Abstract
Regulated disassembly of actin filaments is involved in several cellular processes that require dynamic rearrangement of the actin cytoskeleton. Actin-interacting protein (AIP) 1 specifically enhances disassembly of actin-depolymerizing factor (ADF)/cofilin-bound actin filaments. In vitro, AIP1 actively disassembles filaments, caps barbed ends, and binds to the side of filaments. However, how AIP1 functions in the cellular actin cytoskeletal dynamics is not understood. We compared biochemical and in vivo activities of mutant UNC-78 proteins and found that impaired activity of mutant UNC-78 proteins to enhance disassembly of ADF/cofilin-bound actin filaments is associated with inability to regulate striated organization of actin filaments in muscle cells. Six functionally important residues are present in the N-terminal β-propeller, whereas one residue is located in the C-terminal β-propeller, suggesting the presence of two separate sites for interaction with ADF/cofilin and actin. In vitro, these mutant UNC-78 proteins exhibited variable alterations in actin disassembly and/or barbed end-capping activities, suggesting that both activities are important for its in vivo function. These results indicate that the actin-regulating activity of AIP1 in cooperation with ADF/cofilin is essential for its in vivo function to regulate actin filament organization in muscle cells.
INTRODUCTION
Dynamic reorganization of the actin cytoskeleton is required for a number of cellular processes, including cell motility, cytokinesis, and morphogenesis. Cellular actin dynamics are regulated by complex mechanisms involving several actin-binding proteins in a spatially and temporally regulated and tissue-specific manner. The central machinery of rapid actin turnover requires actin nucleation, filament disassembly, and capping barbed ends to limit the number of elongation sites (Cooper and Schafer, 2000; Carlier et al., 2003; Pollard and Borisy, 2003; Nicholson-Dykstra et al., 2005). In vitro, a minimum set of proteins that regulate these processes can reconstitute actin-based motility (Loisel et al., 1999). However, there are multiple kinds of actin nucleating, depolymerizing/severing, and capping proteins, which might be functionally different in vivo. Therefore, comparing in vitro and in vivo functions of actin-regulatory proteins will be very important to understand the mechanism of the regulation of actin cytoskeletal dynamics.
Actin-depolymerizing factor (ADF)/cofilin is one of the essential proteins that accelerate turnover of actin filaments by depolymerizing actin monomers from the pointed ends and severing filaments (Bamburg, 1999; Bamburg et al., 1999; Carlier et al., 1999; Maciver and Hussey, 2002; DesMarais et al., 2005). The activity of ADF/cofilin is regulated by several mechanisms, including phosphorylation/dephosphorylation, pH, phosphoinositides, and competition with other actin-binding proteins (DesMarais et al., 2005). Actin-interacting protein (AIP) 1 is a unique regulator of ADF/cofilin that enhances actin depolymerization in the presence of ADF/cofilin (Ono, 2003). AIP1 is a conserved WD-repeat protein (Amberg et al., 1995), and its involvement in the reorganization of the actin cytoskeleton has been demonstrated in several model systems. For example, AIP1 is required for actin-rich membrane protrusion (Rogers et al., 2003), myofibril assembly (Ono, 2001), and plant development (Ketelaar et al., 2004), and it is involved in endocytosis, cell movement, and cytokinesis (Konzok et al., 1999). AIP1 functionally interacts with ADF/cofilin in vivo as determined in yeast (Iida and Yahara, 1999; Rodal et al., 1999) and Caenorhabditis elegans (Ono, 2001).
In vitro, AIP1 enhances disassembly of ADF/cofilin-bound actin filaments (Aizawa et al., 1999; Okada et al., 1999; Rodal et al., 1999; Mohri and Ono, 2003). However, the mechanism of this action remains unclear because AIP1 exhibits complex biochemical activity. AIP1 caps the barbed ends of ADF/cofilin-bound actin filaments, and this activity has been proposed to promote disassembly by preventing elongation and reannealing (Okada et al., 2002; Balcer et al., 2003). However, direct microscopic observation of the AIP1 activity suggests that AIP1 actively severs or depolymerizes filaments (Ono et al., 2004). Crystal structures of yeast AIP1 (Voegtli et al., 2003) and C. elegans AIP1 (UNC-78) (Mohri et al., 2004) revealed 14 WD-modules that are arranged into two seven-bladed β-propeller domains. Mutagenesis of conserved surface residues identified five clustered residues in the N-terminal propeller domain that are important for the in vitro actin disassembly activity of AIP1 (Mohri et al., 2004). Furthermore, mutations of these residues did not affect the capping activity, suggesting that disassembly and capping are separable functions of AIP1 that can be uncoupled by point mutations. AIP1 also binds to the side of ADF/cofilin-decorated filaments (Okada et al., 2002). However, how these complex activities of AIP1 influence the in vivo actin dynamics are not understood.
Here, we performed a correlative study on in vivo and in vitro activities of various mutants of AIP1 by using C. elegans and found that the activity of AIP1 to enhance actin disassembly is required for organized assembly of actin filaments in striated muscle. The functional residues on C. elegans AIP1 identified in this study are conserved among AIP1 proteins in eukaryotes and may be commonly utilized for the actin-regulating activity of AIP1 in cooperation with ADF/cofilin.
MATERIALS AND METHODS
Nematode Strains and Culture
Wild-type C. elegans strain N2 was obtained from the Caenorhabditis Genetics Center (Minneapolis, MN). unc-78(gk27) (unc-78 null mutant) was provided by the C. elegans Reverse Genetics Core Facility at the University of British Columbia, Vancouver, British Columbia, Canada, and has been described previously (Ono, 2001). unc-78(su223) (Zengel and Epstein, 1980) was provided by Henry Epstein (University of Texas Medical Branch, Galveston, TX). unc-78(e1221) (Waterston et al., 1980) was provided by Pamela Hoppe (Western Michigan University, Kalamazoo, MI). Nematodes were grown at 20°C as described previously (Brenner, 1974).
Construction of Expression Vectors
The 1.4-kb upstream region of the unc-78 gene was amplified by PCR by using Pfx DNA polymerase (Invitrogen, Carlsbad, CA) with 5′-GATCCCGCGGCCTCTTATCAAGCGAAGAACTCG as a forward primer and 5′-GATCGGTACCGTTTGAGAAAATTCCGACATTCTGC as a reverse primer, digested with SacII and KpnI at the sites introduced in the primer sequences (underlined), and ligated with the 5′ end of the green fluorescent protein (GFP)-coding sequence at the Sac II-Kpn I sites of a GFP-expression vector pPD117.01 (provided by Andrew Fire, Stanford University, Stanford, CA). Subsequently, the full-length UNC-78 cDNA (yk185g6; provided by Yuji Kohara, National Institute of Genetics, Mishima, Japan) was digested with EcoRI and XhoI and ligated in-frame with the 3′ end of GFP at the EcoRI-Nhe I sites of the plasmid.
Transgenic Nematodes
The expression vectors for GFP-UNC-78 (20 μg/ml) were mixed with 80 μg/ml pET-32a (Novagen, Madison, WI) as carrier DNA and injected into the distal arm of the hermaphroditic gonad as described previously (Mello and Fire, 1995). Transformants were selected by expression of GFP as observed by fluorescence microscopy, and at least three transgenic lines with extrachromosomal arrays were established for each expression vector.
Western Blot
Thirty adult worms were picked and lysed in 20 μl of SDS-lysis buffer (2% SDS, 80 mM Tris-HCl, 5% β-mercaptomethanol, 15% glycerol, and 0.05% bromophenol blue, pH 6.8), heated at 97°C for 2 min, homogenized by brief sonication, and heated again at 97°C for 2 min. The samples were resolved by SDS-PAGE by using a 12% acrylamide gel and transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Billerica, MA). The membrane was blocked in 5% nonfat milk in phosphate-buffered saline containing 0.1% Tween 20 for 30 min and incubated with anti-UNC-78 antibody (Mohri and Ono, 2003) for 1 h followed by treatment with peroxidase-labeled goat anti-mouse IgG (Pierce Chemical, Rockford, IL). The reactivity was detected with a SuperSignal chemiluminescence reagent (Pierce Chemical). The membrane was treated with a buffer containing 2% SDS, 100 mM β-mercaptoethanol, and 62.5 mM Tris-HCl, pH 6.8, at 50°C for 30 min to remove bound probes and reprobed with mouse monoclonal anti-actin antibody (C4; MP Biomedicals, Irvine, CA) as a loading control.
Site-directed Mutagenesis
Mutagenesis was performed with a QuikChange mutagenesis kit (Stratagene, La Jolla, CA) by using pGEX-UNC-78 or pGFP-UNC-78 as a template. To make a quadruple mutant, four mutations were sequentially added by separate mutagenesis reactions in the order of F192A, F182A, D168A, and E126A. The entire UNC-78 coding region was sequenced to confirm the presence of introduced mutations and the absence of PCR-induced errors.
Pelleting Assay for F-Actin Binding and Depolymerization
Rabbit muscle actin (Pardee and Spudich, 1982), C. elegans actin (Ono, 1999), and UNC-60B (Ono and Benian, 1998) were purified as described previously. Glutathione S-transferase (GST)-UNC-78 with or without mutation was bacterially expressed and purified as described previously (Mohri et al., 2004). An F-actin copelleting assay was performed as described previously (Mohri et al., 2004) with modifications. Briefly, 10 μM F-actin was incubated with or without 10 or 20 μM UNC-60B and 0.1–2.0 μM GST-UNC-78 in F-buffer (0.1 M KCl, 2 mM MgCl2, 1 mM dithiothreitol, and 20 mM HEPES-NaOH, pH 7.5) for 30 min at room temperature and centrifuged at 80,000 rpm (285,000 × g) for 20 min in a Beckman TLA-100 rotor. The supernatants and pellets were adjusted to the same volumes and analyzed by SDS-PAGE. Gels were stained with Coomassie brilliant blue R-250 (National Diagnostics, Atlanta, GA) and scanned by a UMAX PowerLook III scanner at 300 dpi, and the band intensity was quantified by Scion Image Beta 4.02 (Scion, Frederick, MD). In some experiments, the conditions for centrifugation were changed to 56,000 rpm (140,000 × g) for 20 min in TLA-100 or 14,000 rpm (18,000 × g) for 10 min in a Beckman Microfuge (Beckman Coulter, Fullerton, CA).
Assay for Barbed End Elongation
A spectroscopic assay to examine actin elongation from filament ends was performed as described previously using rabbit muscle actin as seeds (Yamashiro et al., 2005). Briefly, 10 μM F-actin from rabbit muscle was mixed for 30 s with or without 10 μM UNC-60B and with or without 1 μM UNC-78 variants in F-buffer and used as seeds for polymerization of pyrene-labeled rabbit muscle G-actin. The kinetics of actin polymerization was measured by the increase in the fluorescence of pyrene and the initial rate in the presence of UNC-60 proteins was compared with that of actin alone.
Light Scattering Assay
F-actin (5 μM) was mixed with GST-UNC-78 and/or UNC-60B in F-buffer, and light scattering at an angle of 90° and a wavelength of 500 nm was measured with an LS50B fluorescence spectrophotometer (PerkinElmer Life and Analytical Sciences, Boston, MA). Slit width was set at 2.5 nm for both excitation and emission.
Fluorescence Microscopy
Immunofluorescent staining of adult worms was performed as described previously (Finney and Ruvkun, 1990) with anti-UNC-60B antibody (Ono et al., 1999) or anti-UNC-78 antibody (Mohri and Ono, 2003) and anti-actin antibody (C4; MP Biomedicals) as primary antibodies, and Cy3-labeled goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) and Alexa 647-labeled goat anti-mouse antibody (Invitrogen) as secondary antibodies. Staining of worms with tetramethylrhodamine-phalloidin was performed as described previously (Ono, 2001). Live worms expressing GFP-fusion proteins were anesthetized by 0.1% tricaine and 0.01% tetramisole and mounted on 2% agarose pads as described previously (Ono and Ono, 2004).
Samples were viewed by epifluorescence using a Nikon Eclipse TE2000 inverted microscope with a CFI Plan Fluor ELWD 40× objective (dry; numerical aperture [N.A.] 0.60) or a CFI Plan Apo 60× objective (oil; N.A. 1.4). Images were captured by a SPOT RT monochrome charge-coupled device camera (Diagnostic Instruments, Sterling Heights, MI) and processed by the IPLab imaging software (Scanalytics, Rockville, MD) and Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA).
Motility Assay
Worm motility was quantified as described previously (Epstein and Thomson, 1974). Briefly, adult worms were placed in M9 buffer. Then, one beat was counted when a worm swung its head to either right or left. The total number of beats in 30 s was recorded.
RESULTS
Green Fluorescent Protein-tagged UNC-78/AIP1 Rescues the unc-78 Null Phenotype
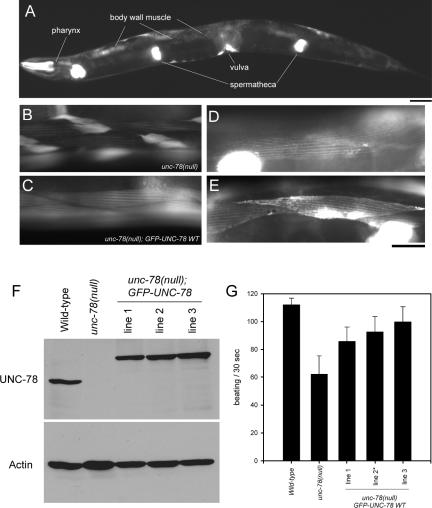
In C. elegans, AIP1 is encoded by the unc-78 gene that is essential for organized assembly of actin filaments into striated myofibrils in the body wall muscle (Ono, 2001). The unc-78 null mutant shows severe disorganization of muscle actin filaments and impaired worm motility because of defective muscle (Ono, 2001) and was used in this study as a genetic background to determine functionality of mutant UNC-78/AIP1 proteins. To construct transgenic animals, we first tested whether the promoter of the unc-78 gene has activity to drive expression of a transgene in a similar pattern to the endogenously expressed UNC-78 protein. The 1.4-kb upstream sequence of the unc-78 gene was fused to GFP and unc-78 cDNA, and the construct was injected into an unc-78-null background [unc-78(gk27)] to make transgenic animals. Expression of GFP-UNC-78 was detected in the pharynx, body wall muscle, spermatheca, and vulva (Figure 1A), which is the same pattern as the endogenously expressed UNC-78 protein detected by immunofluorescent staining (Mohri and Ono, 2003). Importantly, organization of the actin filaments in body wall muscle was restored in the transgenic worms (Figure 1, B and C). In cells expressing GFP-UNC-78, actin is organized in a striated manner, and aggregates of actin were not found (Figure 1, compare B with C). Moreover, the GFP-UNC-78 fusion protein localized in a striated pattern in live animals (Figure 1, D and E), suggesting strongly that our previous observation that endogenously expressed UNC-78 protein colocalizes with actin to the myofibrils (Mohri and Ono, 2003) is not due to an artifact of fixation.
Figure 1.
Transgenic expression of GFP-UNC-78 rescues the unc-78 null phenotype. (A) Expression pattern of GFP-UNC-78. The expression construct was introduced in an unc-78 null mutant, and the fluorescence of GFP was examined. GFP-UNC-78 was expressed in the pharynx, body wall muscle, vulva, and spermatheca. Expression in the body wall muscle was weaker than that in other tissues and underexposed in the micrograph. Bar, 50 μm. (B and C) Rescue of actin disorganization in the unc-78 mutant by GFP-UNC-78 as observed by phalloidin staining. Actin filaments were highly disorganized in the body wall muscle of the unc-78(null) mutant (B), whereas transgenic expression of GFP-UNC-78 restored striated organization of the actin filaments (C). (D and E) Striated localization of GFP-UNC-78 in the body wall muscle in live animals. Bright signals are due to strong expression of GFP-UNC-78 in spermatheca or vulva. Bar, 20 μm for B–E. (F) Protein levels of endogenous UNC-78 and transgenically expressed GFP-UNC-78 as demonstrated by Western blot probed with anti-UNC-78 antibody. Actin was examined as a loading control. GFP-UNC-78 is expressed at comparable levels to endogenous UNC-78 in wild-type. (G) Worm motility of wild-type, unc-78(null), and transgenic strains. Expression of GFP-UNC-78 partially restored motility. Asterisk indicates the strain used for microscopic analysis. Data are means ± SD, n = 10.
The transgenes were maintained as extrachromosomal arrays that are inherited independently from the chromosomes (Mello et al., 1991). Because each array has different characteristics in the level of gene expression, we isolated three independent strains and characterized their phenotypes. Western blot analysis showed that GFP-UNC-78 was expressed at comparable levels to endogenously expressed UNC-78 in wild-type (Figure 1F). All three strains showed significantly improved worm motility compared with the unc-78-null mutant, but the motility was not as fast as wild type (Figure 1G). Many of the transgenic worms had a few cells with no expression of GFP-UNC-78, probably because of loss of the extrachromosomal arrays, and these cells have disorganized actin filaments (our unpublished data). Such mosaic expression of the transgene is a likely explanation for incomplete restoration of worm motility. From these observations, we concluded that GFP-UNC-78 is functional and behaves normally in the muscle cells and that the unc-78 promoter and the GFP-UNC-78 fusion protein are appropriate tools for genetic manipulation and functional analysis of the UNC-78 protein in vivo.
Residues of UNC-78 That Are Important for Actin Filament Disassembly Are Required for Organized Actin Assembly
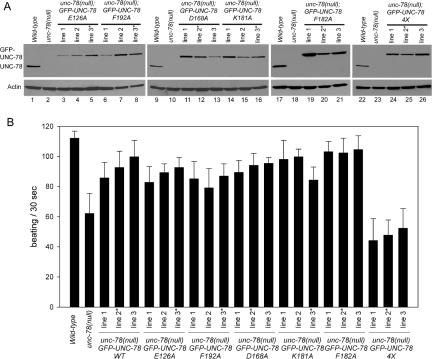
We previously identified five functional residues of UNC-78 (E126, D168, K181, F182, and F192) that are important for disassembly of ADF/cofilin-bound actin filaments in vitro (Mohri et al., 2004). To determine whether these residues are essential for myofibril assembly in vivo, they were converted to alanine in GFP-UNC-78, and the mutants were expressed in the unc-78-null background and tested for their ability to rescue the phenotype. We isolated three independent transgenic strains for each mutant and examined the expression levels of GFP-UNC-78 and worm motility (Figure 2). The levels of expression were variable among the transgenic strains (Figure 2A). Surprisingly, all five single mutants rescued worm motility to similar levels to wild-type GFP-UNC-78 (Figure 2B). Apparently, there was no good correlation between the expression level and the extent of rescue. Therefore, we selected one strain for each mutant that expresses a similar level of GFP-UNC-78 to endogenous UNC-78 in wild-type (selected strains are marked by asterisks in Figure 2) and further examined actin filament organization and localization of GFP-UNC-78 and UNC-60B in body wall muscle cells.
Figure 2.
GFP-UNC-78 with a quadruple mutation (E126A, D168A, F182A, and F192A) fails to rescue motility defects of the unc-78(null) mutant. (A) Protein levels of endogenous UNC-78 and transgenically expressed GFP-UNC-78 with mutations as demonstrated by Western blot probed with anti-UNC-78 antibody. Actin was examined as a loading control. (B) Worm motility of wild-type, unc-78(null), and transgenic strains. Expression of GFP-UNC-78 with single mutations restored motility to similar levels as wild-type GFP-UNC-78. However, expression of a quadruple (4X: E126A, D168A, F182A, and F192A) mutant had a negative effect. Strains that were used in the subsequent microscopic analysis are indicated by asterisks. Data are means ± SD, n = 10.
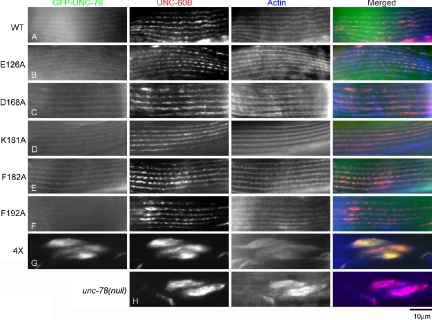
Consistently with the restoration of worm motility, organization of the actin filaments was remarkably improved in the transgenic worms expressing GFP-UNC-78 with single mutations nearly as well as wild type (Figure 3, A–F, and Supplemental Figure 1, A–F). GFP-UNC-78 with single mutations localized to the diffuse cytoplasm and the myofibrils in a striated pattern as observed for wild type (Figure 3, A–F). In the unc-78-null mutant, UNC-60B, the muscle-specific ADF/cofilin, was mislocalized to the actin aggregates (Figure 3H), whereas expression of wild-type GFP-UNC-78 rescued this phenotype to normal localization of UNC-60B to the myofibrils (Figure 3A). UNC-60B also colocalized with actin to the myofibrils in the transgenic worms expressing GFP-UNC-78 with single mutations (Figure 3, B–G). In vitro, the E126A, D168A, F182A, and F192A mutations impair the actin-filament disassembly activity of UNC-78, whereas the K181A mutation enhances the activity (Mohri et al., 2004). However, these in vivo experiments indicate that each single mutant retains functionally sufficient activity to promote organized assembly of actin filaments in muscle cells.
Figure 3.
GFP-UNC-78 with a quadruple mutation (E126A, D168A, F182A, and F192A) fails to rescue defects in the actin organization and the localization of UNC-60B (ADF/cofilin) in the unc-78(null) mutant. Adult unc-78(null) worms that express GFP-UNC-78 were stained with anti-UNC-60B and anti-actin antibodies, and their localization in the body wall muscle was observed. Wild-type (A) or single mutants (B–F) rescued the striated localization of UNC-60B, whereas the 4X mutant did not (G). The unc-78(null) mutant without transgenic expression is shown in H. Bar, 10 μm.
These five residues are clustered on a surface of the N-terminal β-propeller domain, and the in vitro actin disassembly activity of the single mutants is reduced but not abolished (Mohri et al., 2004). Therefore, we reasoned that these residues may consist of a functional surface and work in a partially redundant manner for actin disassembly. We made a quadruple mutant (4X) of GFP-UNC-78 that has four mutations (E126A, D168A, F182A, and F192A), which are biochemically “loss-of-function” mutations, and we tested its ability to rescue the unc-78-null phenotype. The 4X GFP-UNC-78 mutant failed to restore worm motility, and interestingly, its expression worsened motility (Figure 2). This mutant did not restore the actin organization in muscle, and the actin filaments remained highly disorganized and aggregated (Figure 3G and Supplemental Figure 1G). The 4X GFP-UNC-78 mutant and UNC-60B were associated with the aggregates in the body wall muscle (Figure 3H). These results indicate that the quadruple mutation disrupted the essential function of UNC-78 and suggest that these four clustered residues are functionally important for myofibril assembly in vivo.
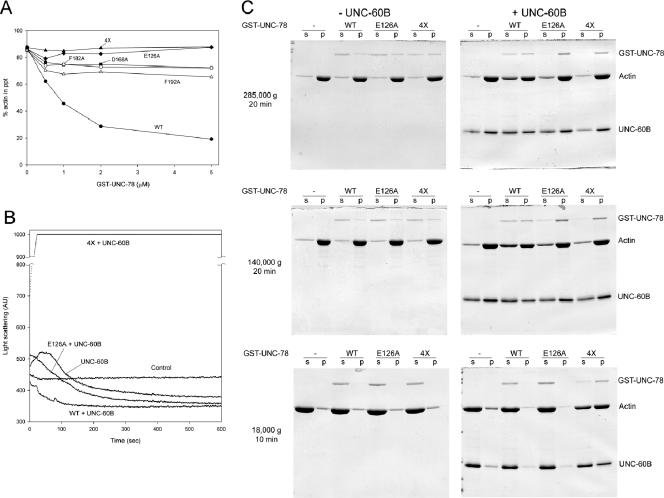
To determine the biochemical difference between the quadruple mutant and each of the single mutants, the activity of the quadruple UNC-78 mutant was characterized in vitro. A sedimentation assay indicated that the 4X mutant and E126A had nearly no UNC-60B-dependent actin disassembly activity (Figure 4A), whereas D168A, F182A, and F192A showed weak disassembly activity (Figure 4A). In addition, the 4X mutant as well as E126A cosedimented with UNC-60B-bound actin filaments (Figure 4C). Thus, the sedimentation assay demonstrated that D168A, F182A, and F192A maintained reduced disassembly activity, but it failed to distinguish the difference between the 4X and E126A mutants. However, their activities were clearly different in the light scattering assay, in which E126A slowly decreased light scattering, whereas the 4X mutant drastically increased light scattering (Figure 4B). E126A binds to UNC-60B-bound actin filaments in a pelleting assay at a 1:2 M ratio (Mohri et al., 2004) and could theoretically contribute to increasing the mass of the filaments. Nonetheless, the decrease in light scattering in the presence of E126A suggests that the filaments were shortened by severing. In contrast, the 4X mutant remains bound to the filaments resulting in the increase in light scattering due to filament bundling through dimerization of the GST moiety. The 4X mutant is most likely defective with the disassembly activity, but it is not conclusive because of its bundling effect. However, it would be reasonable to interpret that the clear difference between 4X and E126A in the light scattering assay (Figure 4B) indicates that E126A has stronger severing activity than 4X and that E126A-severed (and possibly bundled) filaments are short enough to decrease light scattering.
Figure 4.
The 4X UNC-78 mutant is defective in actin filament disassembly. (A) F-actin pelleting assays were performed with 10 μM F-actin, 20 μM UNC-60B, and various concentrations of wild-type or mutant GST-UNC-78 proteins at 285,000 × g for 20 min. Wild-type (black circles) decreased pelletable actin, and D168A (black squares), F182A (white circles), and F192A (white triangles) exhibited reduced activity, whereas E126A (black diamonds) and 4X (black triangles) failed to decrease pelletable actin under these conditions. (B) Light scattering assays were performed with 5 μM F-actin, 5 μM UNC-60B, with or without 1 μM wild type, E126A, or 4X GST-UNC-78. Control experiments were done with F-actin alone. Wild type enhanced actin disassembly and/or severing and rapidly decreased light scattering. E126A exhibited reduced activity and slowly decreased the light scattering signal. In contrast, 4X rapidly increased the signal above the measurable range (arbitrary units [AU] 1000). In the absence of UNC-60B, wild type, E126A, and 4X did not alter light scattering compared with the control (our unpublished data). (C) F-actin pelleting assays were performed with 10 μM F-actin, with or without 10 μM UNC-60B and with or without 1 μM wild-type or mutant GST-UNC-78 proteins at three different centrifugal forces as indicated on the left. The supernatants (s) and pellets (p) were examined by SDS-PAGE. At 285,000 × g for 20 min, only wild type increased actin in the supernatant in the presence of UNC-60B. At 140,000 × g for 20 min, E126A slightly increased actin in the supernatant, but not as much as wild type. In contrast, at 18,000 × g for 10 min, only 4X caused sedimentation of actin in the presence of UNC-60B, suggesting that F-actin bundles were formed.
To further confirm these activities, actin filaments were incubated with UNC-60B and wild-type or mutant GST-UNC-78 and spun at different centrifugal forces (Figure 4C). At 140,000 × g, E126A, but not 4X, slightly increased the amount of actin in the supernatant (Figure 4C), indicating that short filaments were generated by E126A. In addition, the 4X mutant, but not wild-type or E126A, caused sedimentation of actin at low-speed centrifugation only in the presence of UNC-60B (Figure 4C), suggesting that actin bundles were formed by the 4X mutant, again, likely because of dimerization of the GST moiety. Wild-type or E126A may also bundle UNC-60B-bound actin filaments, but such bundles are probably too short to sediment at 18,000 × g. These biochemical results show that the simultaneous mutation of the four residues strongly impairs the filament disassembly activity but not the F-actin-binding activities.
unc-78 Mutations Identified through a Genetic Screen Impair Protein Folding or Filament Disassembly Activity
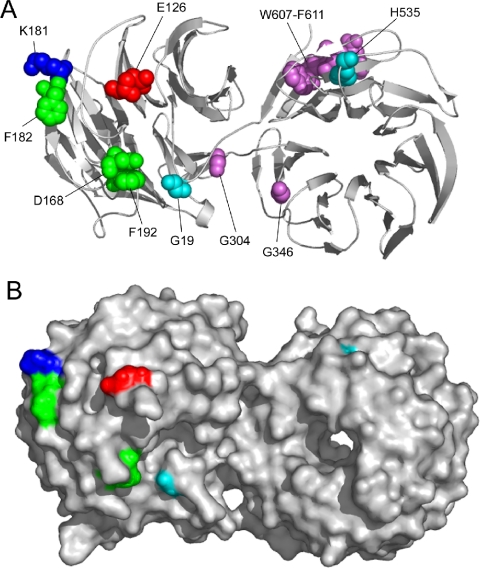
We previously identified three missense mutations (G304E, G346E, and H535Y) and one nonsense mutation (W607-stop) (Ono, 2001) from mutant unc-78 alleles that cause uncoordinated movement and defects in the actin organization in body wall muscle (Waterston et al., 1980; Zengel and Epstein, 1980). Additionally, in this study, we identified a new missense mutation (G19E) from unc-78(su223) that was isolated by Zengel and Epstein (1980). These residues are spatially distinct from the biochemically identified functional residues (Figure 5A). Therefore, they are strong candidates for new functional sites of UNC-78.
Figure 5.
Structure of UNC-78 and its functional surface residues. A ribbon diagram (A) and a surface model (B) of the structure of UNC-78 (Protein Data Bank code 1PEV) (Mohri et al., 2004) are shown. Mutations at green residues (D168, F182, and F192) result in simple reduction in filament disassembly activity, whereas a mutation at E126 (red) reduces the disassembly activity but enhanced filament binding (Mohri et al., 2004). These four residues were mutated in the 4X mutant in this study. A mutation at K181 enhances the activity (Mohri et al., 2004). Purple residues are mutated in unc-78 mutants (Ono, 2001), but mutations at these residues compromised the solubility of the protein. Light blue residues were also mutated in unc-78 mutants, and the effects of the mutations were characterized in this study. Molecular models were generated with PyMOL (DeLano Scientific, South San Francisco, CA).
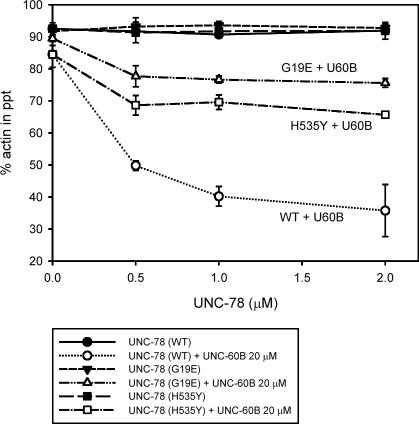
To determine the effects of these mutations on the activity of UNC-78, mutant forms of GST-UNC-78 were bacterially expressed and tested for their activity in vitro. Three mutants (G304E, G346E, and W607-stop) were insoluble after expression in Escherichia coli (our unpublished data). In the crystal structure, G304 and G346 are not exposed on the surface (Figure 5B), and the premature stop codon at position 607 removes the C-terminal five amino acids that associate with the N terminus to conclude the β-propeller (Figure 5A), suggesting that these mutations altered protein folding and/or conformation. The G19E and H535Y mutants were expressed as soluble proteins and successfully purified for subsequent characterization. In sedimentation assays, these two mutants had much weaker activity to disassemble actin filaments in an UNC-60B–dependent manner than wild type (Figure 6). G19E exhibited weaker disassembly activity than H535Y (Figure 6). These mutants had similar activity on C. elegans actin (our unpublished data).
Figure 6.
Effects of the G19E and H535Y mutations on the biochemical properties of UNC-78. F-actin pelleting assays were performed with 10 μM F-actin with or without 20 μM UNC-60B and various concentrations of wild-type or mutant GST-UNC-78 proteins at 285,000 × g for 20 min. Both mutants exhibited reduced disassembly activity. G19E had weaker activity than H535Y. The data are means ± SD of three experiments.
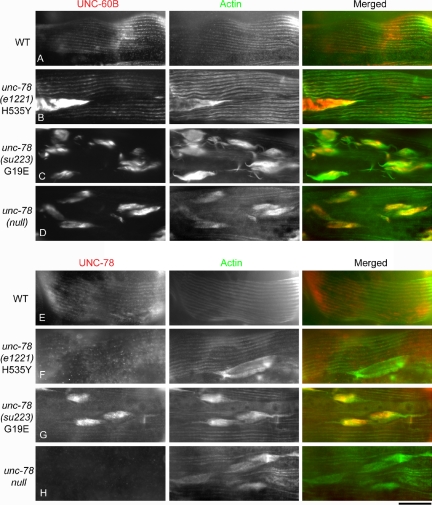
The biochemical defects of these mutants correlated with the severity of the cytoskeletal defects of the unc-78 mutant worms. Worm motility of the G19E mutant was severely impaired to a similar level to the unc-78-null mutant (our unpublished data). In contrast, the H535Y mutant moved nearly as fast as wild type as reported previously (Ono, 2001). The actin filaments in body wall muscle of the G19E mutant was also disorganized as severely as the null mutant (Figure 7C). The H535Y mutant had relatively small actin aggregates only in a subset of muscle cells (Figure 7B). Western blot analysis of the mutant UNC-78 proteins showed that G19E was expressed at a comparable level to wild type, whereas H535Y was expressed at a much lower level than wild type (Figure 8), suggesting that H535Y is not stable or not efficiently expressed. UNC-60B (ADF/cofilin) colocalized with actin to the myofibrils in wild type (Figure 7A), whereas it mislocalized to the actin aggregates in the unc-78-null mutant (Figure 7D). UNC-60B also mislocalized to the actin aggregates in both G19E and H535Y mutants. However, in the H535Y mutant, the phenotype was weak and striated localization of UNC-60B was also observed (Figure 7, B and C). Immunolocalization of the UNC-78 protein showed that wild-type UNC-78 localized to the striated myofibrils (Figure 7E). However, the H535Y mutant was mostly diffuse in the cytoplasm and weakly localized to the striated myofibrils (Figure 7F), and the G19E mutant strongly localized to the actin aggregates and weakly to the myofibrils (Figure 7G). The weak association of H535Y with actin-containing structures is likely due to its low protein level (Figure 8), because H535Y protein coprecipitated with F-actin to a similar extent to wild-type in vitro (our unpublished data). These results further support the significance of the activity of UNC-78 to enhance disassembly of UNC-60B-bound actin filaments for its in vivo function in myofibril assembly.
Figure 7.
Effects of the G19E and H535Y mutations on myofibril organization. Adult worms were stained with anti-UNC-60B and anti-actin antibodies (A–D) or with anti-UNC-78 and anti-actin antibodies (E–H), and the body wall muscle is shown in the figure. Bar, 20 μm.
Figure 8.
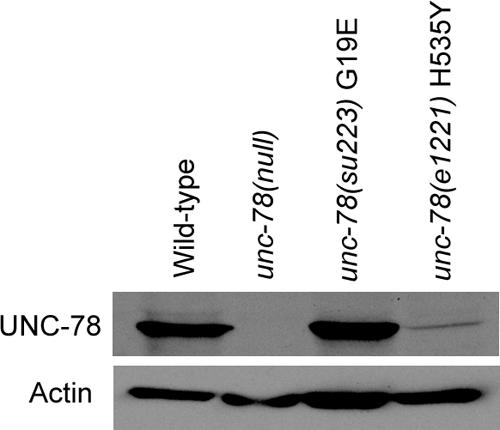
The H535Y mutation reduces the UNC-78 protein level. Thirty adult worms were collected and examined for the UNC-78 protein levels by Western blot. Actin was examined as a loading control.
The Barbed End-capping Activity of Mutant UNC-78 Proteins May Be Impaired
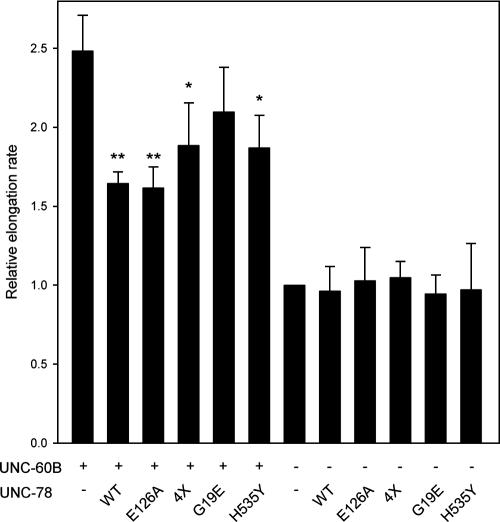
AIP1 caps the barbed ends of ADF/cofilin-bound actin filaments. Studies in yeast suggest that the capping activity of AIP1 is important for its in vivo function (Balcer et al., 2003). Therefore, we examined the barbed end-capping activity of mutant UNC-78 proteins by an actin nucleation assay (Figure 9). UNC-60B severed filaments and increased the number of actin nuclei that enhanced the elongation rate by ∼2.5-fold (Figure 9). Wild-type GST-UNC-78 inhibited the elongation rate by capping the barbed ends (Figure 9) as reported previously for Xenopus laevis AIP1 (Okada et al., 2002). E126A had similar activity to wild type, whereas 4X, G19E, and H535Y showed apparently weaker activity than wild type. Comparison of the data by t test indicates that wild type, E126A, 4X, and H535Y, but not G19E, significantly decreased the elongation rate, suggesting that G19E might be defective with capping activity. The P values for 4X and H535Y were 1 order higher than those for wild type and E126A, suggesting that 4X and H535Y may have partial defects. In the absence of UNC-60B, none of these UNC-78 variants altered the elongation rate (Figure 9). In addition, there was no correlation between the capping defects and disassembly defects. E126A and 4X show strong disassembly defects, whereas the capping activity is unaffected or only mildly reduced. In contrast, G19E has greater disassembly activity but weaker capping activity than E126A or 4X. These data suggest that functionally defective UNC-78 mutants may have impaired capping activity.
Figure 9.
The barbed end-capping activity of mutant UNC-78 proteins may be impaired. F-actin (10 μM) was mixed with or without wild-type or mutant UNC-78 proteins (1 μM) in the presence or absence of 10 μM UNC-60B and used as nuclei at 1.25 μM actin (8-fold dilution) to induce polymerization of pyrene–G-actin. Elongation rates were determined as the initial rates of the increase in the pyrene fluorescence. The data are expressed as relative elongation rates to that of F-actin alone (=1.0) and the means ± SD of three experiments. The data with UNC-78 were compared with those without UNC-78 by the t test to determine whether the elongation rate was significantly reduced. Single asterisks indicate p < 0.05, and double asterisks indicate p < 0.005.
DISCUSSION
In this study, we correlated in vitro and in vivo activities of mutant forms of UNC-78 (AIP1) and found that biochemical defects of UNC-78 mutants in enhancement of ADF/cofilin-dependent actin disassembly are associated with their in vivo defects in actin filament organization in the C. elegans body wall muscle. Single mutations in the residues that are important for the in vitro actin disassembly activity did not completely disrupt its activity or affect the ability to rescue the unc-78 null phenotype. However, a simultaneous mutation of the four residues (E126, D168, F182, and F192) abolished the in vitro actin disassembly activity and its functionality to rescue the unc-78 null phenotype. We also identified two new mutations of UNC-78 that impair both in vitro actin disassembly activity and in vivo function. The G19E mutation caused a reduction in the actin disassembly activity and a defect in the capping activity, and perhaps the combination of these defects resulted in the severe in vivo phenotype in actin organization. These functional residues are separated in the N-terminal and C-terminal propellers and may represent at least two sites for interacting with ADF/cofilin and actin.
Null or loss-of-function phenotypes for AIP1 in several biological systems consistently indicate that AIP1 is required for cellular events that require dynamic rearrangement of the actin cytoskeleton, suggesting that AIP1 is a conserved enhancer of actin filament turnover (Ono, 2003). This conserved function of AIP1 can be explained by its activity to promote disassembly of ADF/cofilin-bound actin filaments. ADF/cofilin by itself severs and depolymerizes actin filaments. However, filament severing by ADF/cofilin increases the number of exposed barbed ends that also enhance polymerization (Ichetovkin et al., 2002; Ghosh et al., 2004). AIP1 can promote actin depolymerization in the presence of ADF/cofilin by actively severing filaments (Ono et al., 2004) and/or capping the barbed ends (Okada et al., 2002; Balcer et al., 2003). The disassembly and capping activities of UNC-78 can be uncoupled by point mutations in evolutionarily conserved residues, suggesting that they are separable activities (Mohri et al., 2004). Both activities are most likely important for the function of AIP1. The 4X and G19E mutants caused very strong actin disorganization in muscle, whereas they showed different biochemical properties. The in vivo phenotypes of 4X and G19E cannot be directly compared because 4X was expressed as a GFP-fusion protein, whereas G19E was expressed endogenously as a nonfusion protein. We showed that GFP-UNC-78 is functional. However, because GFP can dimerize at high concentrations (Kd = ∼100 μM) (Phillips, 1997), GFP-UNC-78–4X could form a dimer when it is locally concentrated in the actin aggregates and bundle actin filaments, which may worsen the phenotype as a secondary effect.
The 4X mutant had a severe disassembly defect and mildly affected capping activity, suggesting the functional significance of the disassembly activity of AIP1 in cytoskeletal reorganization. AIP1 is the only known protein that specifically disassembles ADF/cofilin-bound actin filaments. In contrast, there are a number of barbed end capping proteins in the cell (Weeds and Maciver, 1993; Zigmond, 2004). Thus, the ADF/cofilin-dependent filament disassembly activity may be the unique function of AIP1, but the capping activity could be compensated by other capping proteins within the cell. In support of this idea, in yeast, an aip1-null mutant shows synthetic growth defects in combination with null alleles for capping protein (cap1 and cap2) (Rodal et al., 1999). In contrast, G19E showed a severe capping defect and moderate disassembly activity that is comparable with other functional UNC-78 mutants, suggesting that capping is also an important function. We measured the capping activity by an actin elongation assay to quantify relative numbers of exposed barbed ends. However, the data from this assay contained relatively large deviations, and it is still possible that G19E may have significant capping activity. The relative elongation rate can be reduced when the barbed end is capped or when some filaments are completely disassembled. Although these two phenomena cannot be distinguished in this assay, our results likely represent capping because E126A, which does not increase unpelletable actin in the pelleting assays, shows similar activity to wild type. An actin-pelleting assay in the presence of profilin was used to determine the capping activity of AIP1 (Balcer et al., 2003). However, a recent study has shown that this assay may not accurately measure capping (Clark et al., 2006), probably because depolymerization and severing cannot be distinguished by the pelleting assay. Therefore, more direct methods to determine the capping activity of AIP1 will be needed to understand the functional significance of this activity.
Biochemical analysis of additional mutations of UNC-78 support our previous observations that UNC-78/AIP1 actively disassembles ADF/cofilin-bound actin filaments (Ono et al., 2004) and that the disassembly activity can be uncoupled from the capping activity (Mohri et al., 2004). We found that a simultaneous mutation of four clustered residues is required to eliminate the disassembly activity, suggesting that this activity requires a relatively large surface area of the N-terminal propeller (Figure 6). The quadruple mutant was still capable of F-actin binding and capping, indicating that an actin-binding site exists in the other part of the molecule. G19E is the first mutant with impaired disassembly and capping activities, and this residue is a candidate for a part of a capping site. Gly-19 is also spatially close to the four residues responsible for disassembly and may also be a part of the disassembly site. However, a capping defect of G19E may indirectly affect the disassembly activity. Therefore, the capping site of UNC-78 remains elusive because we have not found any “capping-specific” mutant. In addition, we found that the H535Y mutant caused a defect in the disassembly activity. Although the defect is mild, this is the first functionally defective mutation in the C-terminal propeller, and His-535 is a candidate for a part of a second site responsible for disassembly. In agreement with our data, functional surface residues in the C-terminal propeller of yeast AIP1 have been identified (Clark et al., 2006; Okada and Goode, personal communication). Thus, the C-terminal propeller of AIP1 is very likely to have important function for actin disassembly and/or capping, and further mutational analysis should reveal the mechanism by which AIP1 induces ADF/cofilin-dependent actin disassembly.
Our results demonstrate that morphogenesis of striated myofibrils requires actin filament disassembly by UNC-78 (AIP1) and UNC-60B (ADF/cofilin). Previous studies indicate that UNC-60B is required for organized assembly of actin filaments into the myofibrils (Ono et al., 1999, 2003). UNC-60B by itself enhances filament severing and pointed end depolymerization with only minor effects on the steady-state concentrations of G- and F-actin (Ono and Benian, 1998; Yamashiro et al., 2005). UNC-78 has activity to promote depolymerization and severing (Mohri and Ono, 2003; Ono et al., 2004) to facilitate remodeling of the actin filaments. This study strongly suggests that enhancement of UNC-60B–dependent filament disassembly is the critical and unique function of UNC-78. The mechanism of this function of UNC-78 remains elusive as to whether the enhancement of disassembly is caused by severing or capping, and further biochemical and mutagenesis studies of AIP1 are needed to solve this problem. In muscle cells, the expression of actin is substantially up-regulated during myogenesis and the concentration of actin is very high. This may be the reason why muscle cells require a strong actin disassembly system by ADF/cofilin and AIP1 to achieve assembly of organized myofibrils. In mammalian muscle, a muscle-specific cofilin isoform is expressed (Ono et al., 1994; Thirion et al., 2001; Vartiainen et al., 2002), but it is not known whether mammalian AIP1 (WDR1) (Adler et al., 1999; Fujibuchi et al., 2005) is expressed in muscle. In contrast, cells have to have a system to inhibit the strong activity of ADF/cofilin and AIP1 to maintain the contractile structures. Tropomyosin inhibits ADF/cofilin-dependent actin dynamics in C. elegans body wall muscle (Ono and Ono, 2002) and is a strong candidate for such an inhibitor. However, no direct regulator of the AIP1 activity is currently known. Further studies on actin dynamics in the C. elegans muscle cells may reveal additional regulatory factors that are linked to the functions of ADF/cofilin and AIP1.
Supplementary Material
Acknowledgments
We thank D. Amberg, M. Clark, K. Okada, B. Goode, S. Almo, and D. Sept for helpful discussions and sharing unpublished data. Some nematode strains were provided by Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. This work was supported by Grant MCB-0110464 from the National Science Foundation (to S. O.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–11–1016) on March 8, 2006.
Abbreviations used: ADF, actin-depolymerizing factor; AIP1, actin-interacting protein 1; GFP, green fluorescent protein: GST, glutathione S-transferase.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Adler, H. J., Winnicki, R. S., Gong, T. W., and Lomax, M. I. (1999). A gene upregulated in the acoustically damaged chick basilar papilla encodes a novel WD40 repeat protein. Genomics 56, 59–69. [DOI] [PubMed] [Google Scholar]
- Aizawa, H., Katadae, M., Maruya, M., Sameshima, M., Murakami-Murofushi, K., and Yahara, I. (1999). Hyperosmotic stress-induced reorganization of actin bundles in Dictyostelium cells over-expressing cofilin. Genes Cells 4, 311–324. [DOI] [PubMed] [Google Scholar]
- Amberg, D. C., Basart, E., and Botstein, D. (1995). Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 2, 28–35. [DOI] [PubMed] [Google Scholar]
- Balcer, H. I., Goodman, A. L., Rodal, A. A., Smith, E., Kugler, J., Heuser, J. E., and Goode, B. L. (2003). Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 13, 2159–2169. [DOI] [PubMed] [Google Scholar]
- Bamburg, J. R. (1999). Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15, 185–230. [DOI] [PubMed] [Google Scholar]
- Bamburg, J. R., McGough, A., and Ono, S. (1999). Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 9, 364–370. [DOI] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier, M. F., Le Clainche, C., Wiesner, S., and Pantaloni, D. (2003). Actin-based motility: from molecules to movement. Bioessays 25, 336–345. [DOI] [PubMed] [Google Scholar]
- Carlier, M. F., Ressad, F., and Pantaloni, D. (1999). Control of actin dynamics in cell motility. Role of ADF/cofilin. J. Biol. Chem. 274, 33827–33830. [DOI] [PubMed] [Google Scholar]
- Clark, M. G., Teply, J., Haarer, B. K., Viggiano, S. C., Sept, D., and Amberg, D. C. (2006). A genetic dissection of Aip1p's interactions leads to a model for Aip1p-cofilin cooperative activities. Mol. Biol. Cell 17, 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, J. A., and Schafer, D. A. (2000). Control of actin assembly and disassembly at filament ends. Curr. Opin. Cell Biol. 12, 97–103. [DOI] [PubMed] [Google Scholar]
- DesMarais, V., Ghosh, M., Eddy, R., and Condeelis, J. (2005). Cofilin takes the lead. J. Cell Sci. 118, 19–26. [DOI] [PubMed] [Google Scholar]
- Epstein, H. F., and Thomson, J. N. (1974). Temperature-sensitive mutation affecting myofilament assembly in Caenorhabditis elegans. Nature 250, 579–580. [DOI] [PubMed] [Google Scholar]
- Finney, M., and Ruvkun, G. (1990). The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63, 895–905. [DOI] [PubMed] [Google Scholar]
- Fujibuchi, T., Abe, Y., Takeuchi, T., Imai, Y., Kamei, Y., Murase, R., Ueda, N., Shigemoto, K., Yamamoto, H., and Kito, K. (2005). AIP1/WDR1 supports mitotic cell rounding. Biochem. Biophys. Res. Commun. 327, 268–275. [DOI] [PubMed] [Google Scholar]
- Ghosh, M., Song, X., Mouneimne, G., Sidani, M., Lawrence, D. S., and Condeelis, J. S. (2004). Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304, 743–746. [DOI] [PubMed] [Google Scholar]
- Ichetovkin, I., Grant, W., and Condeelis, J. (2002). Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12, 79–84. [DOI] [PubMed] [Google Scholar]
- Iida, K., and Yahara, I. (1999). Cooperation of two actin-binding proteins, cofilin and Aip1, in Saccharomyces cerevisiae. Genes Cells 4, 21–32. [DOI] [PubMed] [Google Scholar]
- Ketelaar, T., Allwood, E. G., Anthony, R., Voigt, B., Menzel, D., and Hussey, P. J. (2004). The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr. Biol. 14, 145–149. [DOI] [PubMed] [Google Scholar]
- Konzok, A., Weber, I., Simmeth, E., Hacker, U., Maniak, M., and Müller-Taubenberger, A. (1999). DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility. J. Cell Biol. 146, 453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel, T. P., Boujemaa, R., Pantaloni, D., and Carlier, M. F. (1999). Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401, 613–616. [DOI] [PubMed] [Google Scholar]
- Maciver, S. K., and Hussey, P. J. (2002). The ADF/cofilin family: actin-remodeling proteins. Genome Biol. 3, 3007.3001–3007.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, C., and Fire, A. (1995). DNA transformation. Methods Cell Biol. 48, 451–482. [PubMed] [Google Scholar]
- Mello, C. C., Kramer, J. M., Stinchcomb, D., and Ambros, V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri, K., and Ono, S. (2003). Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J. Cell Sci. 116, 4107–4118. [DOI] [PubMed] [Google Scholar]
- Mohri, K., Vorobiev, S., Fedorov, A. A., Almo, S. C., and Ono, S. (2004). Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 279, 31697–31707. [DOI] [PubMed] [Google Scholar]
- Nicholson-Dykstra, S., Higgs, H. N., and Harris, E. S. (2005). Actin dynamics: growth from dendritic branches. Curr. Biol. 15, R346–R357. [DOI] [PubMed] [Google Scholar]
- Okada, K., Blanchoin, L., Abe, H., Chen, H., Pollard, T. D., and Bamburg, J. R. (2002). Xenopus actin-interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. J. Biol. Chem. 277, 43011–43016. [DOI] [PubMed] [Google Scholar]
- Okada, K., Obinata, T., and Abe, H. (1999). XAIP 1, a Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. J. Cell Sci. 112, 1553–1565. [DOI] [PubMed] [Google Scholar]
- Ono, K., and Ono, S. (2004). Tropomyosin and troponin are required for ovarian contraction in the Caenorhabditis elegans reproductive system. Mol. Biol. Cell 15, 2782–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, K., Parast, M., Alberico, C., Benian, G. M., and Ono, S. (2003). Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J. Cell Sci. 116, 2073–2085. [DOI] [PubMed] [Google Scholar]
- Ono, S. (1999). Purification and biochemical characterization of actin from Caenorhabditis elegans: its difference from rabbit muscle actin in the interaction with nematode ADF/cofilin. Cell Motil. Cytoskeleton 43, 128–136. [DOI] [PubMed] [Google Scholar]
- Ono, S. (2001). The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol. 152, 1313–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, S. (2003). Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1, new blades for twisted filaments. Biochemistry 42, 13363–13370. [DOI] [PubMed] [Google Scholar]
- Ono, S., Baillie, D. L., and Benian, G. M. (1999). UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J. Cell Biol. 145, 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, S., and Benian, G. M. (1998). Two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. J. Biol. Chem. 273, 3778–3783. [DOI] [PubMed] [Google Scholar]
- Ono, S., Minami, N., Abe, H., and Obinata, T. (1994). Characterization of a novel cofilin isoform that is predominantly expressed in mammalian skeletal muscle. J. Biol. Chem. 269, 15280–15286. [PubMed] [Google Scholar]
- Ono, S., Mohri, K., and Ono, K. (2004). Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 279, 14207–14212. [DOI] [PubMed] [Google Scholar]
- Ono, S., and Ono, K. (2002). Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J. Cell Biol. 156, 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee, J. D., and Spudich, J. A. (1982). Purification of muscle actin. Methods Enzymol. 85, 164–181. [DOI] [PubMed] [Google Scholar]
- Phillips, G. N., Jr. (1997). Structure and dynamics of green fluorescent protein. Curr. Opin. Struct. Biol. 7, 821–827. [DOI] [PubMed] [Google Scholar]
- Pollard, T. D., and Borisy, G. G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465. [DOI] [PubMed] [Google Scholar]
- Rodal, A. A., Tetreault, J. W., Lappalainen, P., Drubin, D. G., and Amberg, D. C. (1999). Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 145, 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S. L., Wiedemann, U., Stuurman, N., and Vale, R. D. (2003). Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirion, C., Stucka, R., Mendel, B., Gruhler, A., Jaksch, M., Nowak, K. J., Binz, N., Laing, N. G., and Lochmuller, H. (2001). Characterization of human muscle type cofilin (CFL2) in normal and regenerating muscle. Eur. J. Biochem. 268, 3473–3482. [DOI] [PubMed] [Google Scholar]
- Vartiainen, M. K., Mustonen, T., Mattila, P. K., Ojala, P. J., Thesleff, I., Partanen, J., and Lappalainen, P. (2002). The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol. Biol. Cell 13, 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegtli, W. C., Madrona, A. Y., and Wilson, D. K. (2003). The structure of Aip1p, a WD repeat protein that regulates cofilin-mediated actin depolymerization. J. Biol. Chem. 278, 34373–34379. [DOI] [PubMed] [Google Scholar]
- Waterston, R. H., Thomson, J. N., and Brenner, S. (1980). Mutants with altered muscle structure of Caenorhabditis elegans. Dev. Biol. 77, 271–302. [DOI] [PubMed] [Google Scholar]
- Weeds, A., and Maciver, S. (1993). F-actin capping proteins. Curr. Opin. Cell Biol. 5, 63–69. [DOI] [PubMed] [Google Scholar]
- Yamashiro, S., Mohri, K., and Ono, S. (2005). The two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins differently enhance actin filament severing and depolymerization. Biochemistry 44, 14238–14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel, J. M., and Epstein, H. F. (1980). Identification of genetic elements associated with muscle structure in the nematode Caenorhabditis elegans. Cell Motil. 1, 73–97. [DOI] [PubMed] [Google Scholar]
- Zigmond, S. H. (2004). Beginning and ending an actin filament: control at the barbed end. Curr. Top. Dev. Biol. 63, 145–188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.