Abstract
Cohesin maintains sister chromatid cohesion until its Rad21/Scc1/Mcd1 is cleaved by separase during anaphase. DNA topoisomerase II (topo II) maintains the proper topology of chromatid DNAs and is essential for chromosome segregation. Here we report direct observations of mitotic progression in individual HeLa cells after functional disruptions of hRad21, NIPBL, a loading factor for hRad21, and topo II α,β by RNAi and a topo II inhibitor, ICRF-193. Mitosis is delayed in a Mad2-dependent manner after disruption of either or both cohesin and topo II. In hRad21 depletion, interphase pericentric architecture becomes aberrant, and anaphase is virtually permanently delayed as preseparated chromosomes are misaligned on the metaphase spindle. Topo II disruption perturbs centromere organization leading to intense Bub1, but no Mad2, on kinetochores and sustains a Mad2-dependent delay in anaphase onset with persisting securin. Thus topo II impinges upon centromere/kinetochore function. Disruption of topo II by RNAi or ICRF-193 overrides the mitotic delay induced by cohesin depletion: sister centromeres are aligned and anaphase spindle movements occur. The ensuing accumulation of catenations in preseparated sister chromatids may overcome the reduced tension arising from cohesin depletion, causing the override. Cohesin and topo II have distinct, yet coordinated functions in metaphase alignment.
INTRODUCTION
The precise transmission of chromosomes from mother to daughter cells is a fundamental process in cell multiplication. In mitosis, duplicated sister chromatids condense, align to form metaphase plate, and are simultaneously segregated to move to opposing spindle poles. The spindle checkpoint monitors the interaction between microtubules and chromosomes. It restrains chromosome segregation and mitotic exit by negatively regulating polyubiquitination of mitotic cyclin and securin until metaphase chromosome alignment is established. In anaphase, securin and mitotic cyclin are degraded in an ubiquitin-dependent way, leading to chromosome segregation and mitotic exit, respectively. Destruction of securin activates its partner protein, separase, for which it acts as a chaperone, liberating it to cleave hRad21/Scc1, a subunit of cohesin that holds sister chromatids together. These basic principles of chromosome segregation have been evolutionarily conserved from fungi to vertebrates (e.g., Yanagida, 2005).
Cohesin comprises four subunits, two SMC (structural maintenance of chromosome proteins) subunits, SMC1 and SMC3, and two non-SMC proteins Scc3/SA and kleisin (Nasmyth and Haering, 2005). SMCs and kleisin are conserved from bacteria to humans. SMCs consist of very long coiled coil regions that flank a nonstructured hinge region, and the amino and carboxy termini contain ATPase motifs so that the two SMCs that are joined at the hinge are V-shaped with ATPase domains at either end of the V. Kleisin is thought to interact with SMC termini at either end of the V to close the V-shaped structure. Cohesin complexes hold the two sister chromatids together from their loading during DNA replication, until the cleavage of cohesin triggers the resolution of cohesion in anaphase (Nasmyth, 2002). Scc1/Rad21/kleisin is the target for cleavage into nonfunctional portions by separase in eukaryotic cells. There are several models that explain how cohesin acts on chromosome, one of which is so called “embrace” model (Nasmyth, 2005). It presumes that cohesin forms a large ring that holds sister chromatids together and the cleavage of Scc1/Rad21 opens up the cohesin ring to permit the release and separation of sister chromatids. Other models predict distinct nonring based structures (Huang et al., 2005; Nasmyth, 2005; Hirano, 2005). In budding and fission yeasts, Scc2/Mis4 is required to load cohesin onto chromosomes (Ciosk et al., 2000; Tomonaga et al., 2000). The human homologue NIPBL is mutated in Cornelia de Lange syndrome (Krantz et al., 2004; Tonkin et al., 2004) that is manifest as extensive developmental disorders as well as defects in sister chromatid cohesion (Kaur et al., 2005).
Although cohesin loading occurs during the S phase, the major defective phenotypes of cohesion-defective mutants are most apparent during mitosis. Mutation or deletion of cohesion genes in yeasts and higher eukaryotes are remarkably similar, with defects in chromatid cohesion and alignment at the metaphase plate being common features (Guacci et al., 1997; Michaelis et al., 1997; Tomonaga et al., 2000; Sonoda et al., 2001; Vass et al., 2003; Losada et al., 2005). The analysis of temperature-sensitive cohesin mutants in yeast revealed a delay in mitotic progression that was dependent on Mad2 and Bub1 to restrain the entry into anaphase (Stern and Murray, 2001; Biggins and Murray, 2001; Toyoda et al., 2002). In higher eukaryotes, most cohesin dissociates from chromosome in prophase, while any remaining cohesin is cleaved in anaphase (Losada et al., 1998; Sumara et al., 2000; Waizenegger et al., 2000; Hauf et al., 2001). Expression of noncleavable human kleisin, hRad21 leads to defects in chromosome segregation (Hauf et al., 2001). The terminology used in this report is that “segregation” implies anaphase movement of separated chromosomes toward the opposite spindle poles, whereas “separation” means the loss of linkage between sister chromatids (both cohesion and topological linkage such as catenation). Separated sister chromatids may remain in close proximity in the absence of spindle forces; however, these segregated chromosomes are pulled apart by movement toward the spindle poles.
Topo II is essential for mitosis in eukaryotic cells (Holm et al., 1985; Uemura and Yanagida, 1986; Cozzarelli and Wang, 1990). During mitosis topo II may resolve the catenation and other topological links that are generated during replication and in G2-interphase (Sundin and Varshavsky, 1980). In Schizosaccharomyces pombe, active topo II is required for the complete chromosome condensation and segregation of the bulk of the chromosomal DNA during anaphase (Uemura et al., 1986, 1987). Consistently, depletion of topo II by RNAi in higher eukaryotes leads to chromosome mis-segregation alongside slight defects in condensation (Chang et al., 2003; Carpenter and Porter, 2004; Sakaguchi and Kikuchi, 2004). The simultaneous depletion of topo IIα and β generates severe defect in chromosome segregation (Sakaguchi and Kikuchi, 2004). Topo IIα plays a principal role in mitotic chromosome resolution, but topo IIβ shares certain functions with it. Mammalian topo II is known to be broadly distributed along chromosome arms (e.g., Hirano and Mitchison, 1993), but there have been limited studies on its precise role in chromosome segregation. However, topo II mutation and inhibition were recently reported to partly rescue the defects in chromosome alignment induced by depletion of cohesin in yeast and chicken, indicating that cohesin and topo II act in closely related pathways in these organisms (Dewar et al., 2004; Vagnarelli et al., 2004).
Here we examined the role of hRad21, NIPBL, and topo II in chromosome alignment and segregation by investigating the consequences of gene depletion by RNAi on the behavior of sister centromeres during time-lapse analysis of mitosis of a HeLa cell line that stably expresses GFP-fused hMis12 (an essential centromere protein; Goshima et al., 2003; Obuse et al., 2004). To date, such kinetochore behavior has not been investigated for phenotypes arising from defects in hRad21 and topo II depletion or inhibition by ICRF-193, a topo II inhibitor (Ishimi et al., 1992). As the present study is focused on the behavior of chromosomes under spindle checkpoint control, microtubule–kinetochore interaction is also analyzed by visualizing the spindle checkpoint proteins Mad2, Bub1, and BubR1 alongside direct staining of microtubules and kinetochores. The data we present define essential roles for hRad21 and topo II in the correct alignment of centromeres under the surveillance of Mad2-dependent spindle checkpoint control.
MATERIALS AND METHODS
Strains, Media, and Antibodies
HeLa cells were grown at 37°C in a humidified CO2 (5%) incubator in DMEM medium (GIBCO, Rockville, MD) supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% antibiotic-antimycotic. To obtain polyclonal antibodies against human NIPBL and hRad21 proteins, the synthetic peptides of their C-terminal sequences (CRRSSQRISQRITH and CATPGPRFHII, respectively) were used for immunization of rabbits, and polyclonal antisera were obtained. Affinity-purified anti-NIPBL and anti-hRad21 antibodies were used for immunoblot at 1:500 dilutions.
Plasmids and Transfection
A NIPBL cDNA was isolated from a HeLa cDNA library, where a human EST sequence homologous to S. pombe Mis4 (Furuya et al., 1998) was used as a probe. The first codon was determined by 5′-RACE. An hRad21 cDNA plasmid (KIAA0078) was provided from the Kazusa DNA Research Institute. hRad21-myc8 plasmid was constructed by inserting the whole coding region of hRad21 and eight tandem copies of myc antigen in frame into pcDNA3.1/V5His6 (Invitrogen, Carlsbad, CA). Plasmid DNAs were purified using the Endofree Maxi kit (QIAGEN, Chatsworth, CA) and transfected into HeLa cells by the Effectene transfection kit (QIAGEN).
Immunofluorescence Microscopy
Immunofluorescence was performed as previously described (Goshima et al., 2003). Primary antibodies against hMis12, CENP-A, tubulin, CENP-C, Mad2, and cyclin B1 have been described previously (Goshima et al., 2003). Other antibodies were applied as follows; anti-Bub1 and anti-BubR1 (kind gifts from Dr. S. Taylor, 1:500; Taylor et al., 2001), anti-CENP-E and anti-MCAK (Cytoskeleton, Denver, CO; 1:100), anti-Aurora B (BD Transduction Laboratories, Lexington, KY; 1:100), anti-securin (MBL, Nagoya, Japan; 1:20). Slides were observed under a Zeiss Axiophot microscope (Thornwood, NY) equipped with a Plan Apochromat objective lens and an ORCA-ER chilled CCD camera (Hamamatsu, Bridgewater, NJ). Images were acquired by IP Lab software (Scanalytics, Billerica, MA). For detailed observation of microtubule–kinetochore interaction, the DeltaVision microscope system (Applied Precision, Issaquah, WA) equipped with an Olympus IX-70 microscope (Melville, NY) was used. Images were acquired at 0.2-μm steps in the z-axis and deconvolved 15 times by SoftWorx software (Applied Precision). The method of in situ fractionation was performed as described previously (Losada et al., 2005). In short, cells on coverslips were incubated in CSK buffer (10 mM PIPES, pH 6.8, 100 mM NaCl, 300 mM sucrose, 1 mM MgCl2, 1 mM EGTA) supplemented with 0.5% Triton X-100 and 1 mM phenylmethylsulfonyl fluoride for 5 min on ice before paraformaldehyde fixation.
Mitotic Chromosome Spread
For detection of pericentric hRad21-myc8, chromosome spread analysis was performed as described (Goshima et al., 2003; Shimura, M., personal communication). HeLa cells transfected with buffer or siRNA at 0 h were transfected with pcDNA3.1/V5His6 vector or phRad21-myc8 plasmid 24 h later. Nocodazole (100 ng/ml) was added after 48 h, and cells were incubated for a further 4 h. Cells were swollen in a hypotonic solution (75 mM KCl) followed by spreading cells with Cytospin 4 (Thermo, Waltham, MA) at 1000 rpm for 5 min. Spread cells were stained with anti-myc (9E10, 1:100, Santa Cruz Biotechnology, Santa Cruz, CA) and anti-hMis12 antibodies without fixation. After immunostaining, paraformaldehyde fixation was performed followed by staining DNA with Hoechst 33342. For Giemsa staining, nocodazole was added to the cells transfected with buffer or siRNA 30 min before the spreading. Cells were swollen in 75 mM KCl and fixed with cold Carnoy's solution (a 3:1 mixture of methanol and acetic acid). The cell suspension was dropped onto slides in a humid atmosphere. Spread cells were dried, stained with Giemsa (Nakarai, Kyoto, Japan), washed, and mounted for microscopy.
RNAi Method
siRNAs were synthesized for RNAi of NIPBL (5′-GCUUUUGAAUCCUCUAGGATT-3′) and hRad21 (5′-GACCUUAGAAAAAGGAGGATT-3′) by JbioS (Saitama, Japan). The sequences were unique in the human genome database. Transfection of buffer (30 mM HEPES-KOH, pH 7.5, 100 mM potassium acetate, 2 mM Mg acetate) was performed as control. The procedures of RNAi were based on the instruction of Oligofectamine (Invitrogen). When multiple siRNAs were used, 10 pmol of each siRNA was transfected. The sequences of siRNA for Mad2, topo II α and β were previously reported (Martin-Lluesma et al., 2002; Sakaguchi and Kikuchi, 2004). ICRF-193 (Zenyaku Kogyo, Tokyo, Japan) was added after 48 h to give the final concentration of 20 μM and incubated for 30 or 120 min before observation of living cells or fixation, respectively.
Measurement of Cell Proliferation and Mitotic Indices
Cell proliferation was measured directly by counting cell numbers in three identical fields of view for each sample at 24-h intervals. The averages were plotted on the graphs. Mitotic indices were determined by the ratio of round shaped cells and by counting cells with condensed chromosomes stained with Hoechst 33342.
Live Cell Analysis
Live cell analysis was performed as previously described (Haraguchi et al., 1997; Obuse et al., 2004). In short, HeLa cells stably expressing histone H2B-GFP or GFP-hMis12 were used for visualization of chromatin or kinetochores, respectively. Transfected cells grown on glass-based dishes (IWAKI, Tokyo, Japan) were supplemented with 20 mM HEPES (pH 7.4) and 100 ng/ml Hoechst 33342 and observed on the DeltaVision system at 37°C. GFP images were taken at 10-, 20-, or 60-s intervals with exposure time of 0.2 s for H2B-GFP or Hoechst and 1.5 s for GFP-hMis12. Images were deconvolved once by the SoftWorx software running on a SGI O2 computer.
FISH
FISH was performed as described previously with slight modifications (Hoque and Ishikawa, 2002; Nogami et al., 2000). Cells were grown on coverslips and fixed in 3% paraformaldehyde in phosphate-buffered saline (PBS). Fixed cells were successively treated in permeabilization buffer (0.5% Triton X-100, 0.5% saponin, and 20 μg/ml RNase A in PBS), 0.1 N HCl, and 20% glycerol in PBS and stored at –20°C. Procedures after thawing the frozen samples were as described previously (Nogami et al., 2000), with the exception that the denaturation temperatures for probe DNA was 75°C for 5 min and that for the cells 73°C for 4 min. Probes of pBR12 and cCI12–156 were kind gifts from Dr. K. Okumura and were biotinylated by nick translation. CEP 4 SpectrumGreen was purchased from Vysis (Downers Grove, IL).
RESULTS
NIPBL and hRad21 Knockdown Cells Are Mitotically Delayed
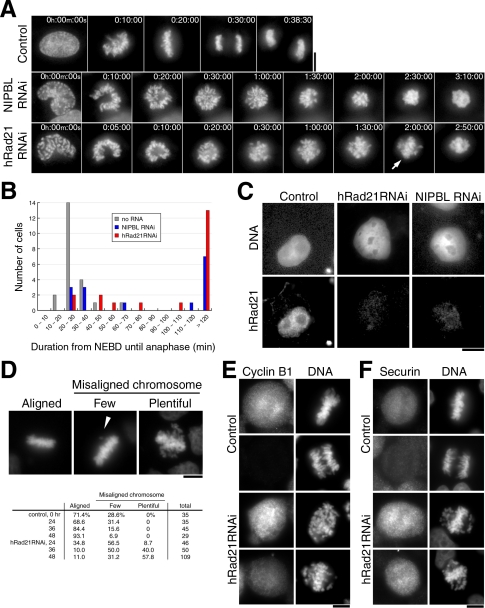
A number of movies were taken for HeLa cells after RNAi of NIPBL or hRad21 using the specific RNA probes (Elbashir et al., 2001; Materials and Methods) in order to examine the timing with which particular phenotypes would emerge. The data presented in Supplement 1, A–C, confirm that hRad21 and NIPBL were depleted to <5% to control levels 1 and 2 d after RNAi, respectively, as reported previously (Atienza et al., 2005; Losada et al., 2005). Initially the HeLa strain that stably expressed histone H2B-GFP (Kanda et al., 1998) was observed by fluorescence microscopy using DeltaVision. In non-RNAi control (Figure 1A, top), the metaphase plate (the congression of condensed chromosomes properly aligned like the plate) was always observed around 20 min after the nuclear envelope breakdown (designated NEBD), followed by the onset of chromosome segregation (anaphase) around 27 min post-NEBD (Supplement 2A for movie).
Figure 1.
hRad21 and NIPBL knockdown cells are mitotically delayed without securin destruction. (A) Time-lapse images taken from movies (Supplement 2) are shown. Top, control no RNAi; middle, NIPBL RNAi (120 h after RNAi); bottom, hRad21 RNAi (104 h after RNAi). Arrow, the partially formed plate. (B) The duration from NEBD until the onset of anaphase was measured in the movies of control non-RNAi, NIPBL, and hRad21 RNAi cells. (C) hRad21 retention in nuclei. Transfected cells were extracted in CSK buffer supplemented with Triton X-100 before fixation. Cells shown are 96 h after transfection. Top, DNA stained with Hoechst 33342; bottom, immunostaining of hRad21. (D) Top, left, normal metaphase; middle, cell showing a few misaligned chromosomes (arrowhead); right, cell showing abundant misaligned chromosomes. Bottom, the frequencies of each appearance measured in control non-RNAi and hRad21 RNAi cells. (E and F) Securin and cyclin B1 were immunostained in control non-RNAi and hRad21 RNAi cells fixed by paraformaldehyde. In hRad21 RNAi cells, mitotic cells containing abundant EMCs are shown. Bars, 10 μm.
In hRad21 knockdown cells (Figure 1A, bottom; Supplement 2B for movie), chromosome condensation occurred, but mitotic progression was considerably delayed after prometaphase. The metaphase plate, one of which is indicated by an arrow in Figure 1A (bottom, 2:00:00), was not obvious as certain chromosomes were misaligned, and condensed chromosomes had still not segregated 2 h 50 min after NEBD. Regular alignment of chromosomes and kinetochores in metaphase is the most prominent feature in normal metaphase (Rieder and Salmon, 1998). A mitotic delay was also prominent in NIPBL knockdown cells (middle panel; Supplement 2C for movie).
The timing with which the onset of anaphase chromosome segregation followed NEBD was measured in movies (Figure 1B; 22, 15, and 20 cells for control no-RNAi, NIPBL, and hRad21 RNAi, respectively). In 75% of control cells that had not been subjected to RNAi, the duration of this period was 20–30 min (the total average, 27.6 min). In hRad21 knockdown cells, however, only 10% of cells showed such duration and in 70% of cells anaphase was not seen after greater than 100 min monitoring. Two classes of cells were present after hRad21 RNAi. In the minor ∼30% class, RNAi was inefficient and the period from NEBD to anaphase ranged from 30 to 70 min, whereas in the majority of cells (∼70%) knockdown was extensive and cells failed to commit anaphase. Similar results were obtained for NIPBL knockdown. Because the mitotic index did not significantly increase in fixed cell preparations, movies were the only way to obtain convincing and quantitative evidence for the mitotic delay. Eventually, after a very long delay, chromosome decondensation was observed in the absence of chromosome segregation (unpublished data).
Chromatin Loading of hRad21 Requires NIPBL
We examined whether, like its yeast homologues, Scc2 and Mis4, human NIPBL functions as a cohesin loading protein (Ciosk et al., 2000; Tomonaga et al., 2000). Retention of hRad21 on chromatin was greatly diminished 48 h after RNAi treatment to deplete NIPBL RNAi (Figure 1C), indicating that, like its yeast counterparts, NIPBL was required for the loading onto chromatin of hRad21. These results are consistent with the frog extracts experiments (Gillespie and Hirano, 2004; Takahashi et al., 2004).
Cells Containing EMCs Increase after hRad21 Knockdown
The degree of chromosome misalignment was measured in fixed cells (Figure 1D). In non-RNAi control, normal metaphase plates in which all the chromosomes were aligned (Aligned) or a few misaligned chromosomes (Few), were the only categories seen. In hRad21 knockdown mitotic cells; however, cells displaying extensively misaligned chromosomes (designated EMCs hereafter) were abundant (Plentiful) as were cells that fell into the class Few. Control non-RNAi never produced EMCs. Note that the horseshoe-shaped figures of prometaphase chromosomes and EMCs were different and could be easily distinguished; EMCs were defined as chromosomes away from a partially formed plate-like chromosome structure. In hRad21 knockdown, 9, 40, and 58% of mitotic cells contained plentiful EMCs 24, 36, and 48 h after RNAi, respectively. The block of anaphase with plentiful EMCs was thus the earliest phenotype so far detected in mitotic cells after hRad21 knockdown.
Securin and Cyclin B1 Are Not Degraded
Next, immunostaining with mouse monoclonal antibodies that recognized cyclin B1 and securin was performed. In the non-RNAi control cells, the signals of securin and cyclin B1 were abolished in all of the anaphase cells (Figure 1, E and F, top). In hRad21 knockdown (Figure 1F, bottom); however, securin was intense in 99.4% (352/354) of cells showing EMCs. Cyclin B1 was also intense but frequently diminished, as only 62.2% (74/119) had strong staining (Figure 1E, bottom). We thus concluded that RNAi cells showing EMCs were arrested in cell cycle progression before commitment to anaphase and securin destruction.
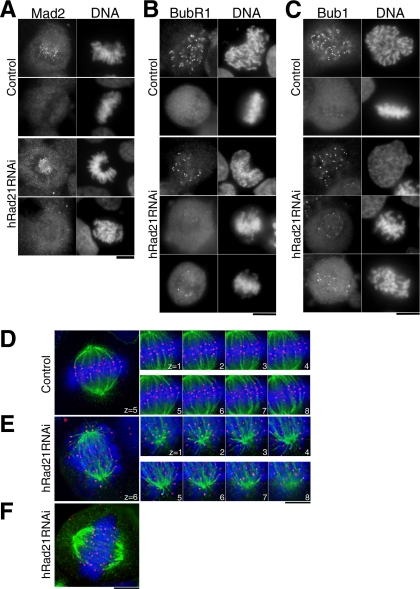
Bub1 Signals, But Not Mad2, Are Bound to EMCs
Mammalian kinetochores that are not attached to microtubules bind the essential spindle checkpoint protein Mad2 (Li and Benezra, 1996; Waters et al., 1998). We therefore asked whether Mad2 was bound to the kinetochores of cells with EMC configurations. In control cells, intense Mad2 signals were seen on prometaphase kinetochores but were missing from metaphase cells (Figure 2A, top panel). Like these controls the Mad2 signal was bound to prometaphase chromosomes of hRad21 knockdown cells, but was also entirely abolished in cells containing EMCs (Figure 2A, bottom panel). Among 190 mitotic cells revealing EMCs, every cell apart from two showed no Mad2 signals, indicating that kinetochores in EMCs were attached to microtubules.
Figure 2.
Bub1, BubR1-positive, and Mad2-negative kinetochores in hRad21 knockdown cells are end-on associated with microtubules. (A–C) Mitotic cells of control non-RNAi and hRad21 RNAi (48 h) cells were stained for DNA with Hoechst 33342, Mad2 (A), BubR1 (B), and Bub1 (C) with antibodies. (D and E) Eight serial sections (0.4-μm intervals) along the z-axis were taken for metaphase in control non-RNAi cells (D) and for hRad21 RNAi cell (E). Red, anti-CENP-C; green, tubulin; blue, DNA. (F) A hRad21 RNAi cell that retained a plate-like structure. Bars, 10 μm.
Bub1 and BubR1 are also spindle checkpoint proteins and bind to microtubule-associated but supposedly discriminate between those at which the microtubule/kinetochore interface does not result in any tension on that kinetochore (Jablonski et al., 1998; Skoufias et al., 2001; Taylor et al., 2001). We examined whether Bub1 and BubR1 were bound to kinetochores in EMCs. In non-RNAi cells, intense BubR1 signals were seen on kinetochores in late prophase and diminished to become virtually invisible by metaphase (Figure 2B, top panel). After hRad21 RNAi, BubR1 was greatly diminished in mitotic cells, although several strong signals were still seen on the periphery of EMCs (bottom panel). Bub1 signals were more intense than those of BubR1 and were particularly strong at the periphery of the EMCs (Figure 2C, bottom panel), while faint Bub1 signals were seen in normal metaphase plates (top panel). These data indicate that incorrect attachment of microtubules to kinetochores, including the type of association resulting into reduced tension, may arise as a consequence of hRad21 depletion.
hRad21 Knockdown Cells Are in a Quasi-Metaphase
To directly examine whether microtubules were associated with kinetochores of EMCs, hRad21 RNAi cells were immunostained by anti-CENP-C to identify the centromeric component of kinetochores (Saitoh et al., 1992) and anti-tubulin antibodies to stain microtubules and observed on a DeltaVision system after the DNA had been stained by Hoechst 33342. In non-RNAi, the paired kinetochores (red) clearly bound to the end of microtubules (green) and faced in opposing directions to opposite spindle poles (Figure 2D). Serial z-sections (0.4-μm intervals) established that all sister kinetochores were paired and microtubules bound these kinetochores in an end-on configuration.
In contrast, sister kinetochores were wide apart, and it was difficult to assign any directionality in their relationship to their partner kinetochore and the spindle poles when hRad21 had been depleted by RNAi. In serial z-section analysis (Figure 2E), nearly all sister kinetochores were bound end-on to microtubules. Of 21 mitotic cells, 13 cells showed the highly scattered EMCs, whereas the remaining 8 cells appeared to have aligned chromosomes (Figure 2F), in which paired centromeres were discernible. The distance between paired kinetochores (the average, 5.0 μm) was approximately twice as great as that separating kinetochores in non-RNAi cells (2.4 μm). Depletion of hRad21 was possibly not complete in a minor fraction of hRad21 knockdown cells. Thus, with respect to microtubule attachment and persistent cyclin B1 and securin signals, cells after hRad21 knockdown were in a quasi-metaphase, although sister kinetochores were preseparated and not properly aligned. MCAK and CENP-E, kinetochore proteins are thought to be required in metaphase to sense attachment/tension and trigger anaphase (Mao et al., 2003; Ohi et al., 2003). We therefore immunostained cells to localize these markers and found that they two were diminished in hRad21 RNAi cells (Supplement 3, A and B). In addition, we confirmed that the localization of aurora B kinase at the inner centromere in metaphase was abolished following hRad21 RNAi (Losada et al., 2005; unpublished data). Similar defects have been reported for INCENP, a partner of aurora B (Sonoda et al., 2001; Vass et al., 2003).
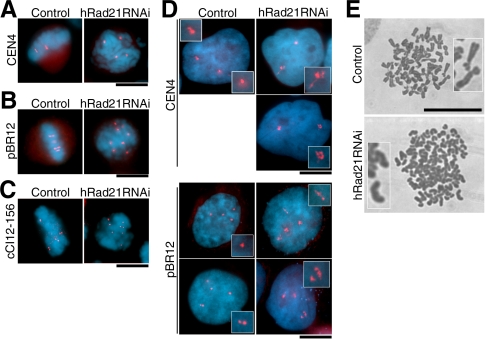
Pericentromeric and Telomeric Repetitive DNAs Are Prematurely Separated
We next used a hybridization approach to visualize the chromosome-specific pericentromeric repeat DNAs of mitotic chromosomes (Lichter et al., 1988). In non-RNAi control cells, the chromosome 4–specific repeat showed the two paired signals on metaphase chromosomes (Figure 3A, left). In hRad21 RNAi, however, four scattered dots were seen in mitosis (right panel), indicating that sister pericentromeric DNAs were prematurely separated. Similar experiments were done using a chromosome 12–specific repeat (pBR12, Baldini et al., 1990). In the HeLa line we used was tetraploid for chromosome 12 so that four paired signals were seen in the control non-RNAi (Figure 3B, left). In hRad21 RNAi cells, eight distantly positioned signals were seen in mitotic cells, as might be expected were cohesion to have failed (right panel). The telomere adjacent probe (cCI12-156, Takahashi et al., 1993; Nogami et al., 2000) specific for chromosome 12 revealed twice as many scattered dots in mitotic cells, than in the control non-RNAi cells (Figure 3C). Thus, although the signals were not preseparated at all in the non-RNAi control, the pericentromeric pBR12 and Cen4 probes and telomeric cCI12-156 were all prematurely separated with a high frequency (∼80%) in mitotic cells after hRad21 RNAi.
Figure 3.
Mitotic and interphase DNAs show aberrant behavior and organization. (A–C) FISH was used for mitotic cells of control non-RNAi and after hRad21 RNAi, using chromosome specific peri-centromeric probes (A and B for chromosomes 4 and 12, respectively) and peri-telomeric probe (C). (D) FISH was applied for interphase cells. (E) Chromosomes were spread and Giemsa stained. Top, control no RNAi; bottom, hRad21 RNAi cells (48 h). Bars, 10 μm.
Aberrant Organization of Pericentric Repetitive DNAs in Interphase Nuclei
In interphase nuclei of control non-RNAi cells, the number of hybridization signals seen for the pericentric repeats was normal (2 and 4, respectively, for the chromosome 4 and 12 pericentromeric probes; Figure 3D, left panels). In interphase of hRad21 RNAi cells (right panels), the same number of signals was found, indicating that within the resolution of this technique, there was no premature separation of sisters. Interestingly, the shapes of signals were extended to a much greater degree than in controls (enlarged insets). Forty-nine of 99 interphase nuclei (49.5%) had one or more aberrant signals with the pBR12 probe, but such an extension was never observed in control non-RNAi and mitotic hRad21 RNAi cells. Note that the pBR12 probe hybridized to around 1.6 Mb within pericentric DNA (Vermeesch et al., 2003), so that the extensions might arise from defects in chromatin organization.
Primary Constriction Is Lost in Giemsa Spread Chromosomes
Spreads of condensed chromosomes were prepared from cells 48 h after treatment with hRad21 RNAi. At ∼136, the number of Giemsa-stained chromosomes in hRad21 RNAi (Figure 3E, bottom) was twice as many as that of control spreads, 66 (top). hRad21 RNAi cells were unlikely to be in anaphase because of the nature of severe preanaphase delay and the use of nocodazole in the spread technique. Control chromosomes were X-shaped, whereas in hRad21 RNAi, chromosomes looked had a rod-like appearance and curiously lacked the primary constriction in centromeric regions. Such abnormal chromosome morphologies were relatively frequent (40%) in hRad21 RNAi cells, but absent from spreads of non-RNAi controls.
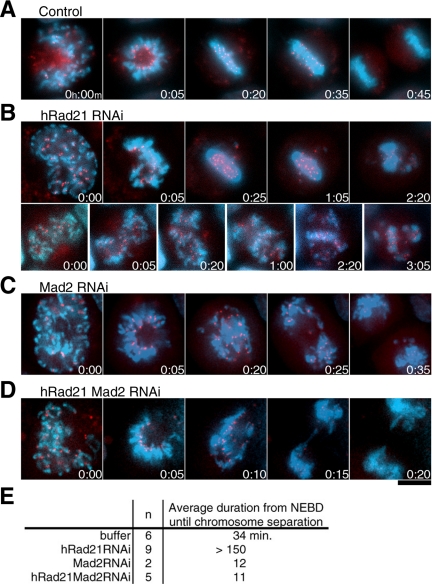
Mitotic Delay in hRad21 RNAi Cells Is Mad2-dependent
To unquestionably establish that the mitotic delay observed upon hRad21 depletion was due to the restraint of mitotic progression by spindle checkpoint, we used multiple RNAi probes to simultaneously deplete multiple proteins. The depletion of all combinations of proteins by multiple RNAi was confirmed by immunoblotting. The RNAi efficiencies were >95% for topo II, >93% for hRad21, and >84% for Mad2. Depletions were done in a HeLa cell line that stably expressed GFP-tagged hMis12 (Goshima et al., 2003; Obuse et al., 2004) in combination with Hoechst 33342 in order to monitor centromeres alongside bulk chromosomal DNA. Movies were taken for control non-RNAi treatment and the combination of hRad21 and Mad2 RNAi (time-lapse micrographs in Figure 4, A–D, and movies in Supplement 4, A–E). The numbers in the micrographs indicate the passage of time (h:min). The average duration for the time from NEBD to the onset of anaphase for control non-RNAi cells, RNAi cells of hRad21, Mad2, and double hRad21 Mad2 are indicated in Figure 4E. A comparison of the results from double hRad21 Mad2 RNAi with single hRad21 RNAi (Supplement 4, B and C) and control non-RNAi (the average, 34 min; Supplement 4A) unequivocally establishes a great shortening in the duration of the period before anaphase chromosome segregation (the average, 11 min: Supplement 4E) in the double hRad21 Mad2 RNAi. For single Mad2 RNAi, the average duration was 12 min (Supplement 4D). Thus, the mitotic delay in hRad21 RNAi was overridden by depletion of the spindle checkpoint protein Mad2. The GFP-hMis12 signals and the bulk DNAs were fully segregated in the double RNAi, albeit along with the occasional presence of lagging chromosomes. Consistently, knockdown of hRad21 and BubR1 compromised the accumulation of cells showing EMCs to produce micronuclei as seen in BubR1 RNAi cells (unpublished data). These data establish that the activation of the spindle checkpoint, rather than any physical restraint, accounts for the delay in commitment to anaphase in hRad21 RNAi cells.
Figure 4.
Sister kinetochore movements are under the Mad2 surveillance. (A–D) A number of movies were taken, and example micrographs are shown. Red, hMis12-GFP; blue, DNA. Elapsed time was shown as h:min. Bar, 10 μm. (A) Control cell. (B) hRad21 RNAi cells. Top, cell displaying a few EMCs. Bottom, cell showing abundant EMCs. No anaphase-like centromere segregation was observed. (C) Mad2 RNAi cell. (D) hRad21 Mad2 double RNAi cell. (E) The average duration (min) from NEBD to the onset of anaphase.
Sister Kinetochores in hRad21 RNAi Cells Experience Tension
The behavior of mitotic kinetochores was closely examined in movies of hRad21 RNAi cells. The intensity of GFP-hMis12 centromere/kinetochore protein (red) highlighted the degree of mitotic progression as it was intense in metaphase but diminished after anaphase (Figure 4A and Supplement 4 movies; T. Kiyomitsu, unpublished result). Two types of kinetochore behavior were noticed. In one type, a few extensively misaligned hMis12 signals were dynamically observed away from the bulk of the DNA, predominantly one to three such signals were seen at any one time (Supplement 4B). The second type had numerous extensively misaligned moving signals (Supplement 4C). Mitotic progression was delayed in both types.
In hRad21 RNAi cells, sister kinetochores visualized by GFP-hMis12 (red) were apparently under a pulling tension that was neither concerted nor bidirectional and did not appear to be sustained or strong enough to induce anaphase segregation of chromatids. An example of such temporal kinetochore “pulling” (indicated by the arrowhead in Figure 5B), seen in all the movies, is represented in the series of movie images (1-min intervals) with the control anaphase movement of hMis12 signals in non-RNAi cells (Figure 5A). Chromosomal arm DNAs (blue) were also pulled out and moved back simultaneously with the kinetochore signals, indicating that the associated arm DNAs were also temporarily pulled out (2–5 μm) from the bulk of chromosome DNA by the force of the spindle.
Figure 5.
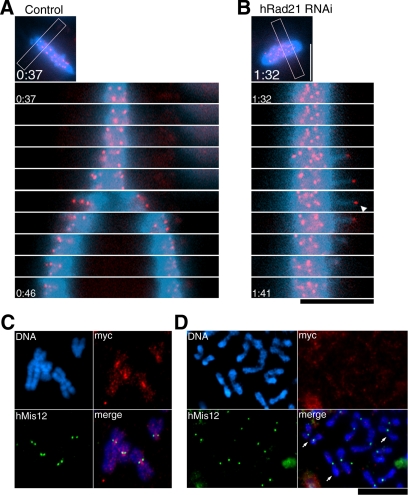
Preseparated centromere signals are under the tension but do not contain detectable hRad21. (A and B) Movie images taken for control non-RNAi (A) and hRad21 RNAi (B) cells are selected for the same chromosome area (indicated with the frame in the small top micrographs) and arranged in a time-lapse order (1-min intervals). GFP-hMis12 signals (red) and chromosome DNA (blue). Elapsed time was shown as h:min. The arrowhead indicates a sister centromere signal. (C and D) Chromosome spread of control non-RNAi (C) and hRad21 RNAi (D). Blue, DNA; red, Myc-tagged hRad21; green, hMis12. Arrows, three remotely paired chromatids. Spread and staining conditions were aimed to maintain the intact structure. Bars, 10 μm.
hRad21 Signals in the Inner Centromeres Are Abolished by RNAi Treatment
We next asked whether residual levels of hRad21 that might physically block chromosome segregation persisted after hRad21 RNAi. To address this question, hRad21-myc8, which permits the visualization of hRad21 in mitotic chromosome (Waizenegger et al., 2000), was expressed by transfection. Resulting HeLa cells were cultured for 24 h, followed by mitotic arrest through nocodazole treatment. A chromosome-spreading procedure that maintained the native chromosome structure was then done. Consistent with a previous report (Hauf et al., 2001), intense hRad21-myc8 signals were seen in the constricted inner centromere regions of metaphase chromosomes, between two hMis12 centromere signals (green, Figure 5C) in non-RNAi controls. In contrast, in hRad21 RNAi cells, the signals of hRad21-myc were not found in the centromeric regions or along the spread of sister chromatids (Figure 5D). In certain spreads, paired chromatids could be seen (indicated by arrows), although they were quite apart and not linked at the centromere regions. Some residual cohesin below the levels of detection might be responsible for such long distance links.
Sister Centromeres Align Correctly in Topo II hRad21 RNAi Cells
RNAi of topo II and topo II hRad21 was done to examine the relationship between topo II and hRad21 in chromosome segregation. As the human genome has two functionally redundant topo II genes (α and β) double RNAi was done (Sakaguchi and Kikuchi, 2004). Both topo IIα and hRad21 were efficiently depleted 48 h after siRNA transfection (Figure 6A). As reported previously, severe phenotypes were observed only when both topo II isotypes were depleted (Sakaguchi and Kikuchi, 2004).
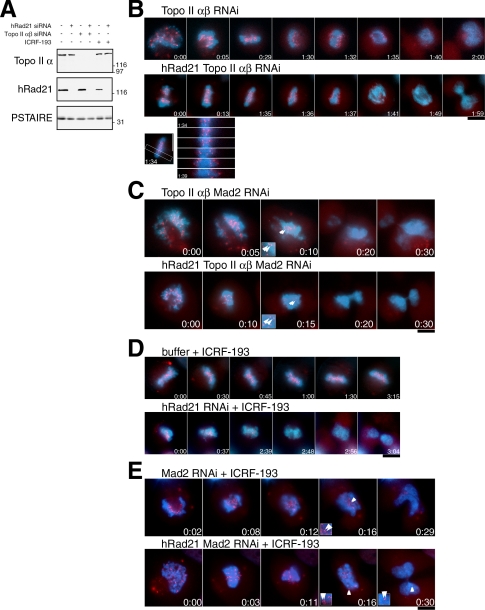
Figure 6.
Defect in topo II induces the Mad2-dependent delay, abolishes EMCs, and causes the tension. (A) Immunoblot of HeLa cell extracts with or without RNAi for topo II α,β and/or hRad21 using antibodies against topo IIα and hRad21. PSTAIRE antibody against Cdc2 was used as a loading control. (B) Time-lapse images of cells (red, hMis12; blue, DNA) after the topo IIα,β double RNAi (top) and the topo IIα,β hRad21 triple RNAi (middle). Time-lapse images for the concerted centromere segregation in the triple RNAi (bottom). (C) Time-lapse images of cells after the topo IIα,β Mad2 triple RNAi (top) and the topo IIα,β hRad21 Mad2 quadruple RNAi (bottom). Enlarged insets were shown to indicate apparent sister separation in comparison with persistent pairing shown in insets in E. Arrowheads, the GFP-hMis12 signals that were considered as sister kinetochores. (D) Movie images of cells treated with the ICRF-193 without or with knockdown of hRad21 (top and bottom, respectively). (E) Time-lapse images of Mad2 single (top) or hRad21 Mad2 double (bottom) RNAi cells treated with ICRF-193. Note that in both cases the pairing of sister kinetochores persisted when the GFP-hMis12 signals started to diminish as shown in insets. Bars, 10 μm.
Analysis of movies revealed a considerable mitotic delay in both double topo II RNAi (Figure 6B, top) and triple topo II hRad21 RNAi (middle) cells with a certain variability in the duration from NEBD till anaphase. The average durations were 87 and 108 min, respectively. Another remarkable result was revealed by signals from the centromere protein GFP-hMis12 signals (red) that apparently showed correct centromere alignment after the delay. Concerted sister centromere segregation occurred, although the bulk of chromosome DNA remained associated in a single mass (movies in Supplement 5, A and B). In the triple hRad21 topo II RNAi, the mode of sister centromere segregation after the delay and alignment was highly reminiscent of the onset of anaphase in non-RNAi control (Figure 6B, bottom). However, undivided chromosomes were either cut by cytokinesis or left in one of the two daughter cells. Our movie analyses confirmed the previous reports that the human double topo II α and β RNAi phenotypes resembled the cut phenotype of fission yeast top2 mutants (Sakaguchi and Kikuchi, 2004). The other striking result was that the EMCs that predominated in hRad21 RNAi were abolished by the additional depletion of topo II in both double and triple RNAi cells, suggesting that EMCs in hRad21 RNAi were produced through topo II activity. Thus, the depletion of topo II overrode the phenotypes of hRad21 knockdown.
The Delayed Mitosis in topo II RNAi Cells Is Dependent on Mad2
We examined whether the mitotic delay in the above double and triple RNAi depletions was Mad2-dependent. When Mad2 and topo II were simultaneously knocked down, the duration of the period from NEBD to diminution of GFP-hMis12 was very short (average, 13 min; Figure 6C, top; Supplement 5C) and was immediately followed by cytokinesis. However, the bulk DNA again remained associated as a single mass in any topo II-depleted RNAi experiment, thus topo II was needed for the physical separation of the bulk of the DNA. A similar phenotype was observed in the quadruple hRad21 topo II Mad2 RNAi (Figure 6C bottom, Supplement 5D); the period up to diminution of GFP-hMis12 was abbreviated (the average, 15 min) and sister centromeres separated to a certain degree, yet the bulk of DNA remained associated as cells underwent cytokinesis (Figure 6C, insets). Therefore, after the Mad2-dependent delay was compromised, some extent of chromatid separation was observed at centromeric regions but not along chromosome arms in both topo II and hRad21 topo II depletions. DNA links such as intertwining or catenation might remain in the arm region of topo II RNAi cells. We found that Topo II BubR1 multiple depletions produced the phenotype similar to that of Topo II Mad2 RNAi (unpublished data).
Topo II Inhibitor Drug Overrides hRad21 Depletion Phenotype and Blocks Every Aspect of Sister Chromatid Separation
ICRF-193 (a topo II inhibitor; Ishimi et al., 1992) was added to hRad21 knockdown cells, and movies taken were analyzed. It was previously shown that ICRF-193 acts on topo II, but not on DNA, and that it inhibits the change in the interaction between topo II and DNA (Roca et al., 1994). ICRF-193 was added 30 and 120 min before live analysis and paraformaldehyde fixation, respectively. The chromosome alignment was similarly observed whether the drug was administered on its own or in combination with hRad21 RNAi (Figure 6D, top and bottom; Supplement 5, E and F). The phenotypes were similar to those described for chicken DT-40 scc1 mutant in the presence of topo II inhibitor (Vagnarelli et al., 2004). However, the phenotypes were distinct from those of topo II RNAi; sister centromeres did not separate after a mitotic delay was finally compromised (the average duration was >180 min; Figure 6E, insets; also see Figure 6C, insets). Such topo II inhibitor-induced mitotic delay in mammalian cells was previously described (Haraguchi et al., 1997; Mikhailov et al., 2002; Skoufias et al., 2004). Consistent with the instigation of a mitotic delay, intense signals of securin and cyclin B1 were observed in metaphase-like cells that were depleted of topo II or treated with ICRF in either a non-RNAi or the hRad21 RNAi background (>98 and >93%, respectively). In contrast, cyclin B1 and securin were not observed in cells showing the cut phenotype (unpublished data). The duration of the time from NEBD until diminution of GFP-hMis12 obtained for a number of specimens is shown in Table 1.
Table 1.
The average duration of mitosis in cells defective in topo II and/or cohesin
| Type of treatment | No. of cells observed | Average duration (min)a |
|---|---|---|
| Non-RNAi control | 6 | 34 |
| Topo II RNAi | 11 | 87 |
| hRad21 Topo II RNAi | 14 | 108 |
| Topo II Mad2 RNAi | 3 | 13 |
| hRad21 Topo II Mad2 RNAi | 9 | 15 |
| Buffer + ICRF-193 | 13 | >180 |
| hRad21 RNAi + ICRF-193 | 11 | >175 |
| Mad2 RNAi + ICRF-193 | 2 | 12 |
| hRad21 Mad2 RNAi + ICRF-193 | 6 | 15 |
The average duration from NEBD until the onset of centromere separation or diminution of GFP-hMis12 signals is shown.
In ICRF-193-treated cells, centromeres were paired but their alignment was disordered, differing from the normal ladder-like figures. This suggested that the drug-inhibited topo II might interfere with the congression of centromeres to the metaphase plate. In hRad21 RNAi plus the drug, the paired signals were often pulled-out from the plate but such excursions were not seen if cells had been subjected to drug treatment alone. This kind of movement, however, never led to anaphase segregation (see Supplement movie 5F), suggesting that depletion of hRad21 loosened the architecture of pericentric and associated arm chromatin, but the paired sister centromeres remained tightly bound.
ICRF-193 blocked sister centromere separation and cytokinesis ensued after a considerable delay. We asked whether these phenotypes were dependent on Mad2. As shown in Figure 6E, the duration from NEBD to diminution of GFP-hMis12 signals (corresponding to anaphase) was very short (∼20 min; Supplement 5, G and H), and was again followed by cytokinesis. Unlike topo II Mad2 RNAi (Figure 6C), there was no sign of sister chromatid separation at all (see insets of Figure 6E), and cytokinesis initiated but often failed. The drug-inhibited topo II thus blocked the every aspect of sister chromosome separation irrespective of the presence of hRad21.
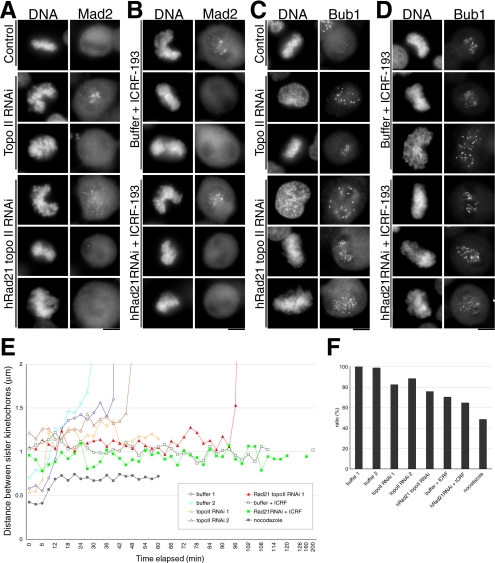
The kinetochore localization of Mad2 and Bub1 was monitored in order to observe the mode of spindle checkpoint activation. The Mad2 signals disappeared (Figure 7, A and B), while Bub1 was quite intense (Figure 7, C and D) in delayed topo II depleted or inhibited cells. Cells defective in both topo II and hRad21 revealed more intense Bub1 signals than cells in which topo II alone was defective. The association between kinetochores and microtubules might be impaired by topo II defects and this impairment appeared to be enhanced by hRad21 RNAi. Topological defects in topo II–depleted or –inhibited cells probably led to structural alterations in the centromere/kinetochore, as reported previously (Rattner et al., 1996). Topo II inhibition caused a disorganization of the metaphase plate and made it difficult to assign sister kinetochores (see Supplement 5, E and F). However, in those instances where we could assign paired kinetochores the distances between sister kinetochores decreased significantly (∼20–30%) in topo II–depleted or –inhibited cells during the delayed metaphase (Figure 7, E and F). This decrease in these distances was confirmed in fixed preparations (unpublished data). Thus, functional topo II might be required for the structure and the function of centromere.
Figure 7.
The mode of microtubule–kinetochore association in topo II–defective cells. (A–D) Kinetochore localization of Mad2 and Bub1 in topo II–deficient cells. Cells treated as indicated were fixed with paraformaldehyde and immunostained by anti-Mad2 (A and B) or anti-Bub1 (C and D) antibodies. Transfected cells were harvested after 48 h with or without an additional incubation for 2 h in the presence of ICRF-193. In each treatment, Mad2 and Bub1 were confirmed to localize in prometaphase or prophase chromosomes, respectively. Bar, 10 μm. (E) Based on movie data, the distances between discernible sister kinetochores were measured and plotted on the graph. Data obtained from nocodazole-treated cells served as the control where no spindle and tension existed. (F) The average distances between discernible sister kinetochores during the metaphase-like stage are shown. The value of buffer-transfected control (buffer 1; 1.45 μm) was set to 100%.
DISCUSSION
Up to anaphase cohesin plays a critical role to hold sister chromatids in metaphase and the loss of its subunit Rad21/Scc1 in anaphase by cleavage then allows chromosome segregation. Although the regulation of topo II during mitosis is barely understood, it may be activated in order to remove topological links (catenation, intertwining) between chromatids during anaphase. Cohesion and catenation may be viewed in a similar light with regard to their formation during DNA replication and dissolution before chromosome segregation. Through the live analyses of human centromere protein hMis12 and arm DNAs we have investigated how hRad21 and topo II are functionally interrelated in metaphase chromosome alignment in HeLa cells, in generating the opposing force that resists the pulling force of the spindle. GFP-hMis12 proves to be an excellent indicator of mitotic progression as well as kinetochores, as bright kinetochore signals diminish after anaphase. RNAi and ICRF-193, respectively, were the tools used for knocking down cohesin and topo II, and inhibiting topo II. We will discuss below the functional relationship between topo II and cohesin in the light of our results and those reported previously for sister chromatid defects in fungi, fly, frog, chicken, and human cells (Guacci et al., 1997; Michaelis et al., 1997; Tomonaga et al., 2000; Sonoda et al., 2001; Hoque and Ishikawa, 2002; Vass et al., 2003; Losada et al., 2005) and topo II (Uemura et al., 1986, 1987; Rattner et al., 1996; Mikhailov et al., 2002; Dewar et al., 2004; Sakaguchi and Kikuchi, 2004; Skoufias et al., 2004; Vagnarelli et al., 2004).
An illustration summarizing present results is shown in Figure 8. Defects in either or both cohesin and topo II activate the Mad2-dependent spindle checkpoint in HeLa cells (indicated by the eye). This is the first full systematic analyses of the dependency between the spindle checkpoint and cohesin and topo II functions. In normal mitoses (A), the presence of cohesion and topological links between chromatids ensures that chromosomes at metaphase promote the Mad2-dependent inhibition of APC/C. After knockdown of cohesin (B), anaphase never occurs in most RNAi cells, as chromosomes are not properly aligned and so activate the spindle checkpoint to restrain anaphase (e.g., a “wait” signal is generated). Ever since the mal-oriented chromosomes were reoriented by spindle fiber tension (Nicklas and Koch, 1969), tension has been considered an important factor in regulating element for controlling the activity of the spindle checkpoint (Stern and Murray, 2001; Indjeian et al., 2005). Although the actual level of tension was not measured, the quasi-metaphase phenotype produced by hRad21 knockdown may be explained by the diminished tension due to preseparation of sister chromatids. This notion is consistent with intense Bub1 signals (but not Mad2) at kinetochores in the delayed, hRad21-knockdown cells. Other less mechanical explanations are that Bub1 may monitor improper microtubule-kinetochore interactions and/or structural alterations of centromeres in hRad21 RNAi cells.
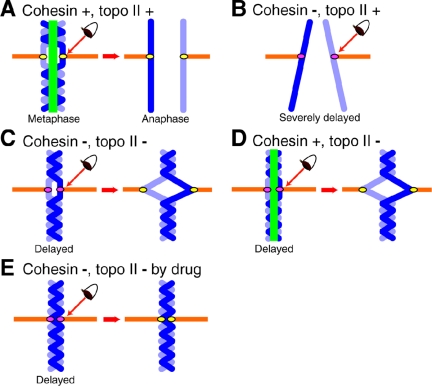
Figure 8.
A model: how cohesin and topo II are implicated in metaphase chromosome alignment. A model illustrating how cohesin and topo II contribute to chromosome alignment in metaphase. (A) Metaphase and anaphase chromosomes of non-RNAi HeLa cells. (B) Preseparated chromosomes in hRad21RNAi. (C) hRad21 Topo II RNAi cells. (D) Topo II RNAi cells. (E) hRad21 RNAi cells treated with ICRF-193. Chromosome, blue; the link regulated by cohesin and topo II, green; spindle, orange. The eye represents surveillance by Mad2-dependent spindle checkpoint control. Kinetochores colored in pink indicate activation of spindle checkpoint, whereas the inactivated checkpoint is indicated by yellow kinetochores. Spindle checkpoint is activated in all the cases except for control non-RNAi cells.
In cohesin- and topo II-knockdown cells (C), Mad2 is localized normally on prometaphase kinetochores, but disappeared from kinetochores in the mitotically delayed cells, after which sister centromeres separated aberrantly. The disappearance of Mad2 can be explained by the fact that the Mad2-dependent delay continues in metaphase, whereas Mad2 localization on kinetochores is abolished (Martin-Lluesma et al., 2002). Kinetochore bound Mad2 was recently reported to activate other Mad2 molecules and so propagate the checkpoint signals (DeAntoni et al., 2005). Although cohesin-knockdown leads to virtually permanent mitotic delay accompanied with chromosome misalignment, cohesin- and topo II-knockdown leads to the Mad2-dependent mitotic delay with enhanced Bub1 kinetochore signals and sister centromere alignment. In addition, the average distance between sister centromeres was shorter than that in normal (buffer control) metaphase chromosomes but longer than in nocodazole-treated chromosomes. In topo II-knockdown (D), similar phenotypes are observed with a shorter delay. In ICRF-193 inhibited cells (E), the delay was virtually permanent, the distance between sister centromeres was rather short (only slightly longer than that in nocodazole-treated chromosome) and sister centromeres never showed any separation.
Our immediate interpretation of these findings is that tensions exerted on metaphase centromeres are reduced in these hRad21- and/or topo II–disrupted cells, and that accumulation or retention of Bub1 on kinetochores by topo II disruption may sustain or engender a Mad2-dependent delay in anaphase entry. As these are the principal novel observations in this study, discussions are first focused on these points. These findings strengthen the connection between topo II and centromere/kinetochore function, although the underlying mechanism remains to be elucidated. The Mad2-dependent delay in topo II disruption is unambiguously established by staining the level of securin in the delayed cells. Topo II disruption- and Mad2-dependent spindle checkpoint activation is not universal phenomenon, however. In fission and budding yeast cells, top2 mutants show no delay in mitosis (Nabeshima et al., 1998; Bhalla et al., 2002), whereas cohesin defects cause the spindle checkpoint activation like that in higher eukaryotic cells (Biggins and Murray, 2001; Toyoda et al., 2002). In yeast top2 mutants, the kinetochore may be normal or fail to transmit the spindle checkpoint signal even though its structure is topologically impaired.
The Earnshaw group (Vagnarelli et al., 2004) showed that treatment with a topo II–inhibiting drug reduced BubR1 staining in scc1/rad21 chicken mutant kinetochores. This is consistent with topo II perturbation correcting an underlying tension defect. In contrast, the present study showed that treatment of human hRad21 RNAi cells with ICRF-193 or joint hRad21 topo II RNAi knockdown actually increased Bub1 kinetochore staining. Despite this, topo II disruption corrected the chromosome alignment defect in hRad21 RNAi and reduced the average time for anaphase onset in Rad21 knockdown cells to 108 from >150 min. These observations seem to be somewhat paradoxical with regard to BubR1 and Bub1 staining, but we have no straightforward explanation. There is already a quite significant difference between the phenotypes of human hRad21 RNAi and the chicken DT-40 rad21 mutant as Mad2 signals are intensely localized on misaligned kinetochores under chicken scc1/rad21-depletion (Sonoda et al., 2001). We never found such intense Mad2-signals in the delayed HeLa cells. These differences may be due to distinctions between the organisms, cell types or the means by which disruption of function was achieved.
Then, how do disruptions of topo II really correct the alignment while Bub1 signals remain on kinetochores? We suggest that centromeric regions experience a lack of tension following topo II disruption, possibly due to a perturbation of kinetochore function that prevents centric regions from being pulled apart. If this is the case, how can one envision that the combined reductions in tension associated with hRad21 and topo II disruption synergize to ameliorate the chromosome alignment defect of Rad21-defective cells? We presume that the combined disruptions of topo II and hRad21 somehow facilitate the alignment of sister centromeres that are under a weak tension over prolonged periods, leading to the inactivation of Mad2. In single hRad21 RNAi cells, sister chromatids are preseparated and never aligned, whereas further topo II disruption produces catenations (intertwining) in the chromosome arm regions, which steadily accumulate the opposing tension, to a point that is sufficient to trigger the inactivation of Mad2. An alternative possibility is that centric regions in topo II–disrupted cells (especially after ICRF-193 treatment) are topologically linked to a point where they cannot be simply drawn far apart by poleward spindle forces. This type of tension problem may be monitored by checkpoint mechanisms, and Bub1 may accumulate on kinetochores. In any case, we presume that misaligned chromosome arising from hRad21 disruption is corrected by topological linking that is generated by disruption of topo II.
The mitotic delay that is induced by disruption of cohesin and/or topo II is abolished by the simultaneous ablation of Mad2. In topo II disruption, the arm DNAs are not separated even after Mad2 depletion, but centric regions do separate once the delay has been overcome. Thus, topo II seems to be differently engaged in centromeres and arms. Consistently, a similar situation is seen with the genomic centromere portions in S. pombe top2 mutant (Uemura et al., 1986; Funabiki et al., 1993). Besides, in budding yeast topo II mutant cells, minichromosomes containing CEN regions are normally segregated (Koshland and Hartwell, 1987). Hence, only the arm portions may be topologically linked upon topo II disruption so that the CEN containing circular plasmids have no restraint for full segregation. This hypothesis may explain the apparent lack of recombination in the centromere regions in human and fission yeast (Nakaseko et al., 1986; Puechberty et al., 1999). Alternatively, topological links present in the centric regions of linear chromosomes may slide away by the pulling force, resulting in centromeric separation in topo II disruption. After ICRF-193 treatment, topological links may become so abundant that sliding hardly occurred, leading to the persistent connection between sister kinetochores.
The ICRF-193–mediated mitotic checkpoint delay in mammalian cells was reported previously with somewhat different results and interpretations. In Mikhailov et al. (2002); DNA damage induced by the drug addition during early mitosis was presumed to cause the delay of anaphase onset via Mad2-dependent spindle checkpoint. Mad2 remained at the kinetochore in the delayed cells. In Skoufias et al. (2004), the Mad2 signal was not found in kinetochore of the delayed cells that were added by the drug upon the release from the nocodazole arrest. Catenation was hypothesized to cause the metaphase arrest, with a surveillance mechanism differing from the regular spindle checkpoint. Our data were obtained for mitotic cells previously treated by ICRF-193, 30 min before the entry into mitosis. Our ICRF-193 results that could not be directly compared with those of two previous reports showed that Mad2 disappeared, whereas Bub1 remained at the centromere, and that the pairs of sister kinetochores in the metaphase plate were not ladder-like: the arrangement of metaphase kinetochore was disorganized. Further studies are definitively needed to resolve these apparently disparate results on the binding mode of spindle checkpoint proteins. Our results and Mikhailov et al. (2002) suggested the centromere/kinetochore role of topo II, which should be focused in the future investigation.
We show that the distance between sister kinetochores in ICRF-193–treated cells is shorter than that of normal metaphase. The intense Bub1 but no Mad2 signals were found on aligned kinetochores. These results may be explained by assuming that the spindle tension exerted on the kinetochore was weak in ICRF-193–treated cells, as the role of topo II in centromere/kinetochore was inhibited from interphase (late G2). We do not consider any novel checkpoint control system but it is of considerable interest if the topological defect could cause the perturbed arrangement of sister kinetochores, which reduced the tension. In Skoufias et al. (2004), ICRF-193 was added in prometaphase immediately after the release from nocodazole so that the centromere/kinetochore role of topo II might not become obvious, and the distances between sister kinetochores were the same as nondrug treatment control. The tension exerted on the kinetochore was normal like metaphase cells untreated with ICRF-193.
The ring model, where cohesin forms a gigantic ring that embraces the duplicated chromosomes within it, was proposed and has been recently discussed in several reports (Hirano, 2005; Huang et al., 2005; Nasmyth, 2005). The model may explain the presence of opposing force in metaphase, and the loss of the ring upon the cleavage of cohesin leads to sister chromatid separation and would enable chromosome segregation by the forces of the spindle. The present result is not entirely harmonious with the ring model: although sister chromatids are indeed preseparated by the depletion of the cohesin subunit Rad21, chromosome segregation is restrained due to the block by activation of the Mad2-dependent spindle checkpoint. Some other factor such as the inactivation of Mad2 is needed to trigger chromosome segregation.
As observed in fungal rad21/scc1 mutants, the present study shows that traverse of interphase after the depletion of hRad21 is required to produce mitotic defects. Indeed, the interphase higher order chromatin DNA is strikingly disordered in repetitive pericentromeric regions after hRad21 RNAi. This could be an important knockdown phenotype, as centric chromatin structures become aberrant in mitosis.
In conclusion, the phenotype of hRad21 knockdown can only be understood through the actions of topo II and the activation of spindle checkpoint. Topo II is required for the ladder-like centromere alignment in metaphase, which is monitored by checkpoint systems, and may be differently engaged in between the centromere and the arm. Functional topo II resolves the catenation and produces the preseparated chromatids in single hRad21 RNAi. Our results thus suggest that topo II and cohesin may equally contribute to regulate sister chromatid association. Further work is definitively needed to understand how cohesin and topo II coordinately function to keep or resolve sister chromatid link and how cohesion and the topological link between sister chromatids are monitored by spindle checkpoint.
Supplementary Material
Acknowledgments
We thank Dr. Takahiro Matsusaka for preparation of polyclonal antibodies against human Rad21 and Mis4/NIPBL, Dr. Katsuzumi Okumura for the FISH probes, Dr. Stephen S. Taylor for anti-Bub1 and anti-BubR1 antibodies, Dr. Kinya Yoda for anti-CENP-A and anti-CENP-C antibodies, Dr. Mari Shimura for an improved spread method to detect hRad21-myc, Dr. Toshiyuki Habu for suggestions on transfection, and Dr. Iain Hagan for valuable suggestions on the writing. We also acknowledge the Kazusa DNA Research Institute for cDNA of hRad21. We declare that none of the authors have a financial interest related to this work. This study was supported by a Specially Promoted Research Grant (Center of Excellence) from the Ministry of Education, Culture, Sports, Science, and Technology. Y. T. was the recipient of the special research fellowship of the Japan Science Promotion Society.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–11–1089) on March 1, 2006.
Abbreviations used: EMC, extensively misaligned chromosome.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Atienza, J. M., Roth, R. B., Rosette, C., Smylie, K. J., Kammerer, S., Rehbock, J., Ekblom, J., and Denissenko, M. F. (2005). Suppression of RAD21 gene expression decreases cell growth and enhances cytotoxicity of etoposide and bleomycin in human breast cancer cells. Mol. Cancer Ther. 4, 361–368. [DOI] [PubMed] [Google Scholar]
- Baldini, A., Rocchi, M., Archidiacono, N., Miller, O. J., and Miller, D. A. (1990). A human alpha satellite DNA subset specific for chromosome 12. Am. J. Hum. Genet. 46, 784–788. [PMC free article] [PubMed] [Google Scholar]
- Bhalla, N., Biggins, S., and Murray, A. W. (2002). Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell 13, 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., and Murray, A. W. (2001). The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15, 3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A. J., and Porter, A. C. (2004). Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIalpha mutant human cell line. Mol. Biol. Cell 15, 5700–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C. J., Goulding, S., Earnshaw, W. C., and Carmena, M. (2003). RNAi analysis reveals an unexpected role for topoisomerase II in chromosome arm congression to a metaphase plate. J. Cell Sci. 116, 4715–4726. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., Shirayama, M., Shevchenko, A., Tanaka, T., Toth, A., and Nasmyth, K. (2000). Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5, 243–254. [DOI] [PubMed] [Google Scholar]
- Cozzarelli, N. R., and Wang, J. C. (eds.) (1990). DNA topology and its biological effect. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- De Antoni, A. et al. (2005). The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15, 214–225. [DOI] [PubMed] [Google Scholar]
- Dewar, H., Tanaka, K., Nasmyth, K., and Tanaka, T. U. (2004). Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature 428, 93–97. [DOI] [PubMed] [Google Scholar]
- Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Funabiki, H., Hagan, I., Uzawa, S., and Yanagida, M. (1993). Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121, 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya, K., Takahashi, K., and Yanagida, M. (1998). Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in the S phase and not destroyed in the G1 phase. Genes Dev. 12, 3408–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, P. J., and Hirano, T. (2004). Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr. Biol. 14, 1598–1603. [DOI] [PubMed] [Google Scholar]
- Goshima, G., Kiyomitsu, T., Yoda, K., and Yanagida, M. (2003). Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160, 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci, V., Koshland, D., and Strunnikov, A. (1997). A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi, T., Kaneda, T., and Hiraoka, Y. (1997). Dynamics of chromosomes and microtubules visualized by multiple-wavelength fluorescence imaging in living mammalian cells: effects of mitotic inhibitors on cell cycle progression. Genes Cells 2, 369–380. [DOI] [PubMed] [Google Scholar]
- Hauf, S., Waizenegger, I. C., and Peters, J. M. (2001). Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293, 1320–1323. [DOI] [PubMed] [Google Scholar]
- Hirano, T. (2005). SMC proteins and chromosome mechanics: from bacteria to humans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, T., and Mitchison, T. J. (1993). Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J. Cell Biol. 120, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, C., Goto, T., Wang, J. C., and Botstein, D. (1985). DNA topoisomerase II is required at the time of mitosis in yeast. Cell 41, 553–563. [DOI] [PubMed] [Google Scholar]
- Hoque, M. T., and Ishikawa, F. (2002). Cohesin defects lead to premature sister chromatid separation, kinetochore dysfunction, and spindle-assembly checkpoint activation. J. Biol. Chem. 277, 42306–42314. [DOI] [PubMed] [Google Scholar]
- Huang, C. E., Milutinovich, M., and Koshland, D. (2005). Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian, V. B., Stern, B. M., and Murray, A. W. (2005). The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science 307, 130–133. [DOI] [PubMed] [Google Scholar]
- Ishimi, Y., Ishida, R., and Andoh, T. (1992). Effect of ICRF-193, a novel DNA topoisomerase II inhibitor, on simian virus 40 DNA and chromosome replication in vitro. Mol. Cell. Biol. 12, 4007–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski, S. A., Chan, G. K., Cooke, C. A., Earnshaw, W. C., and Yen, T. J. (1998). The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma 107, 386–396. [DOI] [PubMed] [Google Scholar]
- Kanda, T., Sullivan, K. F., and Wahl, G. M. (1998). Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8, 377–385. [DOI] [PubMed] [Google Scholar]
- Kaur, M., Descipio, C., McCallum, J., Yaeger, D., Devoto, M., Jackson, L. G., Spinner, N. B., and Krantz, I. D. (2005). Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am. J. Med. Genet. A 138, 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland, D., and Hartwell, L. H. (1987). The structure of sister minichromosome DNA before anaphase in Saccharomyces cerevisiae. Science 238, 1713–1716. [DOI] [PubMed] [Google Scholar]
- Krantz, I. D. et al. (2004). Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet. 36, 631–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., and Benezra, R. (1996). Identification of a human mitotic checkpoint gene: hsMAD2. Science 274, 246–248. [DOI] [PubMed] [Google Scholar]
- Lichter, P., Cremer, T., Borden, J., Manuelidis, L., and Ward, D. C. (1988). Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum. Genet. 80, 224–234. [DOI] [PubMed] [Google Scholar]
- Losada, A., Hirano, M., and Hirano, T. (1998). Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 12, 1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada, A., Yokochi, T., and Hirano, T. (2005). Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J. Cell Sci. 118, 2133–2141. [DOI] [PubMed] [Google Scholar]
- Mao, Y., Abrieu, A., and Cleveland, D. W. (2003). Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell 114, 87–98. [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma, S., Stucke, V. M., and Nigg, E. A. (2002). Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297, 2267–2270. [DOI] [PubMed] [Google Scholar]
- Michaelis, C., Ciosk, R., and Nasmyth, K. (1997). Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91, 35–45. [DOI] [PubMed] [Google Scholar]
- Mikhailov, A., Cole, R. W., and Rieder, C. L. (2002). DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 12, 1797–1806. [DOI] [PubMed] [Google Scholar]
- Nabeshima, K., Nakagawa, T., Straight, A. F., Murray, A., Chikashige, Y., Yamashita, Y. M., Hiraoka, Y., and Yanagida, M. (1998). Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell 9, 3211–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko, Y., Adachi, Y., Funahashi, S., Niwa, O., and Yanagida, M. (1986). Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 5, 1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth, K. (2002). Segregating sister genomes: the molecular biology of chromosome separation. Science 297, 559–565. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. (2005). How might cohesin hold sister chromatids together? Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth, K., and Haering, C. H. (2005). The structure and function of smc and kleisin complexes. Annu. Rev. Biochem. 74, 595–648. [DOI] [PubMed] [Google Scholar]
- Nicklas, R. B., and Koch, C. A. (1969). Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J. Cell Biol. 43, 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogami, M., Nogami, O., Kagotani, K., Okumura, M., Taguchi, H., Ikemura, T., and Okumura, K. (2000). Intranuclear arrangement of human chromosome 12 correlates to large-scale replication domains. Chromosoma 108, 514–522. [DOI] [PubMed] [Google Scholar]
- Obuse, C., Iwasaki, O., Kiyomitsu, T., Goshima, G., Toyoda, Y., and Yanagida, M. (2004). A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 6, 1135–1141. [DOI] [PubMed] [Google Scholar]
- Ohi, R., Coughlin, M. L., Lane, W. S., and Mitchison, T. J. (2003). An inner centromere protein that stimulates the microtubule depolymerizing activity of a KinI kinesin. Dev. Cell 5, 309–321. [DOI] [PubMed] [Google Scholar]
- Puechberty, J. et al. (1999). Genetic and physical analyses of the centromeric and pericentromeric regions of human chromosome 5, recombination across 5cen. Genomics 56, 274–287. [DOI] [PubMed] [Google Scholar]
- Rattner, J. B., Hendzel, M. J., Furbee, C. S., Muller, M. T., and Bazett-Jones, D. P. (1996). Topoisomerase II alpha is associated with the mammalian centromere in a cell cycle- and species-specific manner and is required for proper centromere/kinetochore structure. J. Cell Biol. 134, 1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C. L., and Salmon, E. D. (1998). The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8, 310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca, J., Ishida, R., Berger, J. M., Andoh, T., and Wang, J. C. (1994). Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc. Natl. Acad. Sci. USA 91, 1781–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, H., Tomkiel, J., Cooke, C. A., Ratrie, H., 3rd, Maurer, M., Rothfield, N. F., and Earnshaw, W. C. (1992). CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell 70, 115–125. [DOI] [PubMed] [Google Scholar]
- Sakaguchi, A., and Kikuchi, A. (2004). Functional compatibility between isoform alpha and beta of type II DNA topoisomerase. J. Cell Sci. 117, 1047–1054. [DOI] [PubMed] [Google Scholar]
- Skoufias, D. A., Andreassen, P. R., Lacroix, F. B., Wilson, L., and Margolis, R. L. (2001). Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc. Natl. Acad. Sci. USA 98, 4492–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias, D. A., Lacroix, F. B., Andreassen, P. R., Wilson, L., and Margolis, R. L. (2004). Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol. Cell 15, 977–990. [DOI] [PubMed] [Google Scholar]
- Sonoda, E. et al. (2001). Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell 1, 759–770. [DOI] [PubMed] [Google Scholar]
- Stern, B. M., and Murray, A. W. (2001). Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 11, 1462–1467. [DOI] [PubMed] [Google Scholar]
- Sumara, I., Vorlaufer, E., Gieffers, C., Peters, B. H., and Peters, J. M. (2000). Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 151, 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin, O., and Varshavsky, A. (1980). Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell 21, 103–114. [DOI] [PubMed] [Google Scholar]
- Takahashi, E., Koyama, K., Hitomi, A., and Nakamura, Y. (1993). A high-resolution cytogenetic map of human chromosome 12, localization of 195 new cosmid markers by direct R-banding fluorescence in situ hybridization. Hum. Genet. 92, 405–409. [DOI] [PubMed] [Google Scholar]
- Takahashi, T. S., Yiu, P., Chou, M. F., Gygi, S., and Walter, J. C. (2004). Recruitment of Xenopus Scc2 and cohesin to chromatin requires the prereplication complex. Nat. Cell Biol. 6, 991–996. [DOI] [PubMed] [Google Scholar]
- Taylor, S. S., Hussein, D., Wang, Y., Elderkin, S., and Morrow, C. J. (2001). Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 114, 4385–4395. [DOI] [PubMed] [Google Scholar]
- Tomonaga, T. et al. (2000). Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14, 2757–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin, E. T., Wang, T. J., Lisgo, S., Bamshad, M. J., and Strachan, T. (2004). NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 36, 636–641. [DOI] [PubMed] [Google Scholar]
- Toyoda, Y., Furuya, K., Goshima, G., Nagao, K., Takahashi, K., and Yanagida, M. (2002). Requirement of chromatid cohesion proteins rad21/scc1 and mis4/scc2 for normal spindle-kinetochore interaction in fission yeast. Curr. Biol. 12, 347–358. [DOI] [PubMed] [Google Scholar]
- Uemura, T., Ohkura, H., Adachi, Y., Morino, K., Shiozaki, K., and Yanagida, M. (1987). DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50, 917–925. [DOI] [PubMed] [Google Scholar]
- Uemura, T., and Yanagida, M. (1986). Mitotic spindle pulls but fails to separate chromosomes in type II DNA topoisomerase mutants: uncoordinated mitosis. EMBO J. 5, 1003–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli, P., Morrison, C., Dodson, H., Sonoda, E., Takeda, S., and Earnshaw, W. C. (2004). Analysis of Scc1-deficient cells defines a key metaphase role of vertebrate cohesin in linking sister kinetochores. EMBO Rep. 5, 167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass, S., Cotterill, S., Valdeolmillos, A. M., Barbero, J. L., Lin, E., Warren, W. D., and Heck, M. M. (2003). Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr. Biol. 13, 208–218. [DOI] [PubMed] [Google Scholar]
- Vermeesch, J. R., Duhamel, H., Raeymaekers, P., Van Zand, K., Verhasselt, P., Fryns, J. P., and Marynen, P. (2003). A physical map of the chromosome 12 centromere. Cytogenet. Genome Res. 103, 63–73. [DOI] [PubMed] [Google Scholar]
- Waizenegger, I. C., Hauf, S., Meinke, A., and Peters, J. M. (2000). Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103, 399–410. [DOI] [PubMed] [Google Scholar]
- Waters, J. C., Chen, R. H., Murray, A. W., and Salmon, E. D. (1998). Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 141, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida, M. (2005). Basic mechanism of eukaryotic chromosome segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.