Abstract
Stresses affecting the endoplasmic reticulum (ER) globally modulate gene expression patterns by altering posttranscriptional processes such as translation. Here, we use tunicamycin (Tn) to investigate ER stress-triggered changes in the translation of cytochrome c, a pivotal regulator of apoptosis. We identified two RNA-binding proteins that associate with its ∼900-bp-long, adenine- and uridine-rich 3′ untranslated region (UTR): HuR, which displayed affinity for several regions of the cytochrome c 3′UTR, and T-cell-restricted intracellular antigen 1 (TIA-1), which preferentially bound the segment proximal to the coding region. HuR did not appear to influence the cytochrome c mRNA levels but instead promoted cytochrome c translation, as HuR silencing greatly diminished the levels of nascent cytochrome c protein. By contrast, TIA-1 functioned as a translational repressor of cytochrome c, with interventions to silence TIA-1 dramatically increasing cytochrome c translation. Following treatment with Tn, HuR binding to cytochrome c mRNA decreased, and both the presence of cytochrome c mRNA within actively translating polysomes and the rate of cytochrome c translation declined. Taken together, our data suggest that the translation rate of cytochrome c is determined by the opposing influences of HuR and TIA-1 upon the cytochrome c mRNA. Under unstressed conditions, cytochrome c mRNA is actively translated, but in response to ER stress agents, both HuR and TIA-1 contribute to lowering its biosynthesis rate. We propose that HuR and TIA-1 function coordinately to maintain precise levels of cytochrome c production under unstimulated conditions and to modify cytochrome c translation when damaged cells are faced with molecular decisions to follow a prosurvival or a prodeath path.
In response to environmental stress agents, cells modify the patterns of expressed genes in order to mount adequate responses. In addition to transcriptional events, stress-induced gene expression programs are strongly influenced by posttranscriptional regulatory processes, such as those controlling mRNA turnover and translation (39). In mammalian cells, damaging agents provoke widespread changes in mRNA stability, rendering many mRNAs more stable and others more labile (10), and can also elicit a generalized inhibition of protein biosynthesis (40). In the case of agents that perturb the homeostasis of the endoplasmic reticulum (ER), translational inhibition is effected through the phosphorylation of the alpha subunit of eukaryotic initiation factor 2 (eIF-2α) via the ER-resident kinase PERK (21). With a rapidly growing body of literature on mRNA turnover and translation, it is becoming increasingly clear that these two regulatory paradigms are intimately linked. A central point of convergence between these two processes is the involvement of RNA-binding proteins (RBPs) that associate with mRNAs which are subject to altered mRNA stability and translation (4, 47, 48). These mRNAs frequently bear adenine- and uridine-rich or uridine-rich regions (collectively named AU-rich elements, or AREs) in their 5′- and 3′-untranslated regions (UTRs), through which specialized RBPs govern their half-life and translation rate (4, 7, 51). ARE-RBPs include proteins that promote mRNA decay (including the AU-binding factor 1 [AUF1], tristetraprolin, the K homology splicing regulatory protein, and butyrate response factor 1), proteins that promote mRNA stabilization and modulate translation (such as the Hu proteins HuR, HuB, HuC, and HuD), and proteins that suppress translation, including the T-cell-restricted intracellular antigen 1 (TIA-1) and the TIA-1-related protein TIAR (3, 5, 6, 16, 31, 38, 41, 52).
HuR is predominantly localized in the nucleus, but it can shuttle between the nucleus and the cytoplasm upon cell stimulation. The influence of HuR on target mRNA stabilization and translation depends on its cytoplasmic presence (26, 27). HuR binds target mRNA subsets bearing AREs through its RNA recognition motifs and has been shown to regulate the expression of many target mRNAs, including those that encode c-fos, vascular endothelial growth factor, tumor necrosis factor alpha (TNF-α), β-catenin, c-myc, cyclooxygenase 2, myogenin, MyoD, several cyclins, granulocyte-macrophage colony-stimulating factor, several interleukins, p21, p27, p53, and hsp70. Accordingly, HuR has been directly implicated in regulating various cellular responses, including cell division, carcinogenesis, muscle cell differentiation, replicative senescence, immune cell activation, and stress responsiveness (5, 15).
TIA proteins have been proposed to play general roles as translational repressors in response to environmental stress agents (heat, oxidants, hyperosmolarity, etc.) (1, 2). Under conditions of stress, cells can form discrete cytoplasmic foci called stress granules (SGs). The assembly of SGs can be triggered by the phosphorylation of eIF2α, which effectively prevents the formation of the eIF2-GTP-Met-tRNAi ternary complex. TIA proteins have been postulated to take the place of the ternary complex and recruit untranslated mRNAs into SGs, wherein silent preinitiation complexes comprise small ribosomal subunits and eIF3, eIF4E, and eIF4G but lack eIF2 and eIF5 and do not assemble functional ribosomes (2). TIA proteins possess three RNA recognition motifs and a prion-related domain through which they can self-aggregate within SGs and suppress translation globally, without apparent specificity (14, 23, 43). A related but distinct role for TIA proteins has been theorized whereby TIA proteins specifically mediate the translational silencing of ARE-containing mRNAs, including those that encode TNF-α, matrix metalloproteinase 13, cyclooxygenase 2, the β2-adrenergic receptor, and several other transcripts bearing a recently elucidated TIA-1 motif (8, 9, 16, 19, 32, 50). Given the presence at SGs of RBPs that influence mRNA stability (such as tristetraprolin and HuR) and translation (TIA proteins), these foci have been hypothesized to function as dynamic sites of mRNA triage during stress, where molecular decisions are made regarding the composition of mRNA ribonucleoprotein (RNP) complexes and their subsequent engagement with the translation or degradation machineries (1, 2, 24).
Our previous studies had uncovered global links between mRNA stabilization and translational control during ER stress (22). We obtained systematic evidence that altered mRNA turnover strongly influenced changes in steady-state mRNA levels during the ER stress response and that translation was widely repressed. When the collections of mRNAs present in each turnover and translation group were further contrasted, the most prominently represented group comprised mRNAs that were subject to translational repression and were preferentially stabilized by ER stress (22). To further investigate these observations, we have studied a specific mRNA present in this group, that encoding cytochrome c. We have identified TIA-1 and HuR as RBPs forming specific RNP complexes with the cytochrome c mRNA, have obtained evidence that they potently influence cytochrome c translation, and document the functional consequences of these RNP associations following ER stress.
MATERIALS AND METHODS
Cell culture, treatment, and transfections.
Human HeLa cervical carcinoma cells were cultured in Dulbecco's modified essential medium (Gibco BRL) and treated with tunicamycin (Tn; 2 μg/ml) for the times indicated. Tunicamycin and sodium arsenite were purchased from Sigma. Small interfering RNAs (QIAGEN) targeting TIA-1 (AACACAACAAATTGGCCAGTA) or HuR (AAGAGGCAATTACCAGTTTCA), as well as a control small interfering RNA (siRNA; AATTCTCCGAACGTGTCACGT) were used at 20 nM. The simultaneous reduction of HuR and overexpression of HuR-TAP was performed as previously described (30). Cells were transfected with Oligofectamine (Invitrogen) on day 0 and treated and harvested on day 2. Plasmid pMT2-HA-TIA1 (a gift from P. Anderson and N. Kedersha) was used to overexpress TIA-1; it was used alongside the vector control pMT2.
Synthesis of biotinylated transcripts and biotin pull-down assay.
For in vitro synthesis of biotinylated transcripts, reverse-transcribed total RNA was used as template for PCRs using 5′ oligonucleotides which contained the T7 RNA polymerase promoter sequence [(T7),CCAAGCTTCTAATACGACTCACTATAGGGAGA]. The sequences of all oligonucleotide pairs (forward and reverse, respectively) used to synthesize the DNA templates were as follows: (T7)AGAGAGTGGGGACGTCCGGC and TTACTCATTAGTAGCTTTTTTGAG for fragment CR, (T7)TAATTGGCCACTGCCTTATTT and GATGGCACTCACCATCTTTGTG for fragment a, (T7)CACAAAGATGGTGAGTGCCATC and TCTGTAAGATGTGAGAGGTGTTG for fragment b, (T7)CAACACCTCTCACATCTTACAGA and TTTTGCTCTGTGGTTTTCTTTTT for fragment c, and (T7)CCTCAACGACCACTTTGTCA and GGTTGAGCACAGGGTACTTTATT for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The PCR-amplified products were resolved on agarose gels, and transcripts were purified and used as templates for the synthesis of the corresponding biotinylated RNAs using T7 RNA polymerase and biotin-CTP (45). Biotin pull-down assays were carried out as described elsewhere (45), except that the indicated quantities (2.5 to 20 nM) of recombinant glutathione S-transferase (GST)-TIA-1 and GST-HuR proteins (prepared and used as described in references 32 and 44, respectively) were incubated with a 5- to 50-fold molar excess of biotinylated transcripts for 30 min at room temperature. Complexes were isolated using streptavidin-conjugated Dynabeads (Dynal), and bound proteins in the pull-down material were analyzed by Western blotting using antibodies which recognized HuR or TIA-1 (below).
Polysome analysis.
HeLa cells (5 × 106 per sample) at ∼80% confluence were incubated for 15 min in 0.1 mg/ml cycloheximide and then lifted in 1 ml PEB lysis buffer (0.3 M NaCl, 15 mM MgCl2, 15 mM Tris-HCl, pH 7.6, 1% Triton X-100, 1 mg/ml heparin, and 0.1 mg/ml cycloheximide) by scraping and lysed on ice for 10 min. Nuclei were pelleted at 10,000 × g for 10 min, and the resulting supernatant was fractionated through a 10-to-50% linear sucrose gradient, as described previously (13). The eluted fractions were prepared with a fraction collector (Brandel), and their quality was monitored at 254 nm using a UV-6 detector (ISCO). RNA in each fraction was extracted with 8 M guanidine-HCl. Equal volumes of protein from each fraction were precipitated with deoxycholate and trichloroacetic acid, and the protein pellets were dissolved in Laemmli's sodium dodecyl sulfate (SDS) sample buffer.
Northern and Western blotting.
Northern blot analysis was performed using RNA isolated from whole cells (using standard methodologies) or from each fraction in the sucrose gradients (extracted with 8 M guanidine-HCl). To detect mRNAs encoding cytochrome c and 18S, oligonucleotides GCCTTTGTTCTTATTGGCGGCTGTGTAAGAGTATCCAGGGGCC and ACGGTATCTGATCGTCTTCGAACC, respectively, were end labeled using [α-32P]dATP and terminal transferase. For Western blot analysis, whole-cell (10 μg), cytoplasmic (20 μg), and nuclear (10 μg) lysates were prepared (22), size fractionated by electrophoresis through Tris-HCl gels (Bio-Rad, Life Science), and transferred onto polyvinylidene difluoride membranes. Monoclonal antibodies recognizing HuR and α-tubulin (a control cytoplasmic protein) as well as goat polyclonal anti-TIA-1 and anti-TIAR antibodies and a rabbit polyclonal antibody recognizing hnRNP C1/C2 (a control nuclear protein) were from Santa Cruz Biotechnology. A monoclonal antibody recognizing cytochrome c was from BD Pharmingen, and a polyclonal antibody recognizing cleaved poly(ADP-ribose) polymerase (PARP) was from Cell Signaling Technology. After secondary antibody incubations, signals were detected by enhanced chemiluminescence.
Immunoprecipitation of RNP complexes and reverse transcription-PCR (RT-PCR).
Immunoprecipitation of endogenous RNA-protein complexes was previously described (29). Briefly, cytoplasmic lysates, prepared from either untreated or Tn-treated cells, were divided into two equal parts and incubated (1 h, 4°C) with 100 μl of a 50% (vol/vol) suspension of protein A-Sepharose beads precoated with 30 μg each of mouse immunoglobulin G1 (IgG1; BD Pharmingen), goat IgG, mouse anti-HuR, goat anti-TIAR, or goat anti-TIAR (Santa Cruz Biotechnology). The beads were washed five times with NT2 buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM MgCl2, and 0.05% Nonidet P-40 [NP-40]). For RNA analysis, the beads were incubated with 100 μl NT2 buffer containing 20 U of RNase-free DNase I (15 min, 30°C), washed twice with 1 ml NT2 buffer, and further incubated in 100 μl NT2 buffer containing 0.1% SDS and 0.5 mg/ml proteinase K (15 min, 55°C) to digest the proteins bound to the beads. RNA was extracted using phenol and chloroform and precipitated in the presence of glycoblue. For the analysis of protein-protein and protein-RNA interactions, the beads were incubated in 100 μl of NT2 buffer containing both RNase A and RNase T1. Protein was extracted using Laemmli's SDS sample buffer.
The RNA isolated from immunoprecipitation (IP) material was reverse transcribed using random hexamers, oligo(dT) primer, and SSII reverse transcriptase (Invitrogen). Conditions for quantitative PCR (qPCR) and oligomers to amplify GAPDH and prothymosin α products were described previously (30); oligomers CAACTTTTCACAAAGATGGTGAGTG and GAGGCAAATGAACATGAACACAA were used for the amplification of the cytochrome c PCR product.
Immunofluorescence.
Cells were cultured in 35-mm glass-bottom microwell dishes (MatTek Corporation), fixed for 20 min in phosphate-buffered saline (PBS) containing 2% paraformaldehyde, and incubated for 10 min in ice-cold methanol and for 15 min in PBS containing 0.2% Triton X-100. After incubation for 1 h in blocking buffer (PBS containing 5% horse serum and 0.1% Tween 20), the cell preparations were incubated for 1 h using a 1:500 dilution of mouse anti-HuR (Santa Cruz Biotechnology) and a 1:100 dilution of goat anti-TIA-1 (Santa Cruz Biotechnology) antibodies dissolved in blocking buffer. Following washes with PBS containing 0.1% Tween 20, samples were incubated for 1 h with Alexa Fluor 488 donkey anti-mouse IgG (H+L) and Alexa Fluor 568 donkey anti-goat IgG (H+L) (Molecular Probes) (1 h, 1:500 dilution). After washes with PBS containing 0.1% Tween 20, cells were visualized using a Zeiss Axiovert 200 (Carl Zeiss MicroImaging, Inc.) fluorescence microscope.
Analysis of newly translated protein.
New synthesis of cytochrome c was measured by incubating 106 cells with 1 mCi l-[35S]methionine and l-[35S]cysteine (Easy TagEXPRESS; NEN/Perkin-Elmer, Boston, MA) per 60-mm plate for 20 min, whereupon cells were lysed using RIPA buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS, and 1 mM dithiothreitol). Immunoprecipitations were carried out for 1 h at 4°C using either a monoclonal antibody recognizing cytochrome c or IgG1 (BD Pharmingen). Following extensive washes in TNN buffer (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 5 mM EDTA, 0.5% NP-40), the immunoprecipitated material was resolved by 15% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto polyvinylidene difluoride filters, and visualized with a PhosphorImager (Molecular Dynamics).
RESULTS
ER stress suppresses cytochrome c translation.
Earlier studies had suggested that the expression of cytochrome c was translationally suppressed following exposure to ER stress (22). To directly investigate this process, cervical carcinoma HeLa cells were treated with Tn, an inhibitor of protein glycosylation that potently triggers ER stress. As anticipated, Tn treatment (2 μg/ml) reduced cytochrome c expression levels, as assessed by Western blot analysis (Fig. 1A). This effect was dependent, at least in part, on changes in the translation rate of cytochrome c, since a brief (20-min) incubation of HeLa cells with 35S-labeled amino acids immediately followed by IP analysis using an anti-cytochrome c antibody (employing approaches for measuring new protein translation that were previously described [12, 35, 36]) revealed that the levels of nascent cytochrome c were suppressed by 6 h following exposure to Tn (Fig. 1B). That these experiments measured nascent cytochrome c translation and not protein stability was supported by data indicating that the cytochrome c protein half-life was longer than 6 h (not shown). In addition, the observed reduction in cytochrome c translation was not due to a decrease in cytochrome c mRNA levels, since these actually increased slightly following ER stress, as determined by Northern blot analysis (Fig. 1C). We thus set out to examine the mechanisms underlying the regulation of cytochrome c translation.
FIG. 1.
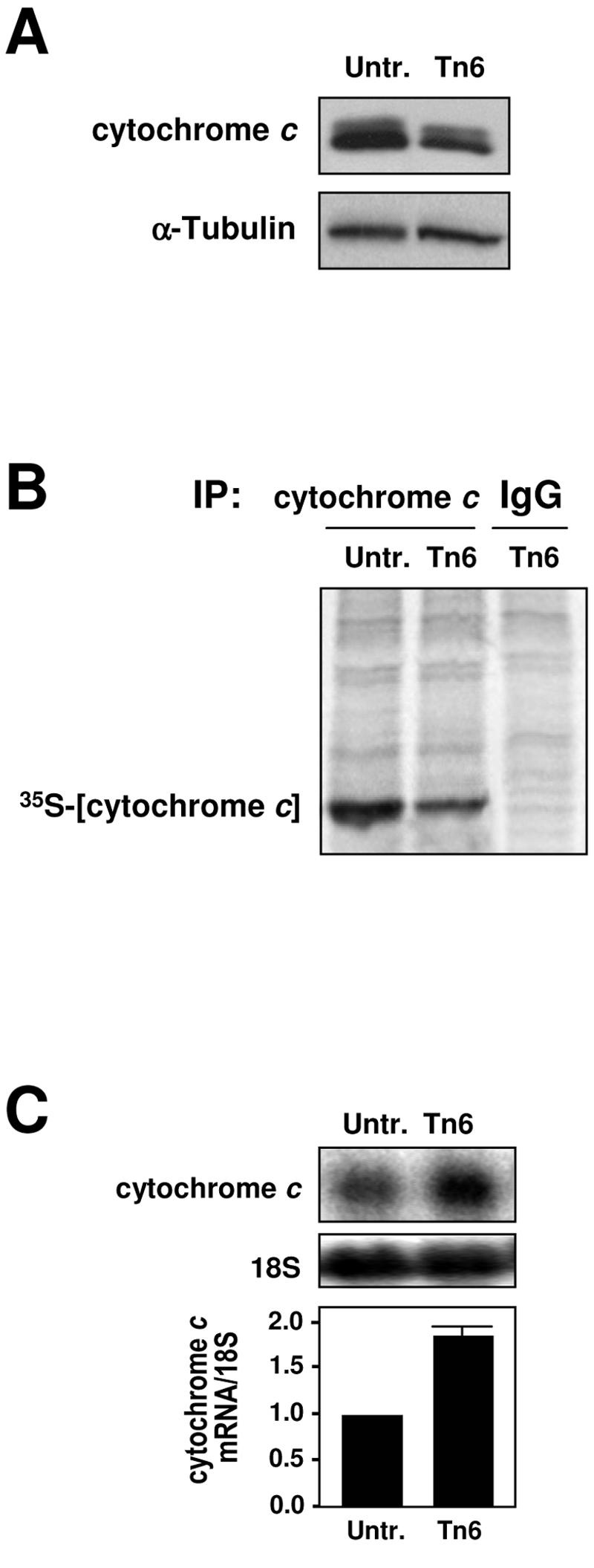
Translational repression of cytochrome c during ER stress. (A) Western blot analysis of cytochrome c expression after continuous treatment of HeLa cells with 2 μg/ml Tn for 6 h. (B) Levels of nascent cytochrome c translation, assessed by incubating untreated cells (Untr.) and cells that had been treated with 2 μg/ml of tunicamycin for 6 h (Tn6) with l-[35S]methionine and l-[35S]cysteine for 20 min. Following IP using either anti-cytochrome c or IgG antibodies, samples were resolved by SDS-PAGE and transferred onto membranes for visualization of the 35S-radiolabeled signals using a PhosphorImager. (C) Northern blot analysis to visualize (top) and quantify (bottom) the expression levels of cytochrome c mRNA in whole-cell lysates (20 μg); 18S rRNA levels served to verify the equal loading of samples.
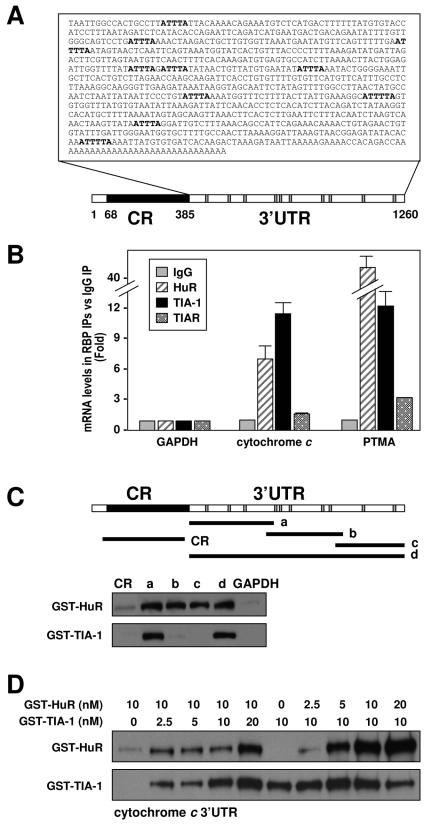
The AU-rich 3′UTR of the cytochrome c mRNA is a target of HuR and TIA-1.
The 3′UTR of the cytochrome c mRNA is relatively long (∼900 bp) and is highly AU rich (Fig. 2A), two features that characterize mRNAs that are the targets of ARE-RBPs. We sought to directly test whether three ARE-RBPs (HuR, TIA-1, and TIAR), which are known to influence translation, were capable of forming a complex with the endogenous cytochrome c mRNA. We immunoprecipitated RNP complexes from cytoplasmic fractions prepared from HeLa cells using antibodies that recognized either HuR, TIA-1, or TIAR and performed RT and real-time PCR (qPCR) analysis to detect endogenous cytochrome c mRNA. The levels of the housekeeping GAPDH mRNA, which was found as a low-level contaminant in all IP samples, were also monitored in order to ascertain any differences in sample input. As shown in Fig. 2B, RBPs HuR and TIA-1 showed significant degrees of binding to the cytochrome c mRNA (7-fold and 11-fold enrichment, respectively) compared to the abundance of cytochrome c mRNA in the control IgG IP samples. As a positive control, we monitored the binding of these proteins to the prothymosin α mRNA, a known target of HuR and TIA-1 (Fig. 2B) (30, 32). By this assay, TIAR did not seem to complex with the cytochrome c mRNA, despite the extensively shared homology between TIAR and TIA-1 (23), the protein-protein interaction reported for TIA-1 and TIAR (14, 23, 43), and the observation that the goat polyclonal anti-TIA-1 antibody employed here recognizes TIAR with very low affinity (14) (data not shown).
FIG. 2.
HuR and TIA-1 bind to cytochrome c mRNA. (A) Schematic of the cytochrome c mRNA and the AU-rich 3′UTR sequence. Gray, location of ATTTA and ATTTTA sequences; CR, coding region. (B) Binding of endogenous HuR, TIA-1, or TIAR to endogenous mRNAs was detected by RT and qPCR assay of material obtained by IP from HeLa cytoplasmic fractions using IgG, anti-HuR, or anti-TIA-1 antibodies. Amplification of mRNAs encoding either GAPDH (a housekeeping gene), prothymosin α (PTMA; a positive control for HuR and TIA-1), and cytochrome c were performed. Changes in the levels of mRNAs associated with each RBP were calculated by measuring their abundance in the IP reaction mixtures using RBP-specific antibodies relative to the levels detected in control IgG IP reaction mixtures. (C) Schematic of the cytochrome c mRNA and the various biotinylated transcripts that were tested in in vitro binding reactions using purified recombinant proteins GST-HuR or GST-TIA-1. Biotin pull-down assays (see Materials and Methods) were employed to assess the binding of purified recombinant HuR and TIA-1 (20 nM each) to the indicated biotinylated transcripts spanning the cytochrome c mRNA. Following pull down, bound proteins were detected by Western blotting using anti-HuR or anti-TIA-1 antibodies. (D) Biotin pull-down assays were performed after incubating transcript d with both HuR and TIA-1 (at the indicated concentrations) in the same binding reaction mixture.
Next, we sought to identify the cytochrome c mRNA regions involved in associating with HuR and TIA-1. To this end, several biotinylated transcripts spanning the mRNA regions shown (Fig. 2C, schematic) were synthesized (see Materials and Methods). Following in vitro binding assays with recombinant GST-HuR or GST-TIA-1 proteins, the interaction between the biotinylated transcripts and the fusion RBPs was assessed by biotin pull-down assay (see Materials and Methods) followed by Western blot analysis. As shown, HuR complexed with nonoverlapping regions of the cytochrome c 3′UTR (but not with the coding region), while TIA-1 preferentially bound the 3′UTR segment proximal to the coding region (Fig. 2C, transcript a). These observations supported a binding scheme on the cytochrome c 3′UTR whereby TIA-1 associated preferentially with the proximal region while HuR was capable of binding to both proximal and distal regions. Accordingly, we postulated that both proteins could bind simultaneously to the cytochrome c 3′UTR, and we carried out additional in vitro binding assays to test this possibility. As shown (Fig. 2D), addition of increasing levels of TIA-1 in the presence of an unchanged HuR concentration caused HuR binding to full-length cytochrome c (transcript d) to increase; conversely, addition of increasing amounts of HuR caused no change in TIA-1 binding (though a reduction was noted at 20 nM HuR). These observations suggest that HuR and TIA-1 may not compete for binding to cytochrome c and, instead, may bind jointly and even synergize in their binding to the cytochrome c mRNA. However, given the variability of this assay, the fact that recombinant proteins were used to study binding, and the observation that no joint binding was observed using RNA electrophoretic mobility shift assays (unpublished), the molecular and functional aspects of these interactions must be investigated using more appropriate systems, particularly through the analysis of the endogenous proteins.
To test if the binding of HuR and TIA-1 to cytochrome c mRNA might be influenced by Tn treatment, IP reactions (similar to those described for Fig. 2B) were performed using cytoplasmic lysates from Tn-treated cells. The cytochrome c mRNA was more abundant in the IP material obtained after TIA-1 IP or HuR IP, compared with its abundance in IgG IPs as well as with the abundance of GAPDH mRNA in these IP materials (Fig. 3). As observed here, whole-cell HuR-cytochrome c mRNA complexes were transiently reduced by 2 h of Tn treatment, whereas whole-cell TIA-1-cytochrome c mRNA complexes remained elevated throughout 6 h of Tn treatment (Fig. 3).
FIG. 3.
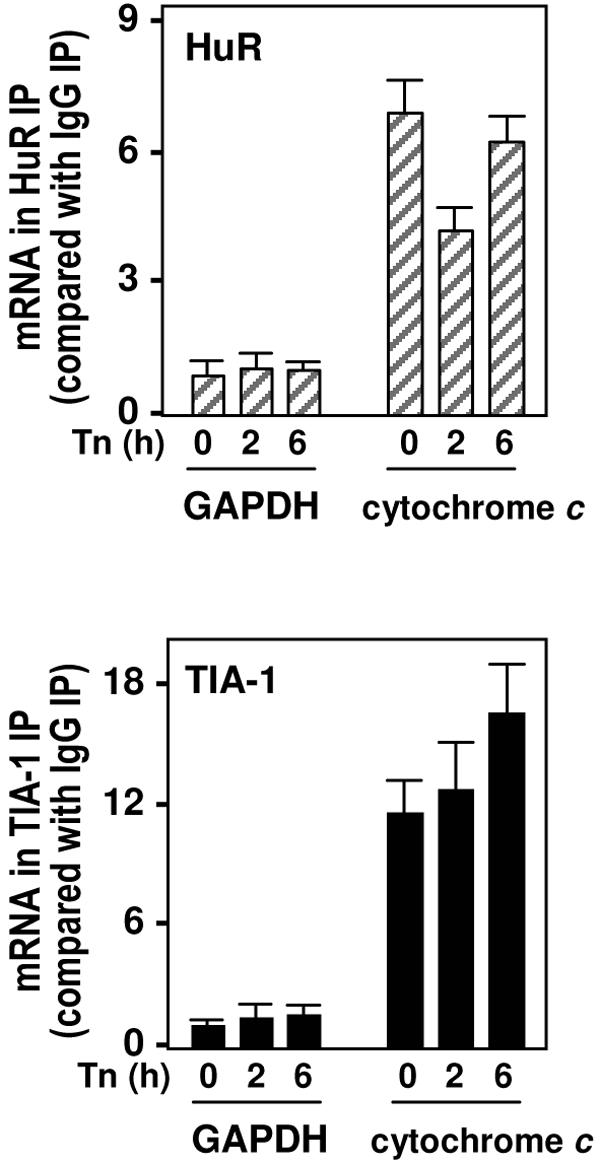
ER stress-dependent binding of endogenous HuR and TIA-1 to endogenous cytochrome c mRNAs. The abundance levels of mRNAs encoding cytochrome c or GAPDH were calculated following IP (with either IgG or with antibodies recognizing HuR [top] or TIA-1 [bottom]) from the cytoplasmic fractions of cells that were either left untreated (0) or treated with Tn for 2 or 6 h; mRNAs present in the IP pellets were quantified by RT and qPCR.
Subcellular distribution of HuR and TIA-1 after Tn treatment.
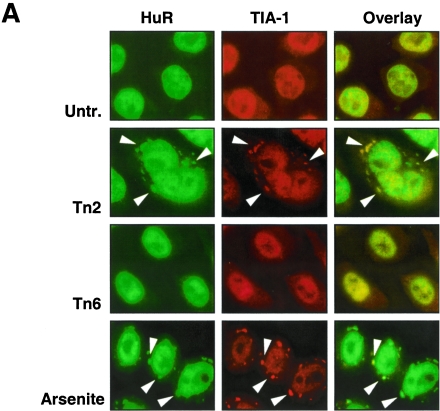
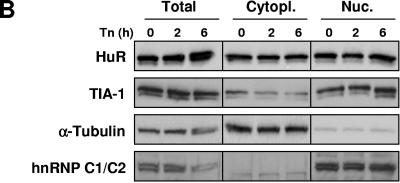
To assess whether Tn treatment caused any changes in the cellular abundance and/or relative distribution of HuR and TIA-1, these RBPs were studied by immunofluorescence in untreated or Tn-treated cultures. As shown in Fig. 4A, HuR and TIA-1 were predominantly nuclear, as reported earlier (26, 42, 44). By 2 h of Tn treatment, HuR and TIA-1 were found to accumulate at several cytoplasmic foci which likely represented SGs. Populations treated with the SG-inducing agent arsenite (Fig. 4A) were included as positive controls; colocalization of these foci with G3BP (an SG marker) (not shown) further supported that they were SGs. Western blot analysis of HuR and TIA-1 levels documented that the overall levels of cytoplasmic HuR and TIA-1 did not change significantly (Fig. 4B).
FIG.4.
Effect of ER stress on the subcellular localization of HuR and TIA-1. (A) At the times indicated following addition of Tn or after a 30-min incubation with 1 mM sodium arsenite followed by 1 h of culture in regular medium (arsenite), the subcellular localizations of HuR (green) and TIA-1 (red) were monitored by immunofluorescence. Overlay: yellow indicates colocalization of HuR and TIA-1 signals. In Tn2 samples, SGs (arrowheads) were observed in >50% of the cells. (B) Western blot analysis of HuR and TIA-1 levels in whole cells (total; 10 μg per lane), cytoplasmic (cytopl.; 20 μg per lane), and nuclear lysates (nuc.; 10 μg per lane) prepared from HeLa cells after Tn treatment for the indicated time periods. The levels of α-tubulin (a cytoplasmic protein) and hnRNP C1/C2 (a nuclear protein) in the same samples were assessed by Western blotting in order to ascertain the quality of the fractionation procedure and to detect loading differences.
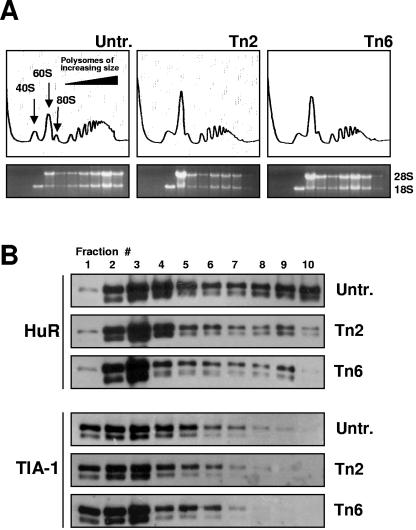
Further analysis of the cytoplasmic distribution of HuR and TIA-1 was carried out by monitoring the relative abundance of HuR and TIA-1 along sucrose density gradients prepared from untreated and Tn-treated cells (Fig. 5A). HuR levels were elevated in high-molecular-weight fractions (9 and 10) of untreated populations, wherein actively translating polysomes are found. HuR levels in Tn-treated cells exhibited a marked shift towards lighter fractions (1 to 8) comprising low-molecular-weight polysomes (fractions 6 to 8), as well as monosomes (5), ribosome subunits (3 and 4), and fractions with no ribosomal material (1 and 2), which had either low or no translational activity (Fig. 5B). Thus, while the association of HuR with the cytochrome c mRNA was restored by 6 h of Tn treatment on a whole-cell level (Fig. 3), HuR-cytochrome c mRNA complexes were reduced in the translating fractions (Fig. 5B and data not shown), thereby contributing to the diminished translation of cytochrome c. In untreated populations, HuR did not appear to associate with polysomes but instead seemed to cosediment with them, as observed after EDTA or puromycin treatment (unpublished). TIA-1 signals were found almost exclusively in the low-molecular-weight fractions of untreated and Tn-treated cells and did not show the distinct shift that was seen with HuR (Fig. 5B).
FIG. 5.
Cytoplasmic association of HuR and TIA-1. (A) Polysomal profiles from cells that either were left untreated (Untr.) or were treated with Tn for 2 or 6 h. From left to right, fractions lacked ribosomes or ribosome subunits (fractions 1 and 2), contained ribosome subunits or single ribosomes (fractions 3 to 5), or spanned polysomes of increasing molecular weights (fractions 6 to 10). The relative abundance levels of 28S and 18S rRNAs were visualized from ethidium bromide-stained agarose gels. (B) Protein from each fraction was prepared for Western blot analysis to detect HuR and TIA-1.
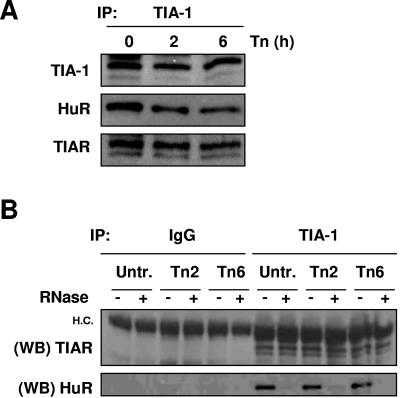
The fact that both proteins shifted towards lighter fractions suggested the possibility that they might be functionally linked through their association with common target mRNAs, akin to the association that was previously described for HuR and AUF1 (29). To test this possibility, TIA-1 was subjected to IP analysis using cytoplasmic lysates, and the presence of HuR was tested in the resulting IP samples. As shown, HuR was readily detectable in the TIA-1 IP material, both before and after Tn treatment (Fig. 6A). TIAR was also detected in these IP analyses, in agreement with previous reports of associations between TIAR and TIA-1 (51) (Fig. 6A). The HuR-TIA-1 interaction likely depends on their association with common target RNAs, since digestion with RNases A and T1 abrogated their colocalization in IP material (Fig. 6B). By contrast, TIA-1-TIAR interactions persisted even after RNase digestion, supporting their previously described protein-protein interaction (Fig. 6B). Together, these data support the views that HuR is found in actively translating cellular fractions but can also be present in fractions with less translation and in translationally silent fractions and that it interacts with TIA-1 via their joint association with common RNA molecules (Fig. 5 and 6).
FIG. 6.
Association of HuR and TIA proteins. (A) IP assays were carried out using cytoplasmic lysates from HeLa cells that were either left untreated or were treated with Tn for the indicated times using an anti-TIA-1 antibody. The presence of TIA-1, HuR, and TIAR in the IP material was monitored by Western blotting. (B) IP reactions were performed in the absence (−) or presence (+) of RNase A and RNase T1, using cytoplasmic lysates and either IgG or anti-TIA-1 antibody. The presence of TIAR and HuR in the IP material was monitored by Western blotting (WB). H.C., heavy immunoglobulin chain.
Knockdown of HuR levels reduces cytochrome c translation, whereas knockdown of TIA-1 levels elevates cytochrome c translation.
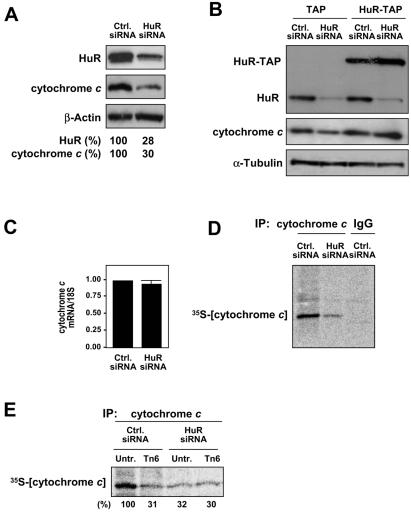
In order to directly test the involvement of HuR in the regulation of cytochrome c expression, siRNA was used. As shown, use of siRNA molecules targeting HuR effectively lowered the HuR concentration to ∼28% of the levels seen in the control populations (Fig. 7A) by 48 h after transfection. This intervention caused a comparable reduction in the levels of cytochrome c (30% of the levels in the control transfection group). Rescue experiments in which HuR was overexpressed as HuR-TAP and the endogenous HuR was silenced using an siRNA targeting the HuR 3′UTR (previously reported [30]) indicated that the HuR effects on cytochrome c abundance were specific (Fig. 7B). As determined by Northern blot analysis (Fig. 7C), HuR knockdown did not significantly alter cytochrome c mRNA levels; this was an important parameter to test, since HuR has been found to stabilize many target mRNAs (5, 44, 45). Instead, the de novo synthesis of cytochrome c protein was markedly reduced after HuR knockdown, both in untreated populations (Fig. 7D) and following Tn treatment (Fig. 7E); no further decrease was observed when HuR-silenced populations were treated with Tn, likely indicating that this basal cytochrome c translation was refractory to the influence by HuR or Tn. Together, these results strongly support a role for HuR as a specific inducer of cytochrome c translation, particularly in untreated cells.
FIG. 7.
Effect of RNAi-mediated HuR knockdown on the expression of cytochrome c. Two days after transfection with either HuR-directed or control siRNA (20 nM each), protein and RNA were collected for analysis. (A) Western blot analysis to monitor the expression levels of HuR, cytochrome c, and the loading control β-actin; signals were quantified, and HuR and cytochrome c levels were normalized to β-actin levels, shown as a percentage of the abundance seen in the control siRNA populations. (B) Cells were cotransfected with siRNA (either control siRNA or siRNA targeting the 3′UTR of the HuR mRNA) and with a plasmid (either the TAP-expressing vector or a TAP-HuR expression plasmid), as previously reported (30); 48 h later the levels of cytochrome c and other proteins were tested by Western blotting. (C) The levels of cytochrome c mRNA and 18S rRNA (loading control) were quantified by Northern blotting and are expressed as the fold difference in control versus HuR siRNA populations. Newly synthesized cytochrome c protein was assessed following a brief (20-min) incubation of siRNA-transfected cells with l-[35S]methionine and l-[35S]cysteine. (D and E) The effect of HuR knockdown on the basal cytochrome c translation (D) or following Tn treatment for 2 h (E) was determined by IP with an anti-cytochrome c antibody; IgG IPs served to assess the specificity of the IP reaction. Samples were resolved by SDS-PAGE and transferred onto membranes for visualization of 35S-radiolabeled signals using a PhosphorImager; nascent cytochrome c signals were quantified and are represented as the percentage of the signal intensity in untreated, control siRNA populations.
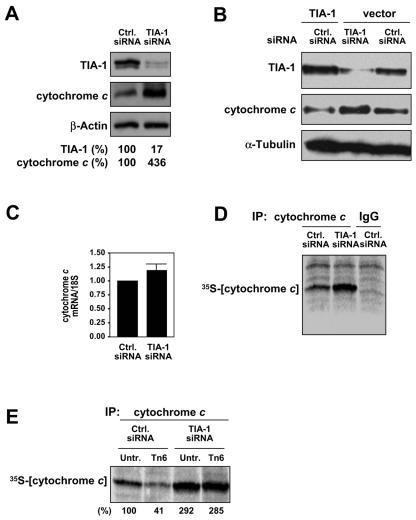
We also investigated the influence of TIA-1 on cytochrome c translation following the siRNA-mediated reduction of TIA-1 levels. As observed by Western blot analysis (Fig. 8A), TIA-1 levels were dramatically suppressed (down to 17% of the levels seen in control populations) by 48 h following transfection of a TIA-1-targeting siRNA, an intervention that caused a strong upregulation (∼4-fold) of cytochrome c levels. Conversely, TIA-1 overexpression reduced cytochrome c levels (Fig. 8B). Cytochrome c mRNA levels, as measured by RT and qPCR analysis, were modestly elevated after silencing TIA-1 (Fig. 8C). In keeping with TIA-1's role as a translational suppressor (32; reviewed in reference 1), the rate of nascent translation of cytochrome c protein was markedly higher in populations with silenced TIA-1 (Fig. 8D). Contrary to what was seen when HuR was silenced, TIA-1 knockdown rescued the Tn-imposed translational repression of cytochrome c (Fig. 8E). These findings indicate that TIA-1 is a suppressor of cytochrome c translation, both in untreated and in Tn-treated cells.
FIG. 8.
Effect of RNAi-mediated TIA-1 knockdown on the expression of cytochrome c. Two days after transfection with either TIA-1-directed or control siRNA (20 nM each), protein and RNA were collected for analysis. (A) Western blot analysis to monitor the expression levels of TIA-1, cytochrome c, and the loading control, β-actin; signals were quantified, and TIA-1 and cytochrome c levels were normalized to β-actin levels and are shown as a percentage of the abundance seen in control siRNA populations. Newly synthesized cytochrome c protein was assessed following a brief (20-min) incubation of siRNA-transfected cells with l-[35S]methionine and l-[35S]cysteine. (B) Cells were cotransfected with siRNA (either control siRNA or TIA-1 siRNA) and with a plasmid (either without an insert or with one expressing TIA-1); 48 h later the levels of cytochrome c and other proteins were tested by Western blotting. (C) The effects of TIA-1 knockdown on the basal cytochrome c mRNA levels were assessed by RT and qPCR and normalized to the levels of GAPDH mRNA. (D and E) The influences of TIA-1 knockdown on cytochrome c translation in untreated cells (D) or following Tn treatment for 2 h (E) were determined by IP with an anti-cytochrome c antibody; IgG IPs served to assess the specificity of the IP reaction. Samples were resolved by SDS-PAGE and transferred onto membranes for visualization of 35S-radiolabeled signals using a PhosphorImager; nascent cytochrome c signals were quantified and are represented as a percentage of the signal intensity in untreated, control siRNA populations.
Reciprocal influence of TIA-1 and HuR expression levels and effects on population growth and survival.
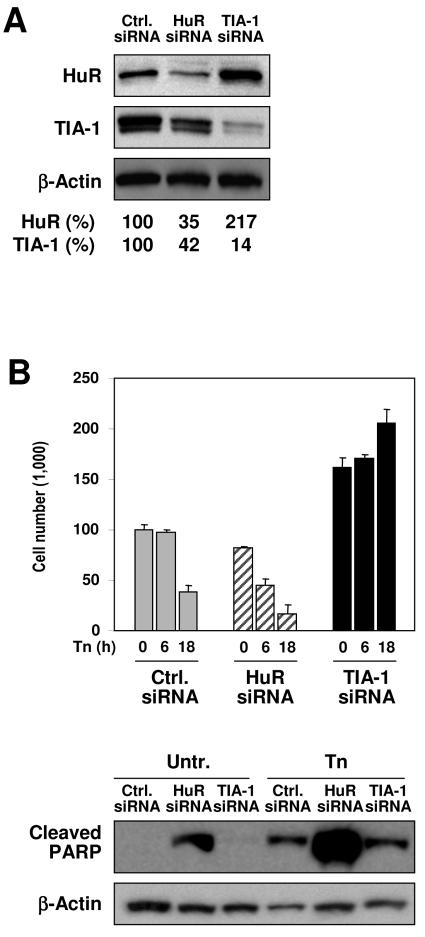
Unexpectedly, the RNA silencing experiments revealed that TIA-1 and HuR influenced each other's expression levels. As shown in Fig. 9A, by 48 h of transfection to knock down HuR, TIA-1 expression levels were also substantially diminished, indicating that HuR might contribute to regulating TIA-1 expression in the cell. In contrast, TIA-1 knockdown caused a marked increase in HuR levels, indicating that TIA-1 may contribute to lowering HuR levels in the cell.
FIG. 9.
Reciprocal influence of HuR and TIA-1 on their expression levels and consequences on population growth and survival. (A) Two days after transfection with siRNA as explained in the legends of Fig. 7 and 8, the expression levels of HuR and TIA-1 (as well as those of the loading control, β-actin) were assessed by Western blot analysis. (B) Following treatment with Tn for 6 or 18 h, cells were collected in order to monitor changes in cell number (graph, reflecting the means and standard errors of the means from three independent experiments). The relative abundance of cleaved PARP, a marker of apoptosis, was monitored after 6 h of Tn treatment; a representative Western blot is shown (20 μg per lane).
Given the central role of cytochrome c in apoptosis, the effects of altering HuR and TIA-1 on cell survival were assessed in HeLa cells that had been left untreated or treated with Tn. Knockdown of HuR significantly lowered the number of remaining cells (Fig. 9B, graph), an effect that was not unexpected, since HuR had been shown to promote cell proliferation (45, 46) and to have an antiapoptotic influence (30). Indeed, increased apoptosis likely contributed to the reduction in size of the HuR siRNA population, given the appearance of cleaved PARP, a well-established marker of apoptosis, in this transfection group (Fig. 9B, Western blot analysis). In striking contrast, knockdown of TIA-1 caused an unanticipated increase in cell numbers; while the downstream mediators of this effect remain to be identified, the observed heightening in the levels of the proliferation-inducing HuR (Fig. 9A) plausibly contributes to this increase. The moderately toxic Tn treatment employed here (2 μg/ml) reduced cell survival in the control transfection group by 18 h, while it significantly lowered cell survival in the HuR siRNA transfection group, associated with a marked increase in cleaved PARP (Fig. 9B). In keeping with the proposed proapoptotic role of TIA-1, its silencing did not appreciably decrease cell viability (instead appearing to promote cell division) and was associated with minimal PARP cleavage (Fig. 9B). Taken together, the influences of HuR and TIA-1 on cell growth and survival following ER stress are consistent with their reported effects on proliferation and survival and with a role of their common target, cytochrome c, in apoptosis, as discussed below.
DISCUSSION
Our earlier studies systematically analyzed possible links between translational inhibition and altered mRNA turnover (22), revealing that the translationally suppressed mRNAs were preferentially stabilized in response to ER stress agents. Since both of these posttranscriptional processes are governed by the formation of RNP complexes, we sought to identify RBPs involved in the regulation of cytochrome c, which is encoded by an mRNA that is stabilized and translationally repressed following ER stress (22). Indeed, IP of RNP complexes indicated that both HuR and TIA-1, but not TIAR, were capable of binding the endogenous cytochrome c mRNA. These associations were verified using recombinant purified proteins and biotin-labeled transcripts and further showed that HuR was capable of binding to proximal and distal regions of the cytochrome c 3′UTR, while TIA-1 bound only the proximal region of the cytochrome c 3′UTR. That these RBPs likely contribute to the translational regulation of cytochrome c mRNA was supported by several experimental pieces of data.
First, whole-cell HuR-cytochrome c mRNA complexes were moderately and transiently reduced by Tn treatment. HuR has been shown to promote the translation of several target mRNAs with which it complexes via their 3′UTR (30, 35) but appears to suppress the translation of mRNAs exhibiting 5′UTR associations (28, 37). While these discrepant roles for HuR remain to be formally examined, the data obtained here support a role for HuR in promoting cytochrome c translation under unstressed conditions: (i) binding of HuR to the cytochrome c mRNA was more abundant in unstressed cells, decreasing when translation was lowered by Tn treatment (Fig. 3); (ii) HuR abundance in the actively translating polysomal fractions (7 to 10) was markedly decreased in the polysomal fractions (Fig. 5B) and consequently its association with cytochrome c mRNA also decreased (not shown); (iii) under conditions of HuR silencing, translation of cytochrome c was reduced (Fig. 7); and (iv) HuR overexpression led to an elevation in cytochrome c abundance. Whether the effect of HuR upon the translation of cytochrome c is linked to an association of HuR ribonucleoprotein complexes with polysomes (Fig. 5) and whether it is subject to posttranslational modifications (e.g., phosphorylation) or to other regulatory processes also remain critical questions to be addressed experimentally.
Second, ribonucleoprotein complexes comprising TIA-1 and cytochrome c mRNA were modestly elevated by 2 h and 6 h in the continuous presence of Tn. The role of TIA-1 as a translational repressor has been characterized in some detail. TIA-1 has been proposed to usurp the ternary complex eIF2-GTP-Met-tRNAi, which is necessary for the formation of a translationally active preinitiation complex, and instead is thought to assemble noncanonical, translationally silent complexes. Under conditions of stress, phosphorylation of eIF2α by one or several kinases (PKR, PERK, GCN2, and HRI) reduces the availability of the ternary complex, resulting in the accumulation of translationally silent preinitiation complexes. The self-aggregating property of TIA-1 (and that of the related protein TIAR) in turn promotes the accumulation of translationally incompetent complexes in SGs. In keeping with our findings that Tn treatment results in the formation of SGs (Fig. 4A), ER stress has been reported to be a potent trigger of eIF2α phosphorylation (17). We devoted a great deal of effort towards investigating the possible colocalization of cytochrome c mRNA (by in situ hybridization) and TIA-1 (by immunofluorescence) in Tn-treated cells. While it was possible to visualize each molecule separately (unpublished), they could not be investigated on the same preparation: to monitor the distribution of the cytochrome c mRNA, the cell preparation had to be digested with proteases, which effectively precluded the subsequent detection of TIA-1; conversely, when TIA-1 was detected first, the integrity and accessibility of the cytochrome c mRNA were compromised, preventing their joint detection on the same preparations.
Although much remains to be elucidated about the regulation of HuR and TIA-1 binding activities, a model emerges from the action of these two RBPs upon the cytochrome c mRNA. TIA-1 and HuR bind the cytochrome c mRNA in unstimulated cells, one promoting its translation and the other inhibiting it. The equilibrium between these two opposing forces shifts following ER stress: Tn causes a reduction in the binding of HuR to the cytochrome c mRNA but does not reduce the association of TIA-1 and, accordingly, the translation-promoting effect of HuR on cytochrome c expression is reduced while the translation-suppressive effect of TIA-1 is accentuated. In in vitro binding assays, HuR and TIA-1 did not appear to compete for binding; instead, the increased binding of TIA-1 seemed to facilitate a greater binding of HuR to the cytochrome c 3′UTR, although increased HuR levels did not influence the binding of TIA-1 to the cytochrome c mRNA (Fig. 2D). While the in vivo relevance of these in vitro complexes remains unclear, this biochemical interaction recapitulates the functional associations recently suggested for HuR and TIA-1 by Katsanou and colleagues (20). Using a mouse model of HuR overexpression, those authors suggested that some of the phenotypic traits (particularly the translation of TNF-α) may be due to a synergistic interaction between HuR and TIA-1 (20). Such complex links between HuR and TIA-1 as well as their joint influence on shared target mRNAs are further highlighted by our findings presented here. As observed, HuR and TIA-1 mutually influence their expression levels, with reductions in HuR causing decreased TIA-1 expression and silencing of TIA-1 leading to elevations in HuR levels (Fig. 9A), suggesting the possible existence of a negative feedback loop involving these RBPs. These regulatory events likely have a significant posttranscriptional component, since TIA-1 binds the HuR mRNA and HuR binds the TIA-1 mRNA (unpublished observations). The influence of HuR and TIA-1 upon each other's expression as well as upon the expression of shared target mRNAs is under investigation in our laboratory (R. Pullmann and M. Gorospe, unpublished data). A systematic dissection of the molecular links and functional interdependence between these two RBPs also awaits further analysis using more suitable systems.
In light of the antiapoptotic influence of HuR (30), it is particularly significant that HuR promoted the translation of cytochrome c. As shown by a number of groups (33, 34, 49), the newly synthesized cytochrome c protein (named apocytochrome c, since it lacks the heme group) both fails to support caspase activation and inhibits the ability of holocytochrome c (the active, heme-conjugated form) to activate caspases. It is likely that the newly translated cytochrome c functions alongside other antiapoptotic effectors, like prothymosin α, to elicit the antiapoptotic program implemented by HuR (30). Likewise, the proapoptotic influence of TIA-1 (11), which remains poorly understood, may rely at least in part on its inhibitory effects on cytochrome c biosynthesis. At later times following Tn treatment, cytochrome c translation resumes (unpublished observations), suggesting that the translational inhibition of this pivotal regulator of apoptosis (18) occurs transiently, presumably during a time of damage assessment, before cells undertake the repair of damaged molecules or engage in apoptotic death. The observation that cytochrome c and prothymosin α share both a functional influence on apoptosis and the presence of regulatory sequences in their 3′UTRs lends support to the posttranscriptional operon model proposed by Keene and colleagues, whereby the expression of functionally related proteins is coordinated through the association of the respective mRNAs with shared RBPs (27). In the system described here, HuR could be envisioned to jointly promote the translation of these two antiapoptotic factors, whereas TIA-1 could simultaneously inhibit their biosynthesis.
It is important to mention that HuR has also been shown to potently increase the stability of many target mRNAs. Unexpectedly, however, HuR did not appear to influence the cytochrome c mRNA stability in this system and instead seemed to influence only the translation of cytochrome c (Fig. 7). Why HuR regulates the stability of certain mRNA subsets and the translation of other mRNA subsets remains an important question for immediate consideration. In the case of the cytochrome c mRNA, additional RNA-binding proteins which influence mRNA turnover were found in our RNP IP studies, most significantly the decay-promoting RBP AUF1 (unpublished observations). Efforts are under way to investigate the possibility that AUF1 is implicated in modulating the stability of the cytochrome c mRNA in Tn-treated cells. A more thorough understanding of the mechanisms influencing cytochrome c mRNA stability will contribute essential information to the processes linking translational suppression with mRNA stabilization (22).
In closing, a recent report by Anderson and colleagues establishes functional and spatio-temporal associations between two cellular aggregates: SGs, which represent sites of silenced translation, and the processing bodies (PBs), which are believed to constitute sites of active mRNA degradation (25). Those authors showed that PBs and SGs are compositionally and morphologically distinct but do share some proteins, such as eIF4E, which shuttle between the two domains. A plausible scenario can thus be envisioned whereby the binding of TIA-1 to cytochrome c mRNA sequesters it within SGs and thereby prevents its degradation at PBs while keeping it from being translated. While additional details of these regulatory paradigms await further experimental demonstration, the findings presented here underscore the complex influence of posttranscriptional gene regulatory processes within the cellular response to harmful stimuli.
Acknowledgments
We are grateful to I. López de Silanes for providing siRNA and to F. E. Indig, M. Juhaszova, and D. V. Dixit for their assistance with the microscopy. We thank P. Anderson, N. Kedersha, and J. Valcarcel for the TIA-1 expression vectors.
This research was supported by the Intramural Research Program of the NIA, NIH.
REFERENCES
- 1.Anderson, P., and N. Kedersha. 2002. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, P., and N. Kedersha. 2002. Stressful initiations. J. Cell Sci. 115:3227-3234. [DOI] [PubMed] [Google Scholar]
- 3.Antic, D., and J. D. Keene. 1997. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am. J. Hum. Genet. 61:273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevilacqua, A., M. C. Ceriani, S. Capaccioli, and A. Nicolin. 2003. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell Physiol. 195:356-372. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell Mol. Life Sci. 58:266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carballo, E., W. S. Lai, and P. J. Blackshear. 1998. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281:1001-1005. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 8.Cok, S. J., S. J. Acton, and A. R. Morrison. 2003. The proximal region of the 3′-untranslated region of cyclooxygenase-2 is recognized by a multimeric protein complex containing HuR, TIA-1, TIAR, and the heterogeneous nuclear ribonucleoprotein U. J. Biol. Chem. 278:36157-36162. [DOI] [PubMed] [Google Scholar]
- 9.Dember, L. M., N. D. Kim, K. Q. Liu, and P. Anderson. 1996. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 271:2783-2788. [DOI] [PubMed] [Google Scholar]
- 10.Fan, J., X. Yang, W. Wang, W. H. Wood III, K. G. Becker, and M. Gorospe. 2002. Global analysis of stress-regulated mRNA turnover using cDNA arrays. Proc. Natl. Acad. Sci. USA 99:10611-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forch, P., and J. Valcarcel. 2001. Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1. Apoptosis 6:463-468. [DOI] [PubMed] [Google Scholar]
- 12.Galban, S., J. Fan, J. L. Martindale, C. Cheadle, B. Hoffman, M. P. Woods, G. Temeles, J. Brieger, J. Decker, and M. Gorospe. 2003. von Hippel-Lindau protein-mediated repression of tumor necrosis factor alpha translation revealed through use of cDNA arrays. Mol. Cell. Biol. 23:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galbán, S., J. L. Martindale, K. Mazan-Mamczarz, I. López de Silanes, J. Fan, W. Wang, J. Decker, and M. Gorospe. 2003. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol. Cell. Biol. 23:7083-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilks, N., N. Kedersha, M. Ayodele, L. Shen, G. Stoecklin, L. M. Dember, and P. Anderson. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorospe, M. 2003. HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle 2:412-414. [PubMed] [Google Scholar]
- 16.Gueydan, C., L. Droogmans, P. Chalon, G. Huez, D. Caput, and V. Kruys. 1999. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem. 274:2322-2326. [DOI] [PubMed] [Google Scholar]
- 17.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, X., and X. Wang. 2004. Cytochrome c-mediated apoptosis. Annu. Rev. Biochem. 73:87-106. [DOI] [PubMed] [Google Scholar]
- 19.Kandasamy, K., K. Joseph, K. Subramaniam, J. R. Raymond, and B. G. Tholanikunnel. 2005. Translational control of β2-adrenergic receptor mRNA by T-cell-restricted intracellular antigen-related protein. J. Biol. Chem. 280:1931-1943. [DOI] [PubMed] [Google Scholar]
- 20.Katsanou, V., O. Papadaki, S. Milatos, P. J. Blackshear, P. Anderson, G. Kollias, and D. Kontoyiannis. 2005. HuR as a negative posttranscriptional modulator of inflammation. Mol. Cell 19:777-789. [DOI] [PubMed] [Google Scholar]
- 21.Kaufman, R. J. 2002. Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 110:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawai, T., J. Fan, K. Mazan-Mamczarz, and M. Gorospe. 2004. Global mRNA stabilization preferentially linked to translational repression during the endoplasmic reticulum stress response. Mol. Cell. Biol. 24:6773-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawakami, A., Q. Tian, X. Duan, M. Streuli, S. F. Schlossman, and P. Anderson. 1992. Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl. Acad. Sci. USA 89:8681-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963-969. [DOI] [PubMed] [Google Scholar]
- 25.Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M. J. Fitzler, D. Scheuner, R. J. Kaufman, D. E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keene, J. D. 1999. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl. Acad. Sci. USA 96:5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keene, J. D. 2001. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl. Acad. Sci. USA 98:7018-7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kullmann, M., U. Gopfert, B. Siewe, and L. Hengst. 2002. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 16:3087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lal, A., K. Mazan-Mamczarz, T. Kawai, X. Yang, J. L. Martindale, and M. Gorospe. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 23:3092-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lal, A., T. Kawai, X. Yang, K. Mazan-Mamczarz, and M. Gorospe. 2005. Anti-apoptotic function of RNA-binding HuR effected through prothymosin α. EMBO J. 24:1852-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loflin, P., C. Y. Chen, and A.-B. Shyu. 1999. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 13:1884-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López de Silanes, I., S. Galbán, J. L. Martindale, X. Yang, K. Mazan-Mamczarz, F. E. Indig, G. Falco, M. Zhan, and M. Gorospe. 2005. Identification and functional outcome of mRNAs associated with TIA-1 ribonucleoprotein complexes. Mol. Cell. Biol. 25:9520-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin, A. G., and H. O. Fearnhead. 2002. Apocytochrome c blocks caspase-9 activation and Bax-induced apoptosis. J. Biol. Chem. 277:50834-50841. [DOI] [PubMed] [Google Scholar]
- 34.Martin, A. G., J. Nguyen, J. A. Wells, and H. O. Fearnhead. 2004. Apocytochrome c inhibits caspases by preventing apoptosome formation. Biochem. Biophys. Res. Commun. 319:944-950. [DOI] [PubMed] [Google Scholar]
- 35.Mazan-Mamczarz, K., S. Galbán, I. López de Silanes, J. L. Martindale, U. Atasoy, J. D. Keene, and M. Gorospe. 2003. RNA-binding HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. USA 100:8354-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazan-Mamczarz, K., T. Kawai, J. L. Martindale, and M. Gorospe. 2005. En masse analysis of nascent translation using microarrays. BioTechniques 39:61-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng, Z., P. H. King, L. B. Nabors, N. L. Jackson, C. Y. Chen, P. D. Emanuel, and S. W. Blume. 2005. The ELAV RNA-stability factor HuR binds the 5′-untranslated region of the human IGF-IR transcript and differentially represses cap-dependent and IRES-mediated translation. Nucleic Acids Res. 33:2962-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min, H., C. W. Turck, J. M. Nikolic, and D. L. Black. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11:1023-1036. [DOI] [PubMed] [Google Scholar]
- 39.Moore, M. J. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309:1514-1518. [DOI] [PubMed] [Google Scholar]
- 40.Ron, D. 2002. Translational control in the endoplasmic reticulum stress response. J. Clin. Investig. 110:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoecklin, G., M. Colombi, I. Raineri, S. Leuenberger, M. Mallaun, M. Schmidlin, B. Gross, M. Lu, T. Kitamura, and C. Moroni. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 21:4709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taupin, J. L., Q. Tian, N. Kedersha, M. Robertson, and P. Anderson. 1995. The RNA-binding protein TIAR is translocated from the nucleus to the cytoplasm during Fas-mediated apoptotic cell death. Proc. Natl. Acad. Sci. USA 92:1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian, Q., M. Streuli, H. Saito, S. F. Schlossman, and P. Anderson. 1991. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell 67:629-639. [DOI] [PubMed] [Google Scholar]
- 44.Wang, W., H. Furneaux, H. Cheng, M. C. Caldwell, D. Hutter, Y. Liu, N. J. Holbrook, and M. Gorospe. 2000. HuR regulates p21 mRNA stabilization by ultraviolet light. Mol. Cell. Biol. 20:760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, W., S. Lin, C. M. Caldwell, H. Furneaux, and M. Gorospe. 2000. HuR regulates cyclin A and cyclin B1 mRNA stability during the cell division cycle. EMBO J. 19:2340-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, W., X. Yang, V. J. Cristofalo, N. J. Holbrook, and M. Gorospe. 2001. Loss of HuR influences gene expression during replicative senescence. Mol. Cell. Biol. 21:5889-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkie, G. S., K. S. Dickson, and N. K. Gray. 2003. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 28:182-188. [DOI] [PubMed] [Google Scholar]
- 48.Wilusz, C. J., and J. Wilusz. 2004. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 20:491-497. [DOI] [PubMed] [Google Scholar]
- 49.Yang, J., X. Liu, K. Bhalla, C. N. Kim, A. M. Ibrado, J. Cai, T.-I. Peng, D. P. Jones, and X. Wang. 1997. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science 275:1129-1132. [DOI] [PubMed] [Google Scholar]
- 50.Yu, Q., S. J. Cok, C. Zeng, and A. R. Morrison. 2003. Translational repression of human matrix metalloproteinase-13 by an alternatively spliced form of T-cell-restricted intracellular antigen-related protein (TIAR). J. Biol. Chem. 278:1579-1584. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, T., V. Kruys, G. Huez, and C. Gueydan. 2002. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem. Soc. Trans. 30:952-958. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, W., B. J. Wagner, K. Ehrenman, A. W. Schaefer, C. T. DeMaria, D. Crater, K. DeHaven, L. Long, and G. Brewer. 1993. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 13:7652-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]