of HIV−, HIVprog, and HIVNP normalized on GAPDH. (c Right) Correlation between TRAIL mRNA expression in LT and HIV-1 plasma viral load (n = 11). (d Left) FasL mRNA expression in LT of HIVprog, HIVNP, and HIV− normalized on GAPDH. (d Right). Correlation between FasL mRNA expression in LT and plasma HIV-1 viral load (n = 11). (e) DR5 (Left) and Fas (Right) mRNA expression in LT of HIVprog, HIVNP, and HIV− controls normalized on CD4 mRNA. LT biopsy data of b and c were compared by the Mann–Whitney test. Horizontal bars indicate median values; upper and lower limits of boxes, 75th and 25th percentiles; vertical lines indicate 90th and 10th percentiles. ∗, P < 0.05 compared with uninfected control.
HIV-1-induced T cell depletion is more extensive in lymphoid organs (23, 30) and gastrointestinal tract (21) than in blood. Because IFN-α regulates TRAIL expression, we compared the above LT samples from HIV−, HIVprog, and HIVNP for expression of TRAIL mRNA. HIVprog expressed significantly higher levels of TRAIL mRNA in LT compared with HIV− (P = 0.05) (Fig. 3c). Because blockers of TRAIL/DR5 pathway did not totally inhibit CD4+ T cell apoptosis (16) and because Fas/FasL is involved in HIV-1-induced CD4+ T cell depletion in LT (30, 31), we also tested LT for FasL mRNA expression. FasL mRNA was significantly increased in HIVprog compared with HIV− (Fig. 3d), similar to our findings for TRAIL mRNA. Importantly, we found that both TRAIL and FasL mRNA expression in LT from HIVNP were comparable with HIV− (Fig. 3 c and d). Because the above finding implicates TRAIL and FasL expression in LT as potential markers for disease progression, we tested the correlation between expression of these apoptotic ligands and plasma viral load. TRAIL mRNA correlated with plasma viral load (R2 = 0.70, P = 0.0001), whereas the correlation between FasL and viral load was not statistically significant (R2 = 0.33, P = 0.06).
TRAIL and FasL induce cell death by binding their respective death receptors DR5 and Fas. Therefore, we tested and found that DR5 and Fas mRNA were both significantly increased in LT from HIVprog, but not from HIVNP, compared with controls (P = 0.05) (Fig. 3e). We previously demonstrated that only CD4+ T cells express DR5 after HIV-1 exposure (16), which suggests that the observed increase in DR5 is mostly by CD4+ T cells. Taken together, the results in Fig. 3 c–e show that both the TRAIL/DR5 and Fas/FasL apoptotic pathways are activated in LT of HIVprog but not in LT of HIVNP or HIV− controls.
Discussion
The findings of this study indicate that, in addition to its known beneficial antiviral activity, type I IFN can exert pathogenic effects by inducing TRAIL expression on CD4+ T cells in patients. We report here that HIV-1 activates the major regulators of IFN-α production (MyD88 and IRF7) by pDCs in vitro and in vivo. IFN-α, MyD88, and IRF7 were increased upon HIV-1-exposure of pDCs from healthy donors and were elevated in the pDCs of HIV+ patients. Our results uniquely demonstrate that pDCs from patients express higher levels of intracellular IFN-α than those from controls, indicating that HIV+ patients’ pDCs can respond to HIV-1.
Our findings that MyD88 and IRF7 are activated in the pDCs of HIV+ patients and after exposure of uninfected pDCs to noninfectious HIV-1 suggests that Toll-like receptor-7 (TLR-7) is involved in IFN-α production. Interestingly, a recent in vitro study demonstrated that HIV-1 activates pDCs to produce IFN-α through TLR-7 (32).
A reduction of circulating pDCs was reported in HIV-1prog disease (6). It is not known whether this reduction is due to cell death or to migration of pDCs to lymphoid organs (2). This report also indicates that pDCs from HIV+ patients express high levels of CCR7 compared with controls, and we confirm that CCR7 expression is increased by exposure to HIV-1 in vitro (17). These results raise the possibility that pDCs migrate to lymphoid tissue after HIV-1 activation. To demonstrate this point, we studied IFN-α expression in LT from HIV+ patients. We found that the number of IFN-α-expressing cells in LT was significantly higher in HIV-1prog than in HIV− and that this number was increased in HIVai patients, possibly because of the higher number of HIV-1 particles in their LT (29). We also found that HIVNP patients did not exhibit IFN-α in their LT.
The major producers of IFN-α are pDCs (1, 4). However, other dendritic cells also produced high levels of IFN-α after viral infection in a murine model (20). Our immunohistochemistry studies demonstrate that IFN-α-expressing cells are localized in the T cell-rich areas of tonsils from HIV+ patients. Because markers of circulating pDCs, CD123 and BDCA-2, may be reduced when the pDCs mature in lymphoid tissue, we could not definitely determine whether the IFN-α-expressing cells that we detected in patients’ tonsils were pDCs. However, our demonstration that IFN-α is expressed in LT after HIV-1 infection in vivo strongly suggests that the IFN-induced immunopathogenic effects that we reported in blood (16, 19) also occurs in lymphoid tissue after HIV-1 infection.
Because apoptosis of CD4+ T cells occurs in lymphoid tissue (21), including tonsils (30), it is crucial to understand the consequences of IFN-α expression in LT. We previously showed that antibodies against IFN-α greatly inhibited TRAIL expression and apoptosis in CD4+ T cells isolated from infected patients’ blood (16). HIV-1-exposed pDCs produced IFN-α, which, in turn, induced TRAIL expression, leading to selective apoptosis of CD4+ T cells (19). Our current analysis of TRAIL expression in LT indicate that untreated patients expressed high levels of TRAIL mRNA compared with controls, which would be expected because of the high levels of IFN-α we found in untreated patients’ LT. Our finding that TRAIL mRNA levels in LT correlates with viral load in HIV+ patients suggests TRAIL as an immunologic marker of HIV-1 disease progression. Interestingly, TCR/CD28-activated human CD4+ T cells undergo Fas-mediated apoptosis upon exposure to IFN-α (33). This finding indicates that IFN-α-mediated apoptosis of CD4+ T cells can occur in the absence of viral infection and is consistent with our data for noninfectious HIV-1. Although increased expression of FasL in HIVprog was observed, FasL mRNA expression in LT did not significantly correlate with viral load.
TRAIL and FasL induce apoptosis by binding to their respective DRs (34, 35). The Fas/FasL apoptotic mechanism was reported to contribute to the rapid and severe depletion of intestinal CD4+ T cells in macaques acutely infected with simian immunodeficiency virus (SIV) (36). We make the observation here that the LT of HIV+ patients express mRNA for TRAIL and FasL and also for their respective receptors DR5 and Fas. Thus, both the TRAIL/DR5 and Fas/FasL mechanisms of CD4+ T cell depletion may contribute to depletion of T helper cells in the LT during HIV-1 disease. HIVNP patients who do not exhibit CD4+ T cell depletion do not express IFN-α, TRAIL/DR5, or Fas/FasL in contrast to HIVprog patients. Therefore, we hypothesize that the lack of HIV-1 particles we observed in LT from HIVNP patients, is responsible for the low level of IFN-α and, consequently, the low level of TRAIL in LT. The absence of high viral titers and of IFN-α may contribute to the high CD4 count seen in HIVNP patients despite infection.
Considered together, these results strongly suggest that IFN-α is involved in HIV-1 pathogenesis. Our observations that LT expressing IFN-α also express TRAIL, FasL, DR5, and Fas and that anti-IFN-α antibodies inhibit apoptosis of CD4+ T cells from patients (16) raise the possibility of considering inhibition of type I IFN as a potential therapeutic approach for reducing CD4+ T cell depletion. This strategy is consistent with a study reporting that immunization against IFN-α generated antibodies, which reduced HIV-1 disease progression (37). Our study highlights a new role for IFN-α in HIV-1 immunopathogenesis.
Materials and Methods
HIV+ Patients.
Blood was collected from 28 HIV+ patients who were enrolled in a U.S. Air Force natural history protocol at Wilford Hall Medical Center, Lackland Air Force Base, and were not receiving antiretroviral therapy. Plasma HIV-1 RNA was measured by quantitative RT-PCR; data are expressed as copies per ml (Amplicor Monitor; Roche Diagnostics; detection limit of 50 copies per ml). Patient and control blood were studied under Institutional Review Board-approved protocols and consent forms from the National Cancer Institute and Wilford Medical Center. Ficoll-purified PBMCs from patients’ blood were cultured for 2 days in medium supplemented with 50% autologous plasma in the absence or presence of anti-IFN-α antibody at 5 μg/ml (BioSource International, Camarillo, CA). Cells were then tested for apoptosis and TRAIL expression.
Tonsil Biopsies.
Twenty-six tonsil biopsies were surgically obtained from HIV− individuals (n = 9), untreated, chronically infected HIV+ patients (n = 6), HIVai patients (n = 6), and untreated HIVNP patients (n = 5). Informed consent was obtained from all subjects, and protocols were approved by the Institutional Review Board (Karolinska University Hospital). HIV+ patients had peripheral CD4 counts (cells per mm3) of 136–470 (mean, 341) and plasma viral loads (copies per ml) of 1,800–405,700 (mean, 155,860). HIVai patients had peripheral CD4 counts of 537–1,318 (mean, 872) and plasma viral loads of 565–3,500,000 (mean, 2,067,1549). HIVNP untreated patients had peripheral CD4 counts of 410–920 (mean, 644) and plasma viral loads ranging from <50 to 4,300 (mean, 980). Viral load values <50 were assigned an arbitrary value of 49 for statistical purposes.
Preparation of Noninfectious AT-2 HIV-1.
HIV-1MN (CXCR4 coreceptor tropic) and HIV-1Ada (CCR5 coreceptor tropic) were inactivated with 1 mM AT-2 for 18 h at 4°C (AT-2 HIV-1), as described in ref. 27. Microvesicles isolated from uninfected cell cultures were used as a negative control (27).
Isolation and Culture of T Cells and pDCs.
In vitro experiments were performed by using PBMCs isolated by density centrifugation from peripheral blood obtained from HIV-1-seronegative blood bank volunteers. Cells were cultured in RPMI medium 1640 (Invitrogen) containing 10% FBS (Sigma) and 1% Pen-Strep-Glut (Invitrogen). The pDCs (CD123+ BDCA-2+) were isolated from healthy donors’ PBMCs by using the BDCA-2 isolation kit (Miltenyi Biotec, Auburn, CA).
Cultures of Leukocytes with HIV-1.
PBMCs or isolated pDCs were cultured with noninfectious AT-2 HIV-1MN or AT-2 HIV-1Ada at 500 ng/ml p24 capsid equivalent overnight. Infectious HIV-1MN and HIV-1Ada were used at the same concentration. We verified infection in the CD4+ cells cultured with infectious HIV-1 and absence of infection in the cells cultured to AT-2 HIV-1 by p24 ELISA of supernatants (Beckman-Coulter) after 3 days of culture.
Detection of MyD88 and IRF7 in pDCs by Flow Cytometry.
Total PBMCs from HIV+ or HIV− donors and isolated pDCs cultured for 24 h with HIV-1MN or AT-2 HIV-1MN cells were incubated for 20 min at room temperature with FITC-conjugated monoclonal antibody anti-human CD123 (MBL International, Woburn, MA), PE-Cy5.5 anti-CD11c (BD Bioscience, San Jose, CA), APC-conjugated monoclonal anti-BDCA-2 antibody (Miltenyi Biotec), or isotype-matched control antibodies (at 5 μg/ml each) (BD Bioscience) in PBS containing 2% mouse serum (Sigma). Cells were washed twice in ice-cold PBS, and intracellular staining was performed by using a Fix and Perm kit (Caltag, Burlingame, CA). Cells were fixed for 20 min at 4°C and then stained for 20 min with rabbit anti-IRF7 or anti-MyD88 antibodies (Santa Cruz Biotechnology). Then cells were stained with phycoerythrin-conjugated goat anti-mouse or goat anti-rabbit antibodies for 20 min. FACS analysis was performed on CD123+ BDCA-2+ CD11c-gated cells.
Immunofluorescence Microscopy.
BDCA2 isolated cells were fixed and labeled for BDCA2 and IFN-α by using the protocol described above for flow cytometry, except an anti-rabbit IFN-α primary antibody (R & D Systems) along with goat anti-rabbit Cy3 secondary antibody (Caltag) combination was used. Cells were washed, allowed to adhere to poly-l-lysine-coated chamber coverglass (MatTek, Ashland, MA), and mounted in staining buffer (Caltag). Confocal images of double immunolabeled cells against BDCA-2 and IFN-α and either untreated or cultured with noninfectious HIV-1 were imaged by using a ×40 C-Apochromat (numerical aperture, 1.2) water immersion lens coupled to a Zeiss LSM510 META confocal microscope equipped with a transmitted light detector for differential interference contrast imaging. The rabbit isotype control antibody followed by anti-rabbit Cy3 was used as the negative control for IFN-α staining (see Fig. 5, which is published as supporting information on the PNAS web site).
Analysis of IFN-α Expression in Lymphoid Tissue by Immunohistochemistry.
Immunohistochemical analysis biopsy samples were cut into 8-μm sections, fixed in 2% formaldehyde, and blocked for endogenous biotin (Vector Laboratories). Anti-human IFN-α monoclonal antibody (R & D Systems) and irrelevant isotype-matched antibody (DAKO) were used. The staining reactions were developed by using diaminobenzidine tetrahydrochloride and hematoxylin and analyzed by using a DMR-X microscope (Leica) and the image analysis system Quantimet Q550IW (Leica Imaging Systems). IFN-α-positive cells were counted manually per digital image, with the total number of cells assessed by in situ imaging. Whole-section scans were performed, and the frequency of positive cells was expressed as the number of positive cells per 104 cells. Double-staining experiments were performed within the T cell-rich area by using anti-human IFN-α polyclonal antibody (BioSource International) in combination with anti-human CD3 or CD4 monoclonal antibodies (DAKO), followed by the appropriate Alexa Fluor-conjugated secondary antibody (Invitrogen). Sections were analyzed and micrographs obtained by using a DMR-X microscope (Leica) coupled to a DFC320 digital camera (Leica).
Real-Time PCR.
Total RNA was extracted from cryopreserved tonsils by using RNAeasy kits (Qiagen, Valencia, CA). RNA was reverse-transcribed by using SuperScript reverse transcriptase (Invitrogen Life Technologies) and random primers (Roche). Real-time PCR and primers for TRAIL and DR5 were conducted as we described in ref. 16. Primer sequences were FasL Forward, TGGCCTTGTGATCAATGAAA; FasL Reverse, GCAGGTTGTTGCAAGATTGA; Fas Forward, TGAAGGACATGGCTTAGAAGTG; and Fas Reverse, GGTGCAAGGGTCACAGTGTT. Data analysis was performed by using sds2.1 software. Standards were obtained by amplification of control sample in PCR using the same primers, reagents, and conditions optimized for real-time analysis. Results are presented as ratios between TRAIL or FasL mRNA and GAPDH mRNA. Because we wanted to observe variations of DR5 and Fas mRNA expression in CD4+ T cells, we normalized Fas and DR5 on CD4 mRNA instead of GAPDH mRNA.
Type I IFN Detection.
Soluble IFN-α levels were detected by ELISA (R & D Systems). Intracellular IFN-α was detected in CD123+ CD11c− BDCA2+ cells by FACS using polyclonal anti-IFN-α antibody (BioSource International).
Statistical Analysis.
Experiments were repeated at least four times. P values were determined by using a two-tailed Student t test. P < 0.05 was considered statistically significant. Univariate distributions of flow cytometric data were performed by probability binning in 300 bins by using flowjo software (38). Gene expression in biopsies was compared with the Mann–Whitney U test. Correlations were determined with Spearman’s rank correlation. A two-tailed P < 0.05 was considered to be significant.
Supplementary Material
Acknowledgments
We thank Dr. J. D. Lifson [Science Applications International Corporation–National Cancer Institute (NCI), Frederick, MD] for providing the microvesicles, infectious HIV-1, and AT-2 HIV-1 particles and Drs. J. C. Grivel and G. Trinchieri [National Institutes of Health (NIH), Bethesda] for reviewing the manuscript. We thank the Fondation pour la Recherche Médicale for its financial support. This research was supported, in part, by the Intramural Program of the Center for Cancer Research, NCI, NIH, and by the NIH Intramural AIDS Targeted Antiviral Program.
Abbreviations
- ai
acutely infected
- AT-2
aldrithiol-2
- DR
death receptor
- HIV−
healthy control
- HIV+
HIV-1-infected
- IRF
IFN regulatory factor
- LT
lymphoid tonsillar tissue
- NP
nonprogressive
- PBMC
peripheral blood mononuclear cell
- pDC
plasmacytoid dendritic cell
- prog
progressive
- TRAIL
TNF-related apoptosis-inducing ligand.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Colonna M., Trinchieri G., Liu Y. J. Nat. Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt B., Ashlock B. M., Foster H., Fujimura S. H., Levy J. A. Virology. 2005;343:256–266. doi: 10.1016/j.virol.2005.09.059. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto J. K., Barre-Sinoussi F., Bolton V., Pedersen N. C., Gardner M. B. J. Interferon. Res. 1986;6:143–152. doi: 10.1089/jir.1986.6.143. [DOI] [PubMed] [Google Scholar]
- 4.Siegal F. P., Kadowaki N., Shodell M., Fitzgerald-Bocarsly P. A., Shah K., Ho S., Antonenko S., Liu Y. J. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 5.Kadowaki N., Antonenko S., Lau J. Y., Liu Y. J. J. Exp. Med. 2000;192:219–226. doi: 10.1084/jem.192.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soumelis V., Scott I., Gheyas F., Bouhour D., Cozon G., Cotte L., Huang L., Levy J. A., Liu Y. J. Blood. 2001;98:906–912. doi: 10.1182/blood.v98.4.906. [DOI] [PubMed] [Google Scholar]
- 7.Kamga I., Kahi S., Develioglu L., Lichtner M., Maranon C., Deveau C., Meyer L., Goujard C., Lebon P., Sinet M., Hosmalin A. J. Infect. Dis. 2005;192:303–310. doi: 10.1086/430931. [DOI] [PubMed] [Google Scholar]
- 8.Muller-Trutwin M., Hosmalin A. Immunol. Cell Biol. 2005;83:578–583. doi: 10.1111/j.1440-1711.2005.01394.x. [DOI] [PubMed] [Google Scholar]
- 9.von Sydow M., Sonnerborg A., Gaines H., Strannegard O. AIDS Res. Hum. Retroviruses. 1991;7:375–380. doi: 10.1089/aid.1991.7.375. [DOI] [PubMed] [Google Scholar]
- 10.Zagury D., Lachgar A., Chams V., Fall L. S., Bernard J., Zagury J. F., Bizzini B., Gringeri A., Santagostino E., Rappaport J., et al. Proc. Natl. Acad. Sci. USA. 1998;95:3851–3856. doi: 10.1073/pnas.95.7.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kayagaki N., Yamaguchi N., Nakayama M., Eto H., Okumura K., Yagita H. J. Exp. Med. 1999;189:1451–1460. doi: 10.1084/jem.189.9.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katsikis P. D., Garcia-Ojeda M. E., Torres-Roca J. F., Tijoe I. M., Smith C. A., Herzenberg L. A. J. Exp. Med. 1997;186:1365–1372. doi: 10.1084/jem.186.8.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeremias I., Herr I., Boehler T., Debatin K. M. Eur. J. Immunol. 1998;28:143–152. doi: 10.1002/(SICI)1521-4141(199801)28:01<143::AID-IMMU143>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 14.Lichtner M., Maranon C., Vidalain P. O., Azocar O., Hanau D., Lebon P., Burgard M., Rouzioux C., Vullo V., Yagita H., et al. AIDS Res. Hum. Retroviruses. 2004;20:175–182. doi: 10.1089/088922204773004897. [DOI] [PubMed] [Google Scholar]
- 15.Miura Y., Misawa N., Maeda N., Inagaki Y., Tanaka Y., Ito M., Kayagaki N., Yamamoto N., Yagita H., Mizusawa H., Koyanagi Y. J. Exp. Med. 2001;193:651–660. doi: 10.1084/jem.193.5.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herbeuval J. P., Grivel J. C., Boasso A., Hardy A. W., Chougnet C., Dolan M. J., Yagita H., Lifson J. D., Shearer G. M. Blood. 2005;106:3524–3531. doi: 10.1182/blood-2005-03-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fonteneau J. F., Larsson M., Beignon A. S., McKenna K., Dasilva I., Amara A., Liu Y. J., Lifson J. D., Littman D. R., Bhardwaj N. J. Virol. 2004;78:5223–5232. doi: 10.1128/JVI.78.10.5223-5232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yonezawa A., Morita R., Takaori-Kondo A., Kadowaki N., Kitawaki T., Hori T., Uchiyama T. J. Virol. 2003;77:3777–3784. doi: 10.1128/JVI.77.6.3777-3784.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbeuval J. P., Hardy A. W., Boasso A., Anderson S. A., Dolan M. J., Dy M., Shearer G. M. Proc. Natl. Acad. Sci. USA. 2005;102:13974–13979. doi: 10.1073/pnas.0505251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diebold S. S., Montoya M., Unger H., Alexopoulou L., Roy P., Haswell L. E., Al-Shamkhani A., Flavell R., Borrow P., Reis e Sousa C. Nature. 2003;424:324–328. doi: 10.1038/nature01783. [DOI] [PubMed] [Google Scholar]
- 21.Brenchley J. M., Schacker T. W., Ruff L. E., Price D. A., Taylor J. H., Beilman G. J., Nguyen P. L., Khoruts A., Larson M., Haase A. T., Douek D. C. J. Exp. Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guadalupe M., Reay E., Sankaran S., Prindiville T., Flamm J., McNeil A., Dandekar S. J. Virol. 2003;77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dyrhol-Riise A. M., Ohlsson M., Skarstein K., Nygaard S. J., Olofsson J., Jonsson R., Asjo B. Clin. Immunol. 2001;101:180–191. doi: 10.1006/clim.2001.5102. [DOI] [PubMed] [Google Scholar]
- 24.Honda K., Ohba Y., Yanai H., Negishi H., Mizutani T., Takaoka A., Taya C., Taniguchi T. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 25.Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., Taniguchi T. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 26.Piatak M., Jr, Saag M. S., Yang L. C., Clark S. J., Kappes J. C., Luk K. C., Hahn B. H., Shaw G. M., Lifson J. D. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 27.Rossio J. L., Esser M. T., Suryanarayana K., Schneider D. K., Bess J. W., Jr, Vasquez G. M., Wiltrout T. A., Chertova E., Grimes M. K., Sattentau Q., et al. J. Virol. 1998;72:7992–8001. doi: 10.1128/jvi.72.10.7992-8001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersson J., Kinloch S., Sonnerborg A., Nilsson J., Fehniger T. E., Spetz A. L., Behbahani H., Goh L. E., McDade H., Gazzard B., et al. J. Infect. Dis. 2002;185:1355–1358. doi: 10.1086/340124. [DOI] [PubMed] [Google Scholar]
- 29.Schacker T., Little S., Connick E., Gebhard K., Zhang Z. Q., Krieger J., Pryor J., Havlir D., Wong J. K., Schooley R. T., et al. J. Infect. Dis. 2001;183:555–562. doi: 10.1086/318524. [DOI] [PubMed] [Google Scholar]
- 30.Badley A. D., Dockrell D. H., Algeciras A., Ziesmer S., Landay A., Lederman M. M., Connick E., Kessler H., Kuritzkes D., Lynch D. H., et al. J. Clin. Invest. 1998;102:79–87. doi: 10.1172/JCI2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dyrhol-Riise A. M., Stent G., Rosok B. I., Voltersvik P., Olofsson J., Asjo B. Clin. Immunol. 2001;101:169–179. doi: 10.1006/clim.2001.5101. [DOI] [PubMed] [Google Scholar]
- 32.Beignon A. S., McKenna K., Skoberne M., Manches O., Dasilva I., Kavanagh D. G., Larsson M., Gorelick R. J., Lifson J. D., Bhardwaj N. J. Clin. Invest. 2005 doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dondi E., Roue G., Yuste V. J., Susin S. A., Pellegrini S. J. Immunol. 2004;173:3740–3747. doi: 10.4049/jimmunol.173.6.3740. [DOI] [PubMed] [Google Scholar]
- 34.Herbeuval J. P., Lambert C., Sabido O., Cottier M., Fournel P., Dy M., Genin C. J. Natl. Cancer Inst. 2003;95:611–621. doi: 10.1093/jnci/95.8.611. [DOI] [PubMed] [Google Scholar]
- 35.Itoh N., Yonehara S., Ishii A., Yonehara M., Mizushima S., Sameshima M., Hase A., Seto Y., Nagata S. Cell. 1991;66:233–243. doi: 10.1016/0092-8674(91)90614-5. [DOI] [PubMed] [Google Scholar]
- 36.Li Q., Duan L., Estes J. D., Ma Z. M., Rourke T., Wang Y., Reilly C., Carlis J., Miller C. J., Haase A. T. Nature. 2005;434:1148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- 37.Gringeri A., Musicco M., Hermans P., Bentwich Z., Cusini M., Bergamasco A., Santagostino E., Burny A., Bizzini B., Zagury D. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1999;20:358–370. doi: 10.1097/00042560-199904010-00006. [DOI] [PubMed] [Google Scholar]
- 38.Roederer M., Treister A., Moore W., Herzenberg L. A. Cytometry. 2001;45:37–46. doi: 10.1002/1097-0320(20010901)45:1<37::aid-cyto1142>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.