Abstract
We describe the molecular characterization and function of vielfältig (vfl), a X-chromosomal gene that encodes a nuclear protein with six Krüppel-like C2H2 zinc finger motifs. vfl transcripts are maternally contributed and ubiquitously distributed in eggs and preblastoderm embryos, excluding the germline precursor cells. Zygotically, vfl is expressed strongly in the developing nervous system, the brain, and in other mitotically active tissues. Vfl protein shows dynamic subcellular patterns during the cell cycle. In interphase nuclei, Vfl is associated with chromatin, whereas during mitosis, Vfl separates from chromatin and becomes distributed in a granular pattern in the nucleoplasm. Functional gain-of-function and lack-of-function studies show that vfl activity is necessary for normal mitotic cell divisions. Loss of vfl activity disrupts the pattern of mitotic waves in preblastoderm embryos, elicits asynchronous DNA replication, and causes improper chromosome segregation during mitosis.
INTRODUCTION
Zinc fingers constitute the most abundant structural motifs in the proteome predicted from the genome sequences of Saccharomyces cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans (Rubin et al., 2000) as well as the draft human genomic sequences (Lander et al., 2001). Zinc finger proteins are best known as transcriptional regulators that participate in a variety of cellular activities such as development, differentiation, and tumor suppression. The most common form of zinc finger domain is the so called C2H2 domain, first identified in the basal transcription factor TFIIIA in Xenopus laevis (Miller et al., 1985) and subsequently found also in DNA-binding transcription factors regulating polymerase II-dependent gene expression (Rosenberg et al., 1985). The three-dimensional structure of the basic C2H2 zinc finger is a small domain composed of a β-hairpin followed by an α-helix, a structure that is held in place by a zinc ion. DNA-binding C2H2 zinc fingers generally occur as tandem arrays with a minimum of two fingers needed to specify the DNA-binding site (Choo and Klug, 1995). In addition to DNA-binding, zinc finger proteins have been implicated in RNA-binding, protein-protein interactions, and lipid binding (Lorick et al., 1999; Bach, 2000; Tucker et al., 2001).
DNA-binding C2H2 zinc finger proteins contain protein-binding domains that provide the basis for the assembly of regulatory complexes involved in chromatin remodeling and transcriptional regulation (Aasland et al., 1995; David et al., 1998). They are frequently expressed in distinct spatial and temporal patterns, and their subcellular localization was found to be regulated in a cell cycle-dependent manner. The entry of mitosis is characterized by the transcriptional shutdown (Prescott and Bender, 1962) that involves inactivation of the general transcription machinery (Segil et al., 1996; Bellier et al., 1997; Akoulitchev and Reinberg, 1998; Long et al., 1998) as well as gene-specific transcription factors during the G2/M transition during mitosis. For some factors, reduced DNA-binding activities have been observed in extracts from mitotic cells (Caelles et al., 1995; Martinez-Balbas et al., 1995; Gottesfeld and Forbes, 1997). For other factors, DNA-binding activities were not affected, but they were displaced from chromatin during mitosis (Martinez-Balbas et al., 1995; Gottesfeld and Forbes, 1997). The reason for the inactivation of gene-specific transcription factors and the mechanism of their inhibition is not known. However, one can speculate that this process is necessary for chromatin condensation. Furthermore, only few factors have been identified that are required for the reinitiation of transcription. Among these factors, the mammalian C2H2 zinc finger protein Ikaros has a function during the reestablishment of gene expression patterns and/or for the heritable silencing of developmentally regulated genes after mitosis (Hahm et al., 1994; Molnar and Georgopoulos, 1994; Georgopoulos, 2002; Smale and Fisher, 2002).
Here, we report the isolation and characterization of the D. melanogaster gene vielfältig (vfl). The gene codes for a member of the C2H2 zinc finger proteom (Chung et al., 2002) that is strongly expressed in mitotically dividing cells. The vfl protein (Vfl) shows a distinct subcellular localization pattern during the cell cycle. Analysis of newly generated vfl mutants, RNA interference (RNAi) injection, and vfl overexpression indicate that vfl is necessary for proper mitosis. Altered levels of vfl activity affect the synchronous early nuclear divisions in the embryo, cause asynchronous DNA replication patterns, and impair chromosome segregation.
MATERIALS AND METHODS
Genetics and Molecular Biology
The vfl gene locus is located on the X-chromosome at position 18F with a single unspliced open reading frame of 4788 base pairs. The expressed sequence tag LD47418 contains the full open reading frame of vfl and was used to generate specific anti-sense digoxigenin-labeled probes according to the instructions of the manufacturer (Roche Diagnostics, Basel, Switzerland) and to generate a pUAST vfl construct. P-elements EP(X)1344 (Rorth, 1996) and PG0353 are positioned in the promotor region, whereas the P-elements PG0225 and PG0427 (Peter et al., 2002) reside in the 5′ untranslated leader sequence of the transcribed region. PG0353 and PG0427 represent strong vfl alleles, showing no or dramatically reduced vfl expression, respectively, whereas PG0225 and EP(X)1344 have only minor effects on expression. EP(X)1344 was used for overexpression experiments.
apGAL4 (Bloomington stock collection, Drosophila Stock Center, Indiana University, Bloomington, IN) controls expression in the ap expression domain, allowing specific expression in the dorsal compartment. The ftzNG-GAL4 (Bloomington stock collection) controls expression in a subset of mitotic dividing neuroblasts, btlGAL4 (Bloomington stock collection) drives high levels of expression in the developing tracheal system and engrailed GAL4 (kindly provided by Andrea Brand, University of Cambridge, Cambridge, United Kingdom) in an epidermal pattern. V32GAL4 (kindly provided by Daniel St. Johnston, University of Cambridge, Cambridge, United Kingdom) contains the DNA-binding domain of GAL4 (amino acids 1-147) with the transactivation domain of VP16 under the expression control of the maternal promotor of α4-tubulin and allows maternal expression in high amounts. For RNAi injection, embryos carrying a His2AVD green fluorescent protein (GFP) (Clarkson and Saint, 1999) were used. All genetic experiments were carried out at 25°C.
Antibodies and Microscopy
Fixations of embryos and third instar imaginal discs, protein detection, and RNA in situ hybridization were performed according to Lehmann and Tautz (1994) and fluorescent detection of RNA according to Knirr et al. (1999). Proteins were either detected using the AB system (Vectastain; Vector Laboratories, Burlingame, CA) or by immunofluorescence by using secondary antibodies directly coupled to Alexa-488 and Alexa-546 (Invitrogen, Carlsbad, CA) or by using the TSA system (PerkinElmer Life and Analytical Sciences, Boston, MA) and documented with a Zeiss Axioplan microscope with Nomarski optics or with a Zeiss LSM confocal microscope. For time-lapse videomicroscopy, images were taken every 30 s on the Zeiss LSM confocal microscope. The following primary antibodies were used: monoclonal antibody (mAb) 22C10 (1:20; Iowa Hybridoma Bank, University of Iowa, Iowa City, IA), anti-Kr (1:250; kindly provided by Pilar Carrera, Universität Bonn, Bonn, Germany), anti-phosphorylated histone H3 (1:500; Upstate Biotechnology, Lake Placid, NY), anti-β-tubulin E7 (1:100; Iowa Hybridoma Bank), or anti-α-tubulin DM1A (Sigma-Aldrich, St. Louis, MO) and anti Crumbs (1:20; Iowa Hybridoma Bank). DNA was stained using Sytox Green (Invitrogen) or Draq5 (Biostatus, Shepshed, Leicestershire, United Kingdom).
Rabbit anti-Vfl antibodies were generated against two peptides corresponding to region 540-554 (TWKSNEARRPKTYNC) and region 923-937 (EQQGTDFSRTTPPQ), respectively (Eurogentec, Seraing, Belgium). After affinity purification using peptides linked to CNBr-activated Sepharose 4B according to the instructions of the manufacturer (Pfizer, New York, NY), embryos were stained (see above). Both affinity-purified antisera (1:200 dilution) showed identical expression and subcellular localization patterns using the TSA detection system (PerkinElmer Life and Analytical Sciences). Furthermore, we used different GAL4 drivers (such as breathless-GAL4 and engrailed-GAL4) to ectopically express vfl cDNA in the developing tracheal system of the embryo and in epidermal stripes, i.e., locations where the gene is normally not expressed. The affinity-purified anti-Vfl antibodies were able to detect ectopically expressed Vfl in the nuclei of the GAL4-expressing cells in a UAS-dependent manner, and the subcellular staining patterns were identical to the patterns of Vfl in cells that express endogenous Vfl. Moreover, injection of the affinity-purified anti-Vfl antibodies was able to cause a phenocopy of the vfl mutant phenotype.
RNAi Synthesis and Injection
To generate double-stranded RNA (dsRNA) of about 1 kb, PCR was performed using the following primers: 5′ sense T7, TAATACGACTCACTATAG GGCACCGTGGGCGTTGACTAT and 3′ sense, GGTTGAGGTGGTGCTGCT and 5′ antisense T7, TAATACGACTCACTATAG TAATACGACTCACTATAG and 3′ antisense, GGCACCGTGGGCGTTGACTAT. After cloning, the added T7 polymerase promotor sequences were used to generate complementary 1-kb RNA transcripts. After phenol/chloroform extraction, identical amounts were used for annealing and adjusted to 2 μg/μl. For the RNA injection, either wild-type embryos or embryos carrying a His2AVD GFP (Clarkson and Saint, 1999) were aged for 0-30 min, and the dsRNA was injected and deposited. As controls, a variety of different dsRNAs were injected, including probes directed against segmentation genes as well as bacterial lacZ. The injected embryos were either investigated using time-lapse imaging (see above) or were aged for additional 2 h and then fixed and stained. Note that no phenotype like the one described in response to vfl dsRNA was observed in response to control dsRNA and that the defects observed after vfl dsRNA injection phenocopied the vfl mutants.
Bromodeoxyuridine (BrdU) Labeling
Embryos were air-dried for 7 min after dechorionation, incubated for 7 min in n-octane, washed in Schneider cell medium, and then incubated for 20 min in 1 mg/ml BrdU (Sigma-Aldrich) in medium. After washing, the embryos were fixed and then incubated for 60 min in 2 N HCl. After washing in phosphate-buffered saline with 0.1% Tween 20, the incorporated BrdU was detected by an anti BrdU-antiserum (Roche Diagnostics) at a dilution of 1:100.
RESULTS
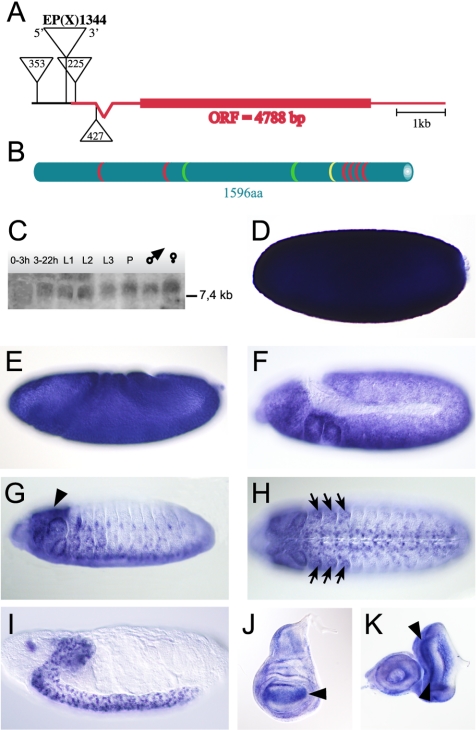
The annotated gene CG12701 of D. melanogaster is located in region 18F of the X-chromosome. Sequence alignment of various isolated cDNA clones and genomic DNA revealed a transcript of 7.7 kbp. It contains a single and unspliced open reading frame of 4788 base pairs and a single intron in the 5′ untranslated region (Figure 1A). The open reading frame translates into a polypeptide of 1596 amino acids. The putative protein contains a nuclear localization signal, two proline-rich domains, and six C2H2-type zinc finger motifs (Rosenberg et al., 1985), four of which are clustered in a C-terminal domain of 110 amino acids (Figure 1B; position 1325-1435). The zinc finger domain is highly similar to the DNA-binding zinc finger domains of the Krüppel-like transcription factors, suggesting that the protein is capable of interacting with DNA (Rosenberg et al., 1985). Sequence homology searches did not reveal a direct vertebrate homologue, but homologues were found in insects such as various Drosophila species, the mosquito Anopheles gambiae and the beetle Tribolium casteneum. For reasons that will be apparent from the mutant phenotype (see below), we refer to the gene as vielfältig (vfl), the German word for versatile or manifold.
Figure 1.
The vfl gene locus, the architecture of the Vfl protein and expression of the vfl transcript. (A) The vfl gene locus is located on the X-chromosome at position 18F with a single unspliced open reading frame of 4788 base pairs. The P-elements EP(X)1344 and PG0353 are positioned in the promotor region, and the P-elements PG0225 and PG0427 are integrated in the 5′ untranslated leader sequence. (B) The vfl gene encodes a 1596 amino acid protein with one nuclear localization signal (yellow), two proline-rich domains (green), and six Krüppel-like C2H2 zinc finger motifs (red), four of which clustered in the carboxy terminus. (C) Northern blot analysis shows expression of a single transcript of more than 7.4 kb throughout Drosophila development. (D) Lateral view of a stage 4 embryo with strong load of maternal contributed mRNA. During gastrulation, maternal mRNA decays (E) and first zygotic expression is detectable at stage 11 in the neuroectoderm and in head segments (F). At embryonic stage 14, vfl mRNA is expressed in the brain (G, lateral view, arrowheads), in the developing imaginal discs (H, arrows), and in cells of the CNS (H, ventral view). (I) At the end of embryogenesis vfl is specifically expressed in cells of the brain and the CNS. (J) In wing imaginal discs of third instars, vfl transcript is enriched in parts of the wing pouch (arrowhead). (K) In eye-antenna imaginal discs, vfl transcript is strongly expressed in two stripes anterior and posterior of the morphogenetic furrow where mitotically active cells are located (arrowheads). Abbreviations in C: L1, first instar; L2, second instar; L3, third instar; and P, pupae.
vfl Is Specifically Expressed in Mitotically Dividing Cells
Expression of vfl RNA was monitored by developmental Northern blot analysis as well as by in situ hybridization on whole mount preparations of embryos and on third instar imaginal discs. Developmental Northern blot analysis revealed that vfl transcripts are present throughout the Drosophila life cycle (Figure 1C). Whole mount in situ hybridization on embryos shows that vfl is strongly expressed during oogenesis. The transcripts are deposited into the egg and remain evenly distributed until blastoderm stage (Figure 1D), excluding the germline progenitor cells (Figure 1D). During gastrulation and germ band extension stages, when maternally derived transcripts become degraded, zygotic expression of the gene is found in the neuroectoderm and in the thoracic segments (Figure 1, E and F). During the subsequent stages of embryogenesis, transcripts remain in cells of the developing central nervous system (CNS), the peripheral nervous system (PNS), and the brain (Figure 1, G-I). Notably, the gene is not expressed in mitotically quiescent or endoreplicative tissues. During larval stages, vfl transcripts are highly enriched in mitotically active zones of imaginal discs such as the developing wing blade area (Figure 1J) or the proliferating cell areas of the eye disk (Wolff and Ready, 1993), i.e., the regions in front of and behind the morphogenetic furrow (Figure 1K). These observations suggest that vfl activity is connected to mitotic events. To study the vfl expression patterns and the subcellular localization of the transcripts in more detail and at higher resolution, we used fluorescence detection of vfl probes and anti-VFL antibodies during the cell cycle of mitotically active cells of the developing embryo.
Differential Subcellular Localization of Vfl during Mitosis
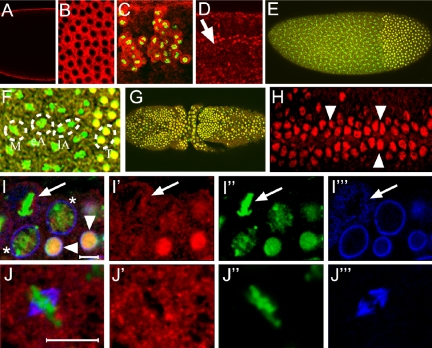
vfl RNA localizes at the apical site of the developing blastoderm cells in proximity of the dividing nuclei (Figure 2, A and B). In the neuroectoderm of embryos, transcripts are also localized at the apical site of the dividing neuroblasts (our unpublished data), suggesting that they are preferentially or exclusively expressed in mitotically active cells. To confirm vfl expression in the mitotically active cells, we combined in situ hybridization analysis with anti phosphorylated histone 3 (H3-P) antibodies, a chromatin marker to visualize mitotically active cells (Schmiesing et al., 2000). The results show that vfl RNA is highly enriched exclusively in the dividing cells of the early developing CNS (Figure 2C).
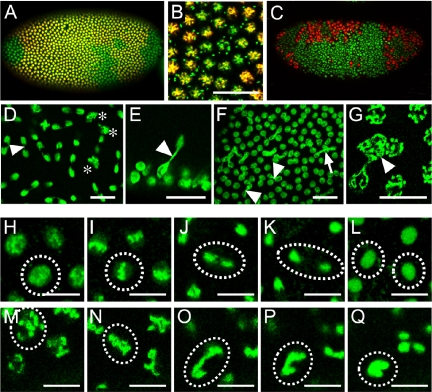
Figure 2.
vfl expression in mitotically dividing tissues and cell cycle dependent Vfl protein localization. (A-C, E-J) Wild-type and enlarged region (D) showing overexpressing vfl in cells of the tracheal system in response to the btlGAL4 driver. (A-C) Enlarged micrographs of embryos showing vfl transcripts (red) after in situ hybridizations with a fluorescent detection of the vfl probe. (A) Note apical localization of the transcripts during nuclear divisions of stage 4 embryos in a longitudinal optical section (A) and in a surface view (B). (C) Neural region of a stage 13 embryo showing a strong enrichment of vfl transcripts (red) in dividing cells of the CNS that are marked by H3-P (green). (D-J) Antibody staining using an anti-Vfl-specific antiserum (D-J, red). (D) Anti-Vfl antibody staining showing ectopic Vfl protein expression in the tracheal system (arrow in an enlargement corresponding to three segment equivalents) in response to btlGAL4 expression. This observation indicates that antibodies recognize the Vfl protein in cells where the protein is normally not expressed. (E) Syncytial blastoderm embryo showing that Vfl protein is associated with chromatin of interphase nuclei as shown by overlap (yellow) of Vfl staining (red) and DNA (green), in the posterior region of these embryos. (F) Enlargement of a mitotic wave border of the same embryo shown in E, indicating that Vfl protein is localized in small granular structures during metaphase (“M”) as well as during early and late anaphases (“eA”, “lA”). Note also chromatin association of the protein from early telophase (“T”) onward. (G) Dorsal view of an embryo during mitosis 15, showing that Vfl is chromatin-associated only in dividing interphase nuclei. (H) Enlarged ventral view of a stage 8 embryo, showing nuclear localization of Vfl in mitotically dividing cells of the forming CNS (arrowheads) and low levels of Vfl protein in cytoplasmic granules in the nondividing epidermal cells. (I) Merged images (I′-I″′) showing Vfl (red), Lamin (blue), and DNA (green) stainings during mitosis 15. Note high amounts of chromatin-associated Vfl in early interphase nuclei (arrowheads) and weaker staining in nuclei that show condensed chromatin (asterisk). Early interphase nuclei are surrounded by a complete nuclear membrane (I″′). In a mitotic nucleus (arrow), Vfl protein dissociates from DNA and forms granular structures not associated with Lamin vesicles (compare I′ and I″′). (J) Merged images (J′-J″′) showing Vfl (red), DNA (green), and α-tubulin (blue) stainings in a single metaphase nucleus. Note that Vfl (J′) is separated from both DNA (J”) and α-tubulin, including the spindle apparatus (see blue structure in J″′). Bar, 5 μm.
To determine the subcellular localization of the vfl protein (Vfl), we generated two sets of anti-Vfl antibodies and used them to stain embryos (see Materials and Methods). To test the specificity of these antibodies, we ectopically expressed vfl using the breathless-GAL4 driver and a UAS-vfl transgene, in the developing tracheal system of the embryo, a tissue that normally does not express vfl. As shown in Figure 2D, affinity-purified anti-Vfl antibodies are able to detect the ectopically expressed Vfl protein in the nuclei of the developing trachea. Furthermore, an engrailed-GAL4 driver caused UAS-dependent Vfl expression in a pattern of epidermal stripes in the embryo (our unpublished data). We subsequently used the antibodies to confirm the in situ pattern of Vfl expression during embryonic development. The protein expression patterns were identical to the patterns of the vfl transcripts, with the exception that no protein could be detected in embryos before mitosis 9 (our unpublished data).
In wild type, Vfl can be detected from mitosis 9-10 onward (Figure 2, E and F). It colocalizes with chromatin during interphase as shown by costaining with Sytox dyes or Histon 2A-GFP fusion gene expression (Clarkson and Saint, 1999). During metaphase and anaphase, Vfl dissociates from chromatin and accumulates in a granular pattern in the nucleoplasm (Figure 2F). Immediately after the chromatide separation in early telophase, when the nuclear membrane starts to be reformed, Vfl is already associated with chromatin (Figure 2F). In addition to this dynamic aspect of the Vfl localization pattern, we also noted a much less pronounced granular distribution of Vfl in the cytoplasm of mitotically inactive cells, suggesting a comparably small storage pool of Vfl in these cells.
During embryonic stage 8/9, Vfl protein is highly enriched in the nuclei of dividing cells of mitotic cycle 15 (Figure 2G) (Foe and Alberts, 1983). The cell cycle-dependent change in subcellular localization is also seen in the dividing neuroblasts on the ventral side of a stage 8 embryo (Figure 2H). In nondividing cells of the epidermis, no Vfl protein could be detected in the nuclei and only small amounts of Vfl protein are maintained in form of the already mentioned cytoplasmic granules (Figure 2H).
During mitosis 15, costaining with anti-lamin antibodies, which mark the nuclear membrane (Figure 2, I-I″′), shows that the majority of Vfl is highly enriched in interphase nuclei. As observed with nuclear divisions in the syncytial blastoderm (Figure 2F), the early telophase nuclei show higher amounts of chromatin-associated Vfl than nuclei at the onset of chromatin condensation (Figure 2I). During mitosis, Vfl is found in a granular pattern in the area of the former nucleus (Figure 2I), which is surrounded by cytoplasmic lamin. At this time, Vfl is neither associated with DNA nor with the spindles as shown by anti-α-tubulin antibody staining (Figure 2, J-J″′). These observations indicate that although Vfl is located in granular structures during meta- and anaphase, it associates with chromatin already in early telophase and remains chromatin bound throughout interphase. In conclusion, the molecular features of Vfl, the strong expression in mitotically active cells as well as the cell cycle-dependence of its subcellular localization pattern are consistent with a role for Vfl during mitosis.
vielfältig Mutations Affect Various Aspects of Drosophila Embryogenesis
To elucidate the function of Vfl activity during mitosis, we generated vfl mutants by making use of P-element insertions that map to the vfl locus. We identified four insertions, including the EP-element EP(X)1344 (Rorth, 1996) and three P-elements (Peter et al., 2002) by inverse PCR analysis (for details, see Peter et al., 2002), where the P-elements reside in the promotor region or in the 5′ untranslated region of vfl (Figure 1A).
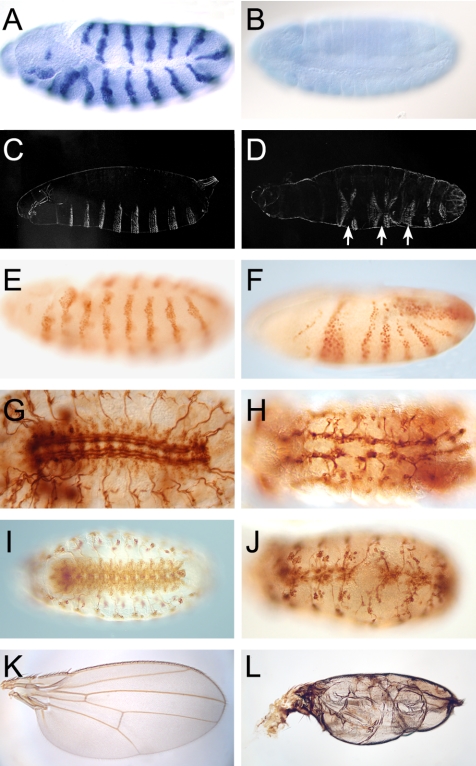
To explore whether the P-element insertions have a mutant effect, we examined the development of hemizygous P-element-bearing individuals, i.e., males that possess only the transposon bearing X-chromosome. In EP(X)1344 and P0225 hemizygous males, the gene seems to be expressed normally and no mutant phenotype was observed (our unpublished data). However, the UAS sequences associated with the EP-element can be used to ectopically express the gene in response to transgene-driven GAL4 activity (Rorth, 1996) (Figure 3A). In contrast, the P-element insertion P0353 as well as P0427 caused the absence of vfl mRNA expression (Figure 3B; our unpublished data). Therefore, both integrations represent vfl loss-of-function alleles.
Figure 3.
vfl loss-of-function and gain-of function phenotypes. (A) Ectopic vfl expression in response to enGAL4 activity driving vfl expression via the EP-element EP(X)1344-associated UAS sequences in a pattern of stripes. (B) Lack of vfl transcripts in a hemizygous l(1)G0427 mutant individual during late embryogenesis, indicating that zygotic expression of vfl is strongly impaired or absent. (C and D). Cuticle preparation of a wild-type (C) and a hemizygous l(1)G0353 mutant larva (D) showing strong segmentation defects (arrows) in the mutant larva. (E and F) Engrailed expression pattern as revealed by anti-Engrailed antibody staining in wild-type (E) and a hemizygous l(1)G0427 mutant individuals (F) at corresponding midstages of embryogenesis, showing that the normally regular Engrailed expression pattern is perturbed as visualized by the fusion of engrailed expressing stripes. (G and H) CNS structure of wild-type (G) and a vfl mutant (H) embryo as revealed by anti-FasII antibody staining during late embryogenesis. Note the strong neuronal and axonal defects. (I and J) mAb 22C10 staining of wild-type embryo (I) and a corresponding embryo that ectopically expressed vfl in a subset of dividing neuroblasts using the ftzNG-GAL4 driver. Note the loss of neurons and axonal fractures. (K and L) Wild-type wing blade (K) and a blistered wing (L) that has been caused by vfl overexpression in the dorsal compartment of the wing imaginal disk in response to the apGAL4 driver.
P0353 and P0427 hemizygous individuals show multiple phenotypic effects in conjunction with embryonic or larval lethality. In P0353 embryos, the phenotypes include segmentation defects (represented by fused segments as shown in Figure 3, C-F), strong abnormalities in the muscle pattern (our unpublished data) and in the PNS and CNS (Figure 3, G and H). The mutant nervous system is characterized by a severe loss of neurons, disrupted axon tracks, and misguided axons (Figure 3, G and H). Similar defects can be observed in mutant embryos, which are hemizygous for the P0427 insertion in which no vfl mRNA expression could be observed (our unpublished data). None of the defects can be seen in individuals when the respective P-element was precisely excised by P-element reversions (for details of the procedure, see Peter et al., 2002), indicating that the inserted P-element is the cause of the mutant phenotype. Moreover, RNAi experiments (see below) caused phenocopies of mutants as described above. These results indicate that loss of vfl (CG12701) expression is the cause of the vfl mutant phenotypes.
The early and multiple pleiotropic loss-of-function defects and the specific expression patterns of vfl in mitotically active cells (see above) suggest that the vfl mutations affect a general cellular process such as mitosis. Overexpression of vfl by the GAL4/UAS system (Brand and Perrimon, 1993) could therefore impair this process as well. To test this proposal, we misexpressed vfl via the UAS-bearing EP(X)1344 insertion in response to specific GAL4-drivers that cause vfl expression in dividing neuroblasts of the nervous system (ftzNG-GAL4) and in the developing wing imaginal discs (ap-GAL4), respectively. Both locations are mitotically active tissues and correspond to sites of vfl expression in wild-type individuals (see above).
The results shown in Figure 3, I-L, indicate that overexpression of Vfl in both the nervous system (Figure 3J) and the dorsal compartment of the wing imaginal discs (Figure 3L) causes strong phenotypes. In the wing imaginal discs, vfl overexpression resulted in adult wing blisters. Blistered wings are generally attributed to improper adherence of the dorsal and ventral wing epithelia and are known to be caused by mutations that interfere with this process, including cell adhesion molecules. Because Vfl is a nuclear protein that is regulated during cell division (see below), we speculate that this mismatch is an indirect consequence of, or is caused by, defective mitotic divisions in response to Vfl overexpression in the dorsal compartment of the wing imaginal disk.
vfl Mutations Affect Size and Alignment of Syncytial Blastoderm Nuclei
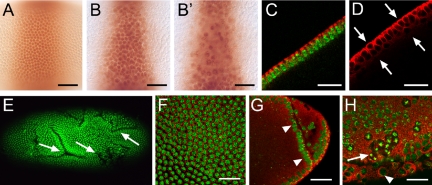
To further explore a possible role of vfl activity during mitosis, we concentrated our efforts on early embryos where, in contrast to the developing wing blade, the mitotic patterns of preblastoderm and blastoderm cells are easily accessible. We used anti-Krüppel antibodies to outline a subset of nuclei in the central regions of syncytial and cellular blastoderm embryos. Figure 4, A and B, shows that nuclei of vfl mutants are abnormally shaped and increased in size. In addition, nuclei fail to become properly aligned in a single layer below the cortical membrane of the preblastoderm embryo (see example in Figure 2B′). The improper alignment of nuclei was also visualized in optical cross sections of embryos that were stained with anti-tubulin antibodies, which label the cytoskeleton (Figure 4, C and D). In addition, we noted deep folds that are uniquely formed by vfl mutant blastoderm embryos as visualized by anti-Crumbs antibody staining, a cellular marker that labels the apical site of the forming blastoderm cells (our unpublished data). These folds remain in pregastrulating embryos (Figure 4E). Double labeling of DNA and cytoskeletal tubulin in wild-type embryos shows the regular pattern of evenly distributed nuclei of about identical size (Figure 4F), whereas in vfl mutant embryos, the region adjacent to folds are marked by enlarged nuclei (Figure 4, G and H). Occasionally, tubulin-surrounded giant cells that contain multiple dividing nuclei were observed (Figure 4H), a finding that indicates incomplete cytokinesis. Collectively, these results indicate that the loss of zygotic vfl activity affects both the size and positioning of the dividing nuclei before cellularization and causes extraepithelial folds during early gastrulation.
Figure 4.
vfl is required for mitotic cytokinesis and polarity formation during early embryogenesis. Embryos were stained with anti-Krüppel (A, B, and B′), anti-α-tubulin (C, D, and F-H, red) antibodies and DNA stain (Sytox green; C and E-H). (A) Nuclei expressing Krüppel are positioned in a single layer in wild-type embryos. (B and B′) Two adjacent optical section of a vfl mutant embryo showing the surface layer of nuclei (B) and a nuclei layer below the surface (B′), which is not observed in wild type. (C and D) Optical cross-sections of anti-α-tubulin-stained wild-type (C) and vfl mutant embryos (D) show that in contrast to the normal arrangement of wild-type nuclei in a single layer (as visualized by DNA [green] and α-tubulin [red] stainings), mutant embryos form an irregular arrangement of nuclei in two layers (arrows). (E) Loss of vfl leads to the formation of ectopic folds as shown by DNA staining of an vfl mutant embryo. (F) Enlarged surface region of a wild-type embryo (DNA in green, anti-α-tubulin in red) showing the regular pattern of cells (F) with a uniform size of nuclei. (G and H) Enlarged regions of vfl mutant embryos showing an extra fold in the posterior region of a gastrulating embryo (G, arrowheads), enlarged nuclei (H, arrowhead) and an area of interconnected nuclei (H, arrow). Bars, 25 μm.
Loss of vfl Activity Affects Chromosome Separation
To explore whether these vfl mutant effects correlate with altered cell cycle progression, we examined the mitotic wave patterns in the early embryo by DNA staining and by using the mitosis marker H3-P (Foe and Alberts, 1983). During the initial syncytial phase of development, the embryo undergoes a series of rapid and synchronous nuclear divisions. Once nuclei align in the cortical region, after nuclear division cycle 10, meta-synchrony of nuclear divisions can be observed along the anterior-posterior axis of the embryo. This phenomenon is referred to as “mitotic wave,” a process that divides the embryo into regions of nuclei that are in the DNA replication phase and other phases of mitosis, respectively (for details, see Supplemental Movie 1; also see Figure 2, E and F). As a result, the wild-type embryo is arranged in areas containing nuclei that are in different cell cycle phases, forming a stereotyped pattern along the anterior-posterior axis (Foe and Alberts, 1983).
Figure 5 shows that the mitotic wave patterns are strongly disrupted in vfl mutant embryos. The defects range from abnormal starting points of the waves and include asynchronous divisions as shown by a “salt-and-pepper” distribution of nuclei that are in different mitotic stages (Figure 5, A-C). Because such defects were only observed in hemizygous mutant embryos, they must be caused by loss of zygotic vfl activity in addition to, or independent of the reduced maternal activity that derives from the heterozygous females. This observation is consistent with the finding that despite the high load of maternal vfl transcripts in eggs and early embryos, Vfl protein can only be detected during nuclear division 9/10 onwards, implying that zygotic vfl expression is already initiated during preblastoderm stage, the stage when first zygotic gene expression occurs in the Drosophila embryo.
Figure 5.
Synchronicity of mitotic divisions and chromosome segregation are affected in syncytial blastoderm stage vfl mutant embryos. vfl mutant blastoderm embryos stained with anti H3-P antibodies (A-C, red) and Sytox green (A-G), vfl dsRNA injected (D-G and M-Q) and wild-type (H-L) embryos carrying H2a-GFP (H-Q). Time-lapse pictures from living embryos (see Supplemental Materials) during one round of mitosis: early prophase (H and M), metaphase (I and N), anaphase (J and O), telophase (K and P), and interphase of the daughter nuclei (L and Q). (A and B) In vfl hemizygous mutant embryos the synchronous mitotic waves are affected (A, enlarged area in B; also see Figure 2, E and F, for the wild-type pattern). (C) In some embryos, irregular areas of nuclei with highly condensed DNA are formed. (D-G) Nuclei of embryos injected with vfl dsRNA form anaphase bridges (D-F, arrowhead) or contain double sets of chromosomes (D, asterisk; G, arrowhead). Note that the two nuclei connected in E (arrowhead) originated from a perpendicular nuclear division. (H-Q) Mitotic phases of H2a-GFP-expressing nuclei (from a time-lapse movie) of a vfl dsRNA-injected embryo show strong chromatin condensation at prophase (H and M), the formation of anaphase chromatin bridges (J and O), and the failure of nuclear separation (L and Q). Bars, 25 μm in (B, D-F) and 5 μm (G-Q).
To see whether the observed defects can be enhanced by an increased loss of maternal gene activity, we aimed to generate homozygous mutant germline clones using the inducible Flipase (FLP)-mediated mitotic recombination technique (Chou et al., 1993). Unfortunately, the vfl gene is positioned beyond the integration point of the proximal-most FRT/ovoD1-element on the X-chromosome (Chou et al., 1993) and thus, for technical reasons, no homozygous vfl mutant germline clones could be established by this approach. To nevertheless generate embryos in which both maternal and zygotic vfl activity is reduced or even absent, we performed RNAi experiments by injecting inhibitory double-stranded RNA (RNAi; Kennerdell and Carthew, 1998). Injections of RNAi specific for the vfl transcript resulted in dose-dependent phenocopies of the vfl mutant segmentation phenotypes (our unpublished data).
We used the RNAi technique to probe the function of the maternally contributed transcript on cell cycle progression in embryos that carried an ubiquitously expressed Histone 2A-GFP transgene (Clarkson and Saint, 1999). Expression of this marker gene visualizes chromatin and makes it possible to follow mitotic division events in living, RNAi-treated embryos. First, defects could be identified before mitosis 10 where few nuclei develop abnormally condensed chromatin and remain connected via small chromatin bridges during late anaphase (Figure 5D). Occasionally, nuclear divisions perpendicular to the surface of the embryo could be observed, resulting in nuclei in a second layer (Figure 5E). During subsequent mitotic cycles, the number of nuclei that remain connected by chromatin bridges was increased, and cases where up to four nuclei are connected by such bridges could be observed (Figure 5F). This finding indicates that nuclei with abnormal chromosome segregation are capable to undergo the next round of mitotic division. In addition, individual nuclei with supernumerary numbers of chromosome arms were observed (Figure 5G).
RNAi injection into embryos that carry a histone 2A-GFP transgene (Clarkson and Saint, 1999) combined with time-lapse recordings allowed us to visualize the course of mitotic waves in vivo under conditions when vfl activity was reduced below the level of maternally derived vfl activity. Comparison of mitotic waves in the wild type (Supplemental Movie 1) with RNAi-injected embryos (Supplemental Movie 2) shows that the experimentally induced loss of vfl activity causes a delay in the front movements of the mitotic wave and eventually blocks the ordered movement, resulting in the salt-and-pepper arrangement of nuclei in different mitotic phases. An enlarged view of nuclei shows that loss of vfl activity does not interfere with nuclear processes such as chromatin condensation or the alignment of chromosomes in the equatorial plate, but impairs the chromosome separation process (Supplemental Movie 3).
Key stages of this process in wild-type and RNAi-injected embryos are shown in Figure 5, H-L and M-Q, respectively. Although the condensed chromatin seems to be abnormally shaped during prophase in RNAi-treated embryos (compare Figure 5, H with M), it can align properly in the equatorial plate (compare Figure 5, I and N). During anaphase, when chromosomes become separated, some of the separating chromosomes remain connected (compare Figure 5, J and O). Subsequently, during telophase, the separation process arrests and the partially separated chromatin fuses again and becomes part of a single enlarged nucleus (compare Figure 5, K and L and P and Q). These observations suggest that mitosis proceeds normally in vfl mutant nuclei, whereas the separation of chromosomes is defective.
Loss of vfl Activity and Overexpression of vfl Impair Proper DNA Replication
Vfl protein colocalizes with chromatin during the interphases. Thus, Vfl is unlikely to participate directly in the separation of chromosomes, but it may instead interfere with an earlier process during telophase or early interphase. Moreover, the DNA-binding properties of Vfl suggest that it functions in a process that involves DNA binding. Thus, Vfl may act in chromatin organization or DNA replication directly or it may function as a transcriptional regulator of genes involved in these processes that are completed before the onset of chromosome segregation. First indications that vfl mutations affect DNA replication and mitosis were based on the time-lapse analysis of embryos injected with vfl RNAi (Supplemental Movie 2). It shows an abnormally delayed mitotic wave during syncytial nuclear division that is followed by an additional round of DNA condensation.
In wild-type embryos, the progression of mitotic waves results in distinct patterns of dividing nuclei in separated regions along the anterior-posterior axis in which DNA replication, DNA condensation, and decondensed interphase DNA can be visualized (Figure 6A). In hemizygous vfl mutant embryos, these processes occur in a noncoordinated manner (Figure 6B; see enlarged Figure 6, C′-C″′). To determine whether DNA replication is affected in the vfl mutant embryos, we performed BrdU incorporation experiments. Figure 6C shows that the levels of BrdU incorporation into replicating DNA vary greatly among the pulse-labeled nuclei in the same region of the mutant embryo and that H3-P-labeled nuclei with condensed chromatin are directly adjacent to nuclei showing high BrdU incorporation along the entire chromatin. Nuclei with high BrdU incorporation do not exert stronger DNA labeling, indicating that these nuclei do not contain an increased number of chromatids. Thus, the salt-and-pepper-like pattern of BrdU incorporation in hemizygous vfl mutant embryos, which reflects the pattern of DNA replication, parallels the distribution of chromosome separation defects in such embryos. The results also indicate that DNA replication is not disrupted because BrdU incorporation can be still be detected. Occasionally, nuclei continue to divide although the chromosome arms failed to be separated, resulting in up to four nuclei that remain connected via chromatin bridges (Figure 5, F and G). Impaired chromosome segregation could therefore be a consequence rather than the cause of the perturbed mitotic division as reflected in altered cell cycle progression and BrdU-labeling patterns.
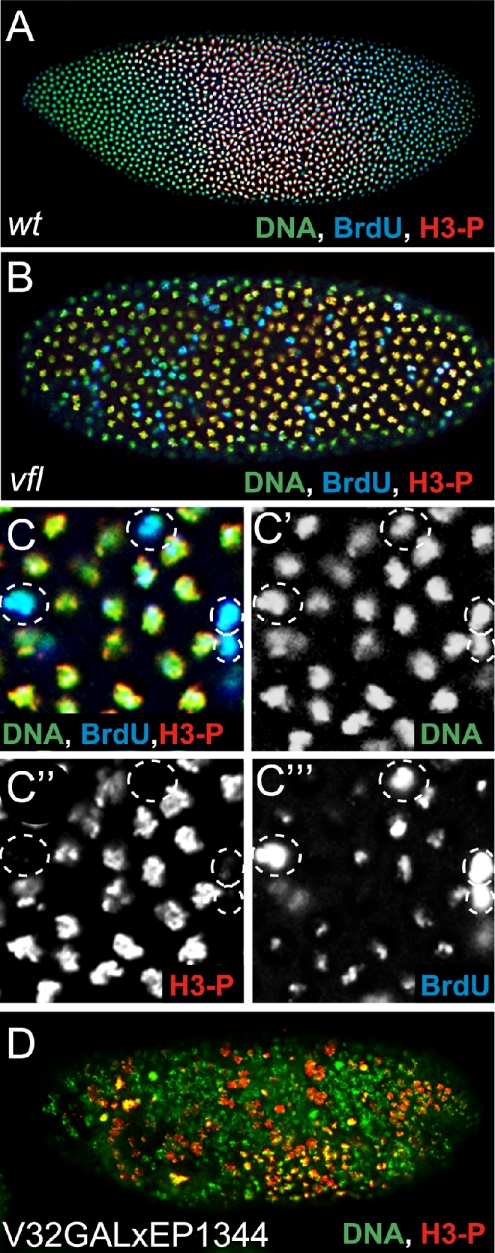
Figure 6.
Synchronicity of DNA replication and mitosis during nuclear divisions depends on the Vfl dosis. Corresponding blastoderm stages of an wild-type embryo (A), a vfl mutant embryo (B, enlarged in C), and an embryo overexpressing vfl using EP(X)1344 and the maternal V32GAL4 driver. Embryos were stained with Sytox green (A-D; green) and anti-H3-P antibodies (A-D; red) and for BrdU incorporation (A-C; blue) to identify nuclei that are in the process of DNA replication. Wild-type blastoderm embryos are divided into regions with DNA-replication (turquoise; overlap of BrdU incorporation and DNA) and regions with condensed DNA (green to yellow; overlap of H3-P and DNA) characteristic for dividing cells (see A). In vfl mutants nuclei with condensed chromatin and nuclei with DNA-replication show a salt-and-pepper-like distribution (B, enlargement in C). C′, C”, and C″′ present the DNA, H3-P, and BrdU staining in single channel display, which are merged in C. Note that high amounts of BrdU incorporation do not correspond to an increased DNA content of the nuclei. (D) In embryos overexpressing Vfl, as in the loss-of-function mutants, the spatial separation of the mitotic domains is lost.
We also asked whether vfl overexpression, which affects development in a vfl mutant-like manner, affects the pattern of DNA replicating cells in the embryos. GAL4/UAS-dependent maternal vfl overexpression in response to the V32GAL4 driver causes irregular early mitotic waves as shown by H3-P labeling (Figure 6D) Thus, both loss-of-function and gain-of-function mutants result in corresponding phenotypes, suggesting that the impaired process is sensitive to the level of vfl activity.
DISCUSSION
We provide evidence that the C2H2 zinc finger protein Vfl participates in mitotic cell division and eventually causes abnormal chromosome segregation during mitosis. This defect results in distinct organismal phenotypes, which are most prominently demonstrated by the lack of synchronous mitotic waves during early Drosophila embryogenesis, as reflected by uncoordinated division patterns and asynchronous DNA-replication cycles. As a result, preblastoderm nuclei are no longer arranged in a single layer at the periphery of the embryo and cause the formation of a number of extra folds during gastrulation. Subsequently, embryos develop an aberrant segmentation pattern, a defective nervous system, and an abnormal muscle pattern in the developing embryo. The mitosis-related phenotype of the vfl mutants is consistent with the notion that vfl transcripts are highly enriched in mitotically active cells and that the protein exerts a dynamic subcellular localization pattern during the cell cycle. From early telophase until the end of interphase, the Vfl protein is chromatin associated. During the DNA-condensation phase, it dissociates from chromatin, remains separated from DNA during metaphase and anaphase, and accumulates in a granular pattern in the area of the disintegrated nucleus once its envelope is dissolved. At this stage, Vfl is neither associated with lamin nor with microtubules or DNA. At the end of mitosis and upon entering the telophase, the granules vanish, and Vfl becomes instantly associated with interphase chromatin. This observation and the canonical DNA-binding domain of Vfl suggest that the protein is active when bound to DNA and inactive when dissociated from chromatin. This proposal implies that the low amounts of Vfl in the cytoplasm of mitotically inactive cells are likely to represent a storage of inactive protein and that Vfl functions once it is chromatin associated during interphase of dividing cells. Vfl may act as a DNA-binding factor that participates directly in chromatin dynamics and accessibility or act indirectly as a transcription factor that regulates the expression of genes involved in these processes. However, because the loss of vfl activity causes notable defects before mitosis 14, i.e., the time point when the embryo switches from maternally controlled nuclear divisions to the zygotic control of mitosis, it seems unlikely that Vfl acts as a transcription factor. Thus, we favor a model in which Vfl functions in DNA replication or participates in some aspects of chromatin dynamics required for proper mitosis.
The BrdU labeling experiments of vfl mutant and wild-type embryos indicate that the coordinated timing of mitosis, including the process of DNA replication, is strongly impaired by altering the normal dose of vfl activity. However, DNA replication is not blocked in the mutants as indicated by the appearance of up to four nuclei that remain associated by chromatin bridges (Figure 5, F and G), indicating that the partially separated nuclei are capable to undergo replication although sister chromatids have failed to separate during the previous anaphase. These observations and the early association of Vfl with early interphase chromatin suggest that the protein could participate in the temporal control of DNA replication. Alternatively, or in addition, the early chromatin association of Vfl could also reflect its requirement during DNA decondensation and/or cohesin loading, two processes that are prerequisite for the initiation of replication (Blow and Tanaka, 2005; Machida et al., 2005).
An interesting aspect of our results concerns the regulation of Vfl localization during mitosis, i.e., association of Vfl with interphase chromatin and with granular structures during all other phases of mitosis. Chromatin condensation during prophase, when Vfl dissociates from chromatin, is accompanied by the cessation of transcription (Prescott and Bender, 1962). This process correlates with inactivation of transcriptional regulators as well as other regulatory chromatin components by removing them from their DNA targets. Such an in vivo mechanism has been recently reported for a mammalian zinc finger protein Ikaros (Hahm et al., 1994; Molnar and Georgopoulos, 1994; Dovat et al., 2002), which plays a key role in the development and the response of the immune system (Cortes et al., 1999; Georgopoulos, 2002; Smale and Fisher, 2002). Ikaros has also been implicated in the regulation of cell cycle progression (Cortes et al., 1999) and was found to dissociate from chromatin during early stages of mitosis (Brown et al., 1997, 1999; Kim et al., 1999) due to a G2/M-specific phosphorylation event (Dovat et al., 2002). We do not know how the dissociation of Vfl from chromatin and its accumulation in the granular structures are achieved mechanistically. We speculate, however, that these structures represent the mitotic containment or a sequestering form for Vfl until cells enter the interphase again. In contrast, cells that do not continue mitosis nevertheless maintain comparatively low levels of Vfl in cytoplasmic granules, likely to represent a small pool of stored and inactive Vfl protein.
Vfl has no direct vertebrate homologue that could be identified by sequence comparison. However, this result has to be taken with caution due to some specifics of zinc finger domains. The C2H2 zinc finger motif is an unusually small, self-folding domain of 25- to 30-amino acid residues. It includes paired cysteines and histidines as zinc-coordinating residues and possesses two short β-strands followed by an α-helix. The DNA-binding properties usually depend on no more than three amino acid residues of the zinc finger loop, the arrangement of C2H2 domains in proteins, and the higher order structure of proteins (Iuchi, 2001). This arrangement and that only a few conserved amino acid residues are required to ensure the sequence-specific DNA binding make it extremely difficult, if not impossible, to predict how the homologous vertebrate C2H2 finger would look. In contrast, we found the protein to be highly conserved in all insects analyzed, including the mosquito An. gambiae and the beetle T. castaneum, which separated from Drosophila for more than 250 and 300 million years ago, respectively (Brown et al., 2003; Kulathinal et al., 2004). Thus, the function of Vfl might be still unrecognized among the more than 2000 C2H2 zinc finger proteins that were annotated in the mouse and human genomes (Lander et al., 2001; Venter et al., 2001; Waterston et al., 2002). Alternatively, vfl may have a function for the insect specific nuclear divisions during syncytial blastoderm stage and therefore may not be conserved in species other than insects.
Loss of vfl activity causes abnormally shaped and enlarged nuclei that fail to become integrated into the cortical arrangement of preblastoderm nuclei. Obviously, these morphological features, in particular the enlargement of the nuclei, cause some space constrictions at the periphery of the embryo, and thus a significant portion of nuclei fail to align properly. The subsequent divisions, which occur in part perpendicular to the normal division axis as frequently observed in such embryos, cause an additional space limitation which forces the epithelial cell layer to form the irregular and variable patterns of folds that were consistently observed in gastrulating embryos. Notably, the positions of the folds vary significantly from embryo to embryo. This finding can be attributed to the fact that the embryos analyzed were most likely not lack-of-function (“null”) mutants and always contained half the normal dose of maternal vfl activity. Furthermore, although the RNAi injections into embryos were done as early as possible after egg deposition, some undetected amounts of protein might have already accumulated in such embryos. This proposal is consistent with the notion of earlier and more severe effects in response to increasing amounts of injected RNAi. Irrespectively of this speculation, the results presented here show that inadequate levels of Vfl interfere with the timing of mitosis and eventually result in impaired chromosome separation. The specific expression of vfl in mitotically active cells, the dose dependence of the protein as demonstrated by gain-of-function and loss-of-function experiments and the shuttling of Vfl between chromatin of interphase nuclei and the granular structures during the other stages of the mitotic cycle suggest that protein function is tightly regulated. The mechanisms of the shuttling, the molecular pathways in which Vfl participates, and the link between Vfl activity and DNA replication remain to be further elucidated.
Supplementary Material
Acknowledgments
We thank our colleagues in the laboratory for various contributions and critical discussions, G. Dowe for sequencing, and U. Jahns-Meyer for transformations. Furthermore, we are grateful to Alf Herzig and Ralf Pflanz for various conceptual contributions. This research was supported the Max-Planck-Society, the SFB271, and the Graduiertenkolleg “Molekulare Genetik der Entwicklung” of the Deutsche Forschungsgemeinschaft (to G. V. and H. J.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-11-1056) on March 8, 2006.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Aasland, R., Gibson, T. J., and Stewart, A. F. (1995). The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20, 56-59. [DOI] [PubMed] [Google Scholar]
- Akoulitchev, S., and Reinberg, D. (1998). The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev. 12, 3541-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach, I. (2000). The LIM domain: regulation by association. Mech. Dev. 91, 5-17. [DOI] [PubMed] [Google Scholar]
- Bellier, S., Chastant, S., Adenot, P., Vincent, M., Renard, J. P., and Bensaude, O. (1997). Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J. 16, 6250-6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow, J. J., and Tanaka, T. U. (2005). The chromosome cycle: coordinating replication and segregation. EMBO Rep. 6, 1028-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415. [DOI] [PubMed] [Google Scholar]
- Brown, K. E., Baxter, J., Graf, D., Merkenschlager, M., and Fisher, A. G. (1999). Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell 3, 207-217. [DOI] [PubMed] [Google Scholar]
- Brown, K. E., Guest, S. S., Smale, S. T., Hahm, K., Merkenschlager, M., and Fisher, A. G. (1997). Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91, 845-854. [DOI] [PubMed] [Google Scholar]
- Brown, S. J., Denell, R. E., and Beeman, R. W. (2003). Beetling around the genome. Genet. Res. 82, 155-161. [DOI] [PubMed] [Google Scholar]
- Caelles, C., Hennemann, H., and Karin, M. (1995). M-phase-specific phosphorylation of the POU transcription factor GHF-1 by a cell cycle-regulated protein kinase inhibits DNA binding. Mol. Cell Biol. 15, 6694-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo, Y., and Klug, A. (1995). Designing DNA-binding proteins on the surface of filamentous phage. Curr. Opin. Biotechnol. 6, 431-436. [DOI] [PubMed] [Google Scholar]
- Chou, T. B., Noll, E., and Perrimon, N. (1993). Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development 119, 1359-1369. [DOI] [PubMed] [Google Scholar]
- Chung, H. R., Schäfer, U., Jäckle, H., and Böhm, S. (2002). Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila. EMBO Rep. 3, 1158-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson, M., and Saint, R. (1999). A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 18, 457-462. [DOI] [PubMed] [Google Scholar]
- Cortes, M., Wong, E., Koipally, J., and Georgopoulos, K. (1999). Control of lymphocyte development by the Ikaros gene family. Curr. Opin. Immunol. 11, 167-171. [DOI] [PubMed] [Google Scholar]
- David, G., Alland, L., Hong, S. H., Wong, C. W., DePinho, R. A., and Dejean, A. (1998). Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene 16, 2549-2556. [DOI] [PubMed] [Google Scholar]
- Dovat, S., Ronni, T., Russell, D., Ferrini, R., Cobb, B. S., and Smale, S. T. (2002). A common mechanism for mitotic inactivation of C2H2 zinc finger DNA-binding domains. Genes Dev. 16, 2985-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe, V. E., and Alberts, B. M. (1983). Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61, 31-70. [DOI] [PubMed] [Google Scholar]
- Georgopoulos, K. (2002). Haematopoietic cell-fate decisions, chromatin regulation and Ikaros. Nat. Rev. Immunol. 2, 162-174. [DOI] [PubMed] [Google Scholar]
- Gottesfeld, J. M., and Forbes, D. J. (1997). Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22, 197-202. [DOI] [PubMed] [Google Scholar]
- Hahm, K., Ernst, P., Lo, K., Kim, G. S., Turck, C., and Smale, S. T. (1994). The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol. Cell Biol. 14, 7111-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi, S. (2001). Three classes of C2H2 zinc finger proteins. Cell Mol. Life Sci. 58, 625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell, J. R., and Carthew, R. W. (1998). Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95, 1017-1026. [DOI] [PubMed] [Google Scholar]
- Kim, J., Sif, S., Jones, B., Jackson, A., Koipally, J., Heller, E., Winandy, S., Viel, A., Sawyer, A., Ikeda, T., Kingston, R., and Georgopoulos, K. (1999). Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10, 345-355. [DOI] [PubMed] [Google Scholar]
- Knirr, S., Azpiazu, N., and Frasch, M. (1999). The role of the NK-homeobox gene slouch (S59) in somatic muscle patterning. Development 126, 4525-4535. [DOI] [PubMed] [Google Scholar]
- Kulathinal, R. J., Bettencourt, B. R., and Hartl, D. L. (2004). Compensated deleterious mutations in insect genomes. Science 306, 1553-1554. [DOI] [PubMed] [Google Scholar]
- Lander, et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- Lehmann, R., and Tautz, D. (1994). In situ hybridization to RNA. Methods Cell Biol. 44, 575-598. [DOI] [PubMed] [Google Scholar]
- Long, J. J., Leresche, A., Kriwacki, R. W., and Gottesfeld, J. M. (1998). Repression of TFIIH transcriptional activity and TFIIH-associated cdk7 kinase activity at mitosis. Mol. Cell Biol. 18, 1467-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick, K. L., Jensen, J. P., Fang, S., Ong, A. M., Hatakeyama, S., and Weissman, A. M. (1999). RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96, 11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida, Y. J., Hamlin, J. L., and Dutta, A. (2005). Right place, right time, and only once: replication initiation in metazoans. Cell 123, 13-24. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas, M. A., Dey, A., Rabindran, S. K., Ozato, K., and Wu, C. (1995). Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83, 29-38. [DOI] [PubMed] [Google Scholar]
- Miller, J., McLachlan, A. D., and Klug, A. (1985). Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4, 1609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar, A., and Georgopoulos, K. (1994). The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell Biol. 14, 8292-8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, A., et al. (2002). Mapping and identification of essential gene functions on the X chromosome of Drosophila. EMBO Rep. 3, 34-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, D. M., and Bender, M. A. (1962). Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell Res. 26, 260-268. [DOI] [PubMed] [Google Scholar]
- Rorth, P. (1996). A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93, 12418-12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, U. B., Preiss, A., Seifert, E., Jäckle, H., and Knipple, D. C. (1985). Production of phenocopies by Krüppel antisense RNA injection into Drosophila embryos. Nature 313, 703-706. [DOI] [PubMed] [Google Scholar]
- Rubin, G. M., Hong, L., Brokstein, P., Evans-Holm, M., Frise, E., Stapleton, M., and Harvey, D. A. (2000). A Drosophila complementary DNA resource. Science 287, 2222-2224. [DOI] [PubMed] [Google Scholar]
- Schmiesing, J. A., Gregson, H. C., Zhou, S., and Yokomori, K. (2000). A human condensin complex containing hCAP-C-hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol. Cell Biol. 20, 6996-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segil, N., Guermah, M., Hoffmann, A., Roeder, R. G., and Heintz, N. (1996). Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 10, 2389-2400. [DOI] [PubMed] [Google Scholar]
- Smale, S. T., and Fisher, A. G. (2002). Chromatin structure and gene regulation in the immune system. Annu. Rev. Immunol. 20, 427-462. [DOI] [PubMed] [Google Scholar]
- Tucker, P., Laemle, L., Munson, A., Kanekar, S., Oliver, E. R., Brown, N., Schlecht, H., Vetter, M., and Glaser, T. (2001). The eyeless mouse mutation (ey1) removes an alternative start codon from the Rx/rax homeobox gene. Genesis 31, 43-53. [DOI] [PubMed] [Google Scholar]
- Venter, J. C., et al. (2001). The sequence of the human genome. Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- Waterston, R. H., et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520-562. [DOI] [PubMed] [Google Scholar]
- Wolff, T., and Ready, D. F. (1993). Pattern formation in the Drosophila retina. In: The Development of Drosophila melanogaster, Vol. 2, ed. M. Bate and A. M. Marinez-Arias, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1277-1325. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.