Abstract
The Ras-GRF1 exchange factor has regulated guanine nucleotide exchange factor (GEF) activity for H-Ras and Rac1 through separate domains. Both H-Ras and Rac1 activation have been linked to synaptic plasticity and thus could contribute to the function of Ras-GRF1 in neuronal signal transduction pathways that underlie learning and memory. We defined the effects of Ras-GRF1 and truncation mutants that include only one of its GEF activities on the morphology of PC12 phaeochromocytoma cells. Ras-GRF1 required coexpression of H-Ras to induce morphological effects. Ras-GRF1 plus H-Ras induced a novel, expanded morphology in PC12 cells, which was characterized by a 10-fold increase in soma size and by neurite extension. A truncation mutant of Ras-GRF1 that included the Ras GEF domain, GRFΔN, plus H-Ras produced neurite extensions, but did not expand the soma. This neurite extension was blocked by inhibition of MAP kinase activation, but was independent of dominant-negative Rac1 or RhoA. A truncation mutant of Ras-GRF1 that included the Rac GEF domains, GRFΔC, produced the expanded phenotype in cotransfections with H-Ras. Cell expansion was inhibited by wortmannin or dominant-negative forms of Rac1 or Akt. GRFΔC binds H-Ras.GTP in both pulldown assays from bacterial lysates and by coimmunoprecipitation from HEK293 cells. These results suggest that coordinated activation of H-Ras and Rac1 by Ras-GRF1 may be a significant controller of neuronal cell size.
INTRODUCTION
The Ras superfamily of GTPases are regulated switches that control many intracellular pathways. The Ras family, which includes H-, K-, and N-Ras and other closely related isoforms, has been particularly associated with the control of proliferation in cells such as fibroblasts and epithelia (Lowy and Willumsen, 1986). This action is thought to be of particular relevance to the common involvement of activated Ras in human cancer (Barbacid, 1987), which can occur by mutational activation (Taparowsky et al., 1982), by inappropriate activation of other elements in the Ras activation pathway, such as the overexpression or aberrant stimulation of growth factor receptors (Malaney and Daly, 2001), or by loss of a deactivating GTPase-activating protein (GAP), such as in type 1 neurofibromatosis (DeClue et al., 1991). Ras proteins are also, however, highly involved in the function of terminally differentiated cells such as neurons of the CNS (Weeber et al., 2002). The Rho family small GTPases, which include Rac1 and many other members, have multiple cellular functions during both cellular differentiation (Beqaj et al., 2002; Sordella et al., 2003) and in the mature phenotype, including regulation of the cytoskeleton and cellular morphology, and coupling to transcription factor pathways (Aznar and Lacal, 2001). There is increasing evidence that the functions of Ras and Rho family small GTPases can be coordinated to produce regulation of cellular phenotypes, with most models suggesting that Ras activation occurs before the activation of Rho proteins (Sarner et al., 2000; Menard and Mattingly, 2003).
Ras superfamily small GTPases function through their cycling between GTP-bound states that can couple to downstream events and GDP-bound states that are conformationally distinct and do not activate those pathways (Macara et al., 1996). The conversion between these states is governed by several groups of enzymes, including the exchange factors (GEFs) that catalyze the release of GDP and subsequent binding of GTP to activate these proteins, and the GAPs, that greatly stimulate the endogenous GTPase activity of Ras proteins and so stimulate their inactivation (Boguski and McCormick, 1993). Physiological control of the switch can reside in regulation of either the relevant GEF or GAP (Bernards and Settleman, 2004), but increasing evidence suggests that the very complex, multidomain structure of the Ras-GEF proteins provides the possibility that they integrate multiple signals to determine the activation state of their target GTPase (Mattingly et al., 1999; Sprang, 2001; Quilliam et al., 2002).
The Ras-GRF1 exchange factor (Shou et al., 1992), which is also termed CDC25Mm (Martegani et al., 1992; Cen et al., 1993), contains both a CDC25 domain that confers exchange factor activity toward Ras (Cen et al., 1993; Wei et al., 1994) and a Dbl homology (DH)/plekstrin homology (PH) region that can act as an exchange factor for Rac1 (Kiyono et al., 1999). It is highly expressed at the synapses of neurons in the CNS (Sturani et al., 1997). There is considerable evidence to link the activation of Ras in general (Manabe et al., 2000; Arendt et al., 2004) and via Ras-GRF1 in particular (Krapivinsky et al., 2003; Schmitt et al., 2005) to molecular and cellular events, such as long-term potentiation and synaptic remodeling, which are thought to underlie memory. Genetic evidence, both from mouse models (Brambilla et al., 1997; Giese et al., 2001; Tian et al., 2004) and human disorders (Weeber et al., 2002), also supports such a link.
The structure and regulation of Ras-GRF1 is complex. The CDC25 domain that confers Ras exchange factor activity is at the far C-terminus of the protein. This activity is controlled by interaction of calcium/calmodulin with the IQ domain near the N-terminus of the protein (Farnsworth et al., 1995), and also by phosphorylation at multiple residues, including Ser-916 (numbering according to the mouse isoform sequence; Mattingly, 1999) and an unknown site of tyrosine phosphorylation by the kinase Ack1 (Kiyono et al., 2000a). Other domains in the N-terminus may also contribute to the regulation of the CDC25 domain, because truncation of the N-terminus has been shown to stimulate Ras exchange factor activity (Baouz et al., 1997). Pathways that stimulate the Ras exchange activity of Ras-GRF1 have been found to be those from G protein–coupled receptors (Shou et al., 1995; Mattingly and Macara, 1996; Zippel et al., 1996), which may require the release of G protein βγ subunits (Mattingly and Macara, 1996), and via direct interactions between Ras-GRF1 and the TrkA nerve growth factor (NGF) receptor (MacDonald et al., 1999; Robinson et al., 2005), and Ras-GRF1 and the NR2B NMDA receptor (Krapivinsky et al., 2003). G protein–coupled agonists, NGF, and NMDA/glycine have all been demonstrated to increase phosphorylation of endogenous Ras-GRF1 at Serine-916 or its equivalent (e.g., Ser-898 in the rat sequence and Ser-927 in the human sequence; Mattingly, 1999; Yang et al., 2003; Norum et al., 2005; Schmitt et al., 2005).
The exchange factor activity toward Rac1 is conferred by the DH/PH domains in the N-terminal half of Ras-GRF1. This activity is also stimulated by overexpression of G protein βγ subunits (Kiyono et al., 1999) or tyrosine phosphorylation by Src (Kiyono et al., 2000b) and is likely to be regulated by G protein–coupled agonists such as lysophosphatidic acid (LPA; Innocenti et al., 1999). Thus there are close parallels between the regulation of the Ras and Rac1 exchange factor activities of Ras-GRF1, but the coordination of these functions has not been defined.
Rat phaeochromocytoma PC12 cells are a widely used model system for neuronal differentiation. Both Ras and Rac activation have been described to produce particular morphological changes and be responsible for parts of the programs induced by stimuli such as NGF (Bar-Sagi and Feramisco, 1985; Guerrero et al., 1986; Yasui et al., 2001; Nusser et al., 2002; Sakai et al., 2004; Robinson et al., 2005). Most work has focused on the mechanisms underlying neurite extension, because this may model events such as neural regeneration and synaptic plasticity. In addition to neuritogenesis, regulation of cell soma size is another relevant, although less studied, neuronal morphology. Increases in soma size of CNS neurons have been reported after constitutive H-Ras activation (Arendt et al., 2004), whereas there is a striking decrease in neuronal soma size in clinical major depression (Rajkowska et al., 1999; Stockmeier et al., 2004). We therefore used PC12 cells as a system to explore the ability of Ras-GRF1 to function as an activator of both Ras and Rac1 and to control cell morphology in a neuronal context.
MATERIALS AND METHODS
Plasmids
The plasmids encoding full-length Ras-GRF1 (residues 1-1262) and the ΔC (residues 1-631) and ΔN (residues 632-1262) truncation mutants fused with triple hemagglutinin-1 (HA) epitope tags at their N-termini have previously been described (Mattingly and Macara, 1996). Plasmids encoding the same inserts with enhanced green fluorescent protein (GFP) replacing the HA1 epitope tags were prepared by subcloning BamHI/EcoRI fragments into the pRK7sGFP vector (Carey et al., 1996). A plasmid encoding the ΔN mutant fused to glutathione S-transferase (GST) has previously been described (Yang et al., 2003). Similar constructs expressing the ΔC mutant or Rac1 were prepared by subcloning the relevant BamHI/EcoRI fragments into pGEX-2T. Plasmids encoding dominant-negative variants of Rac1, Rac1.N17, and of RhoA, RhoA.N19, and of wild-type, constitutively active, and dominant-negative H-Ras fused to triple HA1 tags have previously been described (Mattingly et al., 1994; Mattingly and Macara, 1996; Menard and Mattingly, 2003). Plasmids expressing Myc-tagged H-Ras, K-Ras, and N-Ras were provided by the late Dr. J. Jackson. The plasmid encoding the constitutively activated Src protein, Src.527F, was provided by Dr. D. Flynn. The plasmid encoding triple HA-tagged Sos1 was provided by Dr. M. Czech. The plasmid encoding the HA-tagged, dominant-negative form of Akt (termed Akt-K/M) was provided by Dr. A. Toker. The plasmid encoding the Flag-tagged, dominant-negative form of the MEK1 MAP kinase kinase (mutation K97M) was provided by Dr. G. Tzivion. The plasmid encoding Myc-tagged, constitutively active Rac1 (V12 mutation) was purchased from cdna.org (Rolla, MO). The bacteria transformed with the expression construct for H-Ras were provided by Dr. A. Wolfman.
Cell Culture and Transfection
PC12 cells were cultured in growth media (DMEM [CellGro, Herndon, VA], with 10% fetal bovine serum [Hyclone, Ogden, UT], 5% equine serum, and 100 U/ml penicillin and 100 μg/ml streptomycin [Invitrogen, Carlsbad, CA]) at 37°C under 5% CO2. For transfection, the cells were suspended in room temperature, serum-free DMEM at a cell density of 3 × 106 cells/ml and electroporated (400 V, 500 μF) with 20μg of plasmids in a 0.4-cm gap cuvette. The cells were plated onto Lab-Tek chamber slides (Nalge Nunc, Naperville, IL) that had been precoated with poly-l-lysine (Sigma, St. Louis, MO). The cells were then incubated in either growth media or serum-free media (UltraCulture general purpose medium [Cambrex, Walkersville, MD] plus penicillin and streptomycin) for 48 h before fixation, with additions of drugs or dimethyl sulfoxide (DMSO; Sigma) vehicle as shown. U0126 and SP600125 were purchased from Calbiochem (San Diego, CA) and Stressgen (Vancouver, BC, Canada), respectively. Wortmannin and LPA were from Sigma. HEK-293 cells were transfected by Lipofectamine 2000 (Invitrogen) as previously described (Norum et al., 2005).
Confocal Immunofluorescence
Transfected PC12 cells were fixed and processed for confocal indirect and GFP fluorescence broadly as previously described (Yang et al., 2003), with the following modifications: The primary antibodies used were anti-Myc monoclonal antibody (mAb) 9e10 (1:1000 dilution; Sigma) for detection of cells expressing Myc-tagged Ras or Rac proteins, anti-Ras-GRF1 polyclonal antibody sc224 (1:500 dilution; Santa Cruz Biotechnology, Santa Cruz, CA) to detect untagged Ras-GRF1, anti-Flag mAb M2 (1:200 dilution; Sigma) to detect Flag-tagged MEK1.K97M, and either anti-hemagglutinin-1 (HA1) polyclonal antibody Y-11 (1:150 dilution; Santa Cruz) or 12CA5 mAb (1:500) for detection of cells expressing HA13-tagged proteins. For detection of neurofilaments, mAb RT97 (NICHD Developmental Studies Hybridoma Bank, Iowa City, IA) was used at a 1:100 dilution. The secondary antibodies used were Cy3-coupled anti-mouse antibody (1:300 dilution; Jackson Laboratories, Bar Harbor, ME) and Oregon Green–coupled anti-rabbit antibody (1:300 dilution) or Alexa Fluor 350–coupled goat anti-rabbit antibody (1:300 dilution; Molecular Probes, Eugene, OR). Pictures were taken with a 40× or 63× water immersion lens on a Zeiss LSM310 microscope (Thornwood, NY). For quantification, cells that were identified as positive by fluorescence for the relevant signals were measured using Metamorph software (Universal Imaging, Downingtown, PA). The 30 largest cells recorded for each condition (10 per experiment from 3 independent transfections) were averaged to provide data on cell body area.
SDS-Polyacrylamide Gel Electrophoresis and Western Blotting
SDS-polyacrylamide gel electrophoresis (PAGE) was performed on 10 or 12% gels. Proteins separated by SDS-PAGE were transferred electrophoretically onto nitrocellulose membranes. For antibody detection assays, nitrocellulose membranes were incubated for 1 h at room temperature in blocking buffer (Tris-buffered saline [TBS], pH 7.4, with 0.1% Tween-20 [TBST] containing 5% skim milk powder), washed three times with TBST, and incubated at room temperature for 1 h with primary antibody in TBST with 2% BSA (anti-Myc monoclonal 9B11, 1:1000 dilution [Cell Signaling Technology, Beverly, MA]; anti-HA1 monoclonal 12CA5, 1:5000 dilution; anti-H-Ras, anti-K-Ras, or anti-N-Ras monoclonals, 1:100 dilution [Santa Cruz Biotechnology]; anti-Rac1 monoclonal, 1:2000 dilution [Upstate, Charlottesville, VA]; anti-Ras-GRF1 polyclonal sc244, 1:1000 dilution; anti-GST polyclonal, 1:2000 dilution [Sigma]; or anti-phosphotyrosine polyclonal, 1:1000 dilution [Transduction Laboratories, Lexington, KY]). After washing three times with TBST, membranes were incubated for 1 h with horseradish peroxidase–conjugated secondary antibody (anti-mouse IgG, 1:10,000 dilution or anti-rabbit IgG, 1:20,000 dilution [Santa Cruz]) in blocking buffer. Membranes were then washed five times with TBS. Bound antibodies were detected by enhanced chemiluminescence (Pierce, Rockford, IL).
Pulldown Assays with Proteins Fused to GST
Ras.GTP levels were assayed by pull down with glutathione-Sepharose beads coated with a GST fusion protein with the Ras-binding domain of Raf (Raf.RBD). The protocol was as previously described (Mattingly et al., 2001a) with the following modifications: HEK 293 cells were cotransfected with Myc-tagged H-Ras and Ras-GRF1 or GRF1ΔC for 48 h in presence or absence of serum. Raf.RBD beads were added into cell lysates to pulldown Myc-tagged H-Ras.GTP, which was subsequently detected by Western blotting for the Myc tag.
Rac.GTP levels were assayed by pull down with the Cdc42/Rac1-binding (CRIB) domain of p21-activated kinase 1 (PAK1) fused to GST according to the manufacturer's instructions (Upstate). Briefly, lysates were produced by solubilizing cells in magnesium-containing lysis buffer (Upstate) and then mixed with 5 μg of PAK.PBD agarose beads. The amount of active Rac1 (Rac1-GTP) in each pulldown precipitate was analyzed by Western blot. A volume of the cell lysates equivalent to 5% of that used in the pulldown assay was probed for total Rac1 to determine the total amount of Rac1 protein in each sample.
Escherichia coli transformed with an inducible expression vector for H-Ras (gift from A. Wolfman) were cultured at 37°C, until A600 reached ∼1.0 and then 1 μM IPTG was added for an overnight incubation at room temperature to induce H-Ras expression. The bacteria were harvested and lysed by sonication on ice in 50 mM Tris-Cl (pH 8.0), 150 mM NaCl, 0.2% Na deoxycholate, 0.1% Triton X-100. The lysate was cleared by centrifugation and then split into aliquots and pretreated with 5 mM EDTA, or 5 mM MgCl2 plus 100 μM GTP, or 5 mM MgCl2 plus 100 μM GDP. Pulldown reactions were then initiated by addition of GST.GRFΔN, GST.GRFΔC, or GST.Raf.RBD prebound to glutathione-Sepharose beads to 750 μg of bacterial lysate. After rocking for 1 h at 4°C, the beads were washed three times with TBS containing 250 mM NaCl and 0.25% Triton X-100 and analyzed by Western blotting.
Immunoprecipitation and Coimmunoprecipitation
Transfected cells were solubilized in lysis buffer (100 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.5% Triton X-100) supplemented with 0.5 mM phenylmethylsulfonyl fluoride, 25 μg/ml aprotinin, and 25 μg/ml leupeptin. After centrifugation to remove insoluble material, immunoprecipitates were prepared by addition of anti-Myc mAb 9B11 (2 μg) or anti-HA mAb 12CA5 (2 μg) that had been prebound to protein G-Sepharose beads (Sigma) and washed extensively with lysis buffer. Immunoprecipitated material was then subjected to Western blotting. For coimmunoprecipitation, HA-tagged GRF1ΔC and Myc-tagged H-Ras wild type or its dominant-negative variant H-Ras.N17 or dominant-active variant H-Ras.V12 were expressed in HEK 293 cells. Myc-tagged H-Ras and its variants were immunoprecipitated from cell lysates with anti-Myc mAb 9B11 (2 μg). Coimmunoprecipitated proteins were detected on Western blot with anti-HA polyclonal antibody Y11. All immunoprecipitation experiments were only taken as valid if repeated at least three times with similar results.
GDP/GTP Exchange Assay
In vitro GDP/GTP exchange assays were performed as described (Mattingly and Macara, 1996; Hardt et al., 1998). Briefly, HEK 293 cells transfected with various combinations of plasmids were serum-starved for 24 h. Cells were rinsed twice with cold phosphate-buffered saline, solubilized in RIPA buffer (Mattingly et al., 1999) followed by 10 s of sonication, and centrifuged for 20 min. HA-tagged Ras-GRF1 and its truncated mutant GRFΔC were immunoprecipitated with anti-HA antibody 12CA5. Immunoprecipitates were washed twice with RIPA buffer and twice with exchange buffer (20 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 0.5 mM DTT, 100 mM NaCl, 200 μg/ml BSA) and then incubated with Rac1 that had been preloaded with [3H]GDP to assay for exchange factor activity. Rac1 was prepared from GST-Rac1 by thrombin cleavage, with removal of the thrombin by benzamidine-Sepharose (Sigma; Carey et al., 1996). To load Rac1 with [3H]GDP, 60 pmol of Rac1 were incubated in 20 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 1 mM DTT, 100 mM NaCl, 40 μg/ml BSA, and 1 μM[3H]GDP for 45 min at 37°C. After this incubation, GTP and MgCl2 were added to a final concentration of 1 mM and 10 mM, respectively, to terminate the loading reaction. Immunoprecipitates were incubated with the Rac1.[3H]GDP in exchange buffer at 30°C. At the indicated times, aliquots of the reaction were removed in duplicates and passed through nitrocellulose filters (HAWP025, Millipore, Billerica, MA). Filters were washed twice with a cold wash buffer and radioactivity that remained on the filter was quantified in a liquid scintillation counter (Mattingly and Macara, 1996).
RESULTS
Coexpression of Ras-GRF1 and H-Ras in PC12 Cells Induces a Novel Cellular Morphology
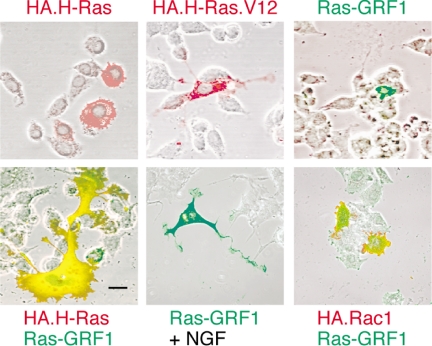
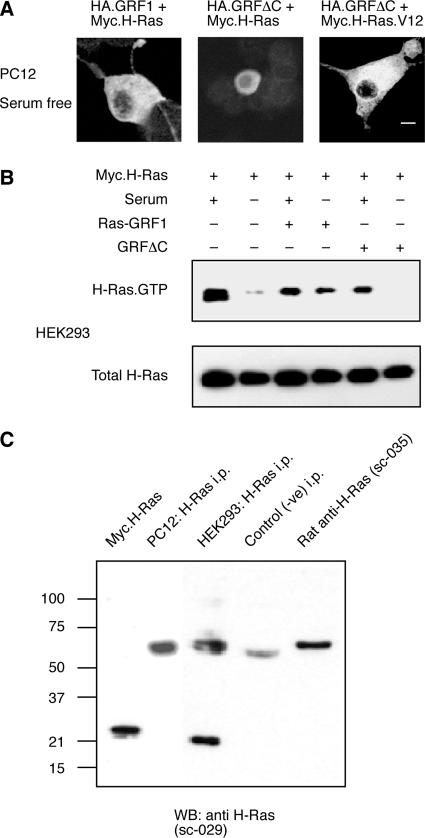
PC12 cells are well known to respond to the continuous activation of Ras, such as occurs after expression of a constitutively activated Ras.V12 protein, by the induction of a morphological differentiation that is characterized by extension of neurites (Bar-Sagi and Feramisco, 1985; Guerrero et al., 1986). We therefore tested whether the overexpression of the neuronal Ras-GRF1 exchange factor would be sufficient to induce differentiation of these cells. The results (Figure 1) showed that the PC12 cells did produce neurites in response to the expression of activated H-Ras.V12, but not in response to wild-type H-Ras overexpression, as would be expected from previous studies. The cells did not, however, show much discernible effect after overexpression of Ras-GRF1. The cells expressing Ras-GRF1 were still competent to differentiate, however, as they produced neurites in response to NGF treatment. Coexpression of Ras-GRF1 and H-Ras in the same cell did produce a morphological transformation that was characterized by the extension of some processes that appeared similar to neurites, but more strikingly was characterized by a great expansion of the size of the soma.
Figure 1.
Effects of Ras-GRF1 expression on the morphology of PC12 cells. PC12 cells were transfected to express the indicated constructs for 48 h. Expression of Ras-GRF1 was detected by green indirect polyclonal immunofluorescence. Expression of HA-tagged H-Ras, H-Ras.V12, or Rac1 were detected by red indirect monoclonal immunofluorescence for the HA tag. To compare morphology with untransfected cells, the fluorescence results are overlaid with a phase-contrast image of the field. Where indicated, the cultures were treated with 40 ng/ml NGF (Genentech) to induce neurite extension (bottom middle). Scale bar, 50 μm. Pictures shown are typical of results from a minimum of three independent experiments. The proportion of cells cotransfected with both H-Ras and Ras-GRF1 (bottom left) that exhibited the novel, expanded morphology was ∼25%. This morphology was never detected after transfection with either H-Ras or Ras-GRF1 alone.
Colocalization of H-Ras and Ras-GRF1 was prominent in an internal, extranuclear region (Figure 1) that may be consistent with the functional colocalization of these proteins that has previously been reported to occur on the endoplasmic reticulum (Arozarena et al., 2004). Reproducibly, the edge of the cell was positive for H-Ras but not Ras-GRF1 (i.e., red), suggesting that Ras-GRF1 may not colocalize with H-Ras at the plasma membrane, at least under these conditions. Ras-GRF1 has also been shown to be an exchange factor for another small GTPase, Rac1 (Kiyono et al., 1999). We therefore tested whether the combination of Ras-GRF1 plus Rac1 had any effect on PC12 cell morphology, but it did not (Figure 1).
Approximately 25% of the cells that were positive for expression of both H-Ras and Ras-GRF1 produced the novel expanded phenotype, which was never recorded in cells transfected to express either protein alone. The remainder of the cells produced a mix of results that included little morphological change (∼20%), clear neurite extension without expansion of the soma (∼20%), intermediate degrees of differentiation (∼25%), and disorganized results that were interpreted as representing cell death (∼10%). With the exception of the latter phenotype, which was often observed with the brightest immunofluorescence and so was presumably due to the highest levels of expression, there was no direct correlation observed between level of transfection and the variation in the phenotypes.
To test the relationship between expression level and phenotype in a simpler system that only requires expression from a single plasmid, we further investigated the established effect of constitutively activated H-Ras to induce neuritogenesis. The results showed that even H-Ras.V12 expression did not produce a uniform response. A representative field is shown top left in Supplementary Figure 1 that includes four cells that are positive for expression of H-Ras.V12 by indirect confocal immunofluorescence. Two of these cells, indicated by arrowheads and including the brightest cell, show no morphological effect, whereas the other two have clear neurite extensions (indicated by arrows).
Ras-GRF1 Induced Neurite Extension Requires H-Ras and ERK MAP Kinase, But Is Independent of Rac1 and PI3K
We have previously reported that the GRFΔN truncation mutant, and other further truncation mutants that preserve the CDC25 domain of the protein, can induce neurite extension from PC12 cells when coexpressed with H-Ras (Yang et al., 2003). In these cases there was no expansion of the soma. To investigate the pathway through which this neurite extension was induced, GRFΔN as a fusion protein with GFP was coexpressed with Myc-tagged H-Ras and dominant-negative Rac1 or RhoA proteins tagged with HA to allow simultaneous detection of the three constructs in the same cell. The results showed that neurite extension induced by GRFΔN occurred even in the presence of dominant-negative Rac1 or RhoA (Figure 2A). These results suggest that GRFΔN/H-Ras can induce neurite extension from PC12 cells through a pathway that is independent of Rac1 or RhoA.
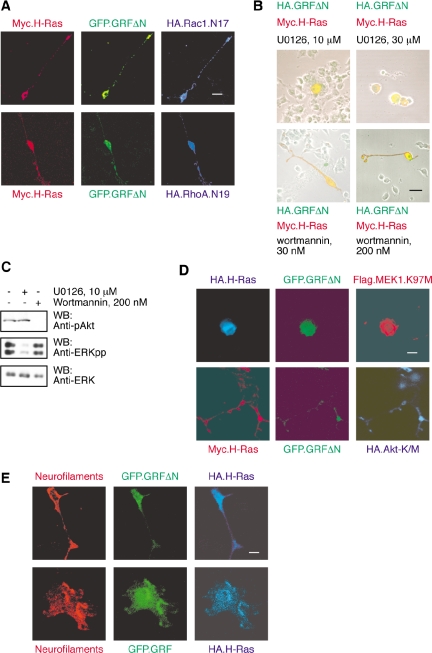
Figure 2.
GRFΔN/H-Ras–induced neurite extension is dependent on ERK activation but not PI3-Kinase or Rho proteins. (A) Triple transfections were performed in PC12 cells, using GFP.GRFΔN (green fluorescent detection), Myc.H-Ras (red indirect immunofluorescent detection), and a dominant-negative variant of either Rac1 (HA.Rac1.N17) or RhoA (HA.RhoA.N19; blue indirect immunofluorescent detection). The result shown is of two single, triply transfected cells that are representative of multiple cells from three independent experiments. Scale bar, 50 μm. Neurite outgrowth of PC12 cells was unaffected by the presence of either dominant-negative Rac1 (top row) or RhoA (bottom row). (B) PC12 cells transfected with HA.GRFΔN (green detection) and Myc.H-Ras (red detection) were maintained in culture medium in the presence of U0126, a MEK inhibitor, or wortmannin, a PI3K inhibitor, for 48 h. U0126 at 10 μM was sufficient to inhibit neurite extension of PC12 cells (top panels). In contrast, neurite outgrowth was not blocked by wortmannin (bottom panels). The results are representative of three independent experiments. (C) Lysates were prepared from PC12 cells that had been treated for 1 h with DMSO vehicle, or 10 μM U0126, or 200 nM wortmannin and subjected to Western blotting for phosphorylated Akt, dually phosphorylated ERK MAP kinases and total ERK as previously described (Mattingly et al., 2001b; Menard et al., 2005). (D) Triple transfections were performed using GFP.GRFΔN (green fluorescent detection), HA- or Myc-tagged H-Ras (blue and red indirect immunofluorescent detection), and dominant-negative variants of either MEK1 (Flag.MEK1.K97M) or Akt (HA.Akt-K/M) (red and blue indirect immunofluorescent detection). The result shown is of two representative, triply transfected cells. Scale bar, 50 μm. Neurite outgrowth from PC12 cells was completely blocked by dominant-negative MEK1 (top row) but unaffected by dominant-negative Akt (bottom row). (E) Cotransfections were performed in PC12 cells, using GFP.GRFΔN or GFP.GRF1 (green fluorescent detection), and HA.H-Ras (blue indirect immunofluorescent detection). PC12 cells that differentiate in response to coexpression of GFP.GRFΔN plus H-Ras (top row) or GFP.Ras-GRF1 plus H-Ras (bottom row) are positive for expression of neurofilaments (red indirect immunofluorescent detection).
To further investigate the pathway for neurite extension, PC12 cells transfected to coexpress GRFΔN and H-Ras were treated with inhibitors of two major signal transduction pathways that are known to be coupled to Ras activation and that have previously been shown to be required for PC12 cellular differentiation (Gotoh et al., 1990; Jackson et al., 1996). Inhibition of the ERK MAP kinase cascade with the MAP kinase kinase inhibitor U0126 was able to completely block neurite extension (Figure 2B). This inhibition of neurite extension was complete at 10 μM U0126, a concentration that did not have any apparent toxic effect and that was sufficient to substantially reduce activation of the ERK MAP kinases (Figure 2C). Higher concentrations of U0126 were toxic to PC12 cells, as was shown by the substantial loss of cells after treatment with 30 μM U0126. This toxicity may be related to inhibition of the MAP kinase pathway as the inactive structural analogue U0124 was not toxic at the same concentration (unpublished data). Interestingly, cells that expressed Ras-GRFΔN and H-Ras were apparently somewhat more resistant to the toxic effect of U0126, because cells positive for the expression of the constructs were retained when the untransfected cells were lost (Figure 2B). This result suggests that this combination of GRFΔN plus H-Ras may produce a prosurvival signal. Inhibition of the phosphatidylinositol 3-kinase (PI3K) pathway with wortmannin did not inhibit neurite extension in response to expression of GRFΔN and H-Ras (Figure 2B), despite the confirmation that Akt phosphorylation was completely inhibited by this treatment (Figure 2C).
The use of the pharmacological inhibitors suggested that GRFΔN plus H-Ras induced neuritogenesis through a pathway that required activation of ERK MAP kinases but that was independent of PI3K activity. To test this model, we performed triple cotransfections of GRFΔN plus H-Ras with dominant-negative forms of either MEK1 or Akt (Figure 2D). The results show that dominant-negative MEK1 blocks neurite production but dominant-negative Akt does not. Another series of experiments to test the model was performed to determine whether a similar pathway for neuritogenesis was induced by H-Ras.V12 (Supplementary Figure 1). These results showed that neurite outgrowth induced by constitutively activated H-Ras was also independent of wortmannin treatment or expression of dominant-negative Akt. Inhibition of ERK MAP kinase activation, whether by treatment with U0126 or expression of dominant-negative MEK1, greatly reduced the extension of neurites but did not reproducibly eliminate them.
To confirm that the morphological changes induced in the PC12 cells by coexpression of H-Ras with Ras-GRF1 were due to induction of neuronal differentiation, we tested for expression of neurofilaments by confocal immunofluorescence (Figure 2E). Control, untransfected PC12 cells were negative for neurofilament expression, whereas the cells induced to extend neurites by expression of H-Ras plus GRFΔN, and those with increased soma size due to expression of H-Ras plus Ras-GRF1, were strongly positive for neurofilaments.
Expression of GRFΔC Induces Expansion of the Soma of PC12 Cells That Is Dependent on H-Ras
Because coexpression of full-length Ras-GRF1 with H-Ras induced both neurite extension and soma expansion, whereas coexpression of GRFΔN and H-Ras only induced neurite extension, we hypothesized that the N-terminal domains of Ras-GRF1 contained the activity responsible for the increase in size of the cell body. We therefore compared the effects of expression of either GRFΔN or GRFΔC on cellular morphology. The results in Figure 3A showed that, just as expression of full-length Ras-GRF1 alone had little effect on PC12 cell morphology, neither GRFΔN nor GRFΔC induced morphological differentiation. Because both exchange factor activities of Ras-GRF1 have been shown to be stimulated by LPA treatment (Innocenti et al., 1999; Mattingly et al., 1999), we tested whether LPA stimulation of PC12 cells that express either GRFΔN or GRFΔC could induce morphological differentiation, but it did not (Figure 3A). Coexpression of GRFΔC with H-Ras, however, was sufficient to induce expansion of the soma. Quantification of the area of the cell body from cotransfected cells showed that the combination of GRFΔC with H-Ras produced a 9.5-fold increase in soma size that was comparable to the 10-fold increase induced by full-length Ras-GRF1 plus H-Ras (Figure 3B). The combination of GRFΔN plus H-Ras, although it induces the extension of neurites, does not significantly change the size of the cell body.
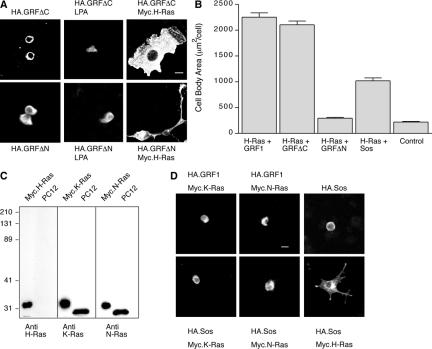
Figure 3.
The expanded PC12 morphology can be induced by cotransfection of GRFΔC and H-Ras. (A) Transfection with either HA.GRFΔC (left, top panel) or HA.GRFΔN (left, bottom panel) alone did not alter cell body size of PC12 cells. Treatment of the cells with 10 μM LPA (middle panels) did not stimulate any morphological differentiation. The combination of HA.GRF1ΔC and Myc.H-Ras produced the expanded morphology (right, top panel). Scale bar, 50 μm. The combination of HA.GRFΔN and Myc.H-Ras produced cells with small soma but long neurite extensions (right, bottom panel). (B) Using the Metamorph software program, we measured cell body areas of transfected cells. We measured 75 cells within each group that were positive for both transfected proteins or else negative for both (as control) from three independent experiments. The largest 10 measurements for each condition from each experiment (total of 30 cells) were averaged to determine cell body area (mean ± SEM). The ability of Sos1 plus H-Ras to increase cell soma size was significantly less than that of Ras-GRF1 plus H-Ras (p < 0.001, by two-way analysis of variance). (C) PC12 cell lysate was separated on 12% SDS-polyacrylamide gel in parallel with positive controls of Myc-tagged Ras proteins expressed in COS-7 cells and transferred onto nitrocellulose membrane. Immunoblotting of the standards with anti-Myc reagents verified their relative loading (Mattingly et al., 2006). The blots were then probed with anti-H-Ras, anti-K-Ras, or anti-N-Ras monoclonal antibodies, as shown. The result showed that the PC12 cells express K-Ras and N-Ras but no detectable H-Ras. (D) Transfection of PC12 cells with Ras-GRF1 plus K-Ras or N-Ras, or with Sos1 alone (top row), or with Sos1 plus K-Ras or N-Ras (bottom left and middle) did not induce morphological changes. Transfection of PC12 cells with Sos1 and H-Ras produced some neurite extension and expansion of the soma (bottom right).
Because Ras-GRF1 had previously been reported to be selective for activation of H-Ras, as opposed to K-Ras or N-Ras, when assayed in situ in NIH-3T3 fibroblasts (Jones and Jackson, 1998) or COS-7 cells (Arozarena et al., 2004), we hypothesized that the reason that expression of Ras-GRF1 or its truncation mutants in PC12 cells did not induce differentiation, whereas in combination with transfected H-Ras they did, could be that endogenous H-Ras was not sufficient. The data in Figure 3C show that although K-Ras and N-Ras were easily detectable in lysates of PC12 cells probed by Western blotting, H-Ras protein could not be detected. Further, overexpression of K-Ras or N-Ras with Ras-GRF1 was not sufficient to induce apparent morphological changes in the PC12 cells (Figure 3D).
Sos1 is a widely expressed protein with dual GEF activities for both Ras and Rac (Nimnual et al., 1998). To test whether similar morphological differentiation of PC12 cells could be induced in the absence of Ras-GRF1, we overexpressed Sos1 in combination with Ras proteins (Figure 3D). Similar to Ras-GRF1, overexpression of Sos1 alone or in combination with K-Ras or N-Ras had minimal effects on the cells. Overexpression of Sos1 with H-Ras produced significant morphological effects, with both an increase in soma size and neurite extension (Figure 3D). Quantification of the effects showed that the increase in soma size was approximately half of that induced by Ras-GRF1 plus H-Ras (Figure 3B).
Expansion of the Soma of PC12 Cells That Is Induced by GRFΔC and H-Ras Is Dependent on Rac1, PI3K, and Akt Activity
The N-terminal half of Ras-GRF1 is missing the CDC25 domain that is known to activate H-Ras, thus the requirement for the combination of GRFΔC plus H-Ras to induce the increase in cell body size was unexpected. To further investigate the requirement of GRFΔC for small GTPases, we coexpressed it as a GFP fusion protein with wild-type or dominant-negative H-Ras, or with Rac1. The results show that GFP.GRFΔC is incapable of affecting cellular morphology when expressed alone (as was shown for HA-tagged GRFΔC in Figure 3A), but induces expansion of the cell body when coexpressed with HA-tagged H-Ras (Figure 4A). Coexpression of Rac1 or of dominant-negative H-Ras was incapable of supporting GRFΔC-induced soma expansion. Because the N-terminal region of Ras-GRF1 (GRFΔC) contains the DH/PH domains that have been demonstrated to be capable of acting as an exchange factor toward Rac1 (Kiyono et al., 1999), we determined whether there was a role for Rac1 in this morphological change. Coexpression of dominant-negative Rac1 with GFP.GRFDC plus H-Ras completely blocked the increase in cell body size (Figure 4B). As a control, we found that coexpression of dominant-negative RhoA did not prevent soma expansion.
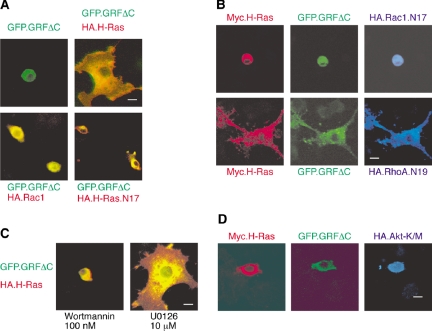
Figure 4.
The expanded PC12 morphology that is induced by GRFΔC/H-Ras is blocked by dominant-negative Rac1 as well as inhibition of PI 3-kinase. (A) PC12 cells were transfected with either GFP.GRFΔC (green fluorescent detection) alone or in combination with HA-tagged H-Ras, H-Ras.N17, or Rac1 (red indirect immunofluorescent detection). The expanded morphology induced by GFP.GRFΔC requires H-Ras (right, top panel). Scale bar, 50 μm. GFP.GRFΔC alone as well as its combination either with a dominant-negative H-Ras variant, H-Ras.N17, or with Rac1 did not induce changed morphology. (B) Triple transfection of GFP.GRFΔC (green detection) and Myc-H-Ras (red detection) plus either HA-Rac1.N17 or HA-RhoA.N19 (blue indirect immunofluorescent detection) was performed in PC12 cells. The expanded morphology induced by GRFΔC/H-Ras, was blocked by Rac1.N17 (top row), but not by RhoA.N19 (bottom row). (C) GFP.GRFΔC and H-Ras exhibit a great expansion of the soma. Expansion of the soma is effectively blocked by wortmannin, a PI3K inhibitor (left panel), but not by U0126, a MEK inhibitor (right panel). (D) Coexpression of dominant-negative HA-Akt-K/M (blue indirect immunofluorescent detection) with GFP.GRFΔC (green detection) and Myc-H-Ras (red detection) blocked expansion of the soma.
To further delineate the pathway through which GRFΔC plus H-Ras induced an increase in cell body area, we again tested for the role of ERK MAP kinases and PI3K. The results showed that expansion of the soma was completely inhibited by wortmannin, whereas it was not inhibited by U0126 (Figure 4C). The requirement for the PI3K pathway was further supported by the observation that coexpression of dominant-negative Akt could block the soma expansion induced by GRFΔC plus H-Ras (Figure 4D). Thus increase in cell body size requires PI3K/Akt but not ERK MAP kinase activity, which is the opposite requirement from that described for neurite extension (Figure 2B).
To test the model that Rac1 activation by GRFΔC may lead to soma expansion, we directly tested whether constitutively activated Rac1 used a similar pathway to increase cell size (Supplementary Figure 2). The results show that Rac1.V12 induces prominent membrane ruffling that increases the area of the cell and that this effect is blocked by wortmannin treatment or expression of dominant-negative Akt, but is independent of dominant-negative MEK1. Jun kinase activation is another prominent signaling pathway downstream of Rac1 activation, including when it is induced by Ras-GRF1 signaling (Innocenti et al., 1999). We tested whether Jun kinase may be involved using the pharmacological inhibitor SP600125 at a concentration of 10 μM, which is at or above that which has been shown to block Jun kinase activation in PC12 cells (Waetzig and Herdegen, 2003; Marek et al., 2004). Treatment with SP600125 did not block either the membrane ruffling induced by Rac1.V12 or the soma expansion induced by Ras-GRF1 plus H-Ras (Supplementary Figure 2A). Similarly, SP600125 did not affect the ability of H-Ras.V12 to induce neuritogenesis (Supplementary Figure 1). The characteristic phenotype induced by Rac1.V12 is apparently dominant over the extension of neurites that is induced by H-Ras.V12 as the coexpression of the two constitutively activated proteins reproducibly produced ruffled cells that lacked prominent neuritis (Supplementary Figure 2A). Similarly, the morphology of cells coexpressing Rac1.V12 with GRFΔN plus H-Ras was indistinguishable from that of cells expressing Rac1.V12 alone (Supplementary Figure 2B).
Constitutive Rac Activation by the GRFΔC Truncation Mutant
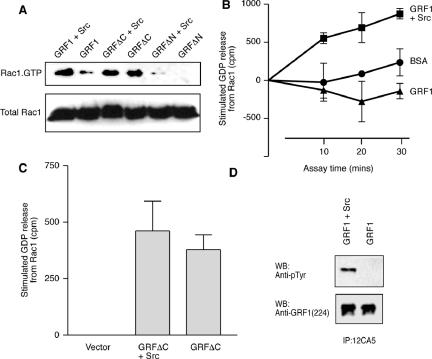
Because dominant-negative Rac1 blocked the increase in cell body size induced by GRFΔC plus H-Ras, and in view of the close correlations found between the morphological differentiation induced by activated Rac1 and the soma expansion induced by Ras-GRF1 plus H-Ras, it was reasonable to assume that activation of endogenous Rac1 by GRFΔC may be required for its effects. To investigate the relative abilities of full-length Ras-GRF1 and GRFΔC to activate Rac1, they were transfected into HEK-293 cells and the level of Rac1.GTP present in cell lysates was assayed. HEK-293 cells allow sufficient efficiency of transfection to allow biochemical assays and some isolates also express endogenous Ras-GRF1 (Norum et al., 2005). Because the Rac exchange factor activity of Ras-GRF1 has previously been shown to be minimal in the absence of tyrosine phosphorylation (Kiyono et al., 2000b), we included cotransfection with a constitutively active Src kinase as a further variable. The results showed that Rac1.GTP levels were increased in cells coexpressing full-length Ras-GRF1 plus active Src, but that GRFΔC increased Rac.GTP levels independent of the presence of Src (Figure 5A). As a control, we also transfected with GRFΔN with or without active Src. This construct is missing the DH/PH domains that are responsible for Rac exchange factor activity. Under those conditions there was a minimal level of Rac1.GTP.
Figure 5.
Activation of Rac1 by GRF1/Src correlates with tyrosine phosphorylation of GRF1. (A) HEK-293 cells were transfected with HA-tagged GRF1, GRFΔC, or GRFΔN, with or without additional activated Src. Activation of the endogenous Rac1 protein was then assayed by determination of the proportion that was bound to GTP. (B) HEK-293 cells were transfected with HA.GRF1 alone or in combination with activated Src. Cell lysates were prepared after serum starvation and anti-HA immunoprecipitates were assayed for their ability to release [3H]GDP that had been prebound to recombinant Rac1 in comparison to incubations with BSA as a control. Data shown are mean ± SEM from four independent experiments conducted in triplicate. (C) HEK-293 cells were transfected with a control, empty vector, or with HA.GRF1ΔC alone or plus activated Src. Anti-HA immunoprecipitates were assayed for 20 min for their ability to release [3H]GDP that had been prebound to recombinant Rac1. Data shown are the change in released GDP relative to that from Rac1 incubated with a control immunoprecipitate and are mean ± SEM from four independent experiments conducted in triplicate. (D) HEK-293 cells transfected with either HA.GRF1 alone or in combination with Src. After serum starvation, cell lysates were prepared in RIPA buffer as described (Yang et al., 2003). HA.Ras-GRF1 was immunoprecipitated and analyzed by Western blot with anti-pTyr antibody (top panel). The blot was then stripped and reprobed with anti-GRF1 antibody sc224 (bottom panel).
To verify whether the levels of Rac1.GTP found in the cell lysates reflected the actual exchange factor activity of Ras-GRF1 and GRFΔC toward Rac1, we immunoprecipitated these proteins using their HA epitope tags and assayed them in vitro against recombinant Rac1. The results showed that Ras-GRF1 immunoprecipitated from HEK-293 cells in the absence of active Src had no measurable exchange factor activity toward Rac1 (Figure 5B). In fact, there was a reproducible effect to slightly stabilize the nucleotide bound to Rac1 in comparison to incubations of Rac1.GDP with BSA as a control. When Ras-GRF1 was immunoprecipitated from cells that had been cotransfected with active Src, then there was significant exchange factor activity toward Rac1 present. Immunoprecipitated GRFΔC exhibited exchange factor activity toward Rac1 whether it had been transfected alone or in the presence of active Src (Figure 5C). Western blotting of the immunoprecipitated Ras-GRF1 confirmed that it was tyrosine phosphorylated when coexpressed with active Src (Figure 5D). We were unable to detect phosphotyrosine on GRFΔC whether it was isolated from cells in the presence or absence of active Src (unpublished data).
Activation of H-Ras Is Required for GRFΔC-induced Cell Body Expansion
The effect of GRFΔC to induce the expansion of the PC12 soma was dependent on the presence of H-Ras and could be blocked by dominant-negative H-Ras. Because GRFΔC is lacking the CDC25 domain to activate H-Ras, we hypothesized that H-Ras may become activated during the assay due to another stimulus. We therefore tested whether GRFΔC plus H-Ras could stimulate differentiation of PC12 cells if serum were omitted from the incubation. The results shown in Figure 6A demonstrated that GRFΔC plus wild-type H-Ras was insufficient to induce any increase in the cell body size in the absence of serum stimulation. Conversely, if GRFΔC was coexpressed with a constitutively active H-Ras.V12 protein, then the cell body increased in size without serum stimulation. Further, full-length Ras-GRF1 plus wild-type H-Ras was also capable of inducing this morphological differentiation in the absence of serum.
Figure 6.
Ras-GRF1 can activate H-Ras and induce expanded PC12 morphology independent of serum; GRF1ΔC requires activation of H-Ras. (A) PC12 cells were transfected with HA.GRF1 in combination with Myc.H-Ras, or with HA.GRF1ΔC in combination either with Myc.H-Ras or Myc.H-Ras.V12 and then cultured for 48 h in the absence of serum. Cells were fixed and subjected to indirect immunofluorescence staining using anti-HA polyclonal antibody Y11 and green detection or anti-Myc mAb 9e10 and red detection. In the absence of serum, GRF1 and H-Ras could still induce cell expansion (left) but GRF1ΔC plus H-Ras could not (center). A combination of GRF1ΔC plus H-Ras.V12 did induce cell expansion (right). Scale bar, 50 μm. (B) HEK-293 cells were transfected with Myc.H-Ras alone, or in combination with Ras-GRF1 or with GRF1ΔC. Ras.GTP pulldown assays were performed on the cell lysates. The active form of the transfected H-Ras was detected by Western blot using anti-Myc 9B11 antibody (top panel). Cell lysates were probed for total transfected H-Ras to determine whether there were equivalent amounts of Myc.H-Ras protein in each sample (bottom panel). (C) Lysates of PC12 and HEK293 cells were immunoprecipitated for H-Ras using a rat anti-H-Ras mAb, followed by rabbit anti-rat polyclonal antibodies and protein A-Sepharose. The immunoprecipitates were analyzed by Western blot in comparison to a Myc-tagged H-Ras positive control, and negative controls of an immunoprecipitation from HEK293 cell lysate that omitted the rat anti-H-Ras antibody and of the rat anti-H-Ras antibody alone.
To verify the relative abilities of Ras-GRF1 and GRFΔC to activate H-Ras, we cotransfected them with Myc-tagged H-Ras into HEK-293 cells and measured their effect on H-Ras.GTP levels. The results showed that H-Ras was activated in cells grown in serum, but not in those incubated without serum (Figure 6B). Only full-length Ras-GRF1, but not GRFΔC, could activate H-Ras in the absence of serum stimulation. Unlike PC12 cells, the HEK-293 cells used in this study also contained detectable, endogenous H-Ras protein (Figure 6C).
Direct Interaction between GRFΔC and H-Ras.GTP
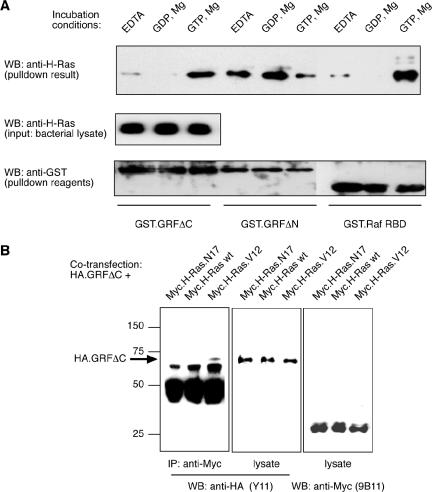
Ras.GTP has recently been shown to bind to the SOS exchange factor's REM (Ras exchange motif) domain and provide feedback activation of CDC25 exchange factor activity (Margarit et al., 2003). Because this establishes a precedent for the interaction of activated Ras with one of its own exchange factors, we decided to test whether there might be a direct interaction between H-Ras.GTP and GRFΔC. The parallel was not close, however, because the REM domain is missing from GRFΔC and we were searching for an interaction that could participate in the activation of the Rac exchange factor activity of its DH/PH domains rather than the Ras exchange factor activity of its CDC25 domain. We therefore prepared lysates of E. coli that expressed H-Ras and incubated them with GST-fusion proteins of GRFΔC, GRFΔN (as a positive control for Ras binding to a CDC25 domain), and Raf-RBD (as a positive control for a Ras effector interaction). The pulldown reactions were performed in the presence of EDTA, to destabilize nucleotide binding to H-Ras, or GDP plus Mg2+, to induce formation of H-Ras.GDP, or GTP plus Mg2+, to induce formation of H-Ras.GTP. The results showed that GRFΔN bound preferentially to H-Ras.GDP and H-Ras in the presence of EDTA, and less to H-Ras.GTP, as would be expected for an exchange factor (Figure 7A). The result for binding to Raf-RBD, a strong preference for binding to H-Ras.GTP over H-Ras.GDP, was also in agreement with expectations. Strikingly, GRFΔC showed an identical preference for selective binding to H-Ras.GTP as for the Raf-RBD effector interaction.
Figure 7.
Ras-GRF1ΔC binds preferentially to GTP-bound H-Ras in a GST-fusion protein pulldown assay and in a coimmunoprecipitation. (A) Lysates of bacteria expressing H-Ras bacteria were pretreated with 5 mM EDTA, 5 mM magnesium plus 100 μM GTP, or GDP, and then incubated with glutathione beads coated with GST.GRFΔC, GST.GRFΔN, or GST.RafRBD. The results shown are for the H-Ras that was detected by Western blotting of the washed pulldown reactions (top panel) or of 5% of the bacterial lysates (middle panel). The membranes were stripped and reprobed with anti-GST antibody to control for the relative loading of the GST fusion proteins onto the pulldown beads (bottom panel). Results shown are representative of three independent experiments. (B) HEK-293 cells were cotransfected with HA.GRFΔC plus Myc.H-Ras, Myc.H-Ras.N17, or Myc.H-Ras.V12. Cell lysates were prepared and blotted for content of Myc-tagged H-Ras proteins (right panel) and HA.GRFΔC (center panel) that confirmed even expression of the constructs. Anti-Myc immunoprecipitates were prepared from cell lysates using the mAb 9B11. HA.GRFΔC was only detected in immunoprecipitations from cells that coexpressed Myc.H-Ras.V12 (left panel).
To further test the potential for activated H-Ras to bind to GRFΔC, we coexpressed Myc-tagged H-Ras wild-type (wt), constitutively active (V12), or dominant-negative (N17) proteins with HA-tagged GRFΔC in HEK-293 cells. Western blotting confirmed that all of the proteins coexpressed and were recovered efficiently after cell lysis (Figure 7B). Immunoprecipitation of the Myc-tagged Ras proteins from the cell lysates demonstrated that GRFΔC only coimmunoprecipitated with the activated H-Ras.V12 protein, and not with H-Ras wt or H-Ras.N17 (Figure 7B).
DISCUSSION
The molecular and cellular basis for memory is still poorly understood, but it is assumed that electrophysiological phenomena, such as long-term potentiation, and morphological characteristics, such as the numbers and shapes of dendritic spines, are useful and relevant experimental systems (Sheng and Kim, 2002). Long-term potentiation has been most actively studied in response to the activation of excitatory glutamate receptors of the NMDA subtype. Activation of these receptors leads to calcium influx, Ras and ERK MAP kinase activation, and subsequent activation of transcription factors such as CREB (Sheng and Kim, 2002). Ras-GRF1 has been identified as a potential key intermediate in this pathway, because it is able to activate Ras and the ERK MAP kinase cascade in response to increases in intracellular calcium (Farnsworth et al., 1995), can directly associate with NMDA receptors and is functionally required for NMDA receptor coupling (Krapivinsky et al., 2003), and becomes phosphorylated at Serine-916 during NMDA receptor-induced long-term potentiation (Schmitt et al., 2005). Genetic support for a role for Ras-GRF1 in memory is provided by the learning deficits that have been found in knockout mice that are deficient in Ras-GRF1 (Brambilla et al., 1997; Giese et al., 2001). Detailed analysis of these Ras-GRF1–deficient mice has revealed abnormalities in long-term potentiation (Brambilla et al., 1997) and neuronal excitability (Tonini et al., 2001), and that there is a developmentally regulated role for Ras-GRF1 to provide signal transduction from NMDA receptors in the adult, but not the neonatal brain (Tian et al., 2004).
A primary role for Ras-GRF1 in signal transduction is presumably to activate H-Ras. In previous studies in NIH-3T3 fibroblasts (Jones and Jackson, 1998) and COS-7 cells (Arozarena et al., 2004), Ras-GRF1 has been shown to selectively activate H-Ras, rather than K-Ras or N-Ras. Our results, which show that Ras-GRF1 is not able to induce morphological changes in PC12 cells in the absence of cotransfected H-Ras, seem to be largely in agreement with these studies. Neither the endogenous nor overexpressed K-Ras and N-Ras were sufficient to allow Ras-GRF1 to induce morphological differentiation, although constitutively activated N-Ras and K-Ras have been reported to be sufficient to induce neurite extension (Guerrero et al., 1986; Brightman et al., 1990). Localization of Ras-GRF1 and H-Ras by indirect immunofluorescent confocal microscopy suggested that they were colocalized in internal, extranuclear regions of the PC12 cells that could be consistent with the endoplasmic reticulum. There was also prominent localization of H-Ras at the apparent plasma membrane, but little colocalization with Ras-GRF1 there. In COS-7 cells, Ras-GRF1 activation of H-Ras on the endoplasmic reticulum required an intact DH domain (Arozarena et al., 2004). In the current study, there was functional cooperation of GRFΔN with H-Ras to enhance cell survival and induce neurite extension from PC12 cells, which would suggest that the N-terminal region is not absolutely required for the interaction.
Activation of H-Ras is significant as that isoform of Ras has also been strongly linked to regulation of long-term potentiation, neuronal morphology, and memory. For example, knockout mice deficient in H-Ras have aberrant NMDA receptor–dependent long-term potentiation (Manabe et al., 2000). Further, transgenic expression of an activated H-Ras.V12 from a synapsin promoter in adult CNS neurons produces an increase in the soma size and dendritic spine density of cortical neurons (Arendt et al., 2004) and was also protective against neural degeneration (Heumann et al., 2000). Soma size of prefrontal and hippocampal neurons is reduced in major depression (Rajkowska et al., 1999; Stockmeier et al., 2004) and so it is important to understand the signaling pathways that regulate this morphology. In the current study we found that Ras-GRF1 plus H-Ras induced an increase in soma size that was blocked by the PI3K inhibitor wortmannin. PI3K has previously been ascribed roles in both NGF-induced differentiation of PC12 cells (Jackson et al., 1996) and in long-term potentiation and synaptic plasticity (Lin et al., 2001). The significance of Ras-induced increases in cell size is not clear, but in addition to the current study on PC12 cells expressing Ras-GRF1 and H-Ras, and the observations on cortical neurons expressing H-Ras.V12 (Arendt et al., 2004), there is also evidence for this phenomenon in hypertrophy of cardiomyocytes (Ramirez et al., 1997). The profound expansion of the soma of PC12 cells that was induced in the current study by Ras-GRF1 and H-Ras does not seem to have previously been reported, although expression of constitutively-activated Rac2 (Daniels et al., 1998) or stimulation of PC12 cells that overexpress the TrkB receptor with brain-derived neurotrophic factor (Iwasaki et al., 1997) have been reported to produce some soma expansion. In the latter case, expansion of the soma took 7 d to occur and was preceded by an apparent thickening of neurite extensions in comparison to those stimulated by NGF treatment (Iwasaki et al., 1997). In our study, soma expansion occurred within 48 h of cotransfection and precursor phenotypes were not identified. Because modification of dendritic shape is thought to be a morphological correlate of long-term potentiation and memory (Carlisle and Kennedy, 2005), it is interesting to speculate whether this modification of PC12 cell morphology may be a parallel phenomenon to that process.
In addition to activation of H-Ras, Ras-GRF1, through its DH/PH2 domains, can also act as a GEF for Rac1. This Rac GEF activity is apparently latent in the intact, unstimulated protein (Kiyono et al., 1999), but is stimulated by overexpression of G protein βγ subunits (Kiyono et al., 1999) or an active Src kinase (Kiyono et al., 2000b) or by stimulation with LPA (Innocenti et al., 1999). Our data support the previous observations that there is minimal Rac GEF activity in Ras-GRF1 under basal conditions and suggest that there may actually be slight stabilization of nucleotide binding to Rac1. Cotransfection with activated Src induced Rac GEF activity that was revealed both in in vitro GEF assays and by increased Rac.GTP loading in HEK293 cells. Our data further show that the GRFΔC truncation protein exhibits constitutive Rac GEF activity in both of these assays. Our data also show, however, that the GRFΔC truncation protein was not sufficient to induce any morphological change upon transfection into PC12 cells. PC12 cells respond to a constitutively activated Rac2 (Daniels et al., 1998) or Rac1 protein (this study) by producing prominent membrane ruffles. The lack of effect on morphology after GRFΔC expression could suggest either that the Rac GEF activity of GRFΔC is not constitutive in PC12 cells or that its constitutive activity does not generate sufficient Rac.GTP levels to reproduce the effects of a constitutively activated Rac1 mutant. To induce expanded morphology of PC12 cells, the GRFΔC truncation protein required the additional presence of activated H-Ras (either H-Ras plus serum stimulation or H-Ras.V12). Thus the expanded morphology of PC12 cells is apparently dependent on activation of both H-Ras and Rac1. It is interesting to note that this dual activation requires coordination as the simple coexpression of activated Rac1 with activated H-Ras (or of activated Rac1 with GRFΔN plus H-Ras) produced a phenotype that was indistinguishable from that induced by Rac1.V12 alone. One possible mechanism for Ras-GRF1 to produce the coordinated activation of H-Ras and Rac1 activation that is required for its characteristic phenotypic effect would be through the directed activation of a particular subcellular pool of Rac1 at the locus of H-Ras activation.
Activation of Rac1 has previously been shown to be critical for both the induction of morphological differentiation of PC12 cells (Daniels et al., 1998; Yasui et al., 2001) and for long-term potentiation and dendritic remodeling in the brain (Luo et al., 1996). Like H-Ras activation, Rac activation in response to NMDA receptors is thought to be a major component in synaptic plasticity (Sheng and Kim, 2002; Weeber et al., 2002). Rac1 is well known for its ability to regulate the actin cytoskeleton, and it is likely that this function may provide a connection to the dynamic remodeling of dendritic spines (Penzes et al., 2001). There are several Rac GEF activities that may explain the activation of Rac in response to NMDA receptor signaling. Both Kalirin-7 and Tiam1 are Rac GEFs that have been localized to postsynaptic densities and modification of their activities can alter dendritic morphology (Penzes et al., 2001; Tolias et al., 2005). In the case of Tiam1, a model was proposed whereby the activation of Ras by Ras-GRF1 in response to NMDA receptor activation would provide Ras.GTP to bind to the RBD of Tiam1 and stimulate its Rac GEF activity (Lambert et al., 2002; Tolias et al., 2005). Both Tiam1 and Ras-GRF1 have previously been shown to bind to the same scaffolding complex (Buchsbaum et al., 2002). Our data suggest that the H-Ras.GTP generated by Ras-GRF1 can act to directly facilitate the Rac GEF activity that is intrinsic to Ras-GRF1 and so would provide a further mechanism to coordinate the activation of H-Ras and Rac1 that may be required for effective synaptic remodeling (Figure 8). We find that H-Ras.GTP can bind to the GRFΔC truncation protein and that this interaction correlates with the induction of a Rac-dependent morphological change. It is therefore surprising that GRFΔC is lacking the REM domain sequence of Ras-GRF1 that is homologous to the region shown to bind Ras.GTP in Sos1 and does not obviously contain sequences homologous to either RBD (Herrmann, 2003) or RA (Ras association) domains (Ponting, 1999) that have been defined to provide interaction with Ras.GTP. The structural basis of the interaction between GRFΔC and H-Ras.GTP will need to be identified.
Figure 8.
The expanded PC12 cell morphology that is induced by Ras-GRF1 is produced by coordinated activation of H-Ras and Rac1. Expansion of the PC12 soma requires activation of both H-Ras and Rac1. Full-length Ras-GRF1 can accomplish both functions, but GRFΔC requires an additional stimulus to activate H-Ras. Activated H-Ras can directly interact with GRFΔC and may provide a mechanistic link for coordination of Rac1 activation at the locus of H-Ras activation.
Supplementary Material
Acknowledgments
We thank Drs. M. Czech, D. Flynn, J. Jackson, A. Toker, G. Tzivion and A. Wolfman, and Genentech, for their generous gifts of reagents; Dr. R. Andrade for useful discussions; and Dr. J. Benjamins for a critical reading of the manuscript. This work was supported by Public Health Service Grant R01 CA-81150 from the National Cancer Institute. Confocal facilities were supported by Center Grants P30 ES06639 and CA22453.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–10–0913) on February 15, 2006.
Abbreviations used: CRIB, Cdc42/Rac1-binding domain; DH, Dbl homology domain; GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; GRFΔC, Ras-GRF1 residues 1-631; GRFΔN, Ras-GRF1 residues 632-1262; GST, glutathione S-transferase; HA, hemagglutinin-1 epitope; LPA, lysophosphatidic acid; NGF, nerve growth factor; PH, plekstrin homology domain; PI3K, phosphatidylinositol 3-kinase; Raf.RBD, Ras-binding domain of Raf; TBST, TBS with Tween-20.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Arendt, T. et al. (2004). Neuronal activation of Ras regulates synaptic connectivity. Eur. J. Neurosci. 19, 2953–2966. [DOI] [PubMed] [Google Scholar]
- Arozarena, I., Matallanas, D., Berciano, M. T., Sanz-Moreno, V., Calvo, F., Munoz, M. T., Egea, G., Lafarga, M., and Crespo, P. (2004). Activation of H-Ras in the endoplasmic reticulum by the RasGRF family guanine nucleotide exchange factors. Mol. Cell. Biol. 24, 1516–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznar, S., and Lacal, J. C. (2001). Rho signals to cell growth and apoptosis. Cancer Lett. 165, 1–10. [DOI] [PubMed] [Google Scholar]
- Baouz, S., Jacquet, E., Bernardi, A., and Parmeggiani, A. (1997). The N-terminal moiety of CDC25(Mm), a GDP/GTP exchange factor of Ras proteins, controls the activity of the catalytic domain. Modulation by calmodulin and calpain. J. Biol. Chem. 272, 6671–6676. [DOI] [PubMed] [Google Scholar]
- Bar-Sagi, D., and Feramisco, J. R. (1985). Microinjection of the Ras oncogene protein into PC12 cells induces morphological differentiation. Cell 42, 841–848. [DOI] [PubMed] [Google Scholar]
- Barbacid, M. (1987). ras genes. Annu. Rev. Biochem. 56, 779–827. [DOI] [PubMed] [Google Scholar]
- Beqaj, S., Jakkaraju, S., Mattingly, R. R., Pan, D., and Schuger, L. (2002). High RhoA activity maintains the undifferentiated mesenchymal cell phenotype, whereas RhoA down-regulation by laminin-2 induces smooth muscle myogenesis. J. Cell Biol. 156, 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards, A., and Settleman, J. (2004). GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 14, 377–385. [DOI] [PubMed] [Google Scholar]
- Boguski, M. S., and McCormick, F. (1993). Proteins regulating Ras and its relatives. Nature 366, 643–654. [DOI] [PubMed] [Google Scholar]
- Brambilla, R. et al. (1997). A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature 390, 281–286. [DOI] [PubMed] [Google Scholar]
- Brightman, M. W., Simpson, D. L., Tao-Cheng, J. H., Bressler, J. P., Okuda, O., and Chang, L. (1990). Some neuronal properties of PC12 cells differentiated by the K-ras oncogene. J. Neurocytol. 19, 776–788. [DOI] [PubMed] [Google Scholar]
- Buchsbaum, R. J., Connolly, B. A., and Feig, L. A. (2002). Interaction of Rac exchange factors Tiam1 and Ras-GRF1 with a scaffold for the p38 mitogen-activated protein kinase cascade. Mol. Cell. Biol. 22, 4073–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey, K. L., Richards, S. A., Lounsbury, K. M., and Macara, I. G. (1996). Evidence using a green fluorescent protein-glucocorticoid receptor chimera that the Ran/TC4 GTPase mediates an essential function independent of nuclear protein import. J. Cell Biol. 133, 985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle, H. J., and Kennedy, M. B. (2005). Spine architecture and synaptic plasticity. Trends Neurosci. 28, 182–187. [DOI] [PubMed] [Google Scholar]
- Cen, H., Papageorge, A. G., Vass, W. C., Zhang, K. E., and Lowy, D. R. (1993). Regulated and constitutive activity by CDC25Mm (GRF), a Ras-specific exchange factor. Mol. Cell. Biol. 13, 7718–7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, R. H., Hall, P. S., and Bokoch, G. M. (1998). Membrane targeting of p21-activated kinase 1 (PAK1) induces neurite outgrowth from PC12 cells. EMBO J. 17, 754–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeClue, J. E., Cohen, B. D., and Lowy, D. R. (1991). Identification and characterization of the neurofibromatosis type 1 protein product. Proc. Natl. Acad. Sci. USA 88, 9914–9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth, C. L., Freshney, N. W., Rosen, L. B., Ghosh, A., Greenberg, M. E., and Feig, L. A. (1995). Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature 376, 524–527. [DOI] [PubMed] [Google Scholar]
- Giese, K. P., Friedman, E., Telliez, J. B., Fedorov, N. B., Wines, M., Feig, L. A., and Silva, A. J. (2001). Hippocampus-dependent learning and memory is impaired in mice lacking the Ras-guanine-nucleotide releasing factor 1 (Ras-GRF1). Neuropharmacology 41, 791–800. [DOI] [PubMed] [Google Scholar]
- Gotoh, Y., Nishida, E., Yamashita, T., Hoshi, M., Kawakami, M., and Sakai, H. (1990). Microtubule-associated-protein (MAP) kinase activated by nerve growth factor and epidermal growth factor in PC12 cells. Identity with the mitogen-activated MAP kinase of fibroblastic cells. Eur. J. Biochem. 193, 661–669. [DOI] [PubMed] [Google Scholar]
- Guerrero, I., Wong, H., Pellicer, A., and Burstein, D. E. (1986). Activated N-ras gene induces neuronal differentiation of PC12 rat pheochromocytoma cells. J. Cell. Physiol. 129, 71–76. [DOI] [PubMed] [Google Scholar]
- Hardt, W. D., Chen, L. M., Schuebel, K. E., Bustelo, X. R., and Galan, J. E. (1998). S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93, 815–826. [DOI] [PubMed] [Google Scholar]
- Herrmann, C. (2003). Ras-effector interactions: after one decade. Curr. Opin. Struct. Biol. 13, 122–129. [DOI] [PubMed] [Google Scholar]
- Heumann, R. et al. (2000). Transgenic activation of Ras in neurons promotes hypertrophy and protects from lesion-induced degeneration. J. Cell Biol. 151, 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti, M., Zippel, R., Brambilla, R., and Sturani, E. (1999). CDC25(Mm)/Ras-GRF1 regulates both Ras and Rac signaling pathways. FEBS Lett. 460, 357–362. [DOI] [PubMed] [Google Scholar]
- Iwasaki, Y., Ishikawa, M., Okada, N., and Koizumi, S. (1997). Induction of a distinct morphology and signal transduction in TrkB/PC12 cells by nerve growth factor and brain-derived neurotrophic factor. J. Neurochem. 68, 927–934. [DOI] [PubMed] [Google Scholar]
- Jackson, T. R., Blader, I. J., Hammonds-Odie, L. P., Burga, C. R., Cooke, F., Hawkins, P. T., Wolf, A. G., Heldman, K. A., and Theibert, A. B. (1996). Initiation and maintenance of NGF-stimulated neurite outgrowth requires activation of a phosphoinositide 3-kinase. J. Cell Sci. 109(Pt 2), 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M. K., and Jackson, J. H. (1998). Ras-GRF activates Ha-Ras, but not N-Ras or K-Ras 4B, protein in vivo. J. Biol. Chem. 273, 1782–1787. [DOI] [PubMed] [Google Scholar]
- Kiyono, M., Kato, J., Kataoka, T., Kaziro, Y., and Satoh, T. (2000a). Stimulation of Ras guanine nucleotide exchange activity of Ras-GRF1/CDC25(Mm) upon tyrosine phosphorylation by the Cdc42-regulated kinase ACK1. J. Biol. Chem. 275, 29788–29793. [DOI] [PubMed] [Google Scholar]
- Kiyono, M., Kaziro, Y., and Satoh, T. (2000b). Induction of rac-guanine nucleotide exchange activity of Ras-GRF1/CDC25(Mm) following phosphorylation by the nonreceptor tyrosine kinase Src. J. Biol. Chem. 275, 5441–5446. [DOI] [PubMed] [Google Scholar]
- Kiyono, M., Satoh, T., and Kaziro, Y. (1999). G protein beta gamma subunit-dependent Rac-guanine nucleotide exchange activity of Ras-GRF1/CDC25(Mm). Proc. Natl. Acad. Sci. USA 96, 4826–4831.10220378 [Google Scholar]
- Krapivinsky, G., Krapivinsky, L., Manasian, Y., Ivanov, A., Tyzio, R., Pellegrino, C., Ben-Ari, Y., Clapham, D. E., and Medina, I. (2003). The NMDA receptor is coupled to the ERK pathway by a direct interaction between NR2B and RasGRF1. Neuron 40, 775–784. [DOI] [PubMed] [Google Scholar]
- Lambert, J. M., Lambert, Q. T., Reuther, G. W., Malliri, A., Siderovski, D. P., Sondek, J., Collard, J. G., and Der, C. J. (2002). Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat. Cell Biol. 4, 621–625. [DOI] [PubMed] [Google Scholar]
- Lin, C. H., Yeh, S. H., Lin, C. H., Lu, K. T., Leu, T. H., Chang, W. C., and Gean, P. W. (2001). A role for the PI-3 kinase signaling pathway in fear conditioning and synaptic plasticity in the amygdala. Neuron 31, 841–851. [DOI] [PubMed] [Google Scholar]
- Lowy, D. R., and Willumsen, B. M. (1986). The ras gene family. Cancer Surv. 5, 275–289. [PubMed] [Google Scholar]
- Luo, L., Hensch, T. K., Ackerman, L., Barbel, S., Jan, L. Y., and Jan, Y. N. (1996). Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature 379, 837–840. [DOI] [PubMed] [Google Scholar]
- Macara, I. G., Lounsbury, K. M., Richards, S. A., McKiernan, C., and Bar-Sagi, D. (1996). The Ras superfamily of GTPases. FASEB J. 10, 625–630. [DOI] [PubMed] [Google Scholar]
- MacDonald, J. I., Verdi, J. M., and Meakin, S. O. (1999). Activity-dependent interaction of the intracellular domain of rat trkA with intermediate filament proteins, the beta-6 proteasomal subunit, Ras-GRF1, and the p162 subunit of eIF3. J. Mol. Neurosci. 13, 141–158. [DOI] [PubMed] [Google Scholar]
- Malaney, S., and Daly, R. J. (2001). The ras signaling pathway in mammary tumorigenesis and metastasis. J. Mammary Gland Biol. Neoplasia 6, 101–113. [DOI] [PubMed] [Google Scholar]
- Manabe, T., Aiba, A., Yamada, A., Ichise, T., Sakagami, H., Kondo, H., and Katsuki, M. (2000). Regulation of long-term potentiation by H-Ras through NMDA receptor phosphorylation. J. Neurosci. 20, 2504–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek, L., Levresse, V., Amura, C., Zentrich, E., Van Putten, V., Nemenoff, R. A., and Heasley, L. E. (2004). Multiple signaling conduits regulate global differentiation-specific gene expression in PC12 cells. J. Cell. Physiol. 201, 459–469. [DOI] [PubMed] [Google Scholar]
- Margarit, S. M., Sondermann, H., Hall, B. E., Nagar, B., Hoelz, A., Pirruccello, M., Bar-Sagi, D., and Kuriyan, J. (2003). Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112, 685–695. [DOI] [PubMed] [Google Scholar]
- Martegani, E., Vanoni, M., Zippel, R., Coccetti, P., Brambilla, R., Ferrari, C., Sturani, E., and Alberghina, L. (1992). Cloning by functional complementation of a mouse cDNA encoding a homologue of CDC25, a Saccharomyces cerevisiae RAS activator. EMBO J. 11, 2151–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly, R. R. (1999). Phosphorylation of serine 916 of Ras-GRF1 contributes to the activation of exchange factor activity by muscarinic receptors. J. Biol. Chem. 274, 37379–37384. [DOI] [PubMed] [Google Scholar]
- Mattingly, R. R., Felczak, A., Chen, C. C., McCabe, M. J., and Rosenspire, A. J. (2001a). Low concentrations of inorganic mercury inhibit Ras activation during T cell receptor-mediated signal transduction. Toxicol. Appl. Pharmacol. 176, 162–168. [DOI] [PubMed] [Google Scholar]
- Mattingly, R. R., Kraniak, J. M., Dilworth, J. T., Mathieu, P., Bealmear, B., Nowak, J. E., Benjamins, J. A., Tainsky, M. A., and Reiners, J. J., Jr. (2006). The mitogen-activated protein kinase/extracellular signal-regulated kinase kinase inhibitor PD184352 (CI-1040) selectively induces apoptosis in malignant Schwannoma cell lines. J. Pharmacol. Exp. Ther. 316, 456–465. [DOI] [PubMed] [Google Scholar]
- Mattingly, R. R., and Macara, I. G. (1996). Phosphorylation-dependent activation of the Ras-GRF/CDC25(Mm) exchange factor by muscarinic receptors and G-protein beta gamma subunits. Nature 382, 268–272. [DOI] [PubMed] [Google Scholar]
- Mattingly, R. R., Milstein, M. L., and Mirkin, B. L. (2001b). Down-regulation of growth factor-stimulated MAP kinase signaling in cytotoxic drug-resistant human neuroblastoma cells. Cell Signal. 13, 499–505. [DOI] [PubMed] [Google Scholar]
- Mattingly, R. R., Saini, V., and Macara, I. G. (1999). Activation of the Ras-GRF/CDC25(Mm) exchange factor by lysophosphatidic acid. Cell Signal. 11, 603–610. [DOI] [PubMed] [Google Scholar]
- Mattingly, R. R., Sorisky, A., Brann, M. R., and Macara, I. G. (1994). Muscarinic receptors transform Nih 3t3 cells through a Ras-dependent signaling pathway inhibited by the Ras-Gtpase-activating protein Sh3 domain. Mol. Cell. Biol. 14, 7943–7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard, R. E., Jovanovski, A. P., and Mattingly, R. R. (2005). Active p21-activated kinase 1 rescues MCF10A breast epithelial cells from undergoing anoikis. Neoplasia 7, 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard, R. E., and Mattingly, R. R. (2003). Cell surface receptors activate p21-activated kinase 1 via multiple Ras and PI3-kinase-dependent pathways. Cell Signal. 15, 1099–1109. [DOI] [PubMed] [Google Scholar]
- Nimnual, A. S., Yatsula, B. A., and Bar-Sagi, D. (1998). Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science 279, 560–563. [DOI] [PubMed] [Google Scholar]
- Norum, J. H., Methi, T., Mattingly, R. R., and Levy, F. O. (2005). Endogenous expression and protein kinase A-dependent phosphorylation of the guanine nucleotide exchange factor Ras-GRF1 in human embryonic kidney 293 cells. FEBS J. 272, 2304–2316. [DOI] [PubMed] [Google Scholar]
- Nusser, N., Gosmanova, E., Zheng, Y., and Tigyi, G. (2002). Nerve growth factor signals through TrkA, phosphatidylinositol 3-kinase, and Rac1 to inactivate RhoA during the initiation of neuronal differentiation of PC12 cells. J. Biol. Chem. 277, 35840–35846. [DOI] [PubMed] [Google Scholar]
- Penzes, P., Johnson, R. C., Sattler, R., Zhang, X., Huganir, R. L., Kambampati, V., Mains, R. E., and Eipper, B. A. (2001). The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron 29, 229–242. [DOI] [PubMed] [Google Scholar]
- Ponting, C. P. (1999). Raf-like Ras/Rap-binding domains in RGS12- and still-life-like signalling proteins. J. Mol. Med. 77, 695–698. [DOI] [PubMed] [Google Scholar]
- Quilliam, L. A., Rebhun, J. F., and Castro, A. F. (2002). A growing family of guanine nucleotide exchange factors is responsible for activation of Ras-family GTPases. Prog Nucleic Acid. Res. Mol. Biol. 71, 391–444. [DOI] [PubMed] [Google Scholar]
- Rajkowska, G., Miguel-Hidalgo, J. J., Wei, J., Dilley, G., Pittman, S. D., Meltzer, H. Y., Overholser, J. C., Roth, B. L., and Stockmeier, C. A. (1999). Morphometric evidence for neuronal and glial prefrontal cell pathology in major depression. Biol. Psychiatry 45, 1085–1098. [DOI] [PubMed] [Google Scholar]
- Ramirez, M. T., Sah, V. P., Zhao, X. L., Hunter, J. J., Chien, K. R., and Brown, J. H. (1997). The MEKK-JNK pathway is stimulated by alpha1-adrenergic receptor and ras activation and is associated with in vitro and in vivo cardiac hypertrophy. J. Biol. Chem. 272, 14057–14061. [DOI] [PubMed] [Google Scholar]
- Robinson, K. N., Manto, K., Buchsbaum, R. J., MacDonald, J. I., and Meakin, S. O. (2005). Neurotrophin-dependent tyrosine phosphorylation of Ras guanine-releasing factor 1 and associated neurite outgrowth is dependent on the HIKE domain of TrkA. J. Biol. Chem. 280, 225–235. [DOI] [PubMed] [Google Scholar]
- Sakai, Y., Hashimoto, H., Shintani, N., Katoh, H., Negishi, M., Kawaguchi, C., Kasai, A., and Baba, A. (2004). PACAP activates Rac1 and synergizes with NGF to activate ERK1/2, thereby inducing neurite outgrowth in PC12 cells. Brain Res. Mol. Brain Res. 123, 18–26. [DOI] [PubMed] [Google Scholar]
- Sarner, S., Kozma, R., Ahmed, S., and Lim, L. (2000). Phosphatidylinositol 3-kinase, Cdc42, and Rac1 act downstream of Ras in integrin-dependent neurite outgrowth in N1E-115 neuroblastoma cells. Mol. Cell. Biol. 20, 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, J. M., Guire, E. S., Saneyoshi, T., and Soderling, T. R. (2005). Calmodulin-dependent kinase kinase/calmodulin kinase I activity gates extracellular-regulated kinase-dependent long-term potentiation. J. Neurosci. 25, 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, M., and Kim, M. J. (2002). Postsynaptic signaling and plasticity mechanisms. Science 298, 776–780. [DOI] [PubMed] [Google Scholar]
- Shou, C., Farnsworth, C. L., Neel, B. G., and Feig, L. A. (1992). Molecular cloning of cDNAs encoding a guanine-nucleotide-releasing factor for Ras p21. Nature 358, 351–354. [DOI] [PubMed] [Google Scholar]
- Shou, C., Wurmser, A., Suen, K. L., Barbacid, M., Feig, L. A., and Ling, K. (1995). Differential response of the Ras exchange factor, Ras-GRF to tyrosine kinase and G protein mediated signals. Oncogene 10, 1887–1893. [PubMed] [Google Scholar]
- Sordella, R., Jiang, W., Chen, G. C., Curto, M., and Settleman, J. (2003). Modulation of Rho GTPase signaling regulates a switch between adipogenesis and myogenesis. Cell 113, 147–158. [DOI] [PubMed] [Google Scholar]
- Sprang, S. (2001). GEFs: master regulators of G-protein activation. Trends Biochem. Sci. 26, 266–267. [DOI] [PubMed] [Google Scholar]
- Stockmeier, C. A., Mahajan, G. J., Konick, L. C., Overholser, J. C., Jurjus, G. J., Meltzer, H. Y., Uylings, H. B., Friedman, L., and Rajkowska, G. (2004). Cellular changes in the postmortem hippocampus in major depression. Biol. Psychiatry 56, 640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturani, E., Abbondio, A., Branduardi, P., Ferrari, C., Zippel, R., Martegani, E., Vanoni, M., and Denis-Donini, S. (1997). The Ras Guanine nucleotide exchange factor CDC25Mm is present at the synaptic junction. Exp. Cell Res. 235, 117–123. [DOI] [PubMed] [Google Scholar]
- Taparowsky, E., Suard, Y., Fasano, O., Shimizu, K., Goldfarb, M., and Wigler, M. (1982). Activation of the T24 bladder carcinoma transforming gene is linked to a single amino acid change. Nature 300, 762–765. [DOI] [PubMed] [Google Scholar]
- Tian, X., Gotoh, T., Tsuji, K., Lo, E. H., Huang, S., and Feig, L. A. (2004). Developmentally regulated role for Ras-GRFs in coupling NMDA glutamate receptors to Ras, Erk and CREB. EMBO J. 23, 1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias, K. F., Bikoff, J. B., Burette, A., Paradis, S., Harrar, D., Tavazoie, S., Weinberg, R. J., and Greenberg, M. E. (2005). The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron 45, 525–538. [DOI] [PubMed] [Google Scholar]
- Tonini, R. et al. (2001). Involvement of CDC25Mm/Ras-GRF1-dependent signaling in the control of neuronal excitability. Mol. Cell Neurosci. 18, 691–701. [DOI] [PubMed] [Google Scholar]
- Waetzig, V., and Herdegen, T. (2003). The concerted signaling of ERK1/2 and JNKs is essential for PC12 cell neuritogenesis and converges at the level of target proteins. Mol. Cell Neurosci. 24, 238–249. [DOI] [PubMed] [Google Scholar]
- Weeber, E. J., Levenson, J. M., and Sweatt, J. D. (2002). Molecular genetics of human cognition. Mol. Interv. 2, 376- 391, 339. [DOI] [PubMed] [Google Scholar]
- Wei, W., Das, B., Park, W., and Broek, D. (1994). Cloning and analysis of human cDNAs encoding a 140-kDa brain guanine nucleotide-exchange factor, Cdc25GEF, which regulates the function of Ras. Gene 151, 279–284. [DOI] [PubMed] [Google Scholar]
- Yang, H., Cooley, D., Legakis, J. E., Ge, Q., Andrade, R., and Mattingly, R. R. (2003). Phosphorylation of the Ras-GRF1 Exchange Factor at Ser916/898 reveals activation of Ras signaling in the cerebral cortex. J. Biol. Chem. 278, 13278–13285. [DOI] [PubMed] [Google Scholar]
- Yasui, H., Katoh, H., Yamaguchi, Y., Aoki, J., Fujita, H., Mori, K., and Negishi, M. (2001). Differential responses to nerve growth factor and epidermal growth factor in neurite outgrowth of PC12 cells are determined by Rac1 activation systems. J. Biol. Chem. 276, 15298–15305. [DOI] [PubMed] [Google Scholar]
- Zippel, R., Orecchia, S., Sturani, E., and Martegani, E. (1996). The brain specific Ras exchange factor CDC25 Mm: modulation of its activity through Gi-protein-mediated signals. Oncogene 12, 2697–2703. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.