Abstract
To maintain genomic stability, reinitiation of eukaryotic DNA replication within a single cell cycle is blocked by multiple mechanisms that inactivate or remove replication proteins after G1 phase. Consistent with the prevailing notion that these mechanisms are redundant, we previously showed that simultaneous deregulation of three replication proteins, ORC, Cdc6, and Mcm2-7, was necessary to cause detectable bulk re-replication in G2/M phase in Saccharomyces cerevisiae. In this study, we used microarray comparative genomic hybridization (CGH) to provide a more comprehensive and detailed analysis of re-replication. This genome-wide analysis suggests that reinitiation in G2/M phase primarily occurs at a subset of both active and latent origins, but is independent of chromosomal determinants that specify the use and timing of these origins in S phase. We demonstrate that re-replication can be induced within S phase, but differs in amount and location from re-replication in G2/M phase, illustrating the dynamic nature of DNA replication controls. Finally, we show that very limited re-replication can be detected by microarray CGH when only two replication proteins are deregulated, suggesting that the mechanisms blocking re-replication are not redundant. Therefore we propose that eukaryotic re-replication at levels below current detection limits may be more prevalent and a greater source of genomic instability than previously appreciated.
INTRODUCTION
Eukaryotic cells must replicate each portion of their genome precisely once per cell cycle to faithfully transmit that genome to succeeding generations. This cell cycle control is enforced at the hundreds to thousands of replication origins where replication is initiated. As part of this regulation, cells must prohibit reinitiation within a single cell cycle at every origin for many successive generations. Even a small or occasional slip in this control will lead to re-replication, which can potentially compromise genome integrity. Hence, the block to reinitiation must be absolutely effective and reliable.
Studies from many laboratories have led to a model for the block to reinitiation that is based on the division of the initiation event into two mutually exclusive stages (reviewed in Bell and Dutta, 2002; Diffley, 2004; Machida et al., 2005). In the first stage, which is restricted to G1 phase, potential origins are selected on chromosomal DNA by assembly of the origin recognition complex (ORC), Cdc6, Cdt1, and the putative replicative helicase, Mcm2-7 into pre-replicative complexes (pre-RCs). In the second stage, which is restricted to S, G2, and M phases, potential origins are activated to initiate DNA replication by two kinases, a cyclin-dependent kinase (CDK) and Cdc7 kinase. Because CDK activity prevents pre-RC assembly in S, G2, and M phases and origins are not activated in G1 phase, passage through the cell cycle is coupled to exactly one round of replication.
Although this model provides a framework for understanding once and only once initiation, it does not explain how the block to reinitiation can be maintained with such high fidelity. This fidelity can be readily incorporated into the model if multiple overlapping mechanisms prevent pre-RC reassembly. In fact, multiple CDK-dependent inhibitory mechanisms that target pre-RC components have been identified in a number of eukaryotic organisms. In budding and fission yeast, CDKs appear to down-regulate ORC through inhibitory phosphorylation of Orc2 and/or Orc6 (Nguyen et al., 2001; Vas et al., 2001) as well as by direct binding to Orc6 (Wilmes et al., 2004). Additionally, CDKs inhibit Cdc6 (or the Schizosaccharomyces pombe ortholog Cdc18) by promoting Cdc6/Cdc18 degradation (Drury et al., 1997, 2000; Jallepalli et al., 1997; Elsasser et al., 1999), by reducing CDC6 transcription (Moll et al., 1991), and by directly inhibiting Cdc6/Cdc18 through phosphorylation (Jallepalli et al., 1997) or binding (Mimura et al., 2004). Finally, CDKs also promote the nuclear exclusion of Mcm2-7 and Cdt1 in budding yeast (Labib et al., 1999; Nguyen et al., 2000; Tanaka and Diffley, 2002), in part by direct phosphorylation of Mcm3 (Liku et al., 2005). In metazoans, CDKs have been implicated in Orc1 degradation, Cdt1 degradation and Cdc6 nuclear exclusion (reviewed in Diffley, 2004). In addition, metazoan cells have a CDK-independent mechanism involving the protein geminin, which binds to Cdt1 and can prevent it from recruiting Mcm2-7 during S, G2, and M phase (reviewed in Blow and Dutta, 2005).
Obtaining clear evidence of re-replication within a single cell cycle has generally required the simultaneous disruption of multiple mechanisms, leading to the presumption that these mechanisms are redundant (Diffley, 2004; Blow and Dutta, 2005). In budding yeast, for example, simultaneous deregulation of ORC phosphorylation, Mcm localization, and Cdc6 protein levels was needed to detect re-replication in G2/M phase (Nguyen et al., 2001). Similarly, disruption of several regulatory mechanisms leads to re-replication in fission yeast (Gopalakrishnan et al., 2001; Vas et al., 2001; Yanow et al., 2001) and in Xenopus replication extracts (McGarry and Kirschner, 1998; Arias and Walter, 2005; Li and Blow, 2005; Yoshida et al., 2005).
In addition to the issue of mechanistic redundancy, the model for the block to re-replication makes predictions that are best examined by a genome-wide analysis of re-replication. First, the re-replication that is induced by deregulating pre-RC assembly should initiate from the potential replication origins used during normal replication. Reinitiation from a few origins has been observed by two-dimensional gel electrophoresis in both budding (Nguyen et al., 2001) and fission (Yanow et al., 2001) yeast, but genome-wide mapping of reinitiation sites is needed to confirm this prediction. Second, deregulation of pre-RC reassembly should be able to induce re-replication throughout the period from S to M phase. Although Cdt1 overexpression has been shown to prolong S phase in Drosophila embryos (Thomer et al., 2004), direct evidence for re-replication within S phase is still lacking. Finally, full deregulation of pre-RC reassembly should allow more than one round of reinitiation and result in rampant re-replication. So far, precise deregulation of replication proteins has led to at most a doubling of genomic DNA content, suggesting that additional inhibitory mechanisms remain to prevent re-replication. A more comprehensive analysis of where re-replication occurs in the genome may provide clues to how re-replication is still inhibited.
We have developed a more sensitive and comprehensive assay for re-replication by adapting and streamlining previously published microarray-based assays for analyzing DNA replication in budding yeast. With this assay we present evidence that reinitiation occurs primarily at a subset of the potential origins normally established for S phase without being strongly affected by the chromosomal determinants that specify the efficiency and timing of these origins in S phase. Our studies suggest that the limited rereplication observed may be due in part to the fewer initiation sites used for re-replication compared with S phase. Additionally, our studies indicate that some of the mechanisms preventing re-replication in G2/M phase also operate in S phase but that the block to re-replication in these two phases is not identical. Finally, we demonstrate that reinitiation from as few as a single origin is detectable when fewer mechanisms are disrupted, consistent with the notion that these mechanisms are not redundant but are each actively maintaining the high fidelity of the block to re-replication.
MATERIALS AND METHODS
Plasmids and Strains
All plasmids are described in Table 1, all strains are described in Table 2, and all oligonucleotides are described in Table 3. Supplementary Methods contains detailed description of plasmid and strain construction.
Table 1.
Plasmids used in this study
| Plasmid | Key features | Source |
|---|---|---|
| pJL737 | ORC6 URA3 | Nguyen et al. (2001) |
| pJL806 | pGAL1 URA3 | Nguyen et al. (2001) |
| pJL1206 | MCM7-(SVNLS)2 URA3 | Nguyen et al. (2001) |
| pJL1488 | pGAL1-Δntcdc6-cdk2A URA3 | This study |
| pJL1489 | pGAL1-Δntcdc6 URA3 | Nguyen et al. (2001) |
| pKI1260 | MCM7-(svnls3A)2 URA3 | Nguyen et al. (2001) |
| pMP933 | ORC2 URA3 | Nguyen et al. (2001) |
| YIp22 | pMET3-HA3-CDC20 TRP1 | Uhlmann et al. (2000) |
| pFA6a | KanMX6 | Wach et al. (1994) |
| pAG25 | NatMX4 | Goldstein et al. (1999) |
| pPP117 | cdc7-1 URA3 | Hollingsworth et al. (1992) |
Table 2.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| YJL310 | leu2-3,112 ura3-52 trp1-289 bar1Δ::LEU2 | Detweiter and Li (1998) |
| YJL3244 | orc2-cdk6A orc6-cdk4A leu2 ura3-52::{pGAL1, URA3} trp1-289 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} | Nguyen et al. (2001) |
| YJL3248 | orc2-cdk6A orc6-cdk4A ura3-52::{pGAL1-Δntcdc6, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} | Nguyen et al. (2001) |
| YJL3249 | orc2-cdk6A orc6-cdk4A ura3-52::{pGAL1-Δntcdc6, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} | This study |
| YJL4486 | ORC2 ORC6 leu2 ura3-52::{pGAL1, URA3} trp1-289 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} | This study |
| YJL4489 | ORC2 ORC6 ura3-52::{pGAL1-Δntcdc6-cdk2A, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} | This study |
| YJL4832 | orc2-cdk6A orc6-cdk4A ura3-52::{pGAL1, URA3} trp1-289 leu2 ade2 ade3 MCM7-2nls3A bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} | This study |
| YJL3240 | orc2-cdk6A orc6-cdk4A ura3-52::{pGAL1-Δntcdc6, URA3} trp1-289 leu2 ade2 ade3 MCM7-2nls3A bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} | This study |
| YJL5038 | his3Δ::KanMX leu2Δ0 met15Δ0 ura3Δ0 bar1Δ::NatMX4 can1Δ::pMFA1-HIS3::pMFα1-LEU2 | This study |
| YJL5493 | orc2-cdk6A orc6-cdk4A leu2 ura3-52::{pGAL1, URA3} trp1-289 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} | This study |
| YJL5834 | ORC2 ORC6 leu2 ura3-52::{pGAL1, URA3} trp1-289 ade2 ade3 MCM7 bar1::LEU2 | This study |
| YJL5787 | ORC2 ORC6 ura3-52::{pGAL1-Δntcdc6-cdk2A, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} Δars316::KanMX6 | This study |
| YJL5858 | ORC2 ORC6 ura3-52::{pGAL1-Δntcdc6-cdk2A, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} Δars317::KanMX6 | This study |
| YJL5861 | ORC2 ORC6 ura3-52::{pGAL1-Δntcdc6-cdk2A, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} Δars318::KanMX4 | This study |
| YJL5816 | ORC2 ORC6 leu2 ura3-52::{pGAL1, URA3} trp1-289 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} cdc7-1 | This study |
| YJL5822 | ORC2 ORC6 ura3-52::{pGAL1-Δntcdc6-cdk2A, URA3} trp1-289 leu2 ade2 ade3 MCM7-2NLS bar1Δ::LEU2 cdc20::{pMET3-HA3-CDC20, TRP1} cdc7-1 | This study |
Table 3.
Oligonucleotides used in this study
| Oligo | Purpose | Sequence |
|---|---|---|
| OJL1596 | ars316Δ | 5′-TTAACTGACAATTCTTTTGAACAAAATTTACACTTCATCAAGAAAGATGCCGGATCCCCGGGTTAATTAA-3′ |
| OJL1597 | ars316Δ | 5′-TGATGACGAAGGATTCGTTGAAGTTGAATGCACACAAAAAAAGCTTGATACATCGATGAATTCGAGCTCG -3′ |
| OJL1639 | ars317Δ | 5′-ATTAAACAATGTTTGATTTTTTAAATCGCAATTTAATACCCGGATCCCCGGGTTAATTAA-3′ |
| OJL1640 | ars317Δ | 5′-ATTTTTATGGAAGATTAAGCTCATAACTTGGACGGGGATCCATCGATGAATTCGAGCTCG-3′ |
| OJL1641 | ars318Δ | 5′-CGATAAAGTTATTATTTAGATTACATGTCACCAACATTTTCGGATCCCCGGGTTAATTAA-3′ |
| OJL1642 | ars318Δ | 5′-AGAGAAAATAGCTATTTACCTCAACATTTAAAGGTATTAACATCGATGAATTCGAGCTCG-3′ |
| OJL1607 | ARS317 probe | 5′-ATCGATTATCTGTTTGGCAGG-3′ |
| OJL1608 | ARS317 probe | 5′-GAATTCAAAGAAGTCAATCTTATG-3′ |
| OJL1452 | bar1Δ | 5′-ATTAAAAATGACTATATATTTGATATTTATATGCTATAAAGAAATTGTACTCCAGATTTCCATCGATGAATTCGAGCTCG-3′ |
| OJL1454 | bar1Δ | 5′-AGTGGTTCGTATCGCCTAAAATCATACCAAAATAAAAAGAGTGTCTAGAAGGGTCATATACGGATCCCCGGGTTAATTAA-3′ |
Yeast Media, Growth, and Arrest
Cells were grown in YEP, synthetic complete (SC), or synthetic (S broth) medium (Guthrie and Fink, 1990) supplemented with 2% dextrose (wt/vol), 2% galactose (wt/vol), 3% raffinose (wt/vol), or 3% raffinose (wt/vol) + 0.05% dextrose (wt/vol). For S phase experiments cells were grown overnight in SDC (YJL5038) or SDC-Met,Ura (YJL3248 and YJL5834) and arrested in G1 phase with 50 ng/ml α factor (all strains were bar1) at 30°C. Cells were released by filtering, washing, and then resuspending in prewarmed 30°C YEPD containing 100 μg/ml pronase, 100 mM hydroxyurea (HU), and 15 μg/ml nocodazole.
To obtain reproducible induction of re-replication, cells were inoculated from a fresh unsaturated culture containing 2% dextrose into a culture containing 3% raffinose + 0.05% dextrose and grown for 12-15 h the night before the experiment. The GAL1 promoter (pGAL1) was induced by addition of 2% galactose and the MET3 promoter (pMET3) was repressed by the addition of 2 mM methionine. All experiments were performed at 30°C except where noted. For induction of re-replication in G2/M phase, cells grown overnight in SRaffC-Met,Ura + 0.05% dextrose were pelleted and resuspended in YEPRaff + 2 mM methionine and 15 μg/ml nocodazole. Once arrested (>90% large budded cells), galactose was added to a final concentration of 2%. In experiments with strains containing cdc7-1, cells were grown and arrested at 23°C. These cultures were split after arresting in G2/M phase and either kept at 23°C or shifted to 35°C for 1 h followed by addition of 2% galactose to both cultures.
For induction of re-replication during the release from G1 phase into a G2/M phase arrest, cells grown overnight in SRaffC-Met,Ura + 0.05% dextrose were arrested with 50 ng/ml α factor (all strains were bar1). Once arrested (>95% small budded cells), galactose was added to a final concentration of 2% for 30 min. Cells were released by filtering, washing, and then resuspending in prewarmed YEPGal + 2 mM methionine, 100 μg/ml pronase, and 15 μg/ml nocodazole. For the induction of re-replication during a release from G1 phase into S phase, cells arrested and released as described above were resuspended in prewarmed YEPGal + 2 mM methionine, 100 μg/ml pronase, and 100 mM HU.
Flow Cytometry
Cells were fixed and stained with 1 μM Sytox Green (Molecular Probes, Eugene, OR) as previously described (Haase and Lew, 1997).
Pulsed-Field Gel Electrophoresis
Pulsed-field gel electrophoresis (PFGE) was performed as described in Green and Li (2005). Probes for ARS305, ARS607, and ARS1413 were prepared as described in Nguyen et al. (2001).
Two-Dimensional Gel Electrophoresis
Neutral-neutral two-dimensional (2-D) gel analysis was performed essentially as described at http://fangman-brewer.genetics.washington.edu. The DNA preparation described there is a slight modification of the one used in Huberman et al. (1987). Modifications to the previous protocols can be found in Supplementary Methods.
Microarray Assay
Microarrays containing 12,034 PCR products representing every ORF and intergenic region were prepared essentially as described (DeRisi et al., 1997; Iyer et al., 2001; see Supplementary Methods). Genomic DNA was prepared, labeled, and hybridized as described in Supplementary Methods.
Data Analysis
Raw Cy5/Cy3 ratios from scanned arrays were normalized to the DNA content per cell based on the flow cytometry data to determine absolute copy number of each DNA segment. Raw values were then binned and smoothed using Fourier convolution smoothing essentially as described (Raghuraman et al., 2001). Peaks in the replication profiles that were both prominent and reproducible among repetitions of an experiment were identified as origins. Details of data analysis (Supplementary Methods) and examples of raw data (Supplementary Figure S1) are contained in Supplementary Information. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO, http:://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE4181.
The “experiment variability” was determined using the equation for calculating one SD. Because there were only two DNA preparations used, each of which was hybridized twice, the trials are not truly independent and thus we call these values “experiment variability” rather than SD.
Scatter Plot
For each pro-ARS (Wyrick et al., 2001), the normalized Cy5/Cy3 ratio of that chromosomal locus during replication or re-replication was determined and plotted. See Supplementary Methods for more details.
RESULTS
A Simplified Microarray CGH Assay for DNA Replication
We have adapted and streamlined existing microarray assays (Raghuraman et al., 2001; Yabuki et al., 2002) to create a rapid and economical genome-wide assay for yeast DNA replication. Our simplified assay uses CGH to directly measure the increase in DNA copy number arising from replication or re-replication. During S phase replication, the copy number of each DNA segment reflects the timing of its replication because the earlier a DNA segment replicates, the greater the proportion of replicating cells containing a duplication of this segment. Origins, which replicate earlier than neighboring regions, can be localized to chromosomal segments where the copy number reaches a local maxima. Thus, use of microarray CGH to monitor copy number changes across the genome can provide a comprehensive view of the location and efficiency/timing of initiation sites during replication and re-replication.
Figure 1A shows a schematic of our microarray CGH replication assay. Genomic DNA from replicating (or rereplicating) and nonreplicating cells is purified and differentially labeled with Cy5 and Cy3. The labeled probes are competitively hybridized to a spotted microarray and the raw Cy5/Cy3 values are normalized such that the average ratio corresponds to the DNA content determined by flow cytometry. Data are smoothed and origins are computationally identified by locating prominent and reproducible peaks in smoothed replication profiles.
Figure 1.
Use of comparative genomic hybridization (CGH) on spotted microarrays to assay DNA replication. (A) Schematic representation of the CGH replication assay. Genomic DNA is purified from nonreplicating and replicating cells, differentially labeled with Cy3 and Cy5, and competitively hybridized to a microarray containing 12,034 ORF and intergenic PCR products. Cy5/Cy3 ratios are normalized so that the average ratio of all elements equals the DNA content of the cells (as determined by flow cytometry). Normalized ratios are plotted against chromosomal position and mathematically smoothed to generate a replication profile. In most cases, two hybridizations are performed from each of two independent experiments. The resulting four replication profiles are averaged into one composite profile, and the locations of origins are identified using a peak finding algorithm. Chromosomal regions lacking data of sufficient quality are represented as gaps in the profiles. (B) CGH replication assay described for A was performed on YJL5038, a wild-type yeast strain in the S288c background. G1 phase genomic DNA was hybridized against S phase genomic DNA obtained 120 min after cells were released from G1 phase into media containing hydroxyurea (HU). The composite replication profile (blue line) plus and minus the “experiment variability” (light gray band; see Materials and Methods) is shown for chromosome X. Positions of origins annotated in the Saccharomyces Genome Database (SGD; Balakrishnan (2006); red triangles) and the centromere (black circle) are marked along the X-axis. Replication profiles derived from Raghuraman et al. (2001) (violet line) and Yabuki et al. (2002) (orange line) are shown for comparison. (C) S phase progression assayed by flow cytometry for experiment described in B at the indicated times after release from G1 phase. DNA content of 1.4 C was used to normalize the S288c replication profile. (D) The S phase replication profile of the re-replication competent OMC strain and the congenic wild-type strain are similar. S phase replication profiles were generated for the OMC strain YJL3248 (MCM7-2NLS orc2-cdk6A orc6-cdk4A pGAL1-Δntcdc6 pMET3-HA3-CDC20) and a congenic wild-type A364a strain YJL5834 (pGAL1) essentially as described in B except S phase cells were harvested, respectively, at 135 min and 180 min after α factor release. The S phase replication profile for the OMC strain (green line) and the A364a strain (black line) for chromosome X is shown. SGD annotated origins (red triangles) and the centromere (black circle) are marked along the X-axis. (E) S phase progression assayed by flow cytometry for experiment described in D at the indicated times after release from G1 phase. DNA contents of 1.35 C and 1.4 C, respectively, were used to normalize the OMC and A364a replication profiles.
Before using the microarray CGH assay to study re-replication, we assessed its reproducibility and its ability to identify known replication origins in the S phase of a wild-type S288c strain (flow cytometry data in Figure 1C). Figure 1B and Supplementary Figure S2 show the mean of the smoothed S phase replication profiles from four hybridizations plus or minus the “experiment variability” (see Materials and Methods) for chromosome X. The small variability demonstrates that this technique is highly reproducible. An overlay of our replication profiles with those generated from previously published data (Raghuraman et al., 2001; Yabuki et al., 2002) shows considerable agreement in both peak positions, which reflects origin locations, and peak heights, which reflects origin timing/efficiency. When our peak finding algorithm was applied to our profiles, we obtained origin numbers (212) comparable to those obtained by Rhaguraman et al. (2001) (332) and Yabuki et al. (2002) (260). Additionally, the alignment of peaks to origins systematically mapped by 2-D gel electrophoresis or ARS plasmid assay was similar to, or better than, published data (Supplementary Table S1). Together, these data confirm that our streamlined assay is reproducible and accurate.
Re-replication Competent Mutant Has a Mostly Normal S Phase
We have previously demonstrated that simultaneous deregulation of three pre-RC components (ORC, Mcm2-7, and Cdc6) leads to limited re-replication in G2/M phase arrested cells (Nguyen et al., 2001). These initiation proteins were deregulated by mutations that make the proteins refractory to CDK regulation. First, the CDK consensus phosphorylation sites of two subunits of the origin recognition complex, Orc2 and Orc6, were mutated, preventing Cdc28/Cdk1 phosphorylation of these subunits (orc2-cdk6A, orc6-cdk4A). Second, two copies of the SV40 nuclear localization signal were fused to MCM7 (MCM7-SVNLS2) to prevent the Cdc28/Cdk1 promoted net nuclear export of the Mcm2-7 complexes. Finally, an extra copy of CDC6, containing a partially stabilizing N-terminal deletion, was placed under control of the galactose inducible promoter (pGAL1-Δntcdc6). This strain re-replicates when Δntcdc6 is induced by addition of galactose and will be referred to as the OMC re-replicating strain in reference to its deregulation of ORC, Mcm2-7, and Cdc6.
A major concern in any genetic analysis of replication control is the possibility that the mutations deregulating replication proteins also disrupt their replication activity. Such a nonspecific perturbation would complicate any interpretation of the resulting phenotype. We and others have previously reported that Δnt-cdc6 expressed under the CDC6 promoter retains full replication initiation function (Drury et al., 2000; Nguyen et al., 2001). To determine whether the mutations deregulating Orc2, Orc6, and Mcm7 in the OMC strain also preserve their initiation function, we compared S phase of the OMC strain (orc2-cdk6A orc6-cdk4A MCM7-2NLS pGAL1-Δntcdc6), when re-replication was not induced, to S phase of the congenic wild-type A364a strain (ORC2 ORC6 MCM7 pGAL1). When cells were harvested at the same point in S phase (Figure 1E), the replication profiles for the two strains showed considerable overlap (Figure 1D, Supplementary Figures S3 and S4), although ORC and Mcm7 mutations cause subtle alterations in the initiation of DNA replication. Because two wild-type strains of different strain backgrounds show nearly identical replication profiles (Supplementary Figures S5 and S6), we believe these differences reflect subtle alterations in the initiation activity of the mutant ORC and Mcm2-7. Nonetheless, we conclude that, overall, the mutant ORC and Mcm2-7 proteins in the OMC strain retain most of their normal initiation activity.
Mapping Reinitiating Origins
A key prediction of the current model for eukaryotic replication control is that pre-RC reassembly and reinitiation should only occur where pre-RCs normally assemble, i.e., the potential origins or pro-ARSs identified by Wyrick et al. (2001). In our previous characterization of re-replication induced at G2/M phase in the OMC strain (orc2-cdk6A orc6-cdk4A MCM7-2NLS pGAL1-Δntcdc6), we observed three active S phase origins reinitiating by 2-D gel electrophoresis (Nguyen et al., 2001). To comprehensively examine this prediction throughout the genome, we performed microarray CGH on the re-replicating DNA from OMC cells. This re-replicating DNA (flow cytometry in Figure 2A) was competitively hybridized against DNA from a congenic non-re-replicating strain that lacks the inducible Δntcdc6 and will be referred to as the OM strain (orc2-cdk6A orc6-cdk4A MCM7-2NLS pGAL1). Another source of non-re-replicating control DNA is OMC DNA from G1 phase cells, and when this was used, virtually identical results were obtained (unpublished data).
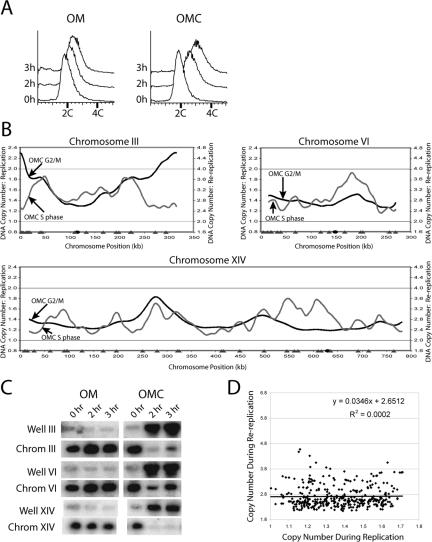
Figure 2.
Re-replication induced during G2/M phase when ORC, Mcm2-7, and Cdc6 are deregulated. (A) G2/M phase re-replication in the OMC strain is readily detectable by flow cytometry. The OMC strain YJL3248 (orc2-cdk6A orc6-cdk4A MCM7-2NLS pGAL1-Δntcdc6 pMET3-HA3-CDC20) and the control OM strain YJL3244 (orc2-cdk6A orc6-cdk4A MCM7-2NLS pGAL1 pMET3-HA3-CDC20) were arrested in G2/M phase. Once arrested, galactose was added, which induced re-replication in the OMC strain. Samples were taken for flow cytometry at the indicated points after galactose addition. The DNA content of 2.7 C at 3 h was used to normalize the OMC re-replication profile in B. (B) Genomic DNA was purified from the OMC strain and the control OM strain after 3 h of galactose induction as described in A and competitively hybridized against each other as described in Figure 1A. The OMC G2/M phase re-replication profiles (black lines, right axis), the OMC S phase replication profiles replotted from Figure 1D (gray lines, left axis), locations of pro-ARSs mapped by Wyrick et al. (2001) (gray triangles) and the centromeres (black circles) are shown for chromosomes III, VI, and XIV. (C) Each chromosome participates when OMC cells are induced to re-replicate in G2/M phase. The OMC strain and the control OM strain from the experiment presented in A were harvested for pulsed-field gel electrophoresis (PFGE) at the indicated times. Southern blots of the gel were probed with fragments containing ARS305 to detect chromosome III, ARS607 to detect chromosome VI, and ARS1413 to detect chromosome XIV. For each chromosome the Southern signal for both the gel well and the normal chromosomal position are shown. (D) Replication timing does not correlate with efficiency of G2/M phase re-replication in the OMC strain. For each of the pro-ARSs defined by Wyrick et al. (2001), the DNA copy number from the OMC G2/M phase re-replication profile in B was plotted versus the DNA copy number from the OMC S phase replication profile in B. Line represents linear regression of plot.
The OMC G2/M phase re-replication profiles are shown in Figure 2B and Supplementary Figure S7. These data confirm that the incomplete re-replication observed by flow cytometry is distributed over all 16 chromosomes, as was first suggested by their limited entry into the gel during PFGE (Nguyen et al., 2001 and Figure 2C). The re-replication profiles also show that individual chromosomes re-replicate very unevenly, with some segments preferentially re-replicating more than others do.
Application of a peak finding algorithm to OMC re-replication profiles identified 106 reinitiating origins. Most of these origins appear to correspond to chromosomal loci that form pre-RCs in G1 phase because more than 80% of the reinitiating origins map to within 10 kb of a pro-ARS identified by Wyrick et al. (2001) as sites of pre-RC binding. The mean distance between the OMC reinitiating origins and the closest Wyrick pro-ARS (Wyrick et al., 2001) is 7.0 kb. This value is highly significant (p < 5 × 10-8) when compared with the mean distances calculated for equivalent numbers of randomly selected chromosomal loci, as the mean distances are tightly distributed around a value of 12.3 kb (Supplementary Figure S8).
Tanny et al. (2006) have analyzed the re-replication profile of a strain similar to our OMC strain containing the additional perturbation of a mutation of an RXL motif in ORC6 that abrogates CDK binding and results in a slightly increased extent of re-replication. Although both articles use slightly different data analysis and presentation, (our profiles are presented to preserve absolute copy number information at the cost of less distinctive peaks), the re-replication profiles are strikingly similar (compare Supplementary Figure S7 to Tanny et al., 2006; Supplementary Figure S2). Like our results, 80% of the 123 re-replication origins identified by Tanny et al. (2006) are within 10 kb of a Wyrick et al. (2001) pro-ARS, further supporting the notion that re-replication occurs at normal sites of pre-RC formation. Overlap of origins identified in both studies is considerable, with 64% of the origins in this study within 10 kb of an origin in Tanny et al. (2006) (20% would be expected by chance). This overlap becomes even more striking, 80% overlap (expected value is also 20%), when the top 40 highest peaks in our analysis are compared with peaks identified in Tanny et al. (2006). Together with our previous confirmation by 2-D gel electrophoresis that ARS305, ARS121, and ARS607 reinitiate (Nguyen et al., 2001), these genomic data suggest that reinitiation primarily occurs at a subset of potential S phase origins.
The efficiency with which these potential origins reinitiate in G2/M phase, however, does not correlate with the efficiency or timing with which they initiate in S phase. For example, only 38% of the active S phase origins reinitiate with enough efficiency to be identified as peaks during re-replication in G2/M phase. Moreover, some regions that normally replicate late in S phase, such as those near the telomeres of chromosome III, re-replicate very efficiently in G2/M phase, apparently from very inefficient or latent S phase origins in those regions. For a systematic comparison of re-replication efficiency versus replication timing of all potential S phase origins, we plotted the re-replication copy number versus the replication copy number for the set of pro-ARSs identified by Wyrick et al. (2001) (Figure 2D). The absence of any significant correlation (R2 = 0.0002) indicates that the efficiency or timing of a replication origin in S phase does not determine its re-replication efficiency during G2/M phase.
Mechanisms That Prevent Re-replication at G2/M Phase Also Act in S Phase
The prevailing model for replication control depicts the prevention of re-replication in S, G2, and M phase as one continuous inhibitory period using a common strategy of preventing pre-RC reassembly. Because CDKs are active throughout this period, the model would predict that mechanisms used by CDKs to regulate replication proteins should prevent re-replication throughout S, G2, and M phase. To determine if CDK regulation of ORC, Mcm2-7, and Cdc6, which prevents re-replication within G2/M phase, also prevents re-replication in S phase, we induced Δntcdc6 in OMC cells (orc2-cdk6A orc6-cdk4A MCM7-2NLS pGAL1-Δntcdc6) as they entered S phase.
OMC cells were arrested in G1 phase with α factor, and half the cells were harvested to obtain G1 phase DNA. The remaining cells were induced to express Δntcdc6 and then released from the G1 arrest into a low concentration of HU to delay their replication and allow us to collect them in S phase. Flow cytometry indicated that the released cells were harvested while still in S phase with a DNA content of 1.4 C (Figure 3A). The S phase and G1 phase DNA were competitively hybridized against the yeast genomic microarray to generate a combined replication/re-replication profile for S phase (Figure 3B and Supplementary Figure S9).
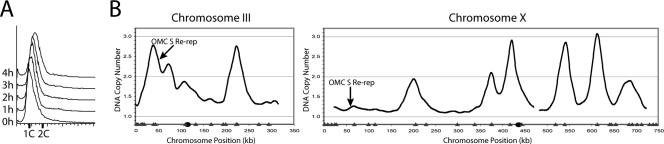
Figure 3.
Deregulation of ORC, Mcm2-7, and Cdc6 can induce re-replication in S phase. (A) Flow cytometry of OMC cells induced to re-replicate in S phase. The OMC strain YJL3249 (orc2-cdk6A orc6-cdk4A MCM7-2NLS pGAL1-Δntcdc6 pMET3-HA3-CDC20) was arrested in G1 phase, induced to express Δntcdc6 by the addition of galactose, then released from the arrest into media containing HU to delay cells from exiting S phase. At 4 h the cells were still in S phase with a DNA content of 1.4 C. This value was used to normalize the re-replication profile in B. (B) OMC cells can reinitiate and re-replicate within S phase. Genomic DNA was isolated at the 0 h (G1 phase) and 4 h (S phase) time points from the OMC strain YJL3249 as described in A and competitively hybridized against each other. The resulting profiles shown for chromosomes III and X reflect copy number increases due to both replication and re-replication. Locations of pro-ARSs mapped by Wyrick et al. (2001) (gray triangles) and the centromeres (black circles) are plotted along the X-axis.
Because normal S phase replication can account for an increase in DNA copy number from 1 to 2, only DNA synthesis beyond this copy number can be unequivocally attributed to re-replication. As seen in Figure 3B and Supplementary Figure S9, many early origins acquired a DNA copy number greater than 2; in some cases reaching values greater than 3. In the same profiles other chromosomal regions had copy numbers significantly below 2, confirming that cells were indeed in the midst of S phase. In fact, early origins reinitiated, whereas forks from their first round of replication were still progressing and before many late origins had fired. Similar re-replication profiles were observed for re-replicating cells synchronously harvested in S phase in the absence of HU (unpublished data). These findings thus directly establish that mechanisms used to prevent re-replication in G2/M phase also act within S phase.
Cell Cycle Position Can Affect the Extent and Location of Re-replication
To determine if the block to re-replication is modulated during progression through the cell cycle, we compared the re-replication profile of OMC cells (orc2-cdk6A orc6-cdk4A MCM7-2NLS pGAL1-Δntcdc6) that were induced to re-replicate through a complete S phase with the profile associated with re-replication in G2/M phase. To obtain the former profile, both OMC and control OM cells (orc2-cdk6A orc6-cdk4A MCM7-2NLS pGAL1) were arrested in G1 phase with α factor followed by addition of galactose to induce Δntcdc6 in the OMC strain. Cells were then released from the G1 arrest, allowed to proceed through S phase, and collected at a G2/M arrest 3 h after the release. DNA prepared from the OMC and OM strains were competitively hybridized to our yeast genomic microarray to obtain a “G1 release” re-replication profile for the OMC cells.
Flow cytometry showed that both the re-replicating OMC and the control OM strain were in the middle of S phase 1 h after the release (Figure 4A). As expected for actively replicating chromosomes (Hennessy and Botstein, 1991), the chromosomes of these strains were retained in the wells during PFGE (Figure 4B). Two hours after the release, S phase was mostly complete in the control OM strain and its chromosomes reentered the gel during PFGE. In the OMC strain, however, the induction of re-replication prevented chromosomes from reentering the PFGE gel at both 2 and 3 h time points. Because significant re-replication could be induced in OMC cells delayed in S phase, we believe that re-replication during the progression through S phase contributed to the re-replication seen in the G1 release experiment.
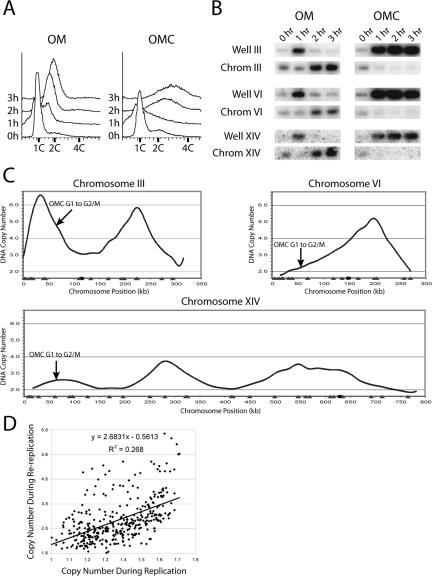
Figure 4.
Re-replication induced upon release from a G1 arrest when ORC, Mcm2-7, and Cdc6 are deregulated. (A) Robust re-replication of OMC cells after G1 release. The OMC strain YJL3248 (orc2-cdk6A orc6-cdk4A MCM7-2NLS pGAL1-Δntcdc6 pMET3-HA3-CDC20) and the control OM strain YJL3244 (orc2-cdk6A orc6-cdk4A MCM7-2NLS pGAL1 pMET3-HA3-CDC20) were arrested in G1 phase, exposed to galactose to induce Δntcdc6 in the OMC strain, and then released from the arrest into G2/M phase. Samples were taken for flow cytometry at the indicated times after release from the α factor arrest. The OMC re-replication profile in C was normalized to the 3 h DNA content of 3.2 C. (B) Cells that were induced to re-replicate in A were harvested for PFGE at the indicated times. Southern blots of the gel were probed for chromosomes III, VI, and XIV as described in Figure 2C. (C) Re-replication profile of the OMC strain after G1 release. Genomic DNA was purified from the OMC strain and the control OM strain 3 h after G1 release. The two DNA preparations were labeled and competitively hybridized against each other to generate the G1 release re-replication profiles shown for chromosomes III, VI, and XIV. Locations of pro-ARSs mapped by Wyrick et al. (2001) (gray triangles) and the centromeres (black circles) are plotted along the X-axis. (D) Re-replication induced in the OMC strain after a G1 release is slightly biased toward early replicating pro-ARSs. For each of the pro-ARSs defined by Wyrick et al. (2001), the DNA copy number from the OMC G1 release re-replication profile in C was plotted versus the DNA copy number from the OMC S phase replication profile in Figure 2B. Line represents linear regression of plot.
Re-replication induced during G1 release of OMC cells was more extensive than re-replication induced in G2/M phase. Despite comparable lengths of induction, flow cytometry reproducibly indicated that the former accumulated a DNA content of 3.2 C, whereas the latter accumulated only 2.7 C (compare 3 h time points in Figure 4A with Figure 2A). More extensive re-replication could also be seen by comparing the re-replication profiles induced during the G1 release (Figure 4C and Supplementary Figure S10) and the G2/M phase arrest (Figure 2B and Supplementary Figure S7). In general the peaks in the G1 release profiles were taller than the G2/M phase profiles, suggesting that more efficient or more rounds of reinitiation can occur when re-replication is induced during S phase. For example, ARS305 reached a copy number of 6.6, indicating it reinitiated a second time, as a single round can only generate a maximum copy number of 4. Overall, multiple rounds of reinitiation were observed on more than half of the chromosomes when re-replication was induced during the G1 release. In contrast, multiple rounds of reinitiation occurred at much fewer loci and to a lesser extent when re-replication was induced in G2/M phase.
A peak finding algorithm identified 87 potential reinitiation sites when re-replication was induced during the G1 release experiment. Of these, 85% were located within 10 kb of a Wyrick pro-ARS Wyrick et al. (2001). These data suggest that re-replication induced during a G1 release occurs from S phase origins of DNA replication.
In addition to the extent of re-replication, another significant difference between re-replication induced during the G1 release and re-replication induced during G2/M phase was their pattern of origin usage. As discussed above, efficiency of re-replication in G2/M phase was not correlated with origin usage during S phase. In contrast, the efficiency of re-replication induced during the G1 release exhibited a modest positive correlation with S phase origin timing (Figure 4D). Although we cannot rule out an intrinsic difference in the reinitiation efficiency of early versus late origins when re-replication is induced during the G1 release, the simplest explanation for this correlation is that earlier replicating origins are cleared of pre-RCs earlier, making them available sooner for reassembly of pre-RCs and reinitiation within S phase.
Limited Re-replication Is Detectable with Fewer Genetic Perturbations
Our previous analysis of budding yeast re-replication failed to detect re-replication when only two pre-RC components were deregulated in G2/M phase (Nguyen et al., 2001). This observation is frequently cited as evidence that eukaryotic replication controls are highly redundant. Both the increased sensitivity of the microarray CGH assay and the enhanced re-replication observed during a G1 release provided opportunities to reexamine whether these controls are indeed redundant in budding yeast.
As a first step, we examined an “OC” strain (orc2-cdk6A orc6-cdk4A pGAL1-Δntcdc6), in which only ORC and Cdc6 are deregulated and compared it with a control “O” strain (orc2-cdk6A orc6-cdk4A GAL1), where only ORC is deregulated. In accordance with our previous results (Nguyen et al., 2001), induction of Δntcdc6 in G2/M phase generated no significant increase in DNA content by flow cytometry (Figure 5A) or chromosome immobilization during PFGE (Figure 5C). Similarly, microarray CGH of DNA prepared from the OC and O strains after 3 h of galactose induction in G2/M phase detected no re-replication on 15 out of 16 chromosomes (Supplementary Figure S11). However, limited re-replication could clearly be observed on both arms of chromosome III (Figure 5E). Thus, the microarray CGH assay can detect re-replication missed by other assays.
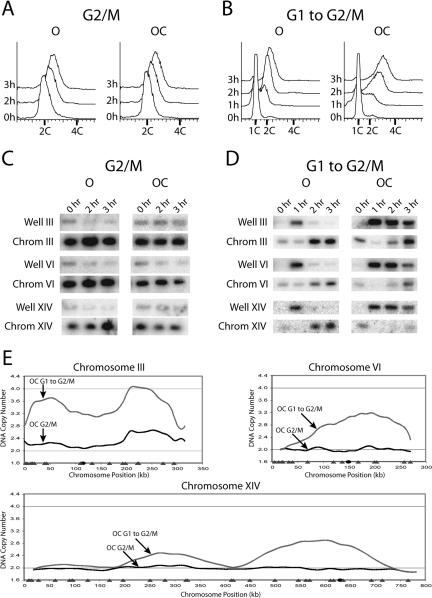
Figure 5.
Re-replication can be induced when only ORC and Cdc6 are deregulated. (A) Re-replication is undetectable by flow cytometry in OC cells in G2/M phase. The OC strain YJL3240 (orc2-cdk6A orc6-cdk4A pGAL1-Δntcdc6 pMET3-HA3-CDC20) and the control O strain YJL4832 (orc2-cdk6A orc6-cdk4A pGAL1 pMET3-HA3-CDC20) were arrested in G2/M phase and induced with galactose as described in Figure 2A. Samples for flow cytometry were taken at the indicated times after galactose addition. The OC G2/M re-replication profile in E was normalized to the 3 h DNA content of 2.0 C. (B) Significant re-replication can be induced in OC cells during a G1 release. The OC strain and the control O strain were induced with galactose and released from a G1 arrest as described in Figure 4A. Samples for flow cytometry were taken at the indicated times after G1 release. The OC G1 release re-replication profile in E was normalized to the 3 h DNA content of 2.6 C. (C) Re-replication is not readily detected by PFGE in OC cells in G2/M phase. Strains that were induced to re-replicate in A were harvested for PFGE at the indicated times. Southern blots of the gel were probed for chromosomes III, VI, and XIV as described in Figure 2C. (D) Some but not all copies of each chromosome participate when OC cells are induced to re-replicate in G2/M phase. Strains that were induced to re-replicate in B were harvested for PFGE at the indicated times. Southern blots of the gel were probed for chromosomes III, VI, and XIV as described in Figure 2C. (E) Cell cycle position significantly affects the extent of re-replication in the OC strain. The OC strain and the control O strain were induced to re-replicate in G2/M phase or during a G1 release as described, respectively, in A and B. For each induction protocol, OC and O strain genomic DNA were prepared and competitively hybridized against each other. Shown for chromosomes III, VI, and XIV are OC G2/M phase re-replication profiles (black lines), OC G1 release re-replication profiles (gray lines), locations of pro-ARSs mapped by Wyrick et al. (2001) (gray triangles), and the centromeres (black circles).
We next asked whether we could detect more re-replication in the OC strain by inducing it during a G1 release. In contrast to the results obtained during a G2/M phase induction, significant re-replication was detected by flow cytometry and PFGE within 2 h of the G1 release (Figure 5, B and D). The re-replication profile of the OC strain induced during a G1 release (Figure 5E and Supplementary Figure S11) showed broad re-replication zones of ∼200-500 kb in width on all chromosomes. These results, along with the re-replication induced during G2/M phase, establish that deregulating just ORC and Cdc6 is sufficient to induce re-replication and thus these inhibitory mechanisms are not truly redundant. The greater amount of re-replication induced during G1 release versus G2/M arrest underscores the dynamic character of the block to re-replication and, in this case, is likely due to the incomplete expulsion of Mcm proteins from the nucleus during S phase.
Microarray CGH Can Detect Re-replication Initiating Primarily from a Single Origin
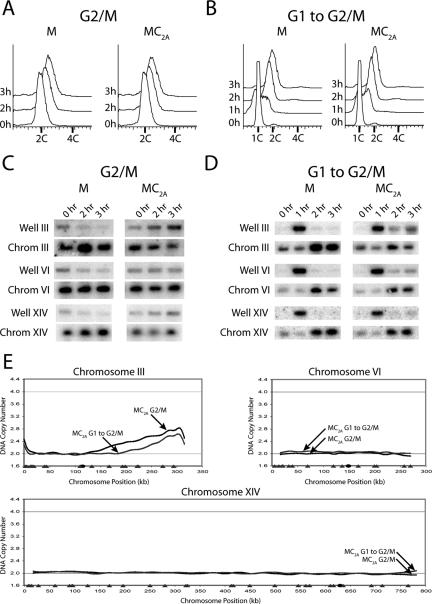
To further investigate the question of redundancy in replication control, we examined the consequences of deregulating just Mcm2-7 and Cdc6. We were not able to detect re-replication in the “MC” strain (MCM7-2NLS pGAL1-Δntcdc6) whether Δntcdc6 was induced in G2/M phase or during a G1 release (unpublished data). Hence, we further deregulated Cdc6 inhibition by mutating the two full CDK consensus phosphorylation sites on Δntcdc6 to generate the MC2A strain (MCM7-2NLS Δntcdc6-cdk2A). These additional mutations increase the stability of Δntcdc6 (Perkins et al., 2001).
Expression of Δntcdc6-cdk2A in the MC2A strain in either G2/M phase or during a G1 release did not cause a detectable increase in DNA content by flow cytometry (Figures 6, A and B). However, PFGE suggested that chromosome III re-replicated in a small subset of MC2A cells when Δntcdc6-cdk2A was induced under either protocol (Figure 6, C and D). Microarray CGH provided definitive evidence that re-replication occurred, in this strain, primarily on the right arm of chromosome III (Figure 6E and Supplementary Figure S12).
Figure 6.
Re-replication occurs primarily on a single chromosome when Mcm2-7 and Cdc6 are deregulated. (A) Re-replication is undetectable by flow cytometry in MC2A cells in G2/M phase. The MC2A strain YJL4489 (MCM7-NLS pGAL1-Δntcdc6-cdk2A pMET3-HA3-CDC20) and the control M strain YJL4486 (MCM7-2NLS pGAL1 pMET3-HA3-CDC20) were arrested in G2/M phase and induced with galactose as described in Figure 2A. Samples for flow cytometry were taken at the indicated times after galactose addition. The MC2A G2/M re-replication profile in E was normalized to the 3 h DNA content of 2.0 C. (B) Re-replication is undetectable by flow cytometry in MC2A cells during a G1 release. The MC2A strain and the control M strain were induced with galactose and released from a G1 arrest as described in Figure 4A. Samples for flow cytometry were taken at the indicated times. The MC2A G1 release re-replication profile in E was normalized to the 3 h DNA content of 2.0 C. (C) A portion of the population of chromosome III molecules participate when MC2A cells are induced to re-replicate in G2/M phase. The strains that were induced to re-replicate in A were harvested for PFGE at the indicated times. Southern blots of the gel were probed for chromosomes III, VI, and XIV as described in Figure 2C. (D) A portion of the population of chromosome III molecules participate when MC2A cells are induced to re-replicate during a G1 release. The strains that were induced to re-replicate in B were harvested for PFGE at the indicated times. Southern blots of the gel were probed for chromosomes III, VI, and XIV as described in Figure 2C. (E) Rereplication in the MC2A strain occurs primarily on chromosome III. The MC2A strain and the control M strain were induced to re-replicate in G2/M phase or during a G1 release as described, respectively, in A and B. For each induction protocol, MC2A and M strain genomic DNA were prepared and competitively hybridized against each other. Shown for chromosomes III, VI, and XIV are MC2A G2/M phase re-replication profiles (black lines), MC2A G1 release re-replication profiles (gray lines), locations of pro-ARSs mapped by Wyrick et al. (2001) (gray triangles) and the centromeres (black circles).
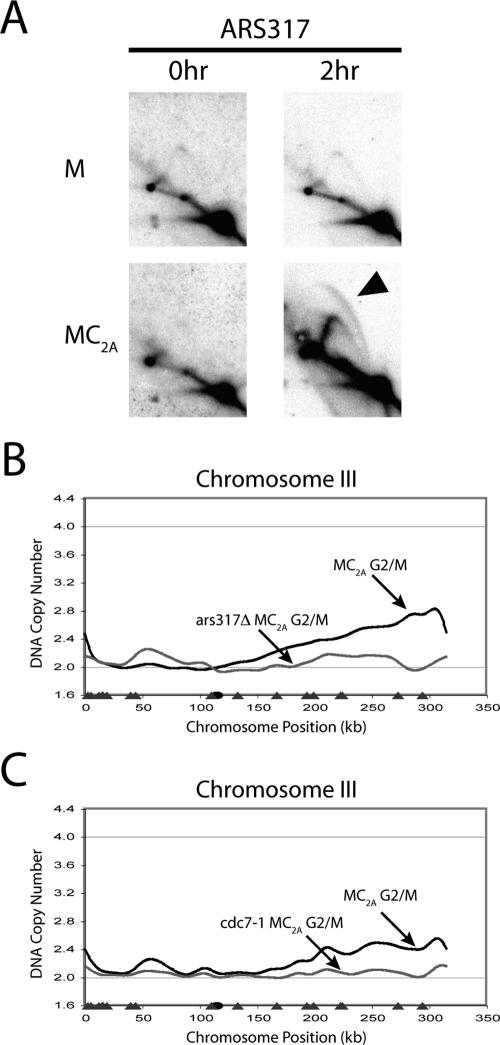
To confirm that the very limited DNA re-replication in the MC2A strain arose from a canonical reinitiation event, we asked whether this re-replication depended on known origins and initiation proteins. Our peak finding algorithm implicated an initiation event at ∼297 kb, close to ARS317, an inefficient S phase origin located at 291 kb. Two-dimensional gel analysis of ARS317 (Figure 7A) detected bubble arcs, indicative of replication initiation, in the MC2A strain but not the control “M” strain (MCM7-2NLS pGAL1). The immediately adjacent origins, ARS316 and ARS318, only displayed fork arcs (unpublished data), suggesting that most of the re-replication on the right arm of chromosome III originates from ARS317. Deletion of ARS317, but not ARS316 or ARS318, in the MC2A strain eliminated the bulk of the re-replication detected by microarray CGH (Figure 7B and unpublished data), demonstrating that re-replication initiates primarily from a single S phase origin.
Figure 7.
The re-replication arising from deregulation of both Mcm2-7 and Cdc6 depends on ARS317 and Cdc7. (A) Reinitiation bubbles are induced at ARS317 when MC2A re-replicates in G2/M phase. The MC2A strain YJL4489 (MCM7-NLS pGAL1-Δntcdc6-cdk2A pMET3-HA3-CDC20) and the control M strain YJL4486 (MCM7-2NLS pGAL1 pMET3-HA3-CDC20) were arrested in G2/M phase and induced with galactose as described in Figure 6A. Genomic DNA was purified from each strain at both 0 and 2 h after induction and subjected to neutral-neutral 2-D gel electrophoresis. Southern blots of the gels were probed with an ARS317 fragment. Black arrow indicates re-replication bubbles. (B) ARS317 sequence is required for the bulk of re-replication induced in MC2A cells. The MC2A-Δars317 strain YJL5858 (MCM7-NLS pGAL1-Δntcdc6-cdk2A pMET3-HA3-CDC20 Δars317) and the control M strain YJL4486 were arrested in G2/M phase and induced with galactose for 3 h as described in Figure 6A. Genomic DNA from the two strains was competitively hybridized against each other to generate the MC2A-Δars317 G2/M phase re-replication profile shown for chromosome III (gray line). The MC2A G2/M phase re-replication profile from Figure 5E is replotted for comparison (black line). The locations of pro-ARSs mapped by Wyrick et al. (2001) (gray triangles), and the centromere (black circle) are plotted along the X-axis. (C) Cdc7 kinase is required for re-replication induced in MC2A cells. The MC2A strain YJL4489, the congenic MC2A-cdc7 strain YJL5821 (MCM7-2NLS pGAL1-Δntcdc6-2A pMET3-HA3-CDC20 cdc7-1), and their respective controls, the M strain YJL4486 and the M-cdc7 strain YJL5816 (MCM7-2NLS pGAL1 pMET3-HA3-CDC20 cdc7-1) were induced with galactose as described in Figure 6A, except the initial arrest was performed at 23°C, and the arrested cells were shifted to 35°C for 1 h, before the addition of galactose. Genomic DNA was isolated 4 h after galactose addition and competitively hybridized (MC2A vs. M and MC2A-cdc7 vs. M-cdc7) as described in Figure 1A. Re-replication profiles for the MC2A (black line) and MC2A-cdc7 (gray line) strains are shown for chromosome III. Locations of pro-ARSs mapped by Wyrick et al. (2001) (gray triangles), and the centromere (black circle) are plotted along the X-axis.
We next asked whether this re-replication is dependent on the essential initiation factor, Cdc7-Dbf4 kinase. Both MC2A and MC2A cdc7-1 strains were induced to re-replicate in G2/M phase under permissive (23°C) and restrictive (35°C) temperatures for the cdc7-1 allele. Microarray CGH demonstrated that both strains re-replicated to a similar extent at 23°C (Supplementary Figure S13), but at 35°C there was little or no re-replication in the MC2A cdc7-1 strain (Figure 7C). Together, the dependence on both ARS317 and Cdc7-Dbf4 indicates that the very limited re-replication induced in the MC2A strain arises primarily from a single bona fide reinitiation event.
DISCUSSION
Use of Microarray CGH as a Routine Genome-wide Assay for Budding Yeast Replication
We have refined previously published genome-wide replication assays for budding yeast and made them more amenable for routine and widespread use in the study of eukaryotic DNA replication. The previous assays required significant effort and cost to generate a single replication profile and were only used to characterize the normal wild-type S phase (Raghuraman et al., 2001; Yabuki et al., 2002). We have obtained comparable replication profiles using a streamlined protocol, collection of a single time point and inexpensive spotted microarrays. Thus, it is feasible to use our streamlined assay to examine the genome-wide replication phenotypes associated with many different genotypes or physiological conditions.
Reinitiation Induced in G2/M Phase Largely Follows the Rules of Origin Selection, But Not the Rules of Origin Activation, That Govern S Phase Replication
We have taken advantage of our microarray CGH assay to perform a genome-wide analysis of eukaryotic re-replication. This comprehensive analysis has allowed us to examine several key tenets of the current model for replication control. One important tenet is that reinitiation that arises from deregulation of ORC, Mcm2-7, and Cdc6 occur from sites of pre-RC formation in S phase. The overall concordance of mapped re-replication origins with pro-ARSs suggests that the reinitiation occurs at sites that normally assemble pre-RCs for S phase replication. Although current limitations of the resolution of microarray data prevent a precise match of replication and re-replication origins, in the few cases where this has been directly tested by 2-D gel electrophoresis or deletion analysis (Figure 7 and Nguyen et al., 2001), we have confirmed that this is, in fact, the case. Thus, the sequence determinants that select potential origins in S phase appear to be conserved during re-replication.
In contrast to the selection of potential origins, the activation of these origins during re-replication in G2/M phase differs considerably from origin activation during replication in S phase. During S phase replication, poorly understood chromosomal determinant specify which potential origins are activated early, which are activated late, and which remain latent. During re-replication in G2/M phase, all three classes are among the 106 origins that reinitiate, and there is no correlation between the time/efficiency pro-ARSs replicate in S phase and the efficiency with which they re-replicate in G2/M phase. These results suggest that the chromosomal determinants governing S phase origin activation are not preserved during G2/M phase re-replication. Such a conclusion is consistent with the finding that the temporal program for origin firing in S phase is lost by G2/M phase and must be reestablished upon entry into each new cell cycle (Raghuraman et al., 1997).
The Block to Re-replication Uses a Common Fundamental Strategy Implemented in a Dynamic Manner Across the Cell Cycle
Another important tenet of the replication control model is that the blocks to re-replication in S, G2, and M phase use the same fundamental strategy of preventing pre-RC reassembly. Deregulating the mechanisms that prevent this reassembly in any of these cell cycle phases should thus lead to re-replication. Studies in human, Drosophila, and Caenorhabditis elegans that deregulate geminin (Melixetian et al., 2004), Cdt1 (Thomer et al., 2004), and Cul-4 (which stabilizes Cdt1; Zhong et al., 2003), respectively, have inferred that re-replication can occur within S phase based on evidence of a prolonged S phase. In this study, we directly demonstrate that cells can reinitiate replication at multiple origins while the first round of replication is still ongoing. Thus, we establish that mechanisms used to prevent re-replication in G2/M phase also prevent re-replication within S phase.
Despite sharing common mechanisms to carry out the same fundamental strategy, the block to re-replication in S phase and G2/M phase are not identical. Two differences are readily apparent when comparing cells re-replicating through S phase during a G1 release with cells re-replicating at a G2/M phase arrest. The first difference is the bias toward reinitiation of early origins that is only observed in the G1 release experiment. The simplest explanation for this bias is suggested by the S phase re-replication profiles, which show reinitiation at early origins occurring before late origins have had a chance to fire. These observations suggest that early origins clear their replication pre-RCs sooner and are more available for pre-RC reassembly during S phase, although other explanations for this bias cannot be ruled out.
The second difference between the G1 release and G2/M phase re-replication is that the amount of re-replication induced during the G1 release was greater than the amount induced in G2/M phase in both the OMC and OC strains. This difference can be observed by flow cytometry but is most striking when G1 release and G2/M phase re-replication profiles are compared. There are a growing number of examples of mechanisms that vary in their efficacy across the cell cycle, such as Cdc6 degradation in budding yeast (Perkins et al., 2001), Cdt1 degradation in Xenopus and humans (Nishitani et al., 2004; Arias and Walter, 2005; Li and Blow, 2005; Yoshida et al., 2005), and geminin inhibition in human cells (Ballabeni et al., 2004). Together these results indicate that the block to re-replication is dynamic with the number and relative contribution of regulatory mechanisms implementing the block changing during the cell cycle.
What Is Limiting Re-replication?
A key difference between re-replication and replication in the OMC strain is that a significantly smaller number of origins initiate efficiently during re-replication (106 vs. 193). This reduction in origin firing likely contributes to the limited re-replication observed in the OMC strain and suggests that additional mechanisms are still restraining reinitiation. Consistent with both notions, additional mechanisms inhibiting ORC (by CDK binding to Orc6; Wilmes et al., 2004) and Cdc6 (by CDK binding to the N-terminus of phosphorylated Cdc6; Mimura et al., 2004) have recently been identified in budding yeast. The latter mechanism is already disrupted in the OMC strain because of the N-terminal deletion of Cdc6. Disrupting the former mechanism in the OMC background moderately enhances re-replication, but this re-replication is still restrained (Wilmes et al., 2004; Tanny et al., 2006), suggesting that still more re-replication controls remain to be identified.
The reduced number of reinitiating pro-ARSs, however, may not be the only factor limiting re-replication. Previous work suggests that a single replication fork should be able to replicate 100-200 kb (Dershowitz and Newlon, 1993; van Brabant et al., 2001). Our re-replicating profiles show that the amount of DNA synthesis associated with many reinitiating origins is significantly reduced 100-200 kb away from these origins (Supplementary Figure S7). These data suggest that re-replicating forks may not be able to progress as far as replicating forks, although a more direct analysis of fork movement will be needed to confirm this hypothesis.
Multiple Nonredundant Mechanisms Work in Combination to Reduce the Probability of Re-replication
We previously showed that we could reliably detect G2/M phase re-replication by flow cytometry in the OMC strain when ORC, Mcm2-7, and Cdc6 are deregulated, but not when only two of the three proteins were deregulated (Nguyen et al., 2001). Since then, there have been many other examples where multiple replication controls had to be disrupted to detect re-replication (reviewed in Diffley, 2004; Blow and Dutta, 2005). These observations have led to the presumption that the eukaryotic replication controls are redundant. We favor an alternative view that replication controls are not redundant and that disruption of one or a few of controls can lead to low levels of re-replication.
Failure to detect this re-replication has been due to the insensitivity of standard replication assays. In support of the view, the more sensitive microarray CGH assay used in this study was able to detect G2/M phase re-replication in the OC and MC2A strains. We did not detect re-replication when only a single mechanism was disrupted, but we note that the microarray CGH assay has its own detection limits and may have difficulty detecting rare or sporadic replication events. The development of even more sensitive single-cell assays that can detect these rare re-replication events may reveal that the chance of re-replication occurring is increased when ORC, Mcm2-7, or Cdc6 is individually deregulated.
Our findings support a model in which the block to rereplication is provided by a patchwork of many mechanisms, each of which contributes to a portion of the block by reducing the probability that re-replication will occur within a cell cycle. The combined action of all these mechanisms is needed to reduce the probability to such low levels that re-replication events become exceedingly rare and virtually prohibited. Successive disruption of these mechanisms does not lead to a sudden collapse of the block after a threshold of deregulation is reached, but instead results in a gradual erosion of the block manifested by incrementally higher frequencies and/or levels of limited re-replication. Because all mechanisms contribute in some way to the block, more than one mechanism or combination of mechanisms can be overridden to generate detectable re-replication. Hence, the fact that disruption of a mechanism is sufficient to induce limited rereplication does not make it the critical or dominant mechanism in the block to re-replication.
Levels of Re-replication Likely to Contribute to Genomic Instability and Tumorigenesis May Not Be Detectable by Most Currently Available Assays
Because genomic instability is associated with, and possibly facilitates, tumorigenesis, there has been much interest in understanding the derangements in DNA metabolism and cell cycle control that can cause genomic instability. Rereplication is a potential source of genomic instability both because it produces extra copies of chromosomal segments and because it generates DNA damage and/or replication stress (Melixetian et al., 2004; Zhu et al., 2004; Archambault et al., 2005; Green and Li, 2005). Re-replication has also been potentially linked to tumorigenesis by the observation that overexpression of Cdt1, which can contribute to re-replication (reviewed in Blow and Dutta, 2005), can transform NIH3T3 cells into tumorigenic cells (Arentson et al., 2002). However, two considerations have raised concerns about the biological relevance of these potential connections. First, if replication controls are highly redundant, the probability that a cell will spontaneously acquire the multiple disruptions needed to induce re-replicate will be extremely small. Second, we and others have shown that cells undergoing overt re-replication experience extensive inviability (Jallepalli et al., 1997; Yanow et al., 2001; Wilmes et al., 2004; Green and Li, 2005) or apoptosis (Vaziri et al., 2003; Thomer et al., 2004), making cell death a more likely outcome than genomic instability or tumorigenesis.
Our results in this study counter the first concern by challenging the concept of redundancy in replication control and showing that very low levels of re-replication can still be observed when fewer controls are disrupted. We also have evidence that lower levels of re-replication induce lower levels of inviability (unpublished data), diminishing the second concern. Consequently, we suggest that re-replication at levels well below current detection limits may occur with greater frequency than previously anticipated and that genomic instability may arise from these low, nonlethal levels of re-replication.
Supplementary Material
Acknowledgments
We thank Adam Carroll, Emily Wang, and Marian Tse for assistance in constructing the microarrays; Hiten Madhani, Bruce Alberts, David Morgan, and David Toczyski for helpful discussions and comments on the manuscript; and Steve Bell for discussion of results before publication. This work was supported by grants to J.J.L. from the Sandler Program in Biological Sciences, the American Cancer Society (RPG-99-169-01-CCG) and the National Institutes of Health (RO1 GM59704). B.R.M. was supported by an National Science Foundation Predoctoral Fellowship (DGE-0202754) and a Department of Defense Breast Cancer Predoctoral Fellowship (W81XWH-04-1-0409). R.J.M. was supported by an NIH Genetics and Cell Biology Training Grant (T32 GM07810).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-11-1043) on February 15, 2006.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Archambault, V., Ikui, A. E., Drapkin, B. J., and Cross, F. R. (2005). Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol. Cell Biol. 25, 6707-6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arentson, E., Faloon, P., Seo, J., Moon, E., Studts, J. M., Fremont, D. H., and Choi, K. (2002). Oncogenic potential of the DNA replication licensing protein CDT1. Oncogene 21, 1150-1158. [DOI] [PubMed] [Google Scholar]
- Arias, E. E., and Walter, J. C. (2005). Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 19, 114-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan, R. et al. (2006). Saccharomyces Genome Database.
- Ballabeni, A., Melixetian, M., Zamponi, R., Masiero, L., Marinoni, F., and Helin, K. (2004). Human geminin promotes pre-RC formation and DNA replication by stabilizing CDT1 in mitosis. EMBO J. 23, 3122-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S. P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333-374. [DOI] [PubMed] [Google Scholar]
- Blow, J. J., and Dutta, A. (2005). Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6, 476-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi, J. L., Iyer, V. R., and Brown, P. O. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680-686. [DOI] [PubMed] [Google Scholar]
- Dershowitz, A., and Newlon, C. S. (1993). The effect on chromosome stability of deleting replication origins. Mol. Cell Biol. 13, 391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley, J. F. (2004). Regulation of early events in chromosome replication. Curr. Biol. 14, R778-R786. [DOI] [PubMed] [Google Scholar]
- Drury, L. S., Perkins, G., and Diffley, J. F. (1997). The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16, 5966-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury, L. S., Perkins, G., and Diffley, J. F. (2000). The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 10, 231-240. [DOI] [PubMed] [Google Scholar]
- Elsasser, S., Chi, Y., Yang, P., and Campbell, J. L. (1999). Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell 10, 3263-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan, V., Simancek, P., Houchens, C., Snaith, H. A., Frattini, M. G., Sazer, S., and Kelly, T. J. (2001). Redundant control of re-replication in fission yeast. Proc. Natl. Acad. Sci. USA 98, 13114-13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, B. M., and Li, J. J. (2005). Loss of rereplication control in Saccharomyces cerevisiae results in extensive DNA damage. Mol. Biol. Cell 16, 421-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G. (eds.) (1990). Guide to Yeast Genetics and Molecular Biology, New York: Academic Press.
- Haase, S. B., and Lew, D. J. (1997). Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol. 283, 322-332. [DOI] [PubMed] [Google Scholar]
- Hennessy, K. M., and Botstein, D. (1991). Regulation of DNA replication during the yeast cell cycle. Cold Spring Harb. Symp. Quant. Biol. 56, 279-284. [DOI] [PubMed] [Google Scholar]
- Huberman, J. A., Spotila, L. D., Nawotka, K. A., el-Assouli, S. M., and Davis, L. R. (1987). The in vivo replication origin of the yeast 2 microns plasmid. Cell 51, 473-481. [DOI] [PubMed] [Google Scholar]
- Iyer, V. R., Horak, C. E., Scafe, C. S., Botstein, D., Snyder, M., and Brown, P. O. (2001). Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409, 533-538. [DOI] [PubMed] [Google Scholar]
- Jallepalli, P. V., Brown, G. W., Muzi-Falconi, M., Tien, D., and Kelly, T. J. (1997). Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev. 11, 2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib, K., Diffley, J. F., and Kearsey, S. E. (1999). G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1, 415-422. [DOI] [PubMed] [Google Scholar]
- Li, A., and Blow, J. J. (2005). Cdt1 downregulation by proteolysis and geminin inhibition prevents DNA re-replication in Xenopus. EMBO J. 24, 395-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liku, M. E., Nguyen, V. Q., Rosales, A. W., Irie, K., and Li, J. J. (2005). CDK phosphorylation of a novel NLS-NES module distributed between two subunits of the Mcm2-7 complex prevents chromosomal rereplication. Mol. Biol. Cell 16, 5026-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida, Y. J., Hamlin, J. L., and Dutta, A. (2005). Right place, right time, and only once: replication initiation in metazoans. Cell 123, 13-24. [DOI] [PubMed] [Google Scholar]
- McGarry, T. J., and Kirschner, M. W. (1998). Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93, 1043-1053. [DOI] [PubMed] [Google Scholar]
- Melixetian, M., Ballabeni, A., Masiero, L., Gasparini, P., Zamponi, R., Bartek, J., Lukas, J., and Helin, K. (2004). Loss of Geminin induces rereplication in the presence of functional p53. J. Cell Biol. 165, 473-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura, S., Seki, T., Tanaka, S., and Diffley, J. F. (2004). Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature 431, 1118-1123. [DOI] [PubMed] [Google Scholar]
- Moll, T., Tebb, G., Surana, U., Robitsch, H., and Nasmyth, K. (1991). The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66, 743-758. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. Q., Co, C., Irie, K., and Li, J. J. (2000). Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr. Biol. 10, 195-205. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. Q., Co, C., and Li, J. J. (2001). Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411, 1068-1073. [DOI] [PubMed] [Google Scholar]
- Nishitani, H., Lygerou, Z., and Nishimoto, T. (2004). Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region. J. Biol. Chem. 279, 30807-30816. [DOI] [PubMed] [Google Scholar]
- Perkins, G., Drury, L. S., and Diffley, J. F. (2001). Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J. 20, 4836-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman, M. K., Brewer, B. J., and Fangman, W. L. (1997). Cell cycle-dependent establishment of a late replication program. Science 276, 806-809. [DOI] [PubMed] [Google Scholar]
- Raghuraman, M. K., Winzeler, E. A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D. J., Davis, R. W., Brewer, B. J., and Fangman, W. L. (2001). Replication dynamics of the yeast genome. Science 294, 115-121. [DOI] [PubMed] [Google Scholar]
- Tanaka, S., and Diffley, J. F. (2002). Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat. Cell Biol. 4, 198-207. [DOI] [PubMed] [Google Scholar]
- Tanny, R. E., MacAlpine, D. M., Blitzblau, H. G., and Bell, S. P. (2006). Genome-wide analysis of re-replication reveals inhibitory controls that target multiple stages of replication initiation. Mol. Biol. Cell 17, 2415-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomer, M., May, N. R., Aggarwal, B. D., Kwok, G., and Calvi, B. R. (2004). Drosophila double-parked is sufficient to induce re-replication during development and is regulated by cyclin E/CDK2. Development 131, 4807-4818. [DOI] [PubMed] [Google Scholar]
- van Brabant, A. J., Buchanan, C. D., Charboneau, E., Fangman, W. L., and Brewer, B. J. (2001). An origin-deficient yeast artificial chromosome triggers a cell cycle checkpoint. Mol. Cell 7, 705-713. [DOI] [PubMed] [Google Scholar]
- Vas, A., Mok, W., and Leatherwood, J. (2001). Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol. Cell Biol. 21, 5767-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri, C., Saxena, S., Jeon, Y., Lee, C., Murata, K., Machida, Y., Wagle, N., Hwang, D. S., and Dutta, A. (2003). A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11, 997-1008. [DOI] [PubMed] [Google Scholar]
- Wilmes, G. M., Archambault, V., Austin, R. J., Jacobson, M. D., Bell, S. P., and Cross, F. R. (2004). Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 18, 981-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick, J. J., Aparicio, J. G., Chen, T., Barnett, J. D., Jennings, E. G., Young, R. A., Bell, S. P., and Aparicio, O. M. (2001). Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294, 2357-2360. [DOI] [PubMed] [Google Scholar]
- Yabuki, N., Terashima, H., and Kitada, K. (2002). Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7, 781-789. [DOI] [PubMed] [Google Scholar]
- Yanow, S. K., Lygerou, Z., and Nurse, P. (2001). Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J. 20, 4648-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K., Takisawa, H., and Kubota, Y. (2005). Intrinsic nuclear import activity of geminin is essential to prevent re-initiation of DNA replication in Xenopus eggs. Genes Cells 10, 63-73. [DOI] [PubMed] [Google Scholar]
- Zhong, W., Feng, H., Santiago, F. E., and Kipreos, E. T. (2003). CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423, 885-889. [DOI] [PubMed] [Google Scholar]
- Zhu, W., Chen, Y., and Dutta, A. (2004). Re-replication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell Biol. 24, 7140-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.