Abstract
Background
SARS coronavirus (SARS-CoV) is the etiologic agent of the severe acute respiratory syndrome. SARS-CoV mainly infects tissues of non-lymphatic origin, and the cytokine profile of those cells can determine the course of disease. Here, we investigated the cytokine response of two human non-lymphatic cell lines, Caco-2 and HEK 293, which are fully permissive for SARS-CoV.
Results
A comparison with established cytokine-inducing viruses revealed that SARS-CoV only weakly triggered a cytokine response. In particular, SARS-CoV did not activate significant transcription of the interferons IFN-α, IFN-β, IFN-λ1, IFN-λ2/3, as well as of the interferon-induced antiviral genes ISG56 and MxA, the chemokine RANTES and the interleukine IL-6. Interestingly, however, SARS-CoV strongly induced the chemokines IP-10 and IL-8 in the colon carcinoma cell line Caco-2, but not in the embryonic kidney cell line 293.
Conclusion
Our data indicate that SARS-CoV suppresses the antiviral cytokine system of non-immune cells to a large extent, thus buying time for dissemination in the host. However, synthesis of IP-10 and IL-8, which are established markers for acute-stage SARS, escapes the virus-induced silencing at least in some cell types. Therefore, the progressive infiltration of immune cells into the infected lungs observed in SARS patients could be due to the production of these chemokines by the infected tissue cells.
Background
For most viruses, the initial encounter with the host takes place in cells of non-lymphatic origin. The outcome of this primary infection can determine the course of disease, and the cytokine response of the infected cell plays a vital part. Type I interferons (IFN-α/β) are potent, antivirally active cytokines which can be produced by most, if not all, body cells in response to virus infection. IFNs not only trigger the synthesis of antivirally active proteins, they also activate the innate immune system and help to shape adaptive immunity [1]. Other virus-induced cytokines and chemokines activate the adaptive immune system and direct the migration of leukocytes [2]. Viruses, on the other hand, have evolved various mechanisms to counteract the host's cytokine response [3], and their ability to induce or inhibit cytokine production in infected cells has direct consequences for the balance between host defense and virus propagation.
SARS coronavirus (SARS-CoV) is the etiologic agent of severe acute respiratory syndrome (SARS), a life-threatening new human disease which recently emerged in China [4-7]. Characteristic SARS symptoms are high fever, myalgia, dry cough and lymphopenia, and in around 30% of cases patients also developed an atypical form of pneumonia [8].
The mechanisms underlying SARS-CoV-mediated pathogenesis remain largely unexplained. Autopsies from deceased patients revealed severe damage of the lungs and lymphatic tissues, accompanied by infiltrations of monocytic cells [9-11]. This may indicate that immunopathogenesis is involved in the severe outcome of the disease, providing the rationale for SARS therapy with immunosuppressant corticosteroids [12]. On the other hand, there is evidence that cell damages could be directly caused by the virus, since SARS-CoV is cytolytic [13] and capable to systemically infect human hosts [14-16]. In addition, virus particles and signs of necrosis were found in affected tissues [11], and high viral loads are predictive of adverse clinical outcome [17]. Interestingly, however, the acute lung injuries and respiratory failure observed in severe cases occured while viral loads were declining [16], again favouring the hypothesis of immune-mediated lung damage.
Virus-induced cytokines not only play a significant role in host defense, but also in immunopathogenesis. Investigations of the cytokine profiles of SARS patients have shown that the proinflammatory cytokines and chemokines IL-6, IL-8 and IP-10 (CXCL10) are strongly upregulated [18-23]. Cell culture studies, by contrast, did not reveal a clear picture of SARS-CoV-induced cytokines. In some cases both IL-8 and IP-10 were upregulated [24], whereas in other cases either only IL-8 [25,26], only IP-10 [27], or no cytokines were induced at all [28]. IL-6 was only moderately upregulated [29], or not detected at all [24-26]. Thus, it is still unclear whether the cytokine storm in SARS patients was directly caused by the virus, i.e. produced by SARS-CoV-infected cells, or whether it is a secondary effect, i.e. the result of strong activation of the immune system.
With one notable exception [24], most studies investigating the cellular cytokine response to SARS-CoV were either based on immune cells [25,27-29] or on Huh7 hepatoma cells inoculated with unphysiologically high amounts of virus [26]. Thus, the overall picture of the cytokine response of non-immune cells, which are most probably the prime targets of SARS-CoV, may still be incomplete. To learn more about it, a human cell line would be needed which, on one hand, can support the complete viral replication cycle, but on the other hand is also able to produce cytokines which are potentially antiviral. However, most cell lines which are permissive for SARS-CoV have lost the ability to synthesize IFNs, the most potent antiviral cytokines [24,30,31]. In this study, we identified an IFN-competent human embryonic kidney (HEK) 293 cell clone which supports the growth of SARS-CoV. Using these cells as well as the established human colon carcinoma cell line Caco-2 [24,31], we investigated the SARS-CoV-induced production of representative cytokines, chemokines and antiviral genes. Our studies revealed that SARS-CoV is capable to suppress the antiviral cytokine response of infected cells to a large extent. Interestingly, however, induction of the chemokines IP-10 and IL-8 escaped suppression by SARS-CoV in Caco-2 cells, but not in HEK 293s. Thus, SARS-CoV efficiently blocks the innate host cell defense at a very early step of infection, buying time to colonize the host. With the possible exception of IP-10 and IL-8, most cytokines detected in SARS patients may therefore be produced by the infiltrating immune cells, and not by the resident tissue cells. These data may help to explain both the rapid rise in virus titers during the initial stage of disease, caused by the suppression of antiviral cytokines, as well as the progressive infiltration of immune cells into the infected lungs, which could be due to the production of chemokines by the infected tissue cells.
Results
Growth of SARS-CoV in different cell lines
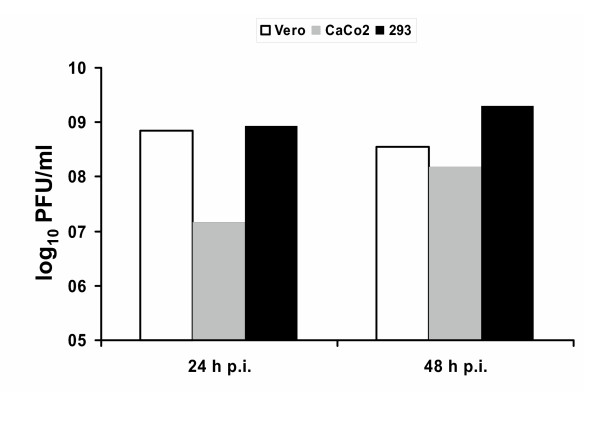
Vero cells, which are standard for growth of SARS-CoV [30,31], lack type I IFN genes [32,33] and therefore are not suitable for cytokine analyses. In search of an appropriate in vitro system, we tested several IFN-competent human cell lines for SARS-CoV growth and identified a low-passage clone of HEK 293 cells [34] as being fully permissive. Fig. 1 shows that titers of HEK 293 cells and Vero cells were comparable and rather high already at 24 h post-infection. Caco-2 cells, by contrast, produce approximately 100-fold less virus at 24 h post-infection, and 10-fold less at 48 h post-infection (Fig. 1). Thus, we considered both the Caco-2 cells and the low passage HEK 293 cells as useful systems for studying the influence of SARS-CoV on the immune system-independent induction of cytokines.
Figure 1.
Virus titers. Simian Vero cells (white bars), human Caco-2 cells (grey bars), and human low-passage HEK 293 cells (black bars) were infected at a multiplicity of infection (MOI) of 5 infectious particles per cell. Virus titers in the supernatants were determined 24 h post-infection and 48 h post-infection by plaque assays.
Interferon genes and their antiviral effectors
To properly assess the cytokine profile of SARS-CoV infection, we compared it with well-characterized cytokine inducers such as Bunyamwera delNSs virus (BdNSs [35]), Sendai virus (SeV) and Newcastle disease virus (NDV). In addition, we deemed it necessary to monitor cytokine synthesis both at 8 h and at 16 h post-infection, since we previously found a striking difference between the early and the late host cell response to SARS-CoV [36].
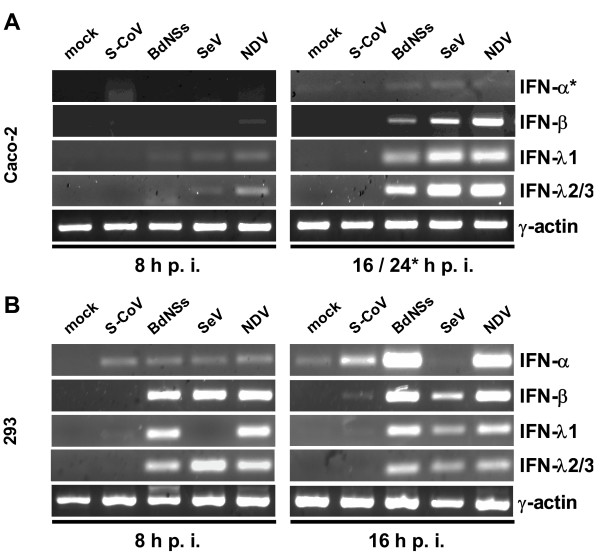
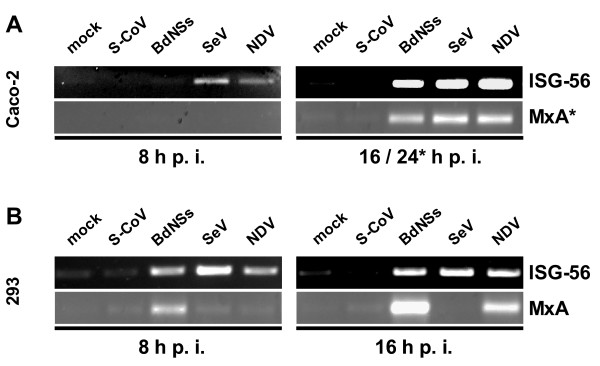
The first set of tested cytokines comprised the classical antiviral cytokines IFN-α and IFN-β [1], and the novel interferons IFN-λ1 and IFN-λ2/3 [37]. To test their induction in cell culture, we infected with 5 plaque-forming units (pfu) of viruses per cell, and analyzed cytokine mRNAs by RT-PCR. As it is shown in (Fig. 2A and 2B), clear signals for all IFNs were detected after infection with the control viruses BdNSs, SeV and NDV. For SARS-CoV, by contrast, only a weak signal for IFN-α was detected in HEK 293 cells, and none for IFN-β or the IFN-λs in either cell line. All preparations contained similar amounts of input RNA, since the γ-actin control mRNA was present in equal amounts (Fig. 2A and 2B, lower panels). It was of interest to see whether virus infection would lead to the upregulation of antiviral, IFN-stimulated genes (ISGs). As specific and sensitive markers we used the ISG56 gene which is induced both by IFNs and by virus infection [38,39] and the MxA gene which is exclusively activated by IFNs [40]. As is evident from Fig. 3, no significant ISG induction occurs for SARS-CoV, whereas the control viruses activated ISG expression. Note that SeV blocks in HEK 293 cells the synthesis of IFN-α (see Fig. 2B, upper right panel) and of MxA (Fig. 3B, lower right panel), most probably because of its ability to inhibit IFN-induced signaling [41]. Curiously, this does not happen in Caco-2 cells (Fig. 3A, lower right panel), suggesting cell type-specific differences in cytokine signaling.
Figure 2.
Interferon production by virus-infected human cells. Caco-2 cells (A) and HEK 293 cells (B) were infected with SARS-CoV or the IFN-inducing control viruses Bunyamwera delNSs (BdelNSs), Sendai virus (SeV), Newcastle disease virus (NDV), or were left uninfected (mock). At 8 h (left panels) or at 16 h (right panels) post-infection, total RNA was isolated and investigated by RT-PCR for the presence of different IFN mRNAs. The cellular γ-actin mRNA served as loading control. Note that for the reliable detection of IFN-α in Caco-2 cells (A, upper right panel) the infection time had to be extended to 24 h.
Figure 3.
Interferon-stimulated genes. RNA samples of Caco-2 cells (A) and HEK 293 cells (B) described in Fig. 2 were investigated by RT-PCR for the presence of ISG56 and MxA mRNAs. As for IFN-α (see Fig. 2A), for detection of MxA mRNA in Caco-2 cells an extended infection period of 24 h was necessary (A, lower right panel).
Taken together, these data demonstrate that, in contrast to the other viruses tested, SARS-CoV suppresses the activation of the antiviral IFNs and the IFN-induced effector genes to a large extent.
Induction of chemokines by infected cells
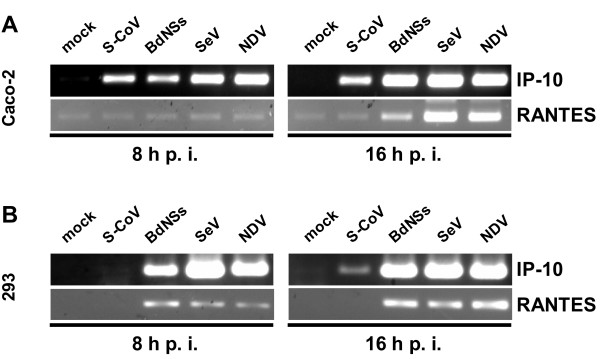
IP-10 and RANTES are potent chemoattractants for activated T cells and NK cells [2]. When we infected Caco-2 cells, significant amounts of IP-10 mRNA were synthesized (Fig. 4A, upper panels). This strong upregulation occurred independently of the virus, suggesting a general response to virus infection. Although IP-10 mRNA levels induced at 16 h p.i. by SARS-CoV are slightly lower than by the other viruses, our data are in good agreement with previous studies [24,27]. Induction of RANTES, by contrast, was restricted to the cytokine-inducing viruses, whereas infection with SARS-CoV had no effect above background levels (Fig. 4A, lower panels). We then tested HEK 293 cells in a similar way. Much to our surprise, IP-10 mRNA was not detectable early after infection with SARS-CoV (Fig. 4B, upper left panel), and only very weakly expressed after longer infection (Fig. 4B, upper right panel).
Figure 4.
Chemokine production. RNA samples of Caco-2 cells (A) and HEK 293 cells (B) described in Fig. 2 were assayed by RT-PCR for IP-10 and RANTES mRNA levels.
RANTES mRNA again was not detectable for SARS-CoV (Fig. 4B, lower panels). All three cytokine-inducing viruses activated IP-10 and RANTES expression in HEK 293 cells as expected (Fig. 4B, upper and lower panels).
Thus, SARS-CoV induces IP-10 gene expression in Caco-2 cells, but not in HEK 293 cells, again suggesting that the cytokine response is dependent on the host cell type. RANTES expression, by contrast, is never induced by SARS-CoV, although the cells respond normally to other viruses.
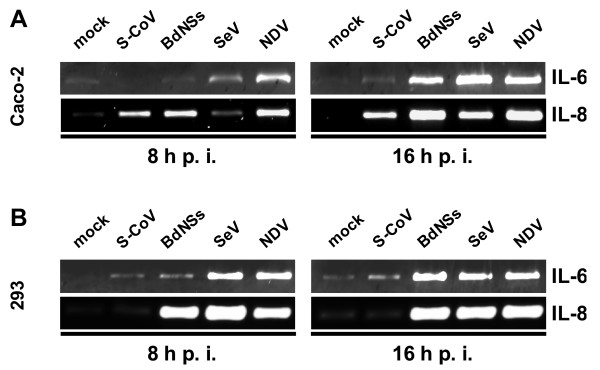
Induction of IL-6 and IL-8
The proinflammatory cytokine IL-6 and the chemokine IL-8 are strongly upregulated in SARS patients [18,19], but from cell culture studies no clear picture emerged [24-26,29]. We investigated IL-6 and IL-8 production by Caco-2 and HEK 293 cells infected with SARS-CoV and compared it with the other RNA viruses. As shown in Fig. 5, IL-6 is induced only weakly by SARS-CoV, independent of the cell line used (Fig. 5A and 5B, upper panels). IL-8, by contrast, is clearly induced by SARS-CoV in Caco-2 cells, but not in HEK 293 cells (Fig. 5A and 5B, lower panels). The control viruses invariably induced both IL-6 and IL-8, demonstrating that the cell lines are capable to produce these cytokines.
Figure 5.
Interleukin production. RNA samples of Caco-2 cells (A) and HEK 293 cells (B) described in Fig. 2 were investigated by RT-PCR for the presence of IL-6 and IL-8 mRNAs.
Thus, SARS-CoV strongly induces IL-8, but not IL-6 in a cell-type dependent manner. This may suggest that the IL-8 detected in SARS patients [18,19] is directly synthesized by infected resident cells, whereas IL-6 is more likely a secondary response mediated by infiltrating immune cells.
Taken together, our data demonstrate that SARS-CoV in general is a weak inducer of cytokines and antiviral genes in non-lymphatic cells. Chemokines like IP-10 and IL-8, however, can be directly upregulated in SARS-CoV in a cell-type-dependent manner.
Discussion
The activation of immune-relevant cytokines and host cell genes by SARS-CoV in cells and patients was the subject of several previous investigations [18,21,22,24-29,42-44]. However, most of the cell culture studies were either based on immune cells which do not represent the majority of infected cells [25,27-29], or on Huh7 hepatoma cells which needed to be infected with 100 pfu per cell, i.e. with unphysiologically high amounts of virus [26]. Moreover, Huh7 cells are known to be deficient in the antiviral cytokine response [45]. Thus, it was not entirely clear whether the patients' cytokine response was caused by virus-infected cells, or whether it was mediated by the activated immune system. Furthermore, it was not systematically investigated how the cytokine induction by SARS-CoV compares to other viruses. Here, we have used three control viruses and two different cells lines to elucidate and compare the induction of cytokines by SARS-CoV. Our results demonstrate that SARS-CoV does not induce significant amounts of IFNs, antiviral genes, RANTES, and IL-6. In agreement with this finding, SARS-CoV-infected macrophages and dendritic cells lack IFN induction [27,29]. IP-10 and IL-8, however, can be activated by SARS-CoV. This suggests that these chemokines, which are reliable markers of acute-stage SARS [18,20,21,23], are not only produced in response to IFN-γ after activation of the immune system as suggested, but may also be directly secreted by infected tissue cells. An upregulation of either IP-10 and/or IL-8 was observed in several studies using SARS-CoV-infected Caco-2 cells [24], macrophages [27], peripheral blood mononuclear cells [25], and dendritic cells [29]. Using HEK 293 cells, by contrast, we found that SARS-CoV is able to downregulate also IP-10 and IL-8 production. Similarly, recent studies showed that peripheral blood monocytes from SARS patients do not produce any cytokines [28]. Thus, chemokine induction by SARS-CoV appears to be highly cell type-specific.
With the exception of IP-10 and IL-8, SARS-CoV is capable to suppress the production of a wide range of cytokines. This is in agreement with our previous finding that SARS-CoV inhibits the crucial cytokine transcription factor IRF-3 [36], providing a possible mechanism for the high potential of this pathogen to suppress the host response. Of note, SARS-CoV is highly sensitive to the antiviral action of IFNs both in vivo and in vitro [46-51], thus explaining why the virus needs to suppress IFN induction in advance.
Conclusion
In the initial phase of SARS, the virus grows exponentially and spreads to different organs, including the lungs [8,14]. Our data may explain this rapid and efficient dissemination of SARS-CoV. By slowing down expression of IFNs and their antiviral genes in the infected tissue cells, the virus buys time during the initial, critical phase of infection in order to grow unhindered in the host. At the same time, however, the virus-induced chemokines IP-10 and IL-8 attract immune cells. Possibly, this mixture of high-level virus replication followed by the invasion of activated immune cells results in a strong inflammatory response, leading to a cytokine storm and the severe and potentially fatal respiratory distress which is the hallmark of full-blown SARS.
Methods
Cells and viruses
Simian VeroE6 cells, human Caco-2 cells and human embryonic kidney (HEK) 293 cells were maintained and grown as described [24,36]. The low-passage HEK 293 cell clone [34] was purchased from Microbix Biosystems, Toronto, Canada. All experiments were performed with HEK 293 cells between passage 38 and 48. The FFM-1 isolate of SARS-CoV was kindly provided by Stephan Becker, University of Marburg, Germany. Bunyamwera delNSs virus [35], Sendai virus and Newcastle disease virus were used as controls.
Plaque assays
Virus plaque assays were performed as described previously [50]. Briefly, Vero cell monolayers were infected with dilutions of supernatants from infected cells, overlaid with soft agar, and allowed to form plaques for 72 h. Then the agar overlay was removed and cells were stained with a solution of 1% crystal violet, 3,6% formaldehyde, 1% methanol, and 20% ethanol.
RT-PCR analyses
Cells were infected for the indicated times, total RNA was extracted and treated with DNase I. For reverse transcription (RT), 1 μg of RNA was incubated with 200 U of Superscript II reverse transcriptase (Invitrogen) and 100 ng random hexanucleotides in 20 μl of 1×RT buffer (Invitrogen) supplied with 1 mM each of the four deoxynucleotide triphosphates, 20 U of RNasin, and 10 mM dithiothreitol. The resulting cDNA was amplified by 35 cycles of PCR, with each cycle consisting of 30 sec at 94°C, 1 min at 58°C (using primer pairs specific for IP-10, IL-6, IL-8 and RANTES) or at 56°C (all other primer pairs), and 1 min at 72°C, followed by 10 min at 72°C. Primer sequences are available from the authors upon request.
List of abbreviations
BdNSs, Bunyamwera delNSs virus; HEK, human embryonic kidney; IFN, interferon; IL, interleukine; ISG, interferon-stimulated gene; NDV, Newcastle disease virus; RANTES, Regulated on activation, normal T cell expressed and secreted; SARS-CoV, SARS coronavirus; SeV, Sendai virus
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
MS carried out the virus growth studies and the RT-PCR analyses, participated in the design of the study, and has given final approval of the version to be published. FW carried out virus infections, participated in the design of the study, and was responsible for drafting and finalizing the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Otto Haller for support and helpful comments, and Martin Michaelis and Peter Staeheli for critically reading the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (grant We 2616/4) and the Sino-German Center for Research promotion (grant GZ Nr. 239 (202/12)).
Contributor Information
Martin Spiegel, Email: martin.spiegel@uniklinik-freiburg.de.
Friedemann Weber, Email: friedemann.weber@uniklinik-freiburg.de.
References
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;4:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins BJ. Chemokines. Blood. 1997;90:909–28. [PubMed] [Google Scholar]
- Weber F, Kochs G, Haller O. Inverse interference: how viruses fight the interferon system. Viral Immunol. 2004;17:498–515. doi: 10.1089/vim.2004.17.498. [DOI] [PubMed] [Google Scholar]
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–76. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–25. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–66. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, van Amerongen G, van Riel D, Laman JD, de Jong T, van Doornum G, Lim W, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–70. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10:S88–97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Wang H, Shen H, Li Z, Geng J, Han H, Cai J, Li X, Kang W, Weng D, et al. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J Pathol. 2003;200:282–9. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang ZW, Zhang LJ, Zhang SJ, Meng X, Li JQ, Song CZ, Sun L, Zhou YS, Dwyer DE. A clinicopathological study of three cases of severe acute respiratory syndrome (SARS) Pathology. 2003;35:526–31. doi: 10.1080/00313020310001619118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, Chu CM, Hui PK, Mak KL, Lim W, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–8. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo JF, Munoz-Fernandez MA. Severe acute respiratory syndrome, a pathological immune response to the new coronavirus – implications for understanding of pathogenesis, therapy, design of vaccines, and epidemiology. Viral Immunol. 2004;17:535–44. doi: 10.1089/vim.2004.17.535. [DOI] [PubMed] [Google Scholar]
- Ng ML, Tan SH, See EE, Ooi EE, Ling AE. Proliferative growth of SARS coronavirus in Vero E6 cells. J Gen Virol. 2003;84:3291–303. doi: 10.1099/vir.0.19505-0. [DOI] [PubMed] [Google Scholar]
- Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, Zou W, Zhan J, Wang S, Xie Z, et al. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202:415–24. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng LF, Wong M, Koh S, Ooi EE, Tang KF, Leong HN, Ling AE, Agathe LV, Tan J, Liu ET, et al. Detection of severe acute respiratory syndrome coronavirus in blood of infected patients. J Clin Microbiol. 2004;42:347–50. doi: 10.1128/JCM.42.1.347-350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, Poon LL, Law KI, Tang BS, Hon TY, Chan CS, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–72. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung IF, Cheng VC, Wu AK, Tang BS, Chan KH, Chu CM, Wong MM, Hui WT, Poon LL, Tse DM, et al. Viral loads in clinical specimens and SARS manifestations. Emerg Infect Dis. 2004;10:1550–7. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KJ, Su IJ, Theron M, Wu YC, Lai SK, Liu CC, Lei HY. An interferon-gamma-related cytokine storm in SARS patients. J Med Virol. 2005;75:185–94. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh PR, Chen PJ, Hsiao CH, Yeh SH, Cheng WC, Wang JL, Chiang BL, Chang SC, Chang FY, Wong WW, et al. Patient data, early SARS epidemic, Taiwan. Emerg Infect Dis. 2004;10:489–93. doi: 10.3201/eid1003.030571. [DOI] [PubMed] [Google Scholar]
- Wang WK, Chen SY, Liu IJ, Kao CL, Chen HL, Chiang BL, Wang JT, Sheng WH, Hsueh PR, Yang CF, et al. Temporal relationship of viral load, ribavirin, interleukin (IL)-6, IL-8, and clinical progression in patients with severe acute respiratory syndrome. Clin Infect Dis. 2004;39:1071–5. doi: 10.1086/423808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CK, Lam CW, Wu AK, Ip WK, Lee NL, Chan IH, Lit LC, Hui DS, Chan MH, Chung SS, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BGoNRPf SARS Dynamic changes in blood cytokine levels as clinical indicators in severe acute respiratory syndrome. Chin Med J (Engl) 2003;116:1283–7. [PubMed] [Google Scholar]
- Xu L, Ran L, Cameron MJ, Persad D, Danesh A, Gold W, Keshavjee S, Brunton J, Loeb M, Kelvin DJ. Proinflammatory gene expression profiles and severity of disease course in SARS patients. International Conference on SARS, one year after the (first) outbreak; Luebeck, Germany. 2004.
- Cinatl J, Jr, Hoever G, Morgenstern B, Preiser W, Vogel JU, Hofmann WK, Bauer G, Michaelis M, Rabenau HF, Doerr HW. Infection of cultured intestinal epithelial cells with severe acute respiratory syndrome coronavirus. Cell Mol Life Sci. 2004;61:2100–12. doi: 10.1007/s00018-004-4222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng LF, Hibberd ML, Ooi EE, Tang KF, Neo SY, Tan J, Murthy KR, Vega VB, Chia JM, Liu ET, et al. A human in vitro model system for investigating genome-wide host responses to SARS coronavirus infection. BMC Infect Dis. 2004;4:34. doi: 10.1186/1471-2334-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BS, Chan KH, Cheng VC, Woo PC, Lau SK, Lam CC, Chan TL, Wu AK, Hung IF, Leung SY, et al. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J Virol. 2005;79:6180–93. doi: 10.1128/JVI.79.10.6180-6193.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CY, Poon LL, Ng IH, Luk W, Sia SF, Wu MH, Chan KH, Yuen KY, Gordon S, Guan Y, et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol. 2005;79:7819–26. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reghunathan R, Jayapal M, Hsu LY, Chng HH, Tai D, Leung BP, Melendez AJ. Expression profile of immune response genes in patients with Severe Acute Respiratory Syndrome. BMC Immunol. 2005;6:2. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law HK, Cheung CY, Ng HY, Sia SF, Chan YO, Luk W, Nicholls JM, Peiris JS, Lau YL. Chemokine upregulation in SARS coronavirus infected human monocyte derived dendritic cells. Blood. 2005. [DOI] [PMC free article] [PubMed]
- Hattermann K, Muller MA, Nitsche A, Wendt S, Donoso Mantke O, Niedrig M. Susceptibility of different eukaryotic cell lines to SARS-coronavirus. Arch Virol. 2005 doi: 10.1007/s00705-004-0461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossel EC, Huang C, Narayanan K, Makino S, Tesh RB, Peters CJ. Exogenous ACE2 expression allows refractory cell lines to support severe acute respiratory syndrome coronavirus replication. J Virol. 2005;79:3846–50. doi: 10.1128/JVI.79.6.3846-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz MO, Ziemin S, Le Beau MM, Pitha P, Smith SD, Chilcote RR, Rowley JD. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc Natl Acad Sci USA. 1988;85:5259–63. doi: 10.1073/pnas.85.14.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emeny JM, Morgan MJ. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J Gen Virol. 1979;43:247–52. doi: 10.1099/0022-1317-43-1-247. [DOI] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Weber F, Bridgen A, Fazakerley JK, Streitenfeld H, Randall RE, Elliott RM. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J Virol. 2002;76:7949–7955. doi: 10.1128/JVI.76.16.7949-7955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M, Pichlmair A, Martinez-Sobrido L, Cros J, Garcia-Sastre A, Haller O, Weber F. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J Virol. 2005;79:2079–86. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- Guo J, Hui DJ, Merrick WC, Sen GC. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. Embo J. 2000;19:6891–9. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SE, Noyce RS, Mossman KL. Innate cellular response to virus particle entry requires IRF3 but not virus replication. J Virol. 2004;78:1706–17. doi: 10.1128/JVI.78.4.1706-1717.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O, Kochs G. Interferon-induced Mx proteins: dynamin-like GTPases with antiviral activity. Traffic. 2002;3:710–7. doi: 10.1034/j.1600-0854.2002.31003.x. [DOI] [PubMed] [Google Scholar]
- Garcin D, Latorre P, Kolakofsky D. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J Virol. 1999;73:6559–65. doi: 10.1128/jvi.73.8.6559-6565.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan ZP, Chen Y, Zhang J, Zhao J, Lang ZW, Meng FK, Bao XL. [Clinical characteristics and mechanism of liver injury in patients with severe acute respiratory syndrome] Zhonghua Gan Zang Bing Za Zhi. 2003;11:493–6. [PubMed] [Google Scholar]
- Yilla M, Harcourt BH, Hickman CJ, McGrew M, Tamin A, Goldsmith CS, Bellini WJ, Anderson LJ. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res. 2005;107:93–101. doi: 10.1016/j.virusres.2004.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li J, Zhan Y, Wu L, Yu X, Zhang W, Ye L, Xu S, Sun R, Wang Y, et al. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect Immun. 2004;72:4410–5. doi: 10.1128/IAI.72.8.4410-4415.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Guerra B, Lee H, Averett DR, Pfeiffer B, Chavez D, Notvall L, Bigger C. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(i)-poly(c), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. J Virol. 2003;77:1092–104. doi: 10.1128/JVI.77.2.1092-1104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Treatment of SARS with human interferons. Lancet. 2003;362:293–4. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J, Jr, Michaelis M, Scholz M, Doerr HW. Role of interferons in the treatment of severe acute respiratory syndrome. Expert Opin Biol Ther. 2004;4:827–36. doi: 10.1517/14712598.4.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans BL, Kuiken T, Martina BE, Fouchier RA, Rimmelzwaan GF, Van Amerongen G, Van Riel D, De Jong T, Itamura S, Chan KH, et al. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat Med. 2004;10:290–3. doi: 10.1038/nm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley LE, Fritz LE, Jahrling PB, Karp CL, Huggins JW, Geisbert TW. Interferon-beta 1a and SARS coronavirus replication. Emerg Infect Dis. 2004;10:317–9. doi: 10.3201/eid1002.030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M, Pichlmair A, Mühlberger E, Haller O, Weber F. The antiviral effect of interferon-beta against SARS-Coronavirus is not mediated by MxA. J Clinical Virol. 2004;30:211–213. doi: 10.1016/j.jcv.2003.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroher U, DiCaro A, Li Y, Strong JE, Aoki F, Plummer F, Jones SM, Feldmann H. Severe acute respiratory syndrome-related coronavirus is inhibited by interferon- alpha. J Infect Dis. 2004;189:1164–7. doi: 10.1086/382597. [DOI] [PMC free article] [PubMed] [Google Scholar]