Abstract
Microtubule-organizing centers (MTOCs) concentrate microtubule nucleation, attachment and bundling factors and thus restrict formation of microtubule arrays in spatial and temporal manner. How MTOCs occur remains an exciting question in cell biology. Here, we show that the transforming acidic coiled coil–related protein Mia1p/Alp7p functions in emergence of large MTOCs in interphase fission yeast cells. We found that Mia1p was a microtubule-binding protein that preferentially localized to the minus ends of microtubules and was associated with the sites of microtubule attachment to the nuclear envelope. Cells lacking Mia1p exhibited less microtubule bundles. Microtubules could be nucleated and bundled but were frequently released from the nucleation sites in mia1Δ cells. Mia1p was required for stability of microtubule bundles and persistent use of nucleation sites both in interphase and postanaphase array dynamics. The γ-tubulin–rich material was not organized in large perinuclear or microtubule-associated structures in mia1Δ cells. Interestingly, absence of microtubules in dividing wild-type cells prevented appearance of large γ-tubulin–rich MTOC structures in daughters. When microtubule polymerization was allowed, MTOCs were efficiently assembled de novo. We propose a model where MTOC emergence is a self-organizing process requiring the continuous association of microtubules with nucleation sites.
INTRODUCTION
Microtubules play an important role in establishment and maintenance of cell polarity and overall intracellular organization. To perform these functions, microtubules are organized into complex arrays by specialized structures called the microtubule-organizing centers (MTOCs). Although distinct higher order microtubule structures can be formed in vitro by combining tubulin and microtubule motors (Nedelec et al., 1997), formation of such arrays is temporally and spatially restricted in living cells where MTOCs concentrate microtubule nucleation, attachment, and bundling factors at specific locations. Thus, positioning of microtubule arrays within a cellular space represents an important aspect of MTOC function.
The fission yeast Schizosaccharomyces pombe is an excellent model for study of MTOC and microtubule dynamics. A relatively large cell size allows detailed dynamic observation of cellular components tagged with fluorescent proteins, its genetics is straightforward, and the genome is fully sequenced and annotated (Wood et al., 2002).
The cellular locations of MTOCs and consequently, architecture of microtubule arrays change in a cell cycle-specific manner (for review, see Hagan, 1998). The spindle pole bodies (SPBs) nucleate and organize spindle microtubules during mitosis. On mitotic exit, the equatorial MTOC (eMTOC) nucleates the postanaphase array (PAA), a specialized symmetrical microtubule structure extending from division site toward the tips of a dividing cell. The eMTOC constricts with the actomyosin ring and eventually disassembles at the end of septation. In interphase, the microtubule-nucleating activity is concentrated at the several nuclear envelope (NE)-bound interphase MTOCs (iMTOCs), the SPB, and satellite sites on microtubules (Sawin et al., 2004; Janson et al., 2005; Zimmerman and Chang, 2005). Establishment of the iMTOCs proceeds synchronously with the disassembly the eMTOC (Zimmerman et al., 2004), but the precise origin of the iMTOCs remains obscure.
A number of S. pombe mutants exhibit microtubule defects that are consistent with abnormalities in MTOC function, such as a decreased number of interphase microtubules and low frequency of microtubule nucleation events. In these cases, the existing microtubules are often longer, probably due to a larger pool of soluble tubulin and microtubule accessory factors. The majority of these mutations are in core components of γ-tubulin ring complexes (γ-TuRCs) (Paluh et al., 2000; Vardy and Toda, 2000; Zimmerman and Chang, 2005) or in γ-tubulin accessory factors (Sawin et al., 2004; Venkatram et al., 2004, 2005; Janson et al., 2005; Samejima et al., 2005; Zimmerman and Chang, 2005) and therefore show pronounced defects in microtubule nucleation.
The S. pombe mutant lacking the homologue of the transforming acidic coiled coil (TACC) protein Mia1p/Alp7p (thereafter referred to as Mia1p) shows multiple microtubule abnormalities. The previous studies concentrated on Mia1p functions in mitosis. It was shown that aster microtubules were either absent or unbalanced (Oliferenko and Balasubramanian, 2002), and the TOG family protein Alp14p was not loaded on spindles and the SPBs resulting in spindle abnormalities (Sato et al., 2004). Interestingly, interphase mia1Δ cells exhibit a reduced number of microtubule bundles that are longer than usual and curve around cell tips, suggesting a possible involvement of Mia1p in interphase MTOC function.
Here, we show that unlike previously described nucleation-deficient mutants, mia1Δ cells can and do nucleate microtubules. However, we find that Mia1p is required for sustaining the attachment of microtubules to nucleation sites and for emergence of the iMTOCs. We propose that Mia1p ensures the persistent use of nucleation sites and allows the existence of stable microtubule bundles. In light of our results, we suggest and test a dynamic model for MTOC assembly.
MATERIALS AND METHODS
Schizosaccharomyces pombe Strains, Antibodies, Reagents, and Constructs
S. pombe strains used in this study and their genotypes are listed in Supplemental Table 1. Media for vegetative growth (EMM2 or YES) and genetic methods were as described in Moreno et al. (1991). Genetic crosses and sporulation were performed on YPD agar plates. The homologous recombination-based method was used to tag endogenous Alp4p with green fluorescent protein (GFP) at its C terminus. The Mto1p-GFP and Rsp1p-GFP strains were gifts from Drs. K. Gould (Vanderbilt University, Nashville, TN) and F. Chang (Columbia University, New York, NY), respectively. For protein purification, Escherichia coli BL21-CodonPlus(DE3)-RIL competent cells (commercially available from Strategene, La Jolla, CA) was used. Dr. Y. Hiraoka (Kansai Advanced Research Center, Kobe, Japan) kindly provided us with the pREP1-α-tubulin-GFP construct. The anti-α-tubulin antibody, TAT-1, was a gift from Dr. K. Gull (University of Oxford, Oxford, United Kingdom). Polylysine used for coating was from Sigma-Aldrich (St. Louis, MO), and the microtubule-destabilizing drug methyl-1-(butylcarbamoyl)-2-benzimidazolecarbamate (Carbendazim; MBC) was from Aldrich Chemical (Milwaukee, WI).
Time-Lapse Fluorescent Microscopy
Time-lapse images were generated on a Zeiss Axiovert 200M microscope equipped with UltraView RS-3 confocal system: CSU21 confocal optical scanner, 12 bit digital cooled Hammamatsu Orca-ER camera (OPELCO, Sterling, VA), and krypton-argon triple line laser illumination source (488, 568, and 647 nm). Still images were collected on a Zeiss Axiovert 200M microscope using Cascade:650 camera (Photometrics/Roper Scientific, Trenton, NJ) and Uniblitz shutter driver (Photonics, Rochester, NY) under the control of MetaMorph software package (Universal Imaging, Sunnyvale, CA). For imaging of microtubule dynamics, S. pombe cells expressing α-tubulin-GFP were grown in appropriate selective medium and placed in sealed growth chambers containing 2% agarose media. For three-dimensional time-lapse imaging, each image stack consisted of 13 sections of 0.5-μm spacing and 15-s intervals between stacks. For single-section time-lapse analyses, images were collected at 5-s intervals. Experiments were carried out at room temperature.
Live Cell Chamber Experiments
Poly-lysine (2 mg/ml) was used to fix cells (which were washed with EMM medium) on the Fisher cover glass with two parallel pieces of double-stick tape mounted on it. The flow chamber was created by overlaying the cover glass with a Matsunami coverslip. Flow-through could be achieved by adding medium on one side and absorbing the liquid from the opposite side by tissue paper. Microtubules were typically depolymerized by 50 μg/ml MBC.
Immunofluorescence Techniques
Cells were fixed with 3.7% formaldehyde and spheroplasted using lysing enzymes and Zymolyase in 1.2 M sorbitol in phosphate-buffered saline (PBS). Permeabilization was performed in 1% Triton X-100 in PBS. PBAL (1 mM sodium azide, 50 μg/ml carbenicillin, 1% bovine serum albumin, and 100 mM lysine hydrochloride in PBS) was used for blocking and for incubation with primary and secondary antibodies. Imaging was done on a Zeiss Axiovert 200M microscope with appropriate sets of filters, and images were generated using Cascade:650 camera and MetaMorph software. Image processing was done in Adobe Photoshop 7.0.
Electron Microscopy Techniques
Cells were rapidly frozen by high-pressure freezing (BAL-TEC HPM-010; Technotrade International, Manchester, NH) and freeze-substituted at –90°C in 0.2% glutaraldehyde plus 0.01% uranyl acetate in acetone for 96 h in an EM-AFS device (Leica, Vienna, Austria). The cells were warmed over 25 h to –40°C and then infiltrated with HM20 (Electron Microscopy Sciences, Hatfield, PA) resin over a period of 5 d. The cells were embedded under UV light at –40°C in HM20 for 3 d and then warmed to room temperature over a 6-h period. Embedded cells were sectioned and immunostained as follows: sections on gold grids (Electron Microscopy Sciences) were floated on blocking buffer (0.02% Tween 20, 0.8% bovine serum albumin, and 0.1% fish gelatin in PBS) for 1 h, immunostained with anti-GFP antibodies overnight at 4°C as described previously, rinsed with PBS plus 0.1% Tween 20 three times, incubated with 10-nm gold goat-anti-rabbit secondary antibodies (BB International, Cardiff, United Kingdom) for 2 h at room temperature, rinsed and fixed with 1.0% glutaraldehyde for 5 min, stained with aqueous uranyl acetate and lead citrate, and imaged in a Philips Tecnai TF20 FEG electron microscope operating at 80 keV.
Mia1p/Microtubule Association Experiments
E. coli BL21 was used to express glutathione S-transferase (GST)-Mia1p. Pelleted bacterial cells were dissolved in the SDS-PAGE sample buffer, separated on SDS-PAGE gel, and then transferred to nitrocellulose filter and incubated in the PEMG buffer [100 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.9, 2 mM EGTA, 1 mM MgSO4, and 1 mM GTP] containing 5% milk and then with taxol-stabilized bovine brain 3X microtubules (50 μg/ml) in the same buffer, followed by washing and immunoblotting with TAT1 anti-tubulin antibodies.
RESULTS
Lack of the Microtubule Binding Protein Mia1p Results in Fewer Interphase Microtubules
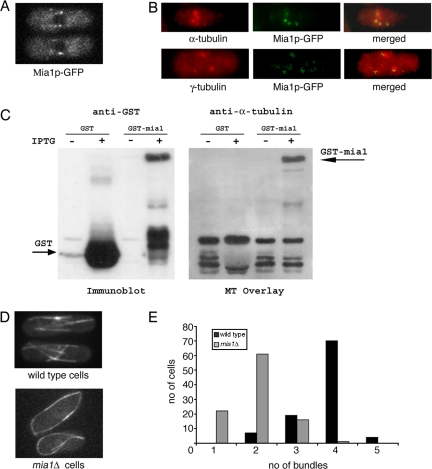
In interphase, S. pombe cells Mia1p-GFP expressed from its native promoter localized in a dotted pattern along microtubules and was particularly concentrated at the regions of microtubule minus ends (Figure 1A; Oliferenko and Balasubramanian, 2002). Interestingly, when microtubules were depolymerized by Carbendazim (MBC) treatment, Mia1p-GFP was visualized as several distinct dots around the NE (Figure 1B) colocalizing with γ-tubulin and short microtubule “stubs.” Far Western analysis using prepolymerized taxol-stabilized microtubules suggested that the recombinant Mia1p-GST could physically associate with microtubules (Figure 1C).
Figure 1.
Lack of the microtubule-binding protein Mia1p results in fewer microtubule bundles in interphase S. pombe cells. (A) Mia1p-GFP is enriched at the minus ends of microtubule bundles in interphase cells. (B) On MBC treatment, Mia1p-GFP localized to distinct dots around the NE coinciding with short microtubule stubs (top) and largely colocalizing with γ-tubulin-rich structures (bottom), as seen from anti-α-tubulin, anti-γ-tubulin, and anti-GFP immunofluorescence images of Mia1p-GFP cells. (C) Filter-immobilized recombinant Mia1p-GST bound taxol-stabilized microtubules in the Far Western assay. Binding was not observed with GST alone. (D) Unlike in the wild-type case, there are fewer microtubule bundles in mia1Δ cells. Shown are single maximum intensity reconstructions of α-tubulin-GFP–expressing wild-type and mia1Δ cells. (E) Histogram of the number of microtubule bundles in wild-type and mia1Δ cells (n = 100 cells).
On average, four antiparallel microtubule bundles run along the axis of the wild-type cell in a characteristic “parallel bars” array (Figure 1, D and E), with microtubule minus ends bundled and anchored at the specific sites on the NE (so-called iMTOCs) and plus ends terminating at the cell tips. In contrast, cells lacking Mia1p usually exhibited two microtubule bundles curving around cell tips (Figure 1, D and E). Because Mia1p was enriched at the sites of microtubule attachment to the NE (iMTOCs), we concluded that it was likely to participate in some aspects of their function.
mia1Δ Cells Exhibit Disorganized and Heterogeneous Microtubule Bundles
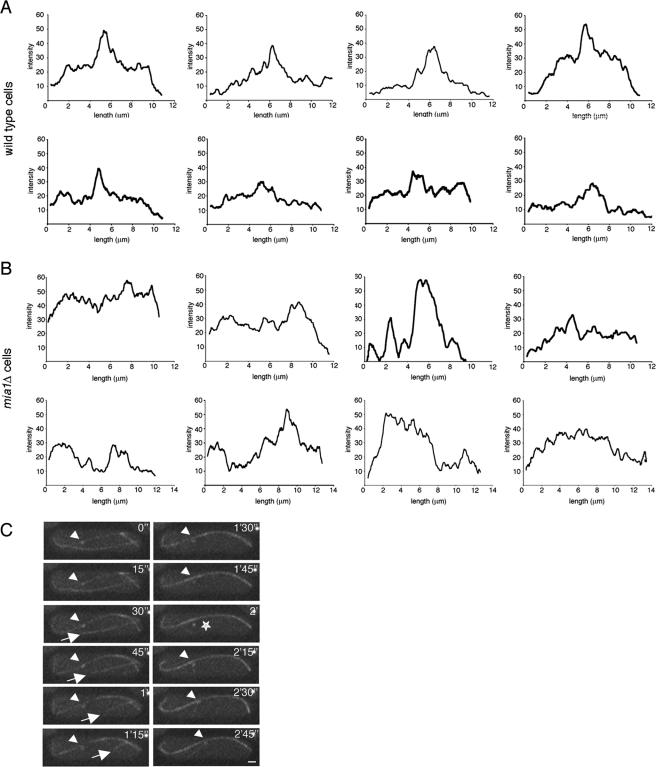
Intensity scan measurements along the length of microtubule bundles in wild-type cells revealed distinct medial zones of overlap (Figure 2A) as published previously (Tran et al., 2001; Loiodice et al., 2005). These double intensity zones are usually positioned near the nucleus and often overlap with the iMTOCs. Using similar techniques, we found no evidence of well defined or properly positioned zones of microtubule overlap in mia1Δ cells (Figure 2B). Indeed, these measurements suggested extreme heterogeneity in architecture of microtubule bundles in the absence of Mia1p. Time-lapse analyses of microtubule and the SPB dynamics in mia1Δ cells revealed presence of free microtubules, not associated with antiparallel bundles and the nuclear surface (Figure 2C and Supplemental Movie 1). When encountering cell tips, microtubules often continued growth and curved around them (Figure 2C and Supplemental Movie 1). We also detected instances of the SPB detachment from the associated microtubule bundles (Figure 2C, star).
Figure 2.
mia1Δ cells have disorganized and heterogeneous microtubule bundles. (A) Intensity scans of individual microtubule bundles in eight wild-type cells. (B) Intensity scans of individual microtubule bundles in eight mia1Δ cells. Note that although there are distinct medial regions of microtubule overlap in wild-type cells (A), mia1Δ cells (B) show heterogeneous patterns of microtubule arrangements. (C) Time-lapse sequence α-tubulin-GFP– and Sid2p-GFP–expressing mia1Δ cell shows presence of free microtubules undergoing catastrophe (indicated by an arrow) and instances of the SPB detachment from microtubule bundles (indicated by a star). The SPB in the sequence is indicated by an arrowhead. Shown are single maximum intensity reconstructions of z-stacks. Numbers refer to the time, in minutes and seconds. Bar, 1 μm.
mia1Δ Cells Are Defective in Attachment of Microtubules to the Nucleation Sites
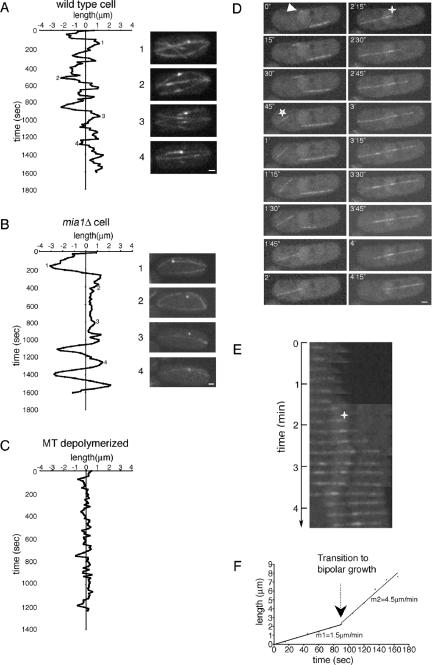
To investigate the phenotypes described above, we followed the SPB behavior and microtubule dynamics in wild-type and mia1Δ cells expressing Sid2p-GFP and α-tubulin-GFP. In wild type, the SPB periodically oscillated along the long axis around the geometric cell center (100%, n = 20 cells, observed for total of 3.5 h; see also Figure 3A and Supplemental Movie 2). This behavior is due to the balanced pushing forces that are exerted by plus ends of microtubules arranged in antiparallel bundles anchored at the nuclear surface (Tran et al., 2001). When microtubules in wild-type cells were depolymerized by MBC, the SPB remained stationary (Figure 3C). In mia1Δ cells, the SPB was either oscillating on much broader scale or abruptly stopped for periods of time (Figure 3B and Supplemental Movie 3). We found that the SPB laterally detached from the associated microtubule bundle during the periods of rest (26%, n = 19 cells, observed for a total of 9.5 h; also see Figure 3B and Supplemental Movie 3).
Figure 3.
Mia1p is required for attachment of microtubules to the SPBs and the NE. (A) Plot of SPB oscillations in α-tubulin-GFP Sid2p-GFP–expressing wild-type cell. Some extreme positions of the SPB (indicated as 1–4) are shown on the right of the graph. (B) Plot of SPB movements in α-tubulin-GFP– and Sid2p-GFP–expressing mia1Δ cell. Note that the SPB detached in the course of an experiment (compare positions 2 and 3 with 1 and 4). (C) In MBC-treated wild-type cell, the SPB remained stationary. Shown are single maximum intensity reconstructions of z-stacks. (D) Time-lapse sequence of α-tubulin-GFP–expressing mia1Δ cell. A microtubule nucleation event (indicated by an arrowhead) is followed by microtubule ejection from the NE (indicated by a star). Shown are single maximum intensity reconstructions of z-stacks. Numbers refer to the time, in minutes and seconds, elapsed from the nucleation event. (E) Kymograph of the growing microtubule from the time-lapse sequence presented in D. Time 0 corresponds to the time of microtubule ejection from the NE (marked by a star). Note that microtubule first grew at the plus end facing the cell tip, pushing its minus end toward the center of the cell. A second nucleation event was followed by formation of the antiparallel bundle (marked by a cross; also see corresponding image in D). The resulting bipolar microtubule bundle continued to grow on both sides until the older bundle underwent catastrophe. (F) Graph of microtubule growth rate obtained from the kymograph.
Interestingly, we found that cells lacking the Mia1p-interacting protein of the MAP215 family, Alp14p, exhibited a different phenotype with respect to the SPB/microtubule detachment. Microtubules remained attached to the SPBs in the course of time-lapse movies of alp14Δ Sid2p-GFP α-tubulin-GFP cells (100%, n = 20 cells, observed for total of 3 h) (Supplemental Figure 1A and Supplemental Movie 4). However, we found that the SPBs hardly oscillated, suggesting that microtubules did not exert sufficient pushing force (Supplemental Figure 1B).
Time-lapse analyses of microtubule dynamics in α-tubulin-GFP expressing mia1Δ cells showed instances of microtubule nucleation at the NE (as shown in Figure 3D and Supplemental Movie 5). In this case, a growing microtubule was ejected from the nucleation site and continued to grow at its plus end, pushing the stable minus end across the cell (Figure 3, D–F). Subsequently, we observed a second nucleation event, as judging by a sudden increase in fluorescence intensity in the vicinity of the minus end (as marked by a cross in Figure 3, D and E). This nucleation event was followed by antiparallel bundling, and the resulting bundle now grew at both plus ends (Figure 3, E and F).
Our analyses so far suggested that nucleation and bundling could occur in mia1Δ cells. However, Mia1p was required for microtubule attachment to the nucleation sites.
Mia1p Is Not Required for Microtubule Nucleation but Is Required for Maintenance of Microtubule Bundles
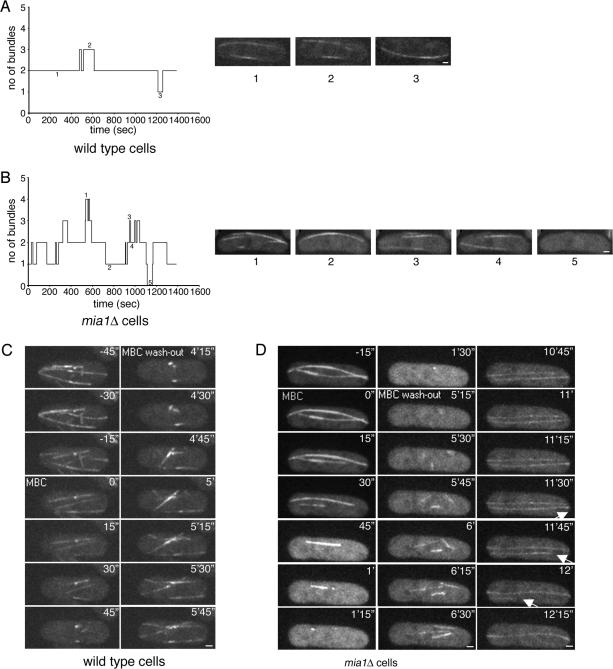
We followed α-tubulin-GFP–expressing wild-type and mia1Δ cells and performed fast (every 5 s) time-lapse analyses of single focal planes. We found that in wild-type cells, microtubule bundles were relatively stable, and we rarely observed the disappearance of preexisting bundles (1.2 events per bundle per hour, n = 8, total time = 3.5 h) (Figure 4A and Supplemental Movie 6). However, in mia1Δ cells the lifetime of microtubule bundles was significantly reduced (bundles disappeared with the rate of 13.6 events per bundle per hour, n = 10, total time = 3.5 h) (Figure 4B and Supplemental Movie 7). We also found that microtubule nucleation in wild-type cells was mainly restricted to preexisting MTOCs (we observed only 4.7 de novo nucleation events per cell per hour, n = 6, total time = 2.5 h). In contrast, microtubules were frequently nucleated from different sites around the NE in the absence of Mia1p (23 events per cell per hour, n = 11, total time = 3.3 h).
Figure 4.
Microtubules can be nucleated from around the nuclear envelope in mia1Δ cells, but the resulting microtubule bundles are not stable. (A) Graph and several still images (1–3) following number of microtubule bundles present in one focal plane of the wild-type cell over a period of time. (B) Graph and several still images (1–5) following number of microtubule bundles present in one focal plane of the mia1Δ cell over a period of time. Data presented in A and B were obtained from time-lapse experiments using α-tubulin-GFP–expressing wild-type and mia1Δ cells (images were taken every 5 s). (C) Wild-type cells expressing α-tubulin-GFP Sid2p-GFP were mounted in the flow chamber allowing the time-lapse analysis of microtubule dynamics during MBC treatment (50 μg/ml) and subsequent washout. Microtubules depolymerized after MBC treatment leaving stable “stubs” at the NE. On washout, microtubules rapidly repolymerized from these “stubs” toward cell periphery. (D) Time lapse analysis of α-tubulin-GFP Sid2p-GFP expressing mia1Δ cell in a similar experimental setup. On washout with fresh medium, we detected efficient nucleation of microtubules from around the NE. At least one microtubule eventually detached and underwent catastrophe (as indicated by an arrow). Shown are single maximum intensity reconstructions of z-stacks. Numbers refer to the time, in minutes and seconds. Bar, 1 μm.
So far our analyses concerned steady-state mia1Δ cells. We decided to perturb the microtubule cytoskeleton by treating cells with MBC and observe microtubule dynamics upon washout of the drug. We performed time-lapse analyses of α-tubulin-GFP Sid2p-GFP–expressing wild-type and mia1Δ cells in a perfusion chamber mounted on a microscope stage. In wild-type cells, microtubules rapidly depolymerized upon addition of 50 mg/ml MBC (within 45 s), leaving stable microtubule “stubs” around the NE (Figure 4C and Supplemental Movie 8). When the drug was washed out with fresh medium, microtubules quickly repolymerized from these “stubs” in a bidirectional manner (Figure 4C and Supplemental Movie 8). MBC-induced depolymerization of microtubules in mia1Δ cells proceeded somewhat slower (∼90 s), but eventually most microtubules disassembled (Figure 4D and Supplemental Movie 9). On washout with fresh medium, we observed microtubule nucleation from several sites around the NE (Figure 4D and Supplemental Movie 9). These microtubules could eventually detach, and we documented one of them undergoing catastrophe and shrinking across the cell body exhibiting a plus-end depolymerization rate of ∼12 mm/min (later time points in Figure 4D and Supplemental Movie 9).
We confirmed that microtubule polymerization could be initiated from the nuclear surface by returning ice-treated wild-type and mia1Δ cells expressing the NE marker Uch2p-GFP and α-tubulin-GFP to the room temperature and performing the fluorescent time-lapse analyses of microtubule regrowth. We found that microtubules could be polymerized from the NE in both wild-type (Supplemental Figure 2A and Supplemental Movie 10) and mia1Δ (Supplemental Figure 2B and Supplemental Movie 11) cells. However, microtubules eventually detached from the NE in cells lacking Mia1p (later time points in Supplemental Figure 2B and Supplemental Movie 11).
Mia1p Is Required for Sustaining Proper Postanaphase Array Dynamics
Dividing fission yeast cells assemble a specialized microtubule arrangement at the division site, called PAA. Microtubules are nucleated symmetrically at the eMTOC with their plus ends reaching toward mother cell tips. eMTOC constricts together with the actomyosin ring and eventually disassembles. We found that Mia1p-GFP localized in the vicinity of the eMTOC both on light microscopy (Figure 5A) and ultrastructural (Figure 5B) levels. We thus set out to examine whether any aspects of the eMTOC/PAA dynamics could be affected in mia1Δ cells. α-Tubulin-GFP–expressing wild-type cells started assembling the medial microtubule foci in late anaphase (Figure 5C, marked with stars in 1′30” time point, and Supplemental Movie 12). These foci persisted and eventually fused into a single ringlike structure that constricted together with the actomyosin ring (Figure 5C and Supplemental Movie 12). We tracked paths of these focal structures over the course of cell division and combined them on a single superimposed image (Figure 5C, C′). Our analysis confirmed that once assembled, these microtubule-nucleating structures remained fairly stable. In contrast, although we detected microtubule nucleation events in the medial region of mia1Δ cells (Figure 5D and Supplemental Movie 13), the resulting microtubule structures were unstable (Figure 5D and Supplemental Movie 13; note the ejection event of a focal structure between 1 min 30 s and 2 min, marked with arrow). Tracking analysis suggested that each structure had a fairly short lifetime and that cells proceeded initiating multiple nucleation events from different regions of the eMTOC (Figure 5D, D′). This behavior resulted in a poorly organized PAA and a precocious loss of microtubules from the medial region (Figure 5D and Supplemental Movie 13).
Figure 5.
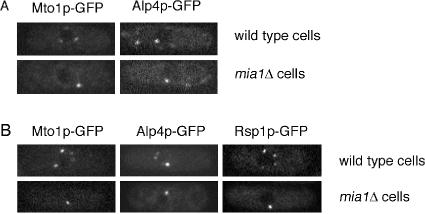
Cells lacking Mia1p do not sustain normal PAA. (A) Mia1p-GFP localizes to the eMTOC at the division site in live cells. (B) Immunoelectron micrograph image of cell expressing Mia1p-GFP using 10-nm gold particles, where Mia1p-GFP is found on both sides of the division site (indicated by stars). Bar, 300 nm. (C) Time-lapse sequence of the dividing α-tubulin-GFP–expressing wild-type cell. Once established, the microtubule-organizing structures are maintained and eventually constrict together with the actomyosin ring. The medial microtubule foci are marked with stars in time point 1′30”. Tracking of these structures from their first appearance to ring constriction suggests that they are relatively persistent (as shown in C′). (D) In contrast, time-lapse sequence of the dividing α-tubulin-GFP–expressing mia1Δ cell suggests that although microtubules can be nucleated from the eMTOC, there are no persistent nucleation sites (compare D′ with C′). Different colors in C′ and D′ represent independent nucleation sites. Ejection event is marked by arrow in time point 1′30”. Shown are single maximum intensity reconstructions of z-stacks. Numbers refer to the time, in minutes and seconds. Bar, 1 μm. (E) Proteins associated with the γ-tubulin complex, Alp4p-GFP, Mto1p-GFP, and Rsp1p-GFP localize to the equatorial ring in the absence of Mia1p.
To probe whether the eMTOC was still present in the absence of Mia1p, we performed live cell fluorescence imaging using several eMTOC markers. We found that localization of Alp4p-GFP, Mto1p-GFP, and Rsp1p-GFP in mia1Δ cells was comparable with wild-type cells (Figure 5E). The eMTOC constriction and disappearance of the equatorial ring material proceeded similarly in mia1Δ and wild-type cells (our unpublished data). We concluded that although Mia1p was not required to localize the γ-tubulin complexes to the actomyosin ring, it was necessary to sustain the continuing use of nucleating sites through the course of cell division.
There Are No Detectable γ-Tubulin–rich Interphase MTOC Structures in Cells Lacking Mia1p
Because microtubules could be nucleated from around the NE in mia1Δ cells, it could be expected that Mia1p executed its function downstream of MTOC components and that those localized normally in the absence of Mia1p. In steady-state interphase, wild-type cells Mto1p-GFP and Alp4p-GFP localized to the SPB and to several bright dots around the nucleus and along microtubules (Figure 6A), likely representing the MTOCs. In mia1Δ cells, we normally detected only the SPB signal together with very faint and even staining along microtubules and around the NE in Mto1p-GFP (Figure 6A).
Figure 6.
Components of the γ-tubulin complex are not found in large structures around the nuclear envelope or on microtubules in interphase mia1Δ cells. (A) Mto1p-GFP and Alp4-GFP localize to the SPB, several large aggregates around the NE, and along microtubules in interphase wild-type cells. In mia1Δ cells, Mto1p and Alp4p-GFP mainly localize to the SPB. Shown are single maximum intensity reconstructions of z-stacks. (B) Components of the iMTOCs are detected as several distinct dots around the nuclear envelope in wild-type but not mia1Δ cells treated with the microtubule-destabilizing drug MBC. Their SPB localization remains unchanged.
When microtubules in wild-type cells expressing fluorescent MTOC markers were depolymerized by addition of MBC, we detected several bright dots around the NE (Figure 6B). One of these is the SPB, and the rest are probably the microtubule attachment sites to the NE, i.e., the iMTOCs. These structures were also rich in γ-tubulin (Figure 1B). We did not detect iMTOCs as visualized by these markers in MBC-treated mia1Δ cells; instead, they only localized to the SPB (Figure 6B).
Because Alp14p was shown to interact with Mia1p (Sato et al., 2004), we addressed the question of γ-tubulin complex distribution in cells lacking this protein. Interestingly, Mto1p-GFP in alp14Δ cells exhibited a strong diffuse signal around the nuclear envelope, in addition to its SPB localization, both in steady-state cells and upon pretreatment with MBC (Supplemental Figure 3, top). This was in marked contrast with cells lacking a distinct microtubule-stabilizing protein, Mal3p (Beinhauer et al., 1997), where Mto1p-GFP could be found in bright puncta around the NE, similar to the wild-type situation (Supplemental Figure 3, bottom).
We concluded that although the microtubule-nucleating complexes were active at the NE of mia1Δ cells, they were not organized into discernible iMTOC structures, probably reflecting the lack of microtubule/NE attachment sites. Thus, it seemed that mia1Δ cells were deficient in establishing the iMTOC structures at the NE.
Microtubules Are Required for Emergence of Interphase MTOC Structures Upon Entry into the New Cell Cycle
We hypothesized that lack of large MTOC structures was related to the deficient microtubule anchorage in the absence of Mia1p. There is a considerable amount of microtubule-nucleating material traveling along microtubules in both plus- and minus-end–directed manner, and it was recently proposed that γ-tubulin complexes nucleate new microtubules as they travel along the preexisting microtubules (Janson et al., 2005). These microtubules could later slide toward the cell center (Carazo-Salas et al., 2005) and eventually get anchored at the NE. We proposed a model where fluctuations in initial distribution and/or activity of γ-tubulin complexes are positively reinforced when additional material is delivered to the nucleating sites via attached microtubules (Figure 7D). Thus, the resulting large structures could serve as dominant microtubule-organizing centers. This positive feedback loop would be disrupted in mia1Δ cells due to the lack of microtubule attachment to the nucleation sites, leading to a defect in γ-tubulin complexes' coalescence into large NE-bound MTOCs (Figure 7D).
Figure 7.
Microtubules are required for emergence of large iMTOC structures upon entry into the new cell cycle. (A) When control Mto1p-GFP mad2Δ cells underwent septation at 18°C, we detected several large Mto1p-GFP–positive structures around the NE of daughter cells. However, Mto1p-GFP was uniformly distributed around the NE of Mto1p-GFP mad2Δ nda3-KM311 cells that septated at the restrictive temperature of 18°C. The SPB localization was comparable in both strains. (B) When Mto1p-GFP–expressing cells exiting mitosis were treated with MBC, septation proceeded normally, but we could not detect formation of the iMTOCs in daughter cells (75%; n = 38 cells). (C) When we prepared daughter cells lacking iMTOC structures in the manner described above and washed out the drug, we observed an emergence of Mto1p-GFP–positive distinct dots around nuclei and along microtubules, presumably the iMTOCs (100%; n = 20 cells). Microtubules were subsequently (3 min) depolymerized to allow better visualization of the emergent structures. In A–C, the single maximum intensity reconstructions of z-stacks are shown. (D) A model of the iMTOC formation in S. pombe cells. Fluctuations in initial distribution and/or activity of γ-tubulin complexes are positively reinforced when additional complexes are delivered to the nucleating sites via attached microtubules. This positive feedback loop is disrupted in mia1Δ cells due to the lack of microtubule attachment to the nucleating sites, leading to a defect in coalescence of γ-tubulin complexes into larger MTOC structures.
This model predicts that emergence of interphase MTOCs in each cell cycle should depend on microtubules. We made use of the cold-sensitive β-tubulin mutation nda3-KM311 and followed the fluorescent MTOC marker Mto1p-GFP through the cell cycle. To allow cell division, we introduced a deletion mutation of the spindle assembly checkpoint gene, mad2Δ (Li and Murray, 1991) into this genetic background. We found that when Mto1p-GFP mad2Δ nda3-KM311 cells underwent septation at the restrictive temperature of 18°C, only one daughter cell inherited the undivided nucleus. Mto1p-GFP was uniformly distributed around the NE and also localized to non-separated SPBs (Figure 7A; 85%; n = 66 cells). In contrast, upon division of control Mto1p-GFP mad2D cells, we detected distinct bright fluorescent structures around the NE of both daughter cells (Figure 7A; 95%; n = 62 cells). This experiment suggested that microtubules were required for enrichment of γ-tubulin complex material in distinct NE-bound MTOCs in each new cell cycle.
To follow this process in real time, we immobilized Mto1p-GFP–expressing wild-type cells in a perfusion chamber mounted on a fluorescence microscope and searched for cells completing mitosis where Mto1p-GFP was localized exclusively to the SPBs and the nascent eMTOC (Figure 7B). We depolymerized microtubules by perfusing medium containing 25 mg/ml MBC and observed the localization of Mto1p-GFP. We found that although eMTOC disassembly and septation proceeded normally, the majority of MBC-treated cells (75%; n = 38 cells) did not exhibit any noticeable MTOC structures around the nuclear envelope; instead, Mto1p-GFP localized exclusively to the SPB (Figure 7B). We also found that when MBC was added just after the onset of the eMTOC/actomyosin ring constriction, we could detect nascent MTOCs around the NE (Supplemental Figure 4), suggesting that iMTOC establishment proceeded simultaneously with the eMTOC disassembly.
Our model also assumes that as long as γ-tubulin complexes and microtubules are present in cells with no previous iMTOCs, these structures should be able to readily self-organize (Figure 7D). We prepared Mto1p-GFP cells lacking iMTOCs as described above (Figure 7B) and allowed microtubule polymerization by washing out MBC. We found that 20 min after MBC washout, 100% of cells (n = 20) exhibited Mto1p-GFP–positive aggregates at the nuclear envelope and spread along the microtubules. We then briefly depolymerized microtubules in these cells to allow a better visualization of the fluorescent material at the NE (Figure 7C). We concluded that γ-tubulin complexes could efficiently self-organize into large interphase microtubule-organizing centers only in the presence of microtubules.
DISCUSSION
In this report, we investigated roles of the fission yeast TACC-related protein Mia1p in organization of interphase microtubule arrays. Based on our findings, we proposed and tested a dynamic model of assembly of the MTOCs at the nuclear surface of interphase S. pombe cells.
There are several clusters of experimentally described mutant phenotypes leading to abnormalities in the fission yeast microtubule cytoskeleton. For example, shorter interphase microtubules might indicate deficiencies in microtubule-stabilizing factors (Beinhauer et al., 1997; Brunner and Nurse, 2000), and disorganized microtubules without obvious overlapping regions could result from the lack of bundling proteins (Loiodice et al., 2005). A separate class of mutants exhibits fewer microtubule bundles that on average are longer than usual. Examples include mutations in γ-tubulin (Gtb1p) (Paluh et al., 2000), the core γ-TuRC components Alp4p and Alp6p (Vardy and Toda, 2000; Zimmerman and Chang, 2005), or γ-TuRC accessory proteins Mto1p (Sawin et al., 2004; Venkatram et al., 2004; Zimmerman and Chang, 2005) and Mto2p (Janson et al., 2005; Samejima et al., 2005; Venkatram et al., 2005). Microtubule nucleation and therefore MTOC function are suppressed in cells lacking either of these factors.
Similarly, we found a decreased number of microtubule bundles in interphase mia1Δ cells (Figure 1, D and E). Microtubules were disorganized (Figure 2B) and curved around cell tips (Figures 1D and 2C); mia1Δ cells were often bent (Radcliffe et al., 1998; Oliferenko and Balasubramanian, 2002). However, unlike previously described mutants, cells lacking Mia1p were not deficient in microtubule nucleation and indeed nucleated microtubules, both in steady state (Figures 3, D and E, and 4B, and Supplemental Movies 5 and 7), and upon experimental perturbations of the microtubule cytoskeleton (Figure 4D, Supplemental Figure 2B, and Supplemental Movies 9 and 11). Indeed, the main abnormality in mia1Δ cells seems to be a faulty attachment of microtubule minus ends to the nucleation sites (Figures 2C, 3, B and D, Supplemental Figure 2B, and Supplemental Movies 1, 3, 5, and 11). We detected instances of lateral loss of microtubule bundles from the NE and the SPBs (Figures 2C and 3B), and microtubule ejection events when minus ends of nucleated microtubules were displaced from the NE before antiparallel bundling (Figure 3, D–F).
Mia1p is related to a large family of TACC proteins implicated in sustaining mitotic centrosome function and spindle integrity and dynamics. Misregulation of TACC gene expression has been linked to development of human cancers (for review, see Raff, 2002). The fruit fly D-TACC participates in spindle pole focusing and organization during both anastral meiotic (Cullen and Ohkura, 2001) and mitotic divisions (Lee et al., 2001). Lack of TAC-1 in one-cell embryos of Caenorhabditis elegans leads to very short, unstable astral and spindle microtubules (Bellanger and Gonczy, 2003; Le Bot et al., 2003; Srayko et al., 2003). TACC family members usually associate with the XMAP215 family of microtubule-stabilizing proteins, and this interaction is functionally significant: mutations in these genes to a certain extent phenocopy each other (Cullen and Ohkura, 2001; Lee et al., 2001; Bellanger and Gonczy, 2003; Le Bot et al., 2003; Srayko et al., 2003).
It was reported that temperature-sensitive allele of the fission yeast XMAP215 protein Alp14p caused shortening of interphase microtubules (Sato et al., 2004), which is consistent with its function in regulating microtubule stability. We found that microtubules remained attached to the SPBs in alp14Δ cells (Supplemental Figure 1A and Supplemental Movie 4), unlike in mia1Δ genetic background. However, it is possible that lack of SPB oscillations in alp14Δ cells could mask the potential detachment phenotype (Supplemental Figure 1B). Detailed studies of the microtubule cytoskeleton in alp14Δ cells will require development of additional tools because only very low levels of α-tubulin expression are tolerated in this genetic background (our unpublished data). In principle, it is possible that Mia1p could function in microtubule attachment to the nucleation sites independently of Alp14p, because localization of Mia1p/Alp7p to the SPBs does not require Alp14p (Sato et al., 2004). However, future studies are needed to address the functional contributions of these two proteins in organizing interphase microtubule arrays.
Another exciting aspect of Mia1p function has been illuminated by lack of large interphase MTOCs in mia1Δ cells. Although microtubule-nucleating activity is present at the NE (Figures 3D and 4D) and along microtubules (Figure 3, E and F) of mia1Δ cells, the γ-tubulin complexes are not organized into discernible structures, unlike in the wild-type case (Figure 6). Interestingly, we noticed that components of the γ-tubulin complex were enriched at the nuclear surface in alp14Δ cells (Supplemental Figure 3). It could be either due to their inability to load on microtubules or due to the general scarcity of microtubules in this genetic background. Interphase alp14Δ cells have very few microtubules, even compared with cells deficient in other microtubule-stabilizing protein, Mal3p (EB1) (Supplemental Figure 1 and Supplemental Movie 4; our unpublished data). It is worth mentioning that mal3Δ cells assembled functional nuclear-bound iMTOCs (Supplemental Figure 3), suggesting that lack of these structures in mia1Δ and alp14Δ cells did not result from general problems with plus end microtubule stability.
We proposed that absence of the iMTOCs in mia1Δ cells could result from a molecular defect of microtubule attachment to the nucleation sites (see model in Figure 7D). In our model, the interphase MTOCs are dynamic structures that are established de novo in each cell cycle after the disassembly of the eMTOC. First, microtubules are nucleated randomly due to variations in density or activity of nascent γ-tubulin complexes at the nuclear surface. However, when newborn microtubules remain anchored, more γ-tubulin complexes together with microtubules nucleated by them (Janson et al., 2005) can be delivered to the original sites of nucleation, resulting in growth of the nucleating structures and eventually restricting the MTOC number. This positive feedback loop would be disrupted in mia1Δ cells due to lack of microtubule attachment to the nucleating sites leading to a defect in coalescence of γ-tubulin complexes into larger iMTOCs (Figure 7D).
These large structures at the NE are best revealed upon depolymerization of microtubules; however, we prefer to view them as functional entities that represent the sites of attachment of microtubule bundles to the NE. One of the more obvious physiological functions for these structures could be ensuring the proper centering of nuclei (Tran et al., 2001). Although nucleation of microtubules can occur elsewhere, either on preexisting microtubules or in cytosol, the resulting microtubules slide toward the nucleus in a Klp2p-dependent manner (Carazo-Salas et al., 2005; Janson et al., 2005) and are likely captured by the NE attachment sites. These attachment sites are thus likely to serve as dominant microtubule-organizing centers contributing to the establishment and maintenance of overall microtubule architecture.
Interestingly, we observed that although we detected instances of microtubule loss from the eMTOC (Figure 5, C and D), all γ-tubulin complex components that we looked at localized to the equatorial MTOC in mia1Δ cells (Figure 5E). This suggested that Mia1p and microtubule attachment were not required for their localization, which is not surprising given that microtubules are not required for the eMTOC formation (Heitz et al., 2001) and Mia1p does not seem to be an integral component of the nucleating complex.
Based on our model for emergence of the interphase MTOCs, we went ahead to show that microtubules are required for their assembly in wild-type cells (Figure 7). When microtubules were absent in dividing mother cells, we could not detect the interphase MTOCs in daughters (Figure 7, A and B). In contrast, when we allowed microtubule polymerization in daughter cells with no previous history of the interphase MTOCs, these structures readily occurred (Figure 7C).
Noncentrosomal MTOCs have been reported in many systems ranging from the nuclear surface of higher plants (Stoppin et al., 1994) to the membranes of animal cells (Keating and Borisy, 1999; Rios et al., 2004). Thus, a common self-organizing mechanism could ensure emergence of the membrane-bound MTOCs. Our model also suggested that one or more minus-end–directed motors transporting γ-tubulin satellites and microtubules nucleated elsewhere could be involved in the process of MTOC assembly. It was recently shown that the Kar3-type kinesin Klp2p is involved in sliding of microtubules toward cell center along preexisting microtubules and focusing of microtubule arrays near the nucleus (Carazo-Salas et al., 2005). Evidence from other systems suggests that the minus-end–directed transport of the microtubule-nucleating machinery contributing to the organization of MTOCs might be a universal occurrence (Kubo et al., 1999; Young et al., 2000). Also, even though the fission yeast iMTOCs do not contain centrioles, our findings could, in principle, be extended to explain the de novo establishment of centriole-containing centrosomes in animal cells. It was shown that centrosomes and centrioles could be assembled de novo in mammalian (Khodjakov et al., 2002) and Chlamydomonas (Marshall et al., 2001) cells. Interestingly, assembly of the pericentriolar material as single spots in mammalian cells depended on microtubules (Khodjakov et al., 2002). Similar dense assemblies of the pericentriolar material including γ-tubulin complexes seemed necessary for the birth of centrioles (Dammermann et al., 2004).
In conclusion, self-organization of γ-tubulin–containing material into large MTOCs could facilitate efficient nucleation, bundling, and intracellular positioning of cytoskeletal arrays.
Supplementary Material
Acknowledgments
We are grateful to P. Tran for help in analyzing and discussing data and to M. Balasubramanian, J. R. McIntosh, and members of the Cell Dynamics and Cell Division groups for discussions and encouragement. M. Balasubramanian and V. Wachtler kindly commented on the manuscript. Many thanks are due to M. Balasubramanian, K. Gull, K. Gould, Y. Hiraoka, J. R. McIntosh, and F. Chang for generous sharing of materials and reagents. This work was supported by National Institutes of Health and Biotechnology Resources grant RR00592 to J. R. McIntosh and intramural funds from the Temasek Life Sciences Laboratory.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–08–0811) on February 15, 2006.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Beinhauer, J. D., Hagan, I. M., Hegemann, J. H., and Fleig, U. (1997). Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J. Cell Biol. 139, 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger, J. M., and Gonczy, P. (2003). TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr. Biol. 13, 1488–1498. [DOI] [PubMed] [Google Scholar]
- Brunner, D., and Nurse, P. (2000). CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102, 695–704. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas, R. E., Antony, C., and Nurse, P. (2005). The kinesin Klp2 mediates polarization of interphase microtubules in fission yeast. Science 309: 297–300. [DOI] [PubMed] [Google Scholar]
- Cullen, C. F., and Ohkura, H. (2001). Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat. Cell Biol. 3, 637–642. [DOI] [PubMed] [Google Scholar]
- Dammermann, A., Muller-Reichert, T., Pelletier, L., Habermann, B., Desai, A., and Oegema, K. (2004). Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 7, 815–829. [DOI] [PubMed] [Google Scholar]
- Hagan, I. M. (1998). The fission yeast microtubule cytoskeleton. J. Cell Sci. 111, 1603–1612. [DOI] [PubMed] [Google Scholar]
- Heitz, M. J., Petersen, J., Valovin, S., and Hagan, I. M. (2001). MTOC formation during mitotic exit in fission yeast. J. Cell Sci. 114, 4521–4532. [DOI] [PubMed] [Google Scholar]
- Janson, M. E., Setty, T. G., Paoletti, A., and Tran, P. T. (2005). Efficient formation of bipolar microtubule bundles requires microtubule-bound {gamma}-tubulin complexes. J. Cell Biol. 169, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating, T. J., and Borisy, G. G. (1999). Centrosomal and non-centrosomal microtubules. Biol. Cell. 91, 321–329. [PubMed] [Google Scholar]
- Khodjakov, A., Rieder, C. L., Sluder, G., Cassels, G., Sibon, O., and Wang, C. L. (2002). De novo formation of centrosomes in vertebrate cells arrested during S phase. J. Cell Biol. 158, 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, A., Sasaki, H., Yuba-Kubo, A., Tsukita, S., and Shiina, N. (1999). Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 147, 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bot, N., Tsai, M. C., Andrews, R. K., and Ahringer, J. (2003). TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr. Biol. 13, 1499–1505. [DOI] [PubMed] [Google Scholar]
- Lee, M. J., Gergely, F., Jeffers, K., Peak-Chew, S. Y., and Raff, J. W. (2001). Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 3, 643–649. [DOI] [PubMed] [Google Scholar]
- Li, R., and Murray, A. W. (1991). Feedback control of mitosis in budding yeast. Cell 66, 519–531. [DOI] [PubMed] [Google Scholar]
- Loiodice, I., Staub, J., Setty, T. G., Nguyen, N. P., Paoletti, A., and Tran, P. T. (2005). Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol. Biol. Cell 16, 1756–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W. F., Vucica, Y., and Rosenbaum, J. L. (2001). Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr. Biol. 11, 308–317. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Nedelec, F. J., Surrey, T., Maggs, A. C., and Leibler, S. (1997). Self-organization of microtubules and motors. Nature 389, 305–308. [DOI] [PubMed] [Google Scholar]
- Oliferenko, S., and Balasubramanian, M. K. (2002). Astral microtubules monitor metaphase spindle alignment in fission yeast. Nat. Cell Biol. 4, 816–820. [DOI] [PubMed] [Google Scholar]
- Paluh, J. L., Nogales, E., Oakley, B. R., McDonald, K., Pidoux, A. L., and Cande, W. Z. (2000). A mutation in gamma-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein pkl1p. Mol. Biol. Cell 11, 1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe, P., Hirata, D., Childs, D., Vardy, L., and Toda, T. (1998). Identification of novel temperature-sensitive lethal alleles in essential beta-tubulin and nonessential alpha 2-tubulin genes as fission yeast polarity mutants. Mol. Biol. Cell 9, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff, J. W. (2002). Centrosomes and cancer: lessons from a TACC. Trends Cell Biol. 12, 222–225. [DOI] [PubMed] [Google Scholar]
- Rios, R. M., Sanchis, A., Tassin, A. M., Fedriani, C., and Bornens, M. (2004). GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell 118, 323–335. [DOI] [PubMed] [Google Scholar]
- Samejima, I., Lourenco, P. C., Snaith, H. A., and Sawin, K. E. (2005). Fission yeast mto2p regulates microtubule nucleation by the centrosomin-related protein mto1p. Mol. Biol. Cell 16, 3040–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M., Vardy, L., Angel Garcia, M., Koonrugsa, N., and Toda, T. (2004). Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol. Biol. Cell 15, 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin, K. E., Lourenco, P. C., and Snaith, H. A. (2004). Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14, 763–775. [DOI] [PubMed] [Google Scholar]
- Srayko, M., Quintin, S., Schwager, A., and Hyman, A. A. (2003). Caenorhabditis elegans TAC-1 and ZYG-9 form a complex that is essential for long astral and spindle microtubules. Curr. Biol. 13, 1506–1511. [DOI] [PubMed] [Google Scholar]
- Stoppin, V., Vantard, M., Schmit, A. C., and Lambert, A. M. (1994). Isolated plant nuclei nucleate microtubule assembly: the nuclear surface in higher plants has centrosome-like activity. Plant Cell 6, 1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, P. T., Marsh, L., Doye, V., Inoue, S., and Chang, F. (2001). A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 153, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy, L., and Toda, T. (2000). The fission yeast gamma-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J. 19, 6098–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatram, S., Jennings, J. L., Link, A., and Gould, K. L. (2005). Mto2p, a novel fission yeast protein required for cytoplasmic microtubule organization and anchoring of the cytokinetic actin ring. Mol. Biol. Cell 16, 3052–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatram, S., Tasto, J. J., Feoktistova, A., Jennings, J. L., Link, A. J., and Gould, K. L. (2004). Identification and characterization of two novel proteins affecting fission yeast gamma-tubulin complex function. Mol. Biol. Cell 15, 2287–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, V., et al. (2002). The genome sequence of Schizosaccharomyces pombe. Nature 415, 871–880. [DOI] [PubMed] [Google Scholar]
- Young, A., Dictenberg, J. B., Purohit, A., Tuft, R., and Doxsey, S. J. (2000). Cytoplasmic dynein-mediated assembly of pericentrin and gamma tubulin onto centrosomes. Mol. Biol. Cell 11, 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, S., and Chang, F. (2005). Effects of γ-tubulin complex proteins on microtubule nucleation and catastrophe in fission yeast. Mol. Biol. Cell 16, 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, S., Tran, P. T., Daga, R. R., Niwa, O., and Chang, F. (2004). Rsp1p, a J domain protein required for disassembly and assembly of microtubule organizing centers during the fission yeast cell cycle. Dev. Cell. 6, 497–509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.