Abstract
NF-κB is critical for determining cellular sensitivity to apoptotic stimuli by regulating both mitochondrial and death receptor apoptotic pathways. The endoplasmic reticulum (ER) emerges as a new apoptotic signaling initiator. However, the mechanism by which ER stress activates NF-κB and its role in regulation of ER stress-induced cell death are largely unclear. Here, we report that, in response to ER stress, IKK forms a complex with IRE1α through the adapter protein TRAF2. ER stress-induced NF-κB activation is impaired in IRE1α knockdown cells and IRE1α−/− MEFs. We found, however, that inhibiting NF-κB significantly decreased ER stress-induced cell death in a caspase-8-dependent manner. Gene expression analysis revealed that ER stress-induced expression of tumor necrosis factor alpha (TNF-α) was IRE1α and NF-κB dependent. Blocking TNF receptor 1 signaling significantly inhibited ER stress-induced cell death. Further studies suggest that ER stress induces down-regulation of TRAF2 expression, which impairs TNF-α-induced activation of NF-κB and c-Jun N-terminal kinase and turns TNF-α from a weak to a powerful apoptosis inducer. Thus, ER stress induces two signals, namely TNF-α induction and TRAF2 down-regulation. They work in concert to amplify ER-initiated apoptotic signaling through the membrane death receptor.
The endoplasmic reticulum (ER) is the organelle where protein folding occurs prior to transport to the extracellular surface or to different intracellular sites. This process depends on molecular chaperones that provide local environments favorable for protein folding. However, under a variety of conditions (ER stress), these folding reactions are compromised, and protein aggregation occurs (21). Persistent protein aggregation eventually causes cell death through apoptosis. ER-resident caspase-12 initiates apoptotic signaling from the ER (26). Mitochondria also play an important role in amplifying apoptotic signaling from the ER (32).
There are at least three known sensors of ER stress in metazoan species, namely, IRE, PERK, and ATF6. PERK mediates global translation attenuation through phosphorylation of the α subunit of eukaryotic translation initiation factor 2 (13). Upon activation, ATF6 is proteolyzed to release a cytosolic fragment that migrates to the nucleus to activate target genes through binding to the ER stress-response element (41). IRE1 is the most conserved ER stress sensor. IRE1α is constitutively expressed in all cells and tissues, whereas IRE1β is restricted to the gut epithelium. In response to ER stress, IRE1 dimerizes, autophosphorylates, and activates its endoribonuclease (RNase) activity. Through site-specific cleavage of HAC1 mRNA (in yeast) and XBP1 mRNA (in mouse), IRE1α converts HAC1 and XBP1 into potent transcription activators (22).
In addition to IRE, PERK, and ATF6, ER retention of adenovirus E3/19K protein activates NF-κB (27). This result suggests that NF-κB may be involved in regulation of ER stress. NF-κB represents a group of structurally related and evolutionarily conserved proteins, with five members in mammals: Rel (c-Rel), RelA (p65), RelB, NF-κB1, and NF-κB2. Regulation of apoptosis is one of the most important functions of NF-κB. It is believed that regulation of NF-κB activity is a key cellular response that modulates the outcome of cells exposed to stress stimulation. NF-κB induces the expression of a number of genes whose products can suppress apoptosis, including inhibitors of apoptosis, c-FLIP, TRAF1, TRAF2, IEX-1, and ferritin heavy chain (20, 29). However, some important proapoptotic genes are also targeted by NF-κB, including Fas ligand (FasL), TRAIL (20), tumor necrosis factor alpha (TNF-α) (37), and p53 (8). So NF-κB is at the crossroads of life and death. Through regulation of the expression of these target genes, NF-κB modulates both mitochondrial and death receptor apoptotic pathways. Considering that the ER has emerged as another apoptosis control point in addition to mitochondrial and death receptor pathways, it is interesting to investigate the role of NF-κB in the regulation of ER stress.
Here, we report that the ER stress sensor IRE1α is required for activation of NF-κB in thapsigargin- or tunicamycin-treated cells. In response to ER stress, IRE1α forms a complex with inhibitor κB kinase (IKK) through TRAF2. Inhibition of NF-κB suppresses ER stress-induced cell death in MCF-7 cells. We also show that expression of TNF-α but not FasL and TRAIL is induced by ER stress in an IRE1α- and NF-κB-dependent manner. Blocking this autocrinely activated TNF receptor 1 (TNFR1) signaling reverses NF-κB-mediated cell death. We further found that ER stress induced a decrease in TRAF2. This inhibits TNF-α-induced activation of NF-κB and c-Jun N-terminal kinase (JNK) and makes cells susceptible to TNF-α-induced cell death. Therefore, our study establishes a novel link between ER stress and the membrane death receptor through IRE1α-NF-κB-TNF-α-TRAF2 and demonstrates cross talk between the ER and the death receptor pathway to initiate apoptotic signaling. The link between the disruption of homeostasis of the ER and induction of TNF-α suggests that the ER can influence the pathogenesis of certain human diseases in addition to its role in unfolded protein aggregation.
MATERIALS AND METHODS
Cell culture.
The human breast cancer cell line MCF-7, human lung cancer cell line H1299, human prostate cancer cell lines PC-3 and DU145, and mouse fibroblast L929 cells were from the American Type Culture Collection. Wild-type and IRE1α−/− mouse embryonic fibroblasts (MEFs) were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS).
Reagents.
Tunicamycin, thapsigargin, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), camptothecin, hydrogen peroxide, and cycloheximide (CHX) were purchased from Sigma-Aldrich. The caspase-8-specific inhibitor Z-IETD-fmk was obtained from Calbiochem. Recombinant TNF-α, TNFR1-Fc, and Fas-Fc were purchased from R&D Systems. Protein A- and protein G-Sepharose were obtained from Amersham Biotech. [γ32P]ATP (3,000 mCi/nmol) and [35S]methionine (>1,000 Ci/mmol) were purchased from NEN.
Western blotting and antibodies.
After treatments as indicated in the experiments below, cells were washed once with phosphate-buffered saline and extracted with sodium dodecyl sulfate (SDS) sample buffer. Equal amounts of protein from each sample were applied to Novex Tris-glycine gels (Invitrogen) and transferred to polyvinylidene difluoride membranes (Millipore). After a blocking step, the membranes were incubated with each primary antibody, followed by incubation with a horseradish peroxidase-conjugated antibody. The protein bands were visualized using an ECL detection system (Amersham Biotech). The following antibodies were used: anti-β-actin and anti-Flag fromSigma; anti-IκBα, anti-IKKβ, anti-phospho-IKKβ, anti-JNK, and anti-phospho-JNK from Cell signaling; anti-RIP (where RIP is receptor-interacting protein) and anti-IKKγ from BD PharMingen; anti-IRE1α and anti-TNFR1 from Santa Cruz Biotechnology; anti-TRAF2 from IMGENEX; anti-RelA from Upstate; and anti-T7 Tag antibody from Novagen.
EMSA.
The NF-κB DNA binding activity was measured using a gel shift assay system (Promega). Nuclear extracts for electrophoretic mobility shift assays (EMSAs) were prepared from treated cells as indicated in the figure legends. For the binding reaction, 5 μg of nuclear extract from each sample was incubated at room temperature for 10 min with reaction buffer, and then 1 μl of [32P]ATP-labeled NF-κB binding consensus oligonucleotide was added to each reaction mixture for an additional 20 min at room temperature. The reaction products were analyzed on a nondenaturing 4% acrylamide gel, which was then dried and subjected to autoradiography.
Plasmids and transfection.
Flag-tagged human IKKα, IKKβ cDNA in CMV4 vector, and a hemagglutinin (HA)-tagged S32A S36A mutant IκBα were cloned into pcDNA3.1. A T7-tagged human IRE1α cDNA was a generous gift from Randal Kaufman (University of Michigan). For stable transfection, the MCF-7 cells were plated at a density of 1.0 × 105 cells/well in six-well plates and transfected with pcDNA3.1m IκBα (S32A S36A) or empty vector (pcDNA3.1) using Lipofectamine Plus (Invitrogen). The stably expressing cells were selected in the presence of G418 (800 μg/ml) for 4 weeks, and the surviving clones were pooled (mass culture). The transfectants were identified by Western blotting using anti-HA antibody. For transient transfection 2.0 × 105 HEK293 cells were plated in each well of the six-well plates. An IKKα or IKKβ construct was transfected alone or cotransfected with the IRE1α expression plasmid using FuGENE6 (Roche) for 48 h before the experiments below (Roche). MEFs were transfected using FuGENE6.
Immunoprecipitation.
For coimmunoprecipitation of IRE1α and IKK, HEK293 cells were transiently transfected with Flag-tagged IKKα or IKKβ alone or cotransfected with T7-tagged IRE1α for 48 h and then collected in lysis buffer (50 mM HEPES at pH 7.6, 250 mM NaCl, 0.1% NP-40, 5 mM EDTA, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin). The lysates were precipitated with anti-Flag antibody and protein G-Sepharose beads by incubation at 4°C for 4 h. The beads were washed five times with lysis buffer, and the bound proteins were analyzed by Western blotting using an anti-T7 antibody. For coimmunoprecipitation of endogenous IRE1α and IKK, MCF-7 cells were treated with tunicamycin for the indicated times. Cell lysates were collected as described above. The lysates were precipitated with an anti-IKKγ antibody and protein A-Sepharose beads and then analyzed by Western blotting with anti-IRE1α and anti-TRAF2 antibodies.
In vitro protein interactions.
[35S]methionine-labeled IKKα, IKKβ, IKKγ, and TRAF2 were produced using TNT-coupled reticulocyte lysate systems (Promega) and then incubated with IRE1α, which was immobilized on beads using a conjugated anti-T7 antibody (Novagen), in binding buffer for 2 h. The mixtures were washed three times with wash buffer before being subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
siRNA knockdown experiments.
The MCF-7 cells plated at a density of 2.0 × 105 cells/well in six-well plates were transfected with the indicated small interfering RNA (siRNA) using Oligofectamine reagent (Invitrogen) for 48 h before treatment. The following siRNAs were used: siRNA pool to human IRE1α, siRNA pool to human TNFR1, a control siRNA (all from Dharmacon), siRNA to human RelA (Upstate), and siRNA to human caspase-8 (Molecula). The siRNA to human TRAF2 duplexes, 5′-CGACAUGAACAUCGCAAGC-3′ (39), was purchased from Dharmacon.
Cell viability assays.
The MTT assay was used to assess cell viability and was performed according to the manufacturer's directions (Sigma). Briefly, cells grown in six-well plates were treated as required. Then, 100 μl of MTT at a concentration of 5 mg/ml was added to each well, and incubation was continued for 2 h. The formazan crystals resulting from mitochondrial enzymatic activity on MTT substrate were solubilized with dimethyl sulfoxide (DMSO). Absorbance was measured at 570 nm using a microplate reader (Molecular Devices). Cell viability was expressed as absorbance relative to that of untreated controls.
Analysis of caspase-8 activity.
MCF-7 cells were treated with thapsigargin or tunicamycin for the indicated times. Cell lysates were used to measure caspase-8 activity using an ApoAlert caspase-8 colorimetric assay kit (BD Clontech). Analyses were performed using a microplate reader at 405 nm (Molecular Devices).
RT-PCR.
Cells were treated with the indicated reagents, and total RNA was extracted by the Trizol method (Invitrogen). It was subjected to reverse transcription-PCR (RT-PCR) in a two-step protocol using SuperScript Reverse Transcriptase (Invitrogen) and Taq polymerase (Applied Biosystems). The number of cycles and annealing temperature were adjusted depending on the genes amplified. Primer sequences are available upon request.
Measurement of TNF-α production.
TNF-α levels were measured in cell supernatants by a human TNF-α chemiluminescent immunoassay kit (R&D Systems).
Adenoviral vector infection and luciferase assay.
An adenoviral vector containing the human TNF-α promoter (−1173 bp) with the 3′ untranslated region of the TNF-α gene was kindly provided by B. M. J. Foxwell (Imperial College School of Medicine, United Kingdom). The cells were plated at a density of 2 × 105 cells/well in six-well plates and exposed to virus at the optimal multiplicity of infection for 1 h in serum-free medium, followed by washing and reculturing in growth medium with 2% FBS for 12 h. After stimulation with thapsigargin or tunicamycin as indicated, luciferase activities were measured using a luciferase assay kit (Promega).
RESULTS
IRE1α is required for ER stress-induced NF-κB activation.
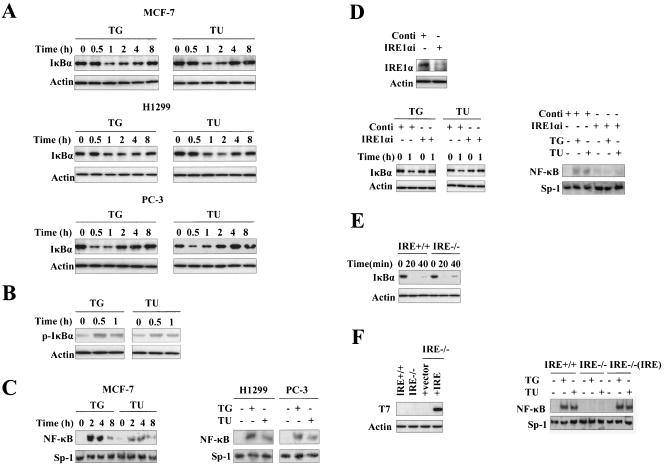
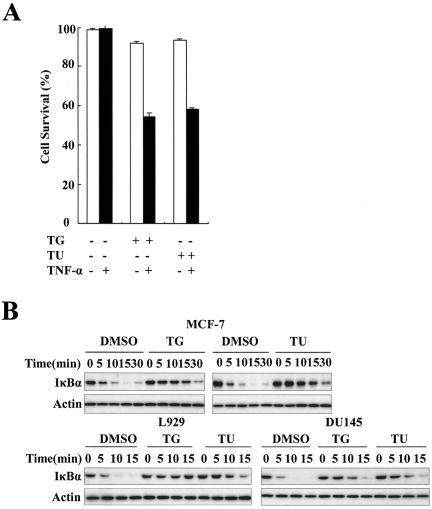
The transcription factor NF-κB is involved in regulation of cellular responses to diverse types of stress, including genotoxic stress (17) and oxidative stress (25). The observation that aberrant accumulation of adenovirus E3/19K protein within the ER activates NF-κB (27) suggests that activation of NF-κB may be an important cellular response to ER stress. To further address the role of NF-κB in the modulation of ER stress, we first studied the effect of two well-known ER stress inducers, thapsigargin and tunicamycin, on the activity of NF-κB in diverse types of mammalian cells. IκB proteins inhibit NF-κB dimers in the cytoplasm, and upstream signaling-mediated phosphorylation and sequent degradation of IκB play a critical role in activating NF-κB (4). As shown in Fig. 1A, I κBα degradation in MCF-7 cells was detected after thapsigargin or tunicamycin treatment for 1 h, and IκBα protein returned to basal levels later. This result is consistent with the fact that IκBα itself is a target gene of NF-κB, so this negative feedback prevents persistent activation of NF-κB (36). Thapsigargin- or tunicamycin-induced degradation of IκBα protein was also observed in H1299 cells and PC-3 cells (Fig. 1A). Consistent with the results of IκBα degradation, the phosphorylation of IκBα protein in MCF-7 cells was detected after thapsigargin or tunicamycin treatment for 0.5 h (Fig. 1B).
FIG. 1.
IRE1α is required for ER stress-induced NF-κB activation. (A) ER stress induces IκBα degradation. MCF-7, H1299, and PC-3 cells were exposed to thapsigargin (2 μM) or tunicamycin (2 μg/ml) for the indicated times, and cell extracts were subjected to SDS-PAGE for Western blotting with anti-IκBα and anti-β-actin antibodies. (B) ER stress induces IκBα phosphorylation. MCF-7 cells were treated with thapsigargin or tunicamycin for the indicated times, and phosphorylated IκBα was examined by Western blotting. (C) ER stress activates NF-κB DNA binding activity. MCF-7, H1299, and PC-3 cells were treated with thapsigargin (2 μM) or tunicamycin (2 μg/ml) for various times as indicated. These concentrations were used in all subsequent experiments. Nuclear extracts were prepared, and 5 μg of nuclear extract from each sample was used to analyze NF-κB DNA binding activity by EMSA with an NF-κB probe. Sp-1 DNA binding activity in each sample was detected as a loading control. (D) Impaired activation of NF-κB by ER stress in IRE1α knockdown cells. MCF-7 cells were transfected with a control siRNA and an siRNA pool specific to human IRE1α for 48 h. Cell lysates were then analyzed for IRE1α level by Western blotting as shown in the upper panel. The same treated MCF-7 cells were subjected to treatment with thapsigargin or tunicamycin for various times as indicated. Whole-cell extracts and nuclear extracts were prepared, and IκBα degradation and NF-κB DNA binding activity were examined by Western blotting and EMSA with an anti-IκBα antibody and an NF-κB probe, respectively. (E) TNF-α-induced NF-κB activation in IRE1α−/− and IRE1α+/+ MEFs. Mouse wild-type and IRE1α−/− fibroblasts were incubated with TNF-α (15 ng/ml) for the indicated times. The levels of IκBα and β-actin were examined by Western blotting. (F) Expression of IRE1α reconstitutes impaired ER stress-induced IκBα degradation and NF-κB activation in IRE1α−/− fibroblast cells. IRE1α−/− MEFs were transfected with a control and a T7-tagged IRE1α vector for 36 h, and cell lysates were analyzed for IRE1α-T7 level by Western blotting as shown in the upper panel. IRE1α+/+, IRE1α−/−, and IRE1α−/− (IRE1α) fibroblast cells were incubated with thapsigargin or tunicamycin as shown in the lower panel. NF-κB activity was measured by EMSA with a NF-κB probe. TG, thapsigargin; TU, tunicamycin; Conti, control siRNA.
To further confirm the evidence of IκBα degradation, we examined the induction of NF-κB activity by EMSA using an NF-κB-specific probe. As expected, an increase in NF-κB DNA-binding activity in response to thapsigargin or tunicamycin was observed in MCF-7, H1299, and PC-3 cells (Fig. 1C). The kinetics of NF-κB activation correlated well with the degradation of IκBα. That different ER stress inducers activated NF-κB in different cell types suggests that ER stress-induced activation of NF-κB is a general response.
In mammals, IRE1, ATF6, and PERK mediate the cellular response to ER stress. A recent paper showed that during ER stress, phosphorylation of the α subunit of eukaryotic initiation factor 2 by PERK is required for activation of NF-κB. However, the mechanism of NF-κB activation does not seem to involve IκB degradation (19). IRE1 induces JNK activation through interaction with TRAF2 in ER stress, which is a pathway similar to that used by cells in response to TNF-α (38), and it is known that in TNF-α signaling, TRAF2 is required for both JNK and NF-κB activation (3). So we focused our research on IRE1.
IRE1 includes IRE1α and IRE1β, and IRE1α is ubiquitously expressed, so we focused on IRE1α. To test our hypothesis, we examined the effect of ablation of endogenous IRE1α on the ER stress induction of NF-κB activity. As shown in the upper panel of Fig. 1D, introduction of a specific siRNA pool to human IRE1α into the MCF-7 cells greatly decreased expression of IRE1α protein compared with a nonspecific control siRNA. More importantly, knockdown of IRE1α significantly inhibited ER stress-induced IκBα degradation and NF-κB DNA binding activity, as shown in the bottom panels of Fig. 1D. Furthermore, we examined ER stress-induced NF-κB activation in wild-type and IRE1α knockout MEF cells. As shown in Fig. 1E, there was little difference in IκBα degradation observed between IRE1α−/− and IRE1α+/+ MEFs when cells were exposed to TNF-α. However, as shown in Fig. 1F, NF-κB DNA binding activity was absent in IRE1α−/− MEFs compared to IRE1α+/+ MEFs when cells were exposed to thapsigargin or tunicamycin. These results suggest that inhibition of NF-κB activation in IRE1α−/− MEFs may be specific to ER stress. To rule out the possibility that some other defects in the ER stress-mediated NF-κB activation were present in the IRE1α−/− MEFs, we tested whether ER stress-induced NF-κB activation could be rescued by ectopic expression of IRE1α. As shown in Fig. 1F, transfection of a human T7-tagged IRE1α cDNA into the IRE1α−/− MEFs reconstituted ER stress-induced activation of NF-κB). These results indicate the essential role of IRE1α in ER stress-induced NF-κB activation.
ER stress induces the formation of IRE1α and IKK complex.
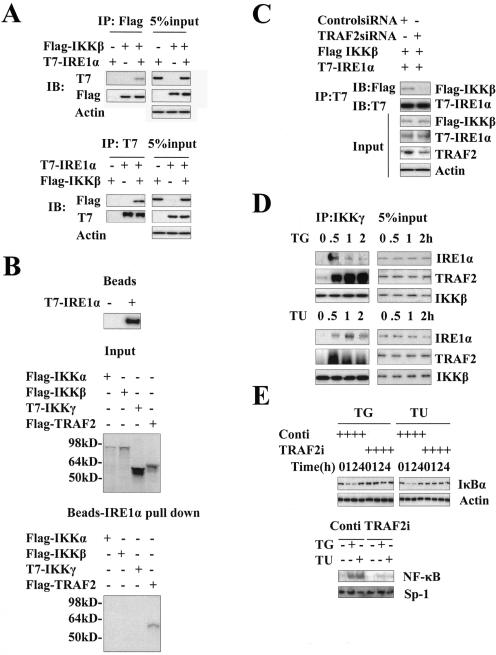
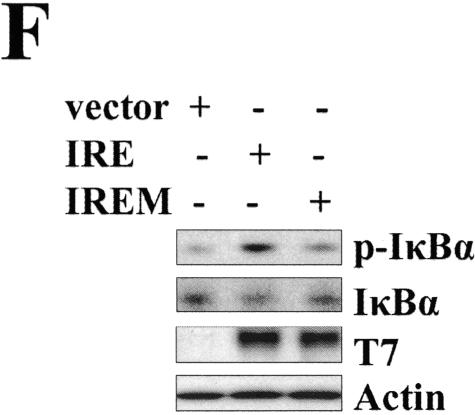
To further investigate the mechanisms by which IRE1α mediates IκBα degradation and NF-κB activation, we examined whether the IKK complex interacts with IRE1α. HEK293 cells were cotransfected with Flag-tagged human IKKβ and T7-tagged human IRE1α cDNA, and then coimmunoprecipitation was tested using the indicated antibodies. As shown in Fig. 2A (top), in the sample in which the IKKβ and IRE1α were cotransfected, IRE1α was clearly detected in the anti-Flag immunoprecipitates by anti-T7 antibody. However, in the control sample, IRE1α was not detected. Portions of the total cell extracts were immunoblotted with anti-β-actin antibody as a control for loading. A reverse coimmunoprecipitation experiment was also conducted; IKKβ was easily detected in the anti-T7 immunoprecipitates by anti-Flag antibody (Fig. 2A, bottom). To further characterize the association between the IKK complex and IRE1α, IRE1α was immobilized by immunoprecipitation with a conjugated T7 antibody as shown in Fig. 2B (top). Input of in vitro translated [35S]methionine-labeled IKKα, IKKβ, IKKγ, or TRAF2 is shown in the middle panel of Fig. 2B, and in vitro binding assays using immobilized IRE1α and in vitro-translated [35S]methionine-labeled IKKα, IKKβ, IKKγ, and TRAF2 are shown on the bottom panel of Fig. 2B. These revealed that IRE1α directly associates with TRAF2 but not with IKKα, IKKβ, or IKKγ. These results suggest that IRE1α does not directly interact with IKK. However, we acknowledge that there is a difference of magnitude between the input values of the four proteins. To check whether TRAF2 is required for formation of the IRE1α-IKK complex, we performed an siRNA knockdown experiment, as shown in Fig. 2C. Reduction of endogenous TRAF2 protein in HEK293 cells dramatically decreased the interaction of cotransfected IKKβ and IRE1α. To test the interaction of endogenous IRE1α and IKK, MCF-7 cells were treated with tunicamycin for different times, and then the cell extracts were collected for immunoprecipitation with an anti-IKKγ antibody. The immunoprecipitates were analyzed by Western blotting with anti-IRE1α and anti-TRAF2 antibody. As shown in Fig. 2D, the interaction between IRE1α, TRAF2, and IKK was undetectable in nontreated cells, whereas in the cells treated with tunicamycin or thapsigargin for 0.5 h, clear bands of IRE1α and TRAF2 were detected. The kinetics of this interaction correlated well with the kinetics of IκBα degradation and NF-κB activation. As a control, the IKKβ level in the immunoprecipitates was measured. The data indicated that the same amount of IKK was precipitated in each sample (Fig. 2D). Consistent with the coimmunoprecipitation results, knockdown of TRAF2 by siRNA significantly inhibited ER stress-induced IκBα degradation and NF-κB activation (Fig. 2E). To further investigate the mechanism by which IRE1α activated the IKK complex, we transiently transfected a wild-type IRE1α and a kinase-defective (K599A) mutant IRE1α into the COS-7 cells. As shown in Fig. 2F, overexpression of wild-type IRE1α alone is enough to activate phosphorylation of IκBα and promote its degradation. However, overexpression of the kinase-dead IRE1α mutant (K599A) (Fig. 2F, IREM) cannot cause phosphorylation and degradation of IκBα. Together, our results suggest that, in response to ER stress, IRE1α forms a complex with IKK through TRAF2 and that kinase activity of IRE1α is required for activating the IKK complex.
FIG.2.
ER stress induces formation of IRE1α and IKK complex. (A) Interaction of IRE1α with IKKβ in vivo. T7-tagged IRE1α and Flag-tagged IKKβ were cotransfected into HEK293 cells. Cell lysates were immunoprecipitated with anti-Flag antibody or T7 antibody, respectively, and then subjected to SDS-PAGE for Western blotting with anti-T7 antibody or anti-Flag antibody. Five percent of the cell extract from each sample was examined with anti-Flag and anti-T7, with anti-β-actin as a control of protein input. (B) Interaction of IRE1α with IKKβ in vitro. Immobilized IRE1α protein was incubated with in vitro translated IKKα, IKKβ, IKKγ, and TRAF2 (top) for 2 h and then subjected to SDS-PAGE and autoradiography (lower panel). (C) Ablation of TRAF2 impairs formation of IKK and IRE1α complex. HEK293 cells were transfected with an siRNA specific to TRAF2 and a control siRNA for 48 h and then cotransfected with T7-tagged IRE1α and Flag-tagged IKKβ for 24 h. Cell lysates were coimmunoprecipitated with anti-T7 antibody, and immunoprecipitates were probed with anti-Flag antibody. (D) ER stress induces the formation of IKK and IRE1α complex. MCF-7 cells were incubated with thapsigargin or tunicamycin for the indicated times. Cell extracts from each sample were immunoprecipitated with an anti-IKKγ antibody. Immunoprecipitates were analyzed by Western blotting with anti-IRE1α and anti-IKKβ antibodies. Five percent of cell extract from each sample was used as a control of protein input. (E) ER stress-induced NF-κB activation is impaired in TRAF2 knockdown cells. After transfection with TRAF2 siRNA and control siRNA for 48 h, MCF-7 cells were subjected to thapsigargin or tunicamycin for the indicated times. Whole-cell extracts and nuclear extracts were prepared, and IκBα degradation (top) and NF-κB DNA binding activity (bottom) were examined by Western blotting and EMSA. (F) Kinase activity is required for IRE1α to activate the IKK complex. COS-7 cells were transfected with T7-tagged wild-type and kinase-defective (K599A) mutant IRE1α. Forty-eight hours later cell extracts were prepared and protein levels of T7, IκBα, phosphorylated IκBα, and actin were detected by Western blotting. TG, thapsigargin; TU, tunicamycin; Conti, control siRNA.
NF-κB mediates ER stress-induced cell death.
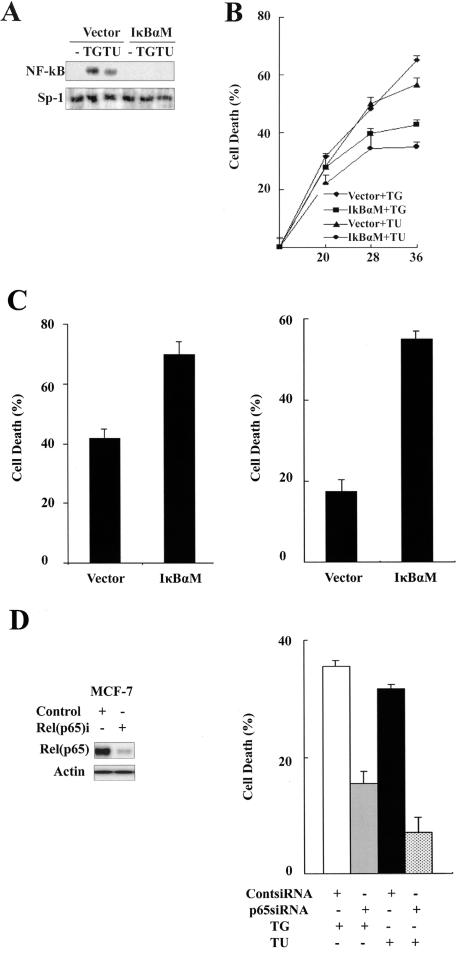
One of most important functions of NF-κB is to regulate cell death and survival. We wished to address the role of NF-κB in ER stress-induced cell death. First, pyrrolidine dithiocarbamate (PDTC), a NF-κB inhibitor, was used to influence thapsigargin- or tunicamycin-induced cell death in MCF-7 cells. A cell viability assay (MTT) showed that PDTC remarkably decreased thapsigargin- or tunicamycin-induced cell death. However, N-acetyl-L-cysteine had no protective effect on ER stress-induced cell death (data not shown). Ubiquitination-mediated IκBα protein degradation depends on phosphorylation of Ser32 and Ser36 of IκBα by the IKK complex (4). To further characterize the role of NF-κB in ER stress, we established stable pools of MCF-7 cells in which a plasmid encoding an HA-tagged S32A and S36A double mutant IκBα was introduced (IκBαM cells). As shown in Fig. 3A, expression of this degradation-resistant IκBα completely inhibited thapsigargin-induced NF-κB activation. The MTT assay showed that IκBαM cells displayed a significantly increased resistance to thapsigargin- and tunicamycin-induced cell death compared with control cells (Fig. 3B). However, IκBαM cells showed increased sensitivity to TNF-α- and hydrogen peroxide-induced cell death (Fig. 3C). These observations suggest that the protective role of mutant IκBα is specific to ER stress.
FIG. 3.
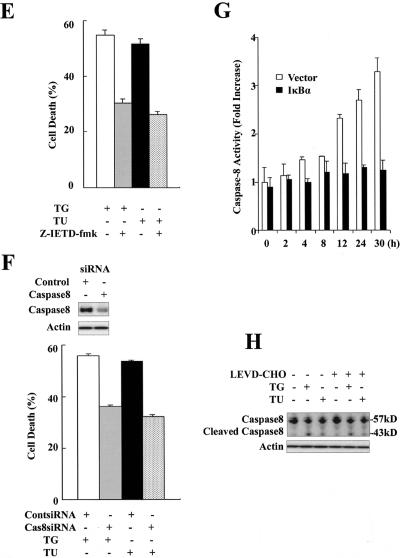
NF-κB mediates ER stress-induced cell death. (A) Expression of mutant IκBα blocks ER stress-induced NF-κB activation. MCF-7 cells were stably transfected with pcDNA3 (empty vector) or pcDNA3 carrying an S32A and S36A double mutant HA-tagged IκBα (IκBαM MCF-7 cells). Pooled control and IκBαM MCF-7 cells were exposed to thapsigargin or tunicamycin for 2 h. NF-κB activity was measured by EMSA as described in the legend of Fig. 1. (B) Increased resistance to ER stress in IκBαM cells. Control and IκBαM MCF-7 cells were incubated with thapsigargin or tunicamycin for 30 h. Cell death was detected by MTT assay. Cell survival was expressed as absorbance relative to that of DMSO-treated controls. (C) Increased sensitivity to TNF-α- and hydrogen peroxide-induced cell death in IκBαM cells. Control and IκBαM MCF-7 cells were exposed to TNF-α (30 ng/ml) for 12 h and H2O2 (500 μM) for 18 h. Cell viability was scored by MTT assay as described in the legend of panel B. (D) Knockdown of RelA inhibits ER stress-induced cell death. MCF-7 cells were transfected with a control siRNA and an siRNA pool specific to RelA for 48 h, and cell lysates were analyzed for RelA level by Western blotting as shown at left. The same treated MCF-7 cells were subjected to treatment with thapsigargin or tunicamycin for 30 h. Cell death was detected by MTT assay (right). (E) Caspase-8 inhibitor Z-IETD-fmk decreases ER stress-induced cell death. MCF-7 cells were exposed to thapsigargin or tunicamycin with or without Z-IETD-fmk (100 μM) for 30 h. Cell death was detected by MTT assay. (F) Knockdown of caspase-8 decreases ER stress-induced cell death. MCF-7 cells were transfected with a control siRNA and an siRNA specific for caspase-8 for 48 h, and caspase-8 levels were detected by Western blotting. The same treated MCF-7 cells were subjected to thapsigargin or tunicamycin for 30 h. Cell death was detected by MTT assay. (G) ER stress activates caspase-8. Control and IκBαM MCF-7 cells were treated with thapsigargin or tunicamycin for the indicated times. Cell lysates were collected and used to measure caspase-8 activity. (H) Caspase-4 is not required for ER stress-induced caspase-8 activation. MCF-7 cells were exposed to thapsigargin or tunicamycin with or without the caspase-4 inhibitor LEVD-CHO (20 μM) for 24 h, and caspase-8 protein level was detected by Western blotting. TG, thapsigargin; TU, tunicamycin; Cont, control.
The p65 subunit (RelA) of NF-κB is believed to play a critical role in inducing target genes of NF-κB. We further confirmed the proapoptotic effect of NF-κB in ER stress by eliminating the p65 protein using specific siRNA (p65-siRNA). As shown in Fig. 3D (left), p65-siRNA largely abolished the endogenous p65 protein in MCF-7 cells. The MTT assay revealed that p65-siRNA significantly reduced thapsigargin- and tunicamycin-induced cell death compared to a control siRNA (Fig. 3D, right). Consistent with the results from the PDTC and IκBα mutant experiments, these data indicate that NF-κB is a major factor in ER stress-induced cell death in MCF-7 cells.
Caspase-8 mediates ER stress-induced cell death.
Apoptosis is primarily mediated by a family of caspases. Different caspases mediate cell death in response to different apoptotic stimuli. Caspase-9 mediates the mitochondrial apoptosis pathway, and caspase-8 is specific to the death receptor apoptosis pathway (5). Recent studies show that caspase-4 in human and caspase-12 in mouse mediate ER stress-induced apoptosis (14, 26). In our previous studies, we found that a broad-spectrum caspase inhibitor zVAD-fmk significantly decreased ER stress-induced cell death, indicating that apoptosis is involved in ER stress (16). To address the exact apoptosis pathway, we tested the ability of different caspase inhibitors to block ER stress-induced cell death in MCF-7 cells. As shown in Fig. 3E, the caspase-8-specific inhibitor Z-IETD-fmk markedly decreased thapsigargin- or tunicamycin-induced cell death. To confirm the results with Z-IETD-fmk, an siRNA specific for human caspase-8 was used. As shown in Fig. 3F (top), caspase-8-siRNA ablated about 70% of endogenous caspase-8 in MCF-7 cells. The MTT assay showed that elimination of caspase-8 dramatically prevented thapsigargin- or tunicamycin-induced cell death compared to control siRNA (Fig. 3F). A caspase-8 activity assay revealed a large increase of caspase-8 activity in control but not IκBαM MCF-7 cells after exposure to tunicamycin (Fig. 3G). Because caspase-4 plays an important role in initiating apoptotic signaling from the ER, it was interesting to examine whether caspase-4 is upstream of caspase-8. As shown in Fig. 3H, LEVD-CHO, a selective caspase-4 inhibitor, did not inhibit thapsigargin- or tunicamycin-induced caspase-8 cleavage. This indicates that caspase-4 and caspase-8 are involved in separate apoptotic signaling in response to ER stress. These results suggest that caspase-8 mediates ER stress-induced cell death in MCF-7 cells and that the membrane death receptor apoptotic pathway may be involved in regulation of ER stress-induced cell death.
ER stress induces TNF-α expression in an NF-κB dependent manner.
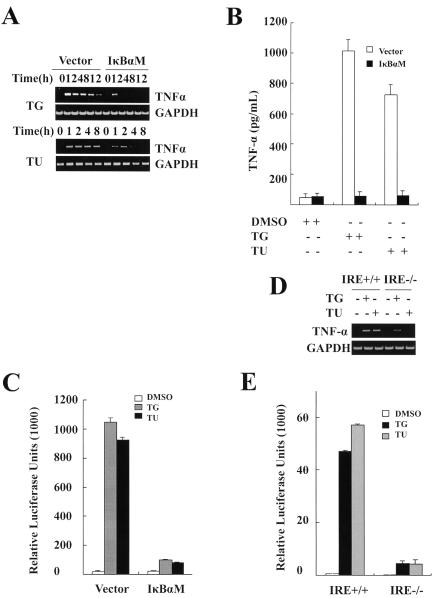
Some targets of NF-κB are important components of the death receptor pathway, for example, FasL, TRAIL, and TNF-α. We hypothesized that ER stress-induced NF-κB activity mediates expression of these targets. To test this hypothesis, an RT-PCR experiment was performed, and we observed that TNF-α mRNA levels rapidly increased when MCF-7 cells (Fig. 4A), MEF cells (Fig. 4D), and P3 and H1299 cells (data not shown) were exposed to thapsigargin or tunicamycin. This suggests that induction of TNF-α transcription in epithelial and mesenchyme-derived cells is a general cellular response to ER stress. However, we did not detect any induction of FasL, TRAIL, and TNF-β in these cells by RT-PCR (data not shown). Induction of TNF-α by ER stress depended on activation of NF-κB. As shown in Fig. 4A, induction of TNF-α in IκBαM cells was dramatically prevented. Enzyme-linked immunosorbent assay results also revealed that the level of secreted TNF-α in the supernatant of medium was very much lower in the IκBαM cells (Fig. 4B). To further address whether the TNF-α promoter is activated in response to ER stress, we infected MCF-7 cells with an adenovirus vector that contained the 5′ sequence of the human TNF-α gene (including a 1.3-kb region upstream of the major transcription start site) upstream of a luciferase coding region. Luciferase assays showed large increases of TNF-α promoter activity after treatment of MCF-7 cells with thapsigargin or tunicamycin compared with treatment with DMSO (Fig. 4C). Similar results were seen in H1299 and PC3 cells (data not shown). ER stress-induced TNF-α promoter activity was remarkably inhibited in IκBαM cells (Fig. 4C). To address the role of IRE1α in ER stress-induced expression of TNF-α, we then examined the TNF-α mRNA level and the TNF-α promoter activity in IRE1α+/+ and IRE1α−/− MEFs. As shown in Fig. 4D and E, absence of IRE1α significantly decreased thapsigargin- or tunicamycin-induced TNF-α transcription and promoter activity. Taken together, our results indicate that ER stress induces expression of TNF-α in an IRE1α- and NF-κB-dependent manner.
FIG. 4.
ER stress induces TNF-α in an IRE1α-NF-κB dependent manner. (A) Induction of TNF-α depends on NF-κB activation. Control and IκBαM MCF-7 cells were incubated with thapsigargin or tunicamycin for the indicated times. Total RNA was analyzed for the expression of TNF-α by RT-PCR. glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. (B) Detection of secreted TNF-α protein. Control and IκBαM MCF-7 cells were incubated with thapsigargin or tunicamycin for the indicated times. Supernatants of cell cultures from each sample were collected, and TNF-α protein levels were determined by enzyme-linked immunosorbent assay. Data are representative of triplicate experiments. (C) ER stress-activated TNF-α promoter activity is inhibited by mutant IκBα. Control and IκBαM MCF-7 cells were incubated with thapsigargin or tunicamycin for 2 h; then TNF-α promoter activity was detected by luciferase assay, and values were normalized based on β-galactosidase activity. (D and E) Impaired TNF-α induction in IRE1α−/− fibroblasts. Wild-type and IRE1α−/− MEFs were exposed to thapsigargin or tunicamycin for various times, as indicated. Total RNA and cell lysates, respectively, were collected and analyzed by RT-PCR for TNF-α expression and by luciferase assay for promoter activity. TG, thapsigargin; TU, tunicamycin.
ER stress-induced NF-κB activation is independent of TNF-α signaling.
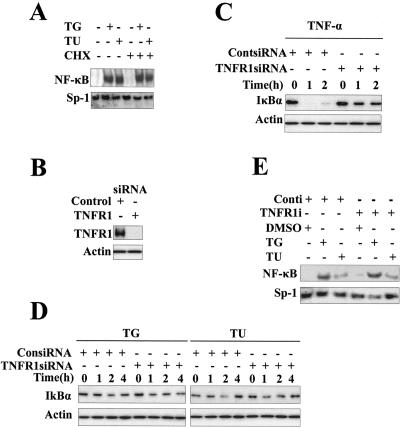
Cytokines like TNF-α and interleukin-1 (IL-1) are strong activators of NF-κB. It has been proposed previously that genotoxic stress may activate NF-κB via cell membrane receptors, such as TNF-α and IL-1 receptors, through the synthesis and release of cytokines (1). To rule out the possibility that ER stress-induced NF-κB activation was caused by the autocrine activity of TNF-α, we tested NF-κB activity in the presence of CHX in response to ER stress. As shown in Fig. 5A, presence of CHX had no effect on tunicamycin- or thapsigargin-induced NF-κB activation in MCF-7 cells. This indicates that ER stress-induced activation of NF-κB does not need de novo protein synthesis, and it is unlikely that newly synthesized TNF-α is responsible for ER stress-induced NF-κB activation. To further exclude the involvement of TNFR1 signaling in ER stress-induced NF-κB activation, a specific siRNA pool to TNFR1 (TNFR1-siRNA) was used to reduce the endogenous level of TNFR1. As shown in Fig. 5B, TNFR1-siRNA almost completely eliminated TNFR1 protein in MCF-7 cells. To determine whether TNFR1 signaling is defective in TNFR1 knockdown cells, TNF-α-induced IκBα degradation was examined. As shown in Fig. 5C, in control MCF-7 cells TNF-α induced rapid and complete IκBα degradation, whereas in the TNFR1 knockdown MCF-7 cells, TNF-α-induced IκBα degradation was dramatically inhibited. These results indicate that TNFR1-siRNA functionally blocked the TNFR1 signaling pathway. We next examined the effect of TNFR1 abolition on ER stress-induced NF-κB activation. As shown in Fig. 5D and E, Western blotting and EMSA revealed similar levels and kinetics of IκBα degradation and NF-κB DNA binding activity between the TNFR1 knockdown cells and control cells when cells were exposed to ER stress. All these data indicate that ER stress-induced NF-κB activation is independent of TNFR1 signaling.
FIG. 5.
ER stress-induced NF-κB activation is independent of TNFR1 signaling. (A) ER stress-induced NF-κB activation does not require de novo protein synthesis. MCF-7 cells were treated with or without CHX (5 μM) for 1 h and then with thapsigargin or tunicamycin for 2 h. Nuclear extracts were prepared for EMSA with an NF-κB probe. (B) Knockdown of TNFR1 by siRNA. MCF-7 cells were transfected with a control siRNA and a specific siRNA pool to TNFR1. Forty-eight hours after transfection, cell lysates were subjected to Western blotting with anti-TNFR1 and anti-β-actin antibodies. (C) Impaired TNF-α-induced NF-κB activation by ablation of TNFR1. After transfection with TNFR1-siRNA and control siRNA (ContsiRNA) for 48 h, MCF-7 cells were subjected to TNF-α (15 ng/ml) for 1 and 2 h. Cell extracts were collected and used to analyze protein levels of IκBα and β-actin by Western blotting with the indicated antibodies. (D and E) ER stress-induced NF-κB is independent of TNFR1 signaling. MCF-7 cells were transfected with the indicated siRNAs as described for panel C. After treatment with thapsigargin or tunicamycin for the indicated times, whole-cell extracts and nuclear extracts were prepared. Cell extracts from each sample were analyzed for IκBα levels by Western blotting. Nuclear extracts were examined by EMSA with an NF-κB probe. TG, thapsigargin; TU, tunicamycin; Conti, control siRNA.
Autocrine action of TNF-α contributes to cell death during ER stress.
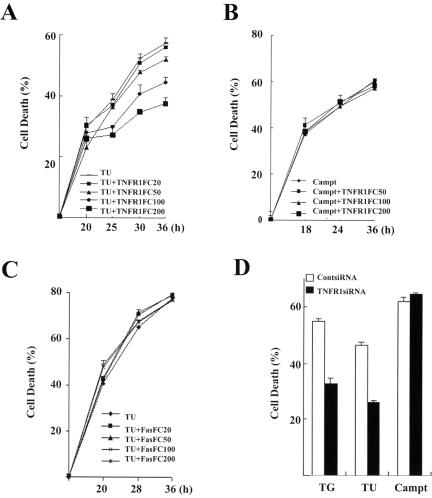
To test whether autocrine TNF-α contributes to ER stress-induced cell death, we first examined the effect of TNFR1-Fc, which binds and neutralizes TNF-α, on thapsigargin- or tunicamycin-induced cell death in MCF-7 cells. As shown in Fig. 6A, the MTT assay revealed that presence of TNFR1-Fc significantly decreased ER stress-induced cell death. However, no significant difference in cell death was observed between TNFR1-Fc-treated and control cells when cells were exposed to the DNA damage agent camptothecin (Fig. 6B). These results suggest that the protective role of TNFR1-Fc is specific to ER stress. To further confirm the specific effect of TNFR1-Fc, we examined the effect of Fas-Fc, which binds and neutralizes FasL. As shown in Fig. 6C, Fas-Fc did not prevent ER stress-induced cell death. Furthermore, we blocked TNFR1 signaling by knockdown of TNFR1 using a specific siRNA (Fig. 5B). As shown in Fig. 6D, ablation of endogenous TNFR1 by TNFR1-siRNA made the cells more resistant to thapsigargin- or tunicamycin-induced cell death compared to control siRNA, and blocking TNFR1 signaling had no effect on camptothecin-induced cell death. Therefore, these results suggest that NF-κB-mediated production of TNF-α contributes to cell death in ER stress in an autocrine manner.
FIG. 6.
Autocrine TNF-α contributes to NF-κB-mediated cell death in ER stress. (A) TNFR1-Fc decreases ER stress-induced cell death. MCF-7 cells were exposed to thapsigargin or tunicamycin with or without TNFR1-Fc at different concentrations for 30 h. Cell death was detected by MTT assay. Data are representative of triplicate experiments. (B) TNFR1-Fc does not affect camptothecin-induced cell death. MCF-7 cells were incubated with camptothecin (1 μM) with or without TNFR1-Fc for 24 h. Cell death was detected by MTT assay as described for panel A. (C) Fas-Fc does not influence ER stress-induced cell death. MCF-7 cells were treated with thapsigargin or tunicamycin in the presence or absence of Fas-Fc for 30 h. Cell death was detected by MTT assay as described for panel A. (D) Elimination of TNFR1 inhibits ER stress-induced cell death. After transfection with TNFR1-siRNA and control siRNA (ContsiRNA) for 48 h, MCF-7 cells were exposed to thapsigargin, tunicamycin, or camptothecin (1 μM) for indicated times. Cell death was scored by MTT assay as described for panel A. Cell survival was expressed as absorbance relative to that of DMSO-treated controls. TG, thapsigargin; TU, tunicamycin; Campt, camptothecin.
ER stress sensitizes cells to TNF-α-induced cell death through down-regulation of TRAF2 expression.
TNF-α is generally considered to be a poor inducer of apoptosis. TNF-α-induced activation of NF-κB mediates expression of c-FLIP, TRAF2, XIAP, and c-IAP1/2, which block TNF-α-induced activation of caspase-8 (20). So, it is of interest to investigate if the cells are more sensitive to TNF-α-induced cell death under ER stress.
We first examined whether ER stress made cells more susceptible to the toxicity of TNF-α. Phase-contrast microscopy revealed that incubation MCF-7 cells with TNF-α for 8 h did not induce obvious cell death, and treatment with thapsigargin or tunicamycin alone for 4 h did not cause cell death after another 8 h. However, exposure to thapsigargin or tunicamycin for 4 h and then TNF-α for another 8 h dramatically decreased cell survival (data not shown). The quantitation of cell death by MTT assay is shown in Fig. 7A. However, preincubation of the cells with thapsigargin did not significantly alter cell death induced by camptothecin or hydrogen peroxide (data not shown). These results suggest that ER stress promotes cellular sensitivity to TNF-α toxicity.
FIG.7.
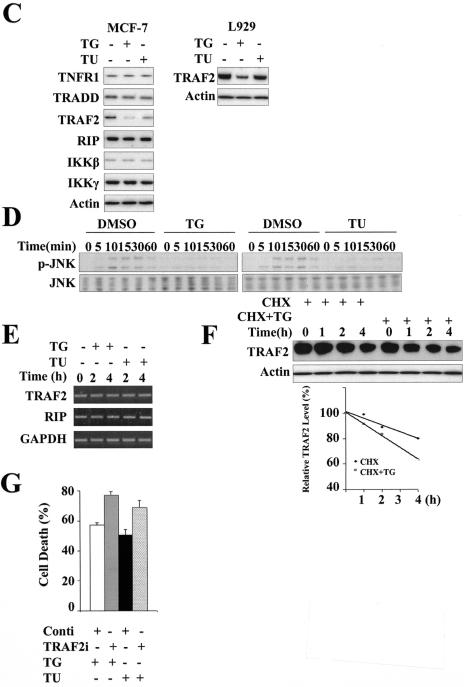
ER stress sensitizes cells to TNF-α-induced cell death. (A) MCF-7 cells were treated with or without thapsigargin or tunicamycin for 4 h. After being washed with medium (Dulbecco's modified Eagle medium + 10% FBS) two times, cells were incubated without or with TNF-α (30 ng/ml). Cell death was quantified by MTT assay. Data are representative of triplicate experiments. (B) ER stress inhibits TNF-α-induced activation of NF-κB. MCF-7, L929, and DU145 cells were treated with DMSO, thapsigargin, or tunicamycin for 4 h and then stimulated with TNF-α (15 ng/ml) for the indicated times. Cell lysates were prepared, and 20 μg of protein from each sample was used for Western blotting with anti-IκBα and anti-β-actin. (C) ER stress induces degradation of TRAF2. MCF-7 and L929 cells were treated with thapsigargin or tunicamycin. Cell lysates were prepared, and 20 μg of protein from each sample was used for Western blotting with anti-TNFR1, anti-TRADD, anti-RIP, anti-TRAF2, anti-IKKβ, and anti-IKKγ. (D) ER stress inhibits TNF-α-induced activation of JNK. The same samples from panel D were used for immunoblotting with anti-JNK and anti-phospho-JNK antibodies. (E) ER stress does not influence transcription of TRAF2. MCF-7 cells were treated with thapsigargin or tunicamycin for 2 and 4 h. Total RNA was collected, and the mRNA level of TRAF2 was measured by RT-PCR. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (F) ER stress decreases stability of TRAF2. L929 cells were incubated with CHX (20 μM) or CHX plus thapsigargin (2 μM) for the indicated time, and then the TRAF2 protein level was examined by Western blotting. (G) Knockdown of TRAF2 protein sensitizes cells to ER stress-induced cell death. After transfection with TRAF2 siRNA or control siRNA for 48 h, MCF-7 cells were subjected to thapsigargin or tunicamycin for the indicated times. Cell death was assessed by an MTT assay. TG, thapsigargin; TU, tunicamycin; Conti, control siRNA.
After TNF-α binding to TNFR1, caspase-8, NF-κB, and JNK are activated. Caspase-8 and JNK are believed to promote cell death, whereas NF-κB inhibits cell death (3). Inhibition of TNF-α-induced activation of NF-κB could unveil its potential proapoptotic ability. As noted above, expression of mutant IκBα made MCF-7 cells remarkably susceptible to the toxicity of TNF-α (Fig. 3C). To address whether ER stress inhibited TNF-α-induced NF-κB activation, IκBα degradation was examined in MCF-7 cells preincubated with thapsigargin or tunicamycin. As shown in Fig. 7B, exposure to ER stress significantly inhibited TNF-α-induced IκBα degradation. We also observed similar results in mouse fibroblast L929 and human prostate cancer DU145 cells. These results indicate that ER stress interrupts TNF-α-induced NF-κB activation and that this is a general cell event. In the TNFR1 signaling pathway, TNFR1, TRADD, RIP, TRAF2, and IKK complex are responsible for NF-κB activation. The levels of these proteins were analyzed by Western blotting in thapsigargin- or tunicamycin-treated MCF-7 cells. The protein abundance of TNFR1, TRADD, RIP, and IKKγ was maintained at the same level as untreated control cells (Fig. 7C, left). However, the protein level of TRAF2 was decreased when cells were exposed to ER stress, and a similar result was observed in L929 cells, as shown in Fig. 7C (right). TRAF2 is also essential for TNF-α-induced activation of JNK. As shown in Fig. 7D, TNF-α-induced phosphorylation of JNK was almost totally absent in thapsigargin- or tunicamycin-treated MCF-7 cells. This is consistent with the results of IκBα degradation and down-regulation of TRAF2 expression. The decreased protein expression of TRAF2 in ER stress is not due to inhibition of transcription or increased turnover of mRNA because RT-PCR data showed that treatment with thapsigargin or tunicamycin did not decrease the level of TRAF2 mRNA (Fig. 7E). A CHX chase experiment showed that thapsigargin promoted TRAF2 degradation (Fig. 7F). Recent evidence indicates that the E3 ubiquitin ligase protein siah2, a homolog of the Drosophila seven in absentia protein, binds and mediates degradation of TRAF2 in several stress situations (10). We wondered whether siah2 also mediated TRAF2 degradation in ER stress. We found that the protein level of TRAF2 decreased in siah2−/− MEFs to the same extent as in siah2+/+ MEFs when cells were exposed to ER stress (data not shown). The presence of MG132, a potent protease inhibitor, also did not maintain the protein level of TRAF2 in ER stress (data not shown). These results indicate that siah2 and protease-mediated degradation are not responsible for the degradation of TRAF2 in ER stress. TRAF2 seems to be a complex regulator in ER stress. First, TRAF2 is required for IRE1α to activate NF-κB in response to ER stress to induce the expression of TNF-α. Second, unavailability of TRAF2 inhibits TNF-α-induced NF-κB activation and sensitizes cells to TNF-α-induced cell death. So it was interesting to explore the exact role of TRAF2 in ER stress. As shown in Fig. 7G, knockdown of TRAF2 by siRNA increased ER stress-induced cell death. This result is consistent with the previous concept that TRAF2 functions as an anti-apoptotic protein. All these results suggest that ER stress inhibits TNF-α-induced activation of NF-κB and sensitizes cells to the toxicity of TNF-α.
DISCUSSION
The ER plays an important role in transcriptional regulation. The ER modulates the activity of transcription factors like XBP1, ATF6, and AP-1. Here we show that, in response to ER stress, IRE1α binds to the IKK complex and then activates NF-κB by promoting degradation of IκBα. Combined with the data that IRE1α directly interacts with TRAF2, but not with IKKα, IKKβ, and IKKγ (Fig. 2), our results indicate that activation of NF-κB and JNK by ER stress may proceed by a pathway similar to that used by cells in response to TNF-α. We further provide evidence that kinase activity is required for IRE1α to activate NF-κB.
The TRAF2 protein seems to play a critical role in activation of NF-κB by different signaling pathways, like TNFR1 (15), PKR (9), CD40, TNFR2 (33), and IL-1 (31). TRAF2 also is a protein which is sensitive to degradation. It has been reported that c-IAP1 (23), siah2 (10), and CD30 (7) mediate its degradation. Therefore, the availability of TRAF2 protein may play an essential role in regulation of the activity of NF-κB in response to stimuli. In our experiments we have observed that there are two different processes of NF-κB activation in response to ER stress. The first one is mediated early by IRE1α while the level of TRAF2 is still high. The second one is induced by autocrine TNF-α. We speculate that there are at least two reasons why ER stress inhibits TNF-α-induced NF-κB activation. The first is that this results from degradation of TRAF2. The second is that in response to ER stress some TRAF2 proteins are recruited by IRE1α.
Based on our observations, TRAF2 plays a complex role in the cellular ER stress response. First, it is required for IRE1α-induced TNF-α expression. Second, impaired TNF-α-induced NF-κB activation due to the unavailability of TRAF2 is critical for cellular sensitivity to TNF-α toxicity. Together, data from siRNA knockdown experiments indicate the protective role of TRAF2 in ER stress. This is consistent with previous results showing that TRAF2 functions as an antiapoptosis protein (24, 40).
It is interesting to observe that ER stress induces early expression of TNF-α in several epithelial-derived cell lines and mesenchyme-derived embryonic fibroblasts. This indicates that induction of TNF-α is a general cellular response to ER stress. TNF-α is a cytokine produced by many cell types in response to inflammation, infection, and other environmental challenges. TNF-α elicits a wide spectrum of organismal and cellular responses. This is consistent with the fact that most cell types constitutively express TNFR1. A number of studies have shown that TNF-α underlies the pathogenesis of some important human diseases, including acute and chronic inflammatory diseases, Crohn's disease (30), insulin resistance in diabetes (2), stroke (11), and neurodegenerative diseases (28).
The physiological significance of ER stress comes from research on neurons, pancreatic β cells, and B cells. The accumulation of amyloid β-precursor proteins in neurons is causally associated with Alzheimer's disease (14, 26). Pancreatic β cells that secrete proteins in large quantities may be at risk for ER stress-induced cell death, which may underlie the pathogenesis of diabetes (12, 34, 35). ER stress pathways also regulate B lymphocytes, which produce large amount of immunoglobulin proteins, and recent data have shown that IRE1α is required for B cell lymphopoiesis (42). Our findings indicate that, in addition to the unfolded protein response, the ER may affect the development of diseases through novel alternative mechanisms. However, how important the role is that TNF-α plays in these processes needs further investigation. Unlike unfolded protein, which can only affect the cell itself, secreted cytokines like TNF-α can affect not only the cell itself through an autocrine mechanism but also the surrounding cells, through a paracrine mechanism, and distant organs through an endocrine mechanism. Therefore, we propose that disruption of the homeostasis of the ER may play an important role in more diseases than previously recognized.
Accumulating evidence suggests that the ER serves as an important apoptotic control point (5). It is believed that the ER can initiate its own apoptotic signaling by activating ER-localized caspase-12 or caspase-4 during ER stress. However, the ER can also initiate apoptosis through cross talk with the mitochondrial apoptotic pathway by inducing the expression of the BH3-only protein PUMA (32). Other studies show that ER stress induces cell death by activating caspase-9 (18), and overexpression of bcl-2 or bcl-xL protein prevents ER stress-induced cell death (14). All these results indicate that mitochondria play an important role in amplifying the apoptotic signaling from the ER. However, there is little known about the role of another important cell death pathway, namely, the membrane death receptor pathway (extrinsic apoptotic pathway), in the modulation of ER stress-induced cell death.
In our experiments, we first provided evidence that ER stress induces a strong expression of TNF-α but not FasL and TRAIL in an NF-κB-dependent manner (Fig. 4). More importantly, autocrine TNF-α promoted cell death by activating caspase-8. Blocking TNFR1 or caspase-8 signaling significantly decreased ER stress-induced cell death (Fig. 3 and 6). That inhibiting caspase-4 had no effect on ER stress-induced cleavage of caspase-8 (Fig. 3) further proves that caspase-8 activation is initiated from IRE1α and that caspase-4 and caspase-8 are on separate apoptotic pathways in ER stress. Our results indicate that the cell surface death receptor pathway is involved in regulation of ER stress-induced cell death, and there is cross talk between the ER and this pathway during apoptotic signaling initiation and amplification.
Unlike FasL and TRAIL, TNF-α is not a strong cell death inducer; TNF-α does not usually trigger apoptosis in TNFR1-bearing cells. It is believed that TRAF2- and RIP-induced activation of NF-κB is a major survival signal in the TNFR1 pathway (20). This result suggests that ER stress inhibits TNF-α-induced activation of NF-κB and sensitizes cells to TNF-α toxicity.
In our experiments, we found that cells exposed to ER stress for a short time are much more susceptible to TNF-α-induced cell death. We provide evidence that ER stress inhibits TNF-α-induced activation of NF-κB and JNK. As mentioned above, decreased levels of TRAF2 may play an important role in this process. However, it is believed that JNK is required for TNF-α-induced cell death. Activation of JNK induces release of Smac from mitochondria. Released Smac binds to IAPs to promote caspase-8 activation (6). We speculate that in ER stress cells can bypass the TNFR1 pathway to provide similar signaling. For example, in response to ER stress IRE1α can activate JNK. Cross talk between ER and mitochondria can also promote the release of Smac.
In this work, we report that ER-resident IRE1α is required for activation of NF-κB in response to ER stress through TRAF2-mediated formation of a complex between IRE1α and IKK. Inhibition of NF-κB suppresses ER stress-induced cell death in MCF-7 cells. The expression of TNF-α is induced by ER stress in an IRE1α- and NF-κB-dependent manner in different cell types. Blocking autocrine TNF-α signaling inhibits NF-κB-mediated cell death. We further found that ER stress down-regulates TRAF2 expression leading to inhibition of TNF-α-induced activation of NF-κB and JNK. This makes cells susceptible to TNF-α-induced cell death. Therefore, our study establishes a novel link between ER and TNF-α through NF-κB, which means that ER may regulate important physiological and pathological processes in addition to promoting protein folding. Our data showing that suppression of caspase-8 inhibits ER stress-induced cell death also indicate that there is cross talk between the ER pathway and the membrane death receptor pathway in initiation and amplification of apoptosis signaling.
Acknowledgments
We thank B. M. J. Foxwell for adenoviral vector containing human TNF-α promoter (−1173bp) with the 3′ untranslated region of the human TNF-α gene and D. W. Ballard for mutant IκB, IKK-α, IKK-β, and IKK-γ plasmids.
Portions of this work were supported by grant DK 42394 from NIH.
REFERENCES
- 1.Bender, K., M. Gottlicher, S. Whiteside, H. J. Rahmsdorf, and P. Herrlich. 1998. Sequential DNA damage-independent and -dependent activation of NF-kappaB by UV. EMBO J. 17:5170-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borst, S. E. 2004. The role of TNF-alpha in insulin resistance. Endocrine. 23:177-182. [DOI] [PubMed] [Google Scholar]
- 3.Chen, G., and D. V. Goeddel. 2002. TNF-R1 signaling: a beautiful pathway. Science 296:1634-1635. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Z. J., L. Parent, and T. Maniatis. 1996. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell 84:853-862. [DOI] [PubMed] [Google Scholar]
- 5.Danial, N. N., and S. J. Korsmeyer. 2004. Cell death: critical control points. Cell 116:205-219. [DOI] [PubMed] [Google Scholar]
- 6.Deng, Y., X. Ren, L. Yang, Y. Lin, and X. Wu. 2003. A JNK-dependent pathway is required for TNFα-induced apoptosis. Cell 115:61-70. [DOI] [PubMed] [Google Scholar]
- 7.Duckett, C. S., and C. B. Thompson. 1997. CD30-dependent degradation of TRAF2: implications for negative regulation of TRAF signaling and the control of cell survival. Genes Dev. 11:2810-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujioka, S., C. Schmidt, G. M. Sclabas, Z. Li, H. Pelicano, B. Peng, A. Yao, J. Niu, W. Zhang, D. B. Evans, J. L. Abbruzzese, P. Huang, and P. J. Chiao. 2004. Stabilization of p53 is a novel mechanism for proapoptotic function of NF-κB. J. Biol. Chem. 279:27549-27559. [DOI] [PubMed] [Google Scholar]
- 9.Gil, J., M. A. Garcia, P. Gomez-Puertas, S. Guerra, J. Rullas, H. Nakano, J. Alcami, and M. Esteban. 2004. TRAF family proteins link PKR with NF-κB activation. Mol. Cell. Biol. 24:4502-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Habelhah, H., I. J. Frew, A. Laine, P. W. Janes, F. Relaix, D. Sassoon, D. D. Bowtell, and Z. Ronai. 2002. Stress-induced decrease in TRAF2 stability is mediated by Siah2. EMBO J. 21:5756-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallenbeck, J. M. 2002. The many faces of tumor necrosis factor in stroke. Nat. Med. 8:1363-1368. [DOI] [PubMed] [Google Scholar]
- 12.Harding, H. P., H. Zeng, Y. Zhang, R. Jungries, P. Chung, H. lesken, D. D. Sabatini, and D. Ron. 2001. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol. Cell 7:1153-1163. [DOI] [PubMed] [Google Scholar]
- 13.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 14.Hitomi, J., T. Katayama, Y. Eguchi, T. Kudo, M. Taniguchi, Y. Koyama, T. Manabe, S. Yamagishi, Y. Bando, K. Imaizumi, Y. Tsujimoto, and M. Tohyama. 2004. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Aβ-induced cell death. J. Cell Biol. 165:347-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu, H., H. B. Shu, M. G. Pan, and D. V. Goeddel. 1996. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 84:299-308. [DOI] [PubMed] [Google Scholar]
- 16.Hu, P., Z. Han, A. D. Couvillon, and J. H. Exton. 2004. Critical role of endogenous Akt/IAPs and MEK1/ERK pathways in counteracting endoplasmic reticulum stress-induced cell death. J. Biol. Chem. 279:49420-49429. [DOI] [PubMed] [Google Scholar]
- 17.Hur, G. M., J. Lewis, Q. Yang, Y. Lin, H. Nakano, S. Nedospasov, and Z. G. Liu. 2003. The death domain kinase RIP has an essential role in DNA damage-induced NF-kappa B activation. Genes Dev. 17:873-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jayanthi, S., X. Deng, P. A. Noailles, B. Ladenheim, and J. L. Cadet. 2004. Methamphetamine induces neuronal apoptosis via cross-talks between endoplasmic reticulum and mitochondria-dependent death cascades. FASEB J. 18:238-251. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, H. Y., S. A. Wek, B. C. McGrath, D. Scheuner, R. J. Kaufman, D. R. Cavener, and R. C. Wek. 2003. Phosphorylation of the α subunit of eukaryotic initiation factor 2 is required for activation of NF-κB in response to diverse cellular stresses. Mol. Cell. Biol. 23:5651-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karin, M., and A. Lin. 2002. NF-κB at the crossroads of life and death. Nat. Immunol. 3:221-227. [DOI] [PubMed] [Google Scholar]
- 21.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 22.Lee, K., W. Tirasophon, X. Shen, M. Michalak, R. Prywes, T. Okada, H. Yoshida, K. Mori, and R. J. Kaufman. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16:452-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, X., Y. Yang, and J. D. Ashwell. 2002. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature 416:345-347. [DOI] [PubMed] [Google Scholar]
- 24.Lin, Y., J. Ryan, J. Lewis, M. A. Wani, J. B. Lingrel, and Z. G. Liu. 2003. TRAF2 exerts its antiapoptotic effect by regulating the expression of Kruppel-like factor LKLF. Mol. Cell. Biol. 23:5849-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martindale, J. L., and N. J. Holbrook. 2002. Cellular response to oxidative stress: signaling for suicide and survival. J. Cell. Physiol. 192:1-15. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa, T., H. Zhu, N. Morishima, E. Li, J. Xu, B. A. Yankner, and J. Yuan. 2000. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature 403:98-103. [DOI] [PubMed] [Google Scholar]
- 27.Pahl, H. L., M. Sester, H. G. Burgert, and P. A. Baeuerle. 1996. Activation of transcription factor NF-κB by the adenovirus E3/19K protein requires its ER retention. J. Cell Biol. 132:511-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perry, R. T., J. S. Collins, H. Wiener, R. Acton, and R. C. Go. 2001. The role of TNF and its receptors in Alzheimer's disease. Neurobiol. Aging 22:873-883. [DOI] [PubMed] [Google Scholar]
- 29.Pham, C. G., C. Bubici, F. Zazzeroni, S. Papa, J. Jones, K. Alvarez, S. Jayawardena, E. De Smaele, R. Cong, C. Beaumont, F. M. Torti, S. V. Torti, and G. Franzoso. 2004. Ferritin heavy chain upregulation by NF-κB inhibits TNFα-induced apoptosis by suppressing reactive oxygen species. Cell 119:529-542. [DOI] [PubMed] [Google Scholar]
- 30.Plevy, S. E., C. J. Landers, J. Prehn, N. M. Carramanzana, R. L. Deem, D. Shealy, and S. R. Targan. 1997. A role for TNF-alpha and mucosal T helper-1 cytokines in the pathogenesis of Crohn's disease. J. Immunol. 159:6276-6282. [PubMed] [Google Scholar]
- 31.Pomerantz, J. L., and D. Baltimore. 1999. NF-κB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 18:6694-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reimertz, C., D. Kogel, A. Rami, T. Chittenden, and J. H. Prehn. 2003. Gene expression during ER stress-induced apoptosis in neurons: induction of the BH3-only protein Bbc3/PUMA and activation of the mitochondrial apoptosis pathway. J. Cell Biol. 162:587-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothe, M., V. Sarma, V. M. Dixit, and D. V. Goeddel. 1995. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science 269:1424-1427. [DOI] [PubMed] [Google Scholar]
- 34.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Sounders, S. Bonner-Weir, and R. J. Kaufman. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed]
- 35.Scheuner, D., D. V. Mierde, B. Song, D. Flamez, J. W. Creemers, K. Tsukamoto, M. Ribick, F. C. Schuit, and R. J. Kaufman. 2005. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat. Med. 11:757-764. [DOI] [PubMed] [Google Scholar]
- 36.Scott, M. L., T. Fujita, H. C. Liou, G. P. Nolan, and D. Baltimore. 1993. The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev. 7:1266-1276. [DOI] [PubMed] [Google Scholar]
- 37.Shakhov, A. N., M. A. Collart, P. Vassalli, S. A. Nedospasov, and C. V. Jongeneel. 1990. Kappa B-type enhancers are involved in lipopolysaccharide-mediated transcriptional activation of the tumor necrosis factor alpha gene in primary macrophages. J. Exp. Med. 171:35-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urano, F., X. Wang, A. Bertolotti, Y. Zhang, P. Chung, H. P. Harding, and D. Ron. 2000. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287:664-666. [DOI] [PubMed] [Google Scholar]
- 39.Wertz, I. E., K. M. O'Rourke, H. Zhou, M. Eby, L. Aravind, S. Seshagiri, P. Wu, C. Wiesmann, R. Baker, D. L. Boone, A. Ma, E. V. Koonin, and V. M. Dixit. 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430:694-699. [DOI] [PubMed] [Google Scholar]
- 40.Yeh, W. C., A. Shahinian, D. Speiser, J. Kraunus, F. Billia, A. Wakeham, J. L. de la Pompa, D. Ferrick, B. Hum, N. Iscove, P. Ohashi, M. Rothe, D. V. Goeddel, and T. W. Mak. 1997. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity 7:715-725. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881-891. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, K., H. N. Wong, B. Song, C. N. Miller, D. Scheuner, and R. J. Kaufman. 2005. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J. Clin. Investig. 115:268-281. [DOI] [PMC free article] [PubMed] [Google Scholar]