Abstract
The PA200 proteasome activator is a broadly expressed nuclear protein. Although how PA200 normally functions is not fully understood, it has been suggested to be involved in the repair of DNA double-strand breaks (DSBs). The PA200 gene (Psme4) is composed of 45 coding exons spanning 108 kb on mouse chromosome 11. We generated a PA200 null allele (PA200Δ) through Cre-loxP-mediated interchromosomal recombination after targeting loxP sites at either end of the locus. PA200Δ/Δ mice are viable and have no obvious developmental abnormalities. Both lymphocyte development and immunoglobulin class switching, which rely on the generation and repair of DNA DSBs, are unperturbed in PA200Δ/Δ mice. Additionally, PA200Δ/Δ embryonic stem cells do not exhibit increased sensitivity to either ionizing radiation or bleomycin. Thus, PA200 is not essential for the repair of DNA DSBs generated in these settings. Notably, loss of PA200 led to a marked reduction in male, but not female, fertility. This was due to defects in spermatogenesis observed in meiotic spermatocytes and during the maturation of postmeiotic haploid spermatids. Thus, PA200 serves an important nonredundant function during spermatogenesis, suggesting that the efficient generation of male gametes has distinct protein metabolic requirements.
The 20S proteasome, or core particle (CP), is a highly conserved complex that mediates protein catabolism in the nucleus and cytoplasm of all eukaryotic cells. The CP is cylindrical in structure with a central channel formed by four heptameric rings, two composed of β subunits sandwiched between two composed of α subunits (8, 29). The β subunits have active sites, positioned towards the inside of the channel, that mediate indiscriminate protein degradation (8, 29). The α-rings serve as gatekeepers, preventing proteins from inadvertently gaining access to the central channel of the CP, where they would be degraded (8, 29).
Access to the CP channel is regulated by proteasome activators that associate with the α heptameric rings at either end of the CP. There are four known proteasome activators, PA700/19S, PA28α/β, PA28γ, and PA200, that are broadly expressed in mammalian cells (1, 8, 33, 37, 42, 43). The 19S proteasome activator, also referred to as the regulatory particle (RP), is composed of approximately 20 subunits and associates with the CP in an ATP-dependent manner. Capping of the CP at one or both ends by the 19S proteasome activator leads to formation of the 26S proteasome, which degrades ubiquitinylated proteins. The remaining proteasome activators, PA28α/β, PA28γ, and PA200, associate with the CP in an ATP-independent manner and stimulate the degradation of peptides, but not whole proteins, in vitro. Notably, the CP can be capped at either end by different proteasome activators, forming hybrid complexes (16, 36, 41).
PA28α and PA28β combine to form PA28α/β hetero-hexameric or hetero-heptameric rings (39, 45). PA28α and PA28β expression is induced by gamma interferon, and the PA28α/β activator has been implicated in regulating the function of the immunoproteasome, a variant of the 20S proteasome that enhances the generation of antigenic peptides for presentation to T lymphocytes during immune responses (20). PA28α and PA28β are encoded by relatively small genes (Psme1 and Psme2, respectively) that are closely linked on mouse chromosome 14 (21). Mice with complete deletions of both the PA28α and PA28β genes have normal immunoproteasome levels and selective defects in the generation and presentation of antigenic peptides (26).
PA28γ homopolymers of six or seven subunits also form a proteasome activator that binds the α-ring of the CP (42). Unlike the 19S and PA28α/β activators, which are distributed throughout the cell, PA28γ appears to be localized primarily to the nucleus (40, 44). PA28γ-deficient mice have a slight reduction in body weight, and PA28γ-deficient fibroblasts exhibit mild cell cycle progression defects not observed in proliferating PA28γ-deficient lymphocytes (5, 25). PA28γ-deficient mice have lower numbers of CD8+ T cells and have a diminished capacity to clear some pathogens, possibly reflecting defects in antigen processing (5).
The PA200 proteasome activator is a 200-kDa protein localized primarily to the nucleus (27, 43). PA200 is broadly expressed in mammalian tissues, existing at the highest levels in testes (43). Although PA200 is distributed throughout the nucleoplasm, it forms discrete foci in mammalian cells upon exposure to ionizing radiation, suggesting that it may be involved in DNA double-strand break (DSB) repair (43). Studies in Saccharomyces cerevisiae have demonstrated that proteasomes localize to the sites of DNA DSBs and function in their repair (23). Whether PA200 serves an active role in DSB repair or simply localizes to DSBs with the proteasome is not known. Recent studies of the yeast PA200 ortholog Blm10 (previously called Blm3), revealed that it binds the CP as a single ring-shaped polypeptide chain and can participate in the formation of hybrid complexes with the 19S activator (36). Yeast with Blm10 null mutations exhibit only a mild increase in sensitivity to the radiomimetic drug bleomycin (36). However, yeast expressing a C-terminal-truncated version of Blm10 exhibit a significant increase in bleomycin sensitivity (36). Thus, if and how PA200 and Blm10 function in the repair of DNA DSBs remain open questions.
In addition to the responses to genotoxic agents such as ionizing radiation and bleomycin, DNA DSBs are also generated under physiologic conditions. In this regard, DNA DSBs are generated during antigen receptor gene assembly in developing B and T lymphocytes and during immunoglobulin class switch recombination in mature B cells (6, 9, 14). Mice deficient in proteins required to repair these DSBs exhibit profound defects in lymphocyte development and/or defects in immunoglobulin class switch recombination (6, 9). In addition, DNA DSBs are generated during meiosis, and successful gametogenesis depends upon the repair of these DSBs (7, 12, 31). Thus, if PA200 is involved in the repair of DNA DSBs, its absence could impact any, or all, of these processes.
Here we have generated and analyzed PA200 null mice. The PA200 gene (Psme4) lies on mouse chromosome 11 and is composed of 48 exons spanning 108 kb. The PA200Δ allele was generated by Cre-mediated recombination after the sequential targeting of loxP sites at the 5′ and 3′ ends of the PA200 gene. PA200Δ/Δ cell lines do not exhibit increased sensitivity to either bleomycin or ionizing irradiation, and PA200Δ/Δ mice do not exhibit measurable lymphocyte developmental defects suggestive of a significant DNA DSB repair defect. However, although PA200Δ/Δ females exhibited normal fertility, PA200Δ/Δ males exhibit a marked reduction in fertility due to pre- and postmeiotic defects in spermatogenesis. Our findings establish a critical and nonredundant function for the PA200 proteasome activator during spermatogenesis.
MATERIALS AND METHODS
Generating PA200 targeting constructs.
pPA2005′LoxP.Neo was generated by cloning the 5′ homology arm (2.5-kb HindIII fragment from a PCR product generated with I15.03 [GCTTGCTCAGGGTGCT] and I15.04 [TTGTTAGTTGTCAGGCTCA]) and the 3′ homology arm (2.4-kb PCR product generated with I16.08 [CCCATCGATATGTTGTCCATACTAACTAAGC] and I16.11 [CGGGATCCTA AAAAGACTATCCTCCTAGTTT]) into pLNTK (10). pPA2003′LoxP.Neo was generated by cloning the 5′ homology arm (6.0-kb EcoRV-ClaI fragment from a PCR product generated using I26.03 [ACTTAATAGAAAACCATGCTGTGAC] and I29.02 [CCGCTCGAGGGCAGTACAGTCTTACTCAA]) and the 3′ homology arm (2.4-kb ClaI-HindIII fragment from a PCR product generated with I26.05 [TACACTTCAAATGAGTGGAA] and I26.06 [GGATGCTCCAATATAAGAAA]) into pLNTK.
ES cell culture.
129/B6 embryonic stem (ES) cells, used for initial gene targetings, were cultured, transfected, and selected as previously described (17). Cre deletions were carried out in vitro using a Cre-expressing adenoviral vector as previously described (17). PA200+/+ and PA200Δ/Δ ES cells were derived through intercrossing PA200Δ/+ mice as previously described (19). For ionizing radiation and bleomycin sensitivity assays, ES cells were plated at various cell numbers 12 h prior to ionizing radiation or incubation in medium containing bleomycin as previously described (35). ES cells were incubated in the indicated concentration of bleomycin for 72 h. At day 7 of culture, cell colonies were counted after staining with 1% crystal violet.
Southern and Northern blot analyses.
Genomic DNA and whole-cell RNA were isolated and prepared for Southern and Northern blotting, respectively, as previously described (17). The probes used were generated by PCR using the following primer pairs: P1, a 0.5-kb ScaI-KpnI fragment of the PCR product generated with I15.01 (CCACTTCTGTGTTTGCCA) and I15.02 (TGGAACACAGATTACGGGAA); P2, a 1.6-kb PCR product generated with I16.01 (CACTCCAGAACCAAGACT) and I16.02 (GATGTCTCCCCAGTGC); P3, a 2.5-kb EcoRV fragment of the PCR product generated with I26.03 and I26.04 (TTCCATCTCCATCTTGGTAG); P4, a 3.2-kb PCR product generated with I26.07 (CATGATCTTGAACCTGTCTT) and I26.08 (TCCTATTTTAAGTCTCTTAGTGT). The PA200 cDNA probe is a 1.2-kb XbaI fragment. The c-myc probe used was a 0.7-kb cDNA fragment generated by PCR with C-MYC-F (AGCAACAGCAGAG) and C-MYC-R (ACAGACACCACATCAATTTC).
Generation of anti-PA200 antiserum and Western blotting.
Anti-PA200 rabbit antiserum was generated by Quality Controlled Biochemicals (Hopkinton, MA) through immunization and boosting of rabbits with a PA200 N-terminal peptide (RLPNSVVRRLHRERFKKPC) linked to keyhole limpet hemocyanin. To obtain whole ES cell lysate for Western blotting, ES cells were resuspended in 1× Laemmli sample buffer, boiled for 5 min, and sonicated. Testes lysate was prepared by homogenizing (Omni tissue homogenizer) in five volumes of S1 buffer (200 mM dithiothreitol, 120 mM Tris-HCl, pH 6.8, 10% protease inhibitor [Sigma]). Sodium dodecyl sulfate (Invitrogen) and bromophenol blue (Mallinckrodt) were added at final concentrations of 2% and 0.1%, respectively. Samples were run on a 10% polyacrylamide Criterion gel system (Bio-Rad) and transferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore). Rabbit anti-PA200 serum (batch 7523) was diluted 1:500 into phosphate-buffered saline-0.05% Tween (TPBS)-1% ovalbumin and detected with horseradish peroxidase-conjugated donkey anti-rabbit F(ab′)2 (Amersham) diluted 1:5,000 into TPBS-1% ovalbumin and ECL detection reagents (Amersham).
Flow cytometric analyses.
Flow cytometric analyses were performed as previously described using a FACScalibur (Becton Dickinson) (17). Antibodies used were phycoerythrin-conjugated anti-CD4, fluorescein isothiocyanate-conjugated anti-CD8, phycoerythrin-conjugated anti-Thy1.2, and fluorescein isothiocyanate-conjugated anti-B220 (Pharmingen).
Immunoglobulin ELISA analyses.
Splenic B cells were purified by depletion of T cells with anti-Thy1.2 magnetic beads (Dynal Biotech) and spun over Lympholyte medium (Cedarlane). Purified B cells were determined to be >90% B220+ by fluorescence-activated cell sorting. B cells were stimulated at 105 cells/ml with medium containing 30 μg/ml lipopolysaccharide (LPS) with or without 100 ng/ml interleukin-4 (IL-4), and supernatants were harvested at day 6. Total immunoglobulin (Ig) of diluted supernatants was captured with goat anti-mouse Ig (Southern Biotechnology) adsorbed onto Maxisorp plates (Nunc). Horseradish peroxidase-conjugated goat anti-mouse γ1 (Southern Biotechnology) was used to detect IgG1. Enzyme-linked immunosorbent assays (ELISAs) were developed with H2O2-activated 2,2′-azinobis(3-ethylbenzthiazoline), the optical density at 405 nm was measured, and antibody titer was determined by comparison to standard curves generated with known amounts of IgG1 (Southern Biotechnology).
Histologic analysis.
PA200+/+ and PA200Δ/Δ male mice were fixed with 1% glutaraldehyde by perfusion prior to removal of testes and epididymides, which were embedded in epoxy resin, sectioned, and stained with toluidine blue prior to light microscopic analyses.
RESULTS
Generation of the PA200Δ allele through loxP targeting and Cre-mediated recombination.
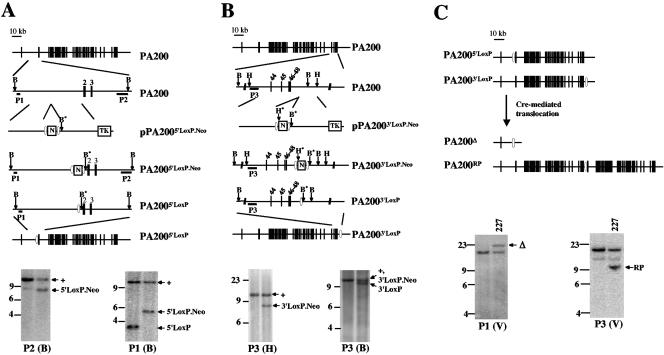
The murine PA200 is a 1,869-amino-acid protein encoded by a gene composed of 45 coding exons spanning 108 kb. Thus, it is unlikely that a single gene targeting could remove a significant portion of the gene. Hence, we chose to delete the majority of the PA200 gene by targeting loxP sites at the 5′ and 3′ ends of the locus followed by Cre-mediated recombination between the two loxP sites.
The loxP-flanked neomycin resistance gene was inserted immediately upstream of exon 2 (generating the PA2005′loxP.Neo allele) using the pPA2005′loxP.Neo targeting vector (Fig. 1A). The PA2005′loxP allele was generated from the PA2005′loxP.Neo allele through Cre-mediated deletion of the neomycin resistance gene (Fig. 1A). The PA2005′loxP allele differs from the wild-type PA200 allele only by the insertion of a single loxP site upstream of exon 2 (Fig. 1A).
FIG. 1.
Generation of the PA200Δ allele. A and B. Schematic of the murine PA200 locus, drawn approximately to scale. The targeting constructs pPA2005′LoxP.Neo and pPA2003′LoxP.Neo are shown, with relevant features indicated, including the neomycin resistance gene (N), flanking loxP sites (open ovals), and thymidine kinase gene (TK). Relevant endogenous and introduced (*) BamHI (B and B*) and HindIII (H and H*) sites are shown. Also shown are schematics of the targeted alleles before (PA2005′LoxP.Neo and PA2003′LoxP.Neo) and after (PA2005′LoxP and PA2003′LoxP) Cre deletion of the neomycin resistance gene. The relative positions of the P1, P2, and P3 probes are shown along with Southern blot analyses of either BamHI- (B) or HindIII- (H) digested ES cell genomic DNA. The bands representing the different targetings are indicated with arrows. The BamHI P3-hybridizing bands from the PA200+ and PA2003′LoxP.Neo alleles are of similar size and are indicated with a single arrow. DNA molecular mass markers (in kb) are also indicated. C. Schematic of Cre-mediated recombination between the PA2005′LoxP and PA2003′LoxP alleles, generating the PA200Δ and PA200RP alleles through a chromosomal translocation. Southern blot analyses of EcoRV (V)-digested ES cell genomic DNA probed with P1 and P3 are shown. The bands generated by the PA200Δ (Δ) and PA200RP (RP) alleles in the PA200-227 (227) ES cell line are indicated.
The pPA2003′loxP.Neo targeting vector was used to target the loxP-flanked neomycin resistance gene downstream of PA200 exon 48 in the PA2005′loxP/+ ES cell line, and two targeted ES cell lines were obtained (Fig. 1B). In these cell lines the loxP-flanked neomycin resistance gene could lie in cis, 90 kb downstream of the 5′ loxP site, on the same chromosome 11. Alternatively, the 5′ loxP site and the loxP-flanked neomycin resistance gene could lie in trans on separate chromosomes 11. Expression of the Cre recombinase in the two ES cell lines readily resulted in clones in which the neomycin resistance gene was deleted (Fig. 1B). However, only one clone (PA200-227) was obtained that appeared to have deleted the entire PA200 gene through Cre-mediated recombination (Fig. 1C, PA200Δ).
This low frequency could result from the 5′ loxP site being in trans with the loxP-flanked neomycin resistance gene. If this were the case, the PA200Δ allele would have been generated through Cre-loxP-mediated interchromosomal recombination between the maternal and paternal chromosomes 11, and this ES cell should contain the reciprocal product of this unequal interchromosomal recombination (Fig. 1C, PA200RP) (46). Indeed, Southern blot analysis of the PA200-227 ES cell line revealed the presence of both the PA200Δ and the PA200RP alleles (Fig. 1C). Thus, the PA200Δ allele was generated through a Cre-loxP-mediated interchromosomal recombination (Fig. 1C). PCR and sequence analyses revealed that this recombination was precise without any alterations of the regions flanking the loxP sites (data not shown). The PA200Δ allele contains only the first coding exon, which could, at most, encode the N-terminal 80 amino acids of the 1,869-amino-acid PA200 protein.
PA200Δ/Δ embryos are viable and born at Mendelian ratios.
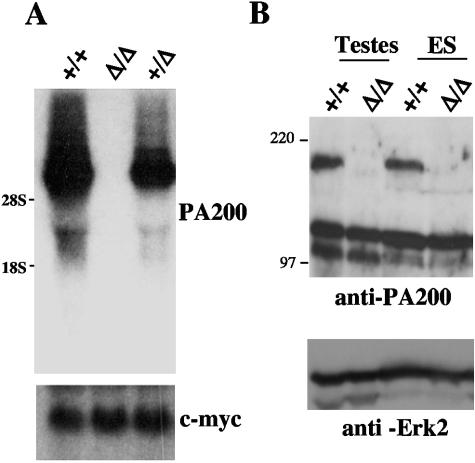
Germ line transmission of the PA200Δ allele was achieved from chimeric mice generated from the PA200-227 ES cell line. Intercrossing of PA200+/Δ mice yielded near-Mendelian ratios of PA200+/+ (26%), PA200+/Δ (46%), and PA200Δ/Δ (29%) offspring (Table 1). There were no overt differences in the appearance or size of neonatal and adult PA200+/+, PA200+/Δ, and PA200Δ/Δ mice (data not shown). Northern blot analysis of PA200Δ/Δ ES cells (see below) did not reveal any PA200 hybridizing mRNA (Fig. 2A). Moreover, we generated a rabbit antiserum to an N-terminal PA200 peptide, and Western blot analysis of whole-cell lysates from PA200Δ/Δ ES cells and PA200Δ/Δ testes using this antiserum failed to reveal any PA200 protein (Fig. 2B). Together, these data demonstrate that PA200 deficiency does not lead to significant defects in either embryonic development or viability of adult mice.
TABLE 1.
Number of PA200+/+, PA200+/Δ, and PA200Δ/Δ offspring from intercrosses of PA2000+/Δ males and females
| Offspring | No. (%) |
|---|---|
| PA200+/+ | 41 (26) |
| PA200+/Δ | 74 (46) |
| PA200Δ/Δ | 46 (29) |
| Total | 161 |
FIG. 2.
PA200 mRNA and protein are not detected in PA200Δ/Δ cells. A. Northern blot analyses of mRNA from PA200+/+, PA200+/Δ, and PA200Δ/Δ ES cells using a PA200 cDNA probe and a c-myc probe as an RNA loading control. B. Western blot analyses of total cell lysates from PA200+/+ and PA200Δ/Δ ES cells and testes using an anti-PA200 antibody and an anti-Erk2 antibody as a protein loading control.
Development of PA200Δ/Δ lymphocytes.
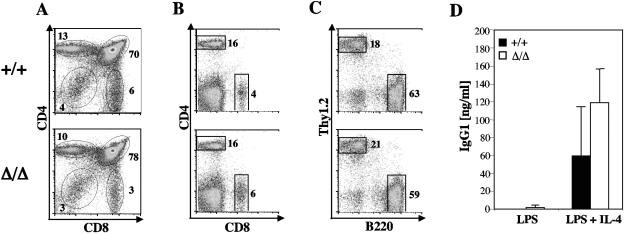
Flow cytometric analyses of thymocytes from PA200Δ/Δ mice did not reveal obvious defects in T-cell development (Fig. 3A and Table 2). In this regard, PA200Δ/Δ and PA200+/+ mice had similar numbers of CD4− CD8− (double-negative) pro-T cells, CD4+ CD8+ (double-positive) pre-T cells, and normal numbers of CD4+ and CD8+ (single-positive) mature T cells in the thymus (Fig. 3A; Table 2). Furthermore, flow cytometric analyses revealed similar numbers of mature CD4+ and CD8+ splenic T cells in PA200Δ/Δ and PA200+/+ mice (Fig. 3B; Table 2). Finally, similar fractions of developing IgM− (pro-B and pre-B) and IgM+ (immature and mature B) cells were found in the bone marrow of both PA200Δ/Δ and PA200+/+ mice, and analyses of splenocytes revealed similar fractions of mature B220+ B cells (Fig. 3C and Table 2).
FIG. 3.
Lymphocyte development and immunoglobulin class switch recombination in PA200Δ/Δ mice. A, B, and C. Flow cytometric analyses of thymocytes (A) and splenocytes (B and C) from PA200+/+ and PA200Δ/Δ mice using either anti-CD4 and anti-CD8 (A and B) or anti-Thy1.2 and anti-B220 (C) antibodies. Gates and percentages are shown. D. Concentration of IgG1 in the supernatant of purified PA200+/+ (filled bars) and PA200Δ/Δ (open bars) B cells stimulated with either LPS or LPS plus IL-4. IgG1 concentrations were determined by ELISA. The mean concentrations and standard deviations from two experiments are shown.
TABLE 2.
Thymocytes, bone marrow B-cell precursors, and mature splenic B and T cells in PA200Δ/Δ and PA200+/+ 6-week-old micea
| Mice (n) | Thymus
|
Bone marrow
|
Spleen
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DN | DP | CD4 SP | CD8 SP | B220low IgM− | B220low IgM+ | B220high IgM+ | CD4+ | CD8+ | B220+ | |
| +/+ (3) | 10 ± 1 | 239 ± 44 | 29 ± 5 | 8 ± 1 | 4.5 ± 1.2 | 0.8 ± 0.2 | 2.1 ± 0.7 | 15 ± 2 | 5 ± 2 | 56 ± 9 |
| Δ/Δ (3) | 11 ± 6 | 279 ± 123 | 28 ± 11 | 7 ± 3 | 5.4 ± 1.4 | 1.4 ± 0.9 | 3.7 ± 1.0 | 20 ± 10 | 8 ± 4 | 85 ± 40 |
Shown are the mean numbers of CD4− CD8− (double negative [DN]), CD4+ CD8+ (double positive [DP]), and CD4+ and CD8+ single positive (SP) thymocytes. Also shown are the mean ± standard deviation numbers of B220low IgM−, B220low IgM+, and B220high IgM+ cells per femur. Finally, the mean ± standard deviation (×10−6) numbers of splenic CD4+ and CD8+ T cells and B220+ B cells are shown.
It is possible that PA200Δ/Δ mice could have subtle defects in lymphocyte development not detectable in the analyses above. Such subtle defects have been revealed through the analysis of mutant and wild-type lymphocytes developing in a competitive fashion in the same host (2). To this end, we generated mixed bone marrow chimera mice using equal numbers of PA200Δ/Δ and PA200+/+ bone marrow cells. Analysis of these chimeric mice revealed no differences in the development of PA200Δ/Δ and PA200+/+ B and T cells (data not shown).
To assess immunoglobulin class switch recombination, PA200+/+ and PA200Δ/Δ B cells were stimulated in vitro with either LPS and IL-4, which stimulate switching from IgM to IgG1, or LPS alone, which does not. Both PA200+/+ and PA200Δ/Δ B cells exhibited similarly robust production of IgG1 when stimulated with LPS and IL-4 but not when stimulated with LPS alone, demonstrating that PA200Δ/Δ B cells can undergo immunoglobulin class switch recombination (Fig. 3D).
PA200Δ/Δ ES cells do not exhibit increased sensitivity to ionizing radiation or bleomycin.
To determine whether PA200 deficiency leads to defects in the repair of DNA DSBs generated by genotoxic agents, PA200+/+ and PA200Δ/Δ ES cell lines were assayed for sensitivity to ionizing radiation and bleomycin as described in Materials and Methods. Three PA200+/+ (X15-1, −4, and −8) and three PA200Δ/Δ (X16-A1, -B3, and -B4) ES cell lines were generated through the intercrossing of PA200+/Δ mice as previously described (19). Thus, the PA200+/+ and PA200Δ/Δ ES cell lines tested were of comparable genetic background and had been cultured for similar periods of time before being tested. The 50% lethal doses (LD50s) for ionizing radiation of the three PA200+/+ and PA200Δ/Δ ES cell lines were not significantly different (Table 3). Moreover, the LD50 doses for bleomycin treatment were also similar in the PA200+/+ and PA200Δ/Δ ES cell lines (Table 3). In contrast, the LD50s for ionizing radiation and bleomycin treatment of cells deficient in XRCC4, an NHEJ protein required for DNA DSB repair, were about half of those observed for the PA200+/+ ES cells, consistent with previous reports (Table 3) (15). Thus, PA200 is not essential for the repair of DNA DSBs generated in ES cell lines upon treatment with ionizing radiation or bleomycin.
TABLE 3.
Sensitivity of PA200+/+, PA200Δ/Δ, and XRCC4−/− ES cell lines to irradiation and bleomycin treatmenta
| Treatment | LD50
|
||||||
|---|---|---|---|---|---|---|---|
| PA200+/+ ES cell lines
|
PA200Δ/Δ ES cell lines
|
XRCC4−/− cells | |||||
| X15-1 | X15-4 | X15-8 | X16-A1 | X16-B3 | X16-B4 | ||
| Bleomycin (ng/ml) | 36 ± 1 | 51 ± 12 | 55 ± 3 | 56 ± 8 | 40 ± 8 | 47 ± 8 | 21 ± 1 |
| X-ray (cGy) | 87 ± 12 | 80 ± 14 | 99 ± 11 | 98 ± 8 | 69 ± 10 | 76 ± 10 | 38 ± 10 |
Shown is the mean dose at which 50% of the plated cells survived (LD50) ± the standard deviation for the three experiments performed.
Loss of PA200 does not increase the mortality of p53-deficient mice.
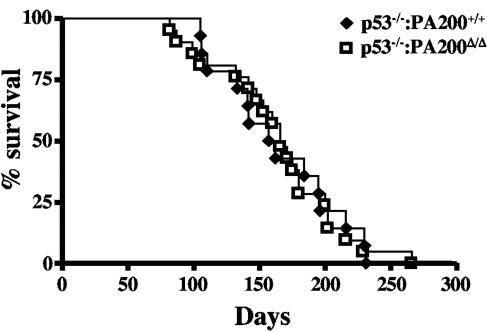
PA200Δ/Δ mice do not exhibit significant survival differences compared to wild-type mice (data not shown). Mice deficient in p53 have increased tumor formation and decreased survival due to genomic instability (18). Coupling p53 deficiency with a DNA DSB repair defect often further increases genomic instability, accelerates tumor formation, and decreases survival in mice (47). Analysis of cohorts of p53−/− PA200+/+ and p53−/− PA200Δ/Δ mice revealed no significant differences in survival (Fig. 4). Thus, mice with a compound loss of both PA200 and p53 do not exhibit decreased survival compared to mice with an isolated loss of p53.
FIG. 4.
Kaplan-Meier survival curves for PA200+/+ p53−/− (filled diamonds; n = 21) and PA200Δ/Δ p53−/− (open squares; n = 14) mice.
Defective spermatogenesis in PA200Δ/Δ mice.
Initial attempts to breed PA200Δ/Δ males yielded few offspring, whereas PA200Δ/+ males yielded litters of relatively normal size at an approximately normal frequency (data not shown). Three 12-week-old PA200+/+ and three 12-week-old PA200Δ/Δ male mice were each bred to two PA200+/+ females for 11 weeks, and the numbers of litters and offspring were monitored. During this period, the PA200+/+ males produced 14 litters with a total of 119 pups, whereas the PA200Δ/Δ males had only 6 litters with a total of 30 pups. Copulatory plugs were generated on a regular basis by PA200Δ/Δ males, suggesting that the reduced number of offspring was not due to a copulatory defect (data not shown). Light microscopic analysis of semen isolated from the epididymides of PA200Δ/Δ males revealed a general decrease in the level of mature spermatozoa (data not shown). In addition, many of the spermatozoa present were malformed and exhibited motility defects (data not shown). Together, these findings suggested that PA200Δ/Δ males have a defect in spermatogenesis.
Mouse spermatogenesis is a highly ordered process that occurs in the seminiferous tubules of the testes, taking 34.5 days to complete (11). The stem cells, or spermatogonia, lie in the periphery of the seminiferous tubule and divide mitotically (11). Once cells enter meiosis, they are referred to as spermatocytes and, after completing the second meiotic division, they become round haploid spermatids. Spermatids undergo a series of morphological changes, collectively referred to as spermiation, leading to the formation of mature spermatozoa. These morphological changes include the generation of the acrosome and tail, reorganization of the mitochondria, and loss of most of the cytoplasm as a residual body. Spermatogenesis occurs in waves across the length of the seminiferous tubule, with new waves starting before maturation of the previous wave is complete. Consequently, cells at several stages of sperm development can be observed at any location of the seminiferous tubule. This heterogeneity in development across the length of the seminiferous tubule allows for the discrimination of 12 histologic stages (I through XII) in mice (11).
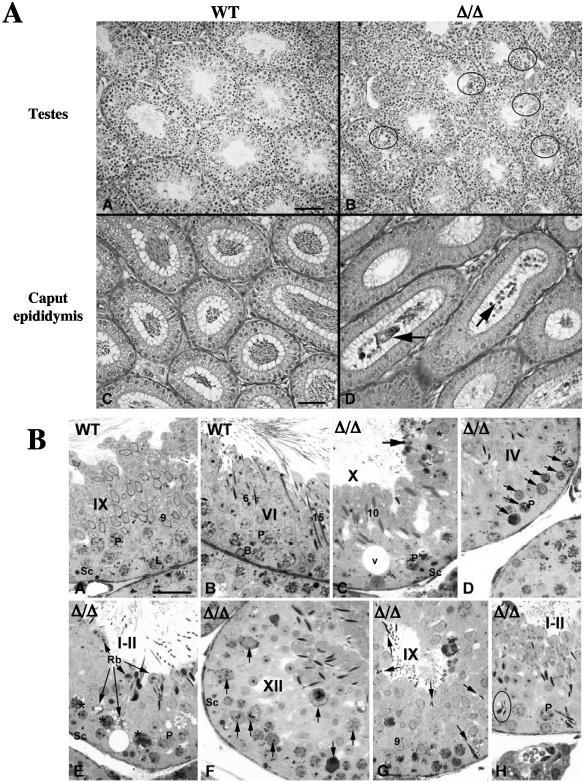
Histologic analysis of PA200Δ/Δ testes revealed multiple defects in spermatogenesis (Fig. 5). PA200Δ/Δ testes have focal areas of abnormal spermatogenesis showing vacuolation, multinucleated giant germ cells, and hypospermatocytogenesis (Fig. 5A, plate B). Evaluation of epididymal lumens from PA200Δ/Δ males revealed a low concentration of normal mature spermatozoa and the presence of immature germ cells not seen in epididymides of PA200+/+ males (Fig. 5A, plates C and D). More-detailed histologic analyses of PA200Δ/Δ testes revealed normally appearing spermatagonia; however, significant defects were noted in both meiotic spermatocytes and postmeiotic spermatids (Fig. 5B). Numerous abnormal pachytene spermatocytes (Fig. 5B, plates C, E and H) and apoptotic or necrotic pachytene spermatocytes (Fig. 5B, plates D and F) were noted in several stages of spermatogenesis. Defects in spermiation were also evidenced by reduced numbers of spermatids, abnormal-appearing spermatids (Fig. 5B, plates C and H), spermatid phagocytosis (Fig. 5B, plate G), and the accumulation of residual bodies (Fig. 5B, plate E) in PA200Δ/Δ testes.
FIG. 5.
Histology of PA200Δ/Δ testes and epidydimides. A. Low magnification of testes (plates A and B; bar, 100 μm) and epididymis (plates C and D; bar, 50 μm) from 5-month-old PA200+/+ (wild type [WT]; plates A and C) and PA200Δ/Δ (plates B and E) mice. Areas showing vacuolation, multinucleated giant germ cells, and hypospermatocytogenesis are indicated (plate B, circles). Immature germ cells in the PA200Δ/Δ caput epididymis are indicated (plate D, arrows). B. Seminiferous tubules from PA200+/+ (WT; plates A and B) and PA200Δ/Δ (Δ/Δ; plates C to H) mice. Bar, 50 μm. Histologic stages of development are indicated with roman numerals. Stages of spermatid maturation are indicated with Arabic numbers. Also indicated are pachytene (P) and leptotene (L) spermatocytes, Sertoli cells (Sc), B-type spermatogonia (B), areas of vacuolation (V), and residual bodies (Rb). The arrows indicate residual bodies (plates C and E), apoptotic and necrotic pachytene spermatocytes (plates D and F), and spermatid phagocytosis (plate G). The asterisk indicates abnormal spermatids (plate C) and abnormal pachetyne spermatocytes (plate E). Abnormal spermatid heads in the seminiferous epithelium (plate H, circle) are shown.
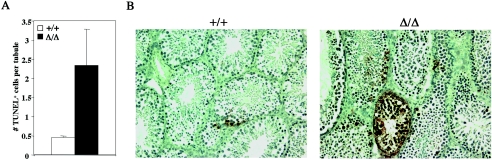
To assess the extent of apoptosis in PA200Δ/Δ testes, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining was carried out (Fig. 6). These analyses revealed an overall increase in the average number of apoptotic TUNEL-positive cells per seminiferous tubule section in PA200Δ/Δ testes (Fig. 6A). Notably, there was significant variation in the number of TUNEL-positive cells in different PA200Δ/Δ tubule sections, suggesting that apoptosis is more pronounced at specific stages of development (Fig. 6B). Together, these findings demonstrate that PA200 deficiency leads to defective spermatogenesis with histological abnormalities observed in both meiotic spermatocytes and during the maturation of postmeiotic haploid spermatids.
FIG. 6.
Increased apoptotic cells in PA200Δ/Δ testes. A. Mean ± standard deviation number of TUNEL-positive cells per seminiferous tubule section in PA200+/+ (open bar; n = 2) and PA200Δ/Δ (filled bar; n = 4) mice. A minimum of 125 seminiferous tubule sections was counted from each mouse. B. TUNEL staining of testes sections from PA200+/+ and PA200Δ/Δ mice. Shown are the sections containing the seminiferous tubule, with the greatest number of TUNEL-positive cells from +/+ and Δ/Δ mice.
DISCUSSION
Generating true null alleles through a single gene-targeting step poses challenges for genes with many exons spanning large genomic regions, such as the PA200 gene. The importance of generating a PA200 null allele in establishing the true function of PA200 is underscored by the observation that yeast expressing a C-terminal-truncated version of Blm10, the PA200 ortholog, exhibit bleomycin sensitivity, whereas true Blm10 null mutants do not (36). Here we have generated a PA200 null allele (PA200Δ) through a gene-targeting approach involving Cre-mediated recombination between loxP sites introduced at either end of the PA200 gene. Notably, the two loxP sites were introduced in trans on separate chromosomes 11, with the PA200Δ allele being generated through Cre-loxP-mediated interchromosomal recombination. Thus, a Cre-loxP-mediated recombination approach can be used to generate null alleles by removing large regions of a gene, whether the loxP sites at either end of the gene lie in cis or trans.
Analysis of PA200-deficient mice and cells revealed little evidence to support the notion that PA200 is essential for the repair of DNA DSBs generated either by genotoxic agents or under physiologic conditions. In this regard, PA200Δ/Δ and PA200+/+ ES cells exhibit similar sensitivity to ionizing radiation and bleomycin. Mice with deficiencies in proteins involved in DNA DSB repair often exhibit increased genomic instability and decreased survival when they have a compound deficiency of the cell cycle checkpoint protein p53. However, mice with a compound deficiency of PA200 and p53 do not exhibit diminished survival compared to those with an isolated deficiency of p53. Lymphocyte development, which depends on the generation and repair of DNA DSBs, also appear unperturbed in PA200Δ/Δ mice, and PA200Δ/Δ B cells undergo efficient immunoglobulin class switch recombination. Finally, although male PA200Δ/Δ mice exhibit fertility defects, it is unlikely that the defects are solely attributable to a requirement for PA200 in the repair of DNA DSBs generated during meiosis (see below). Nevertheless, it is possible that PA200 could have subtle activities in DNA DSB repair not detected by the parameters assessed here in the PA200-deficient mice. In addition, other proteins, such as the nuclear PA28γ proteasome activator, could compensate for PA200 function in PA200-deficient mice (40).
Unexpectedly, PA200Δ/Δ males had significantly impaired fertility due to a decreased production of normal spermatozoa, demonstrating that PA200 serves a critical function in spermatogenesis. DNA DSBs are normally generated and repaired during the first meiotic division of male and female gametogenesis. In this regard, deficiency of Dmc1 or Msh5, which are required for the repair of DSBs generated during meiosis, leads to both male and female sterility (13, 31). The apoptotic pachytene spermatocytes in PA200Δ/Δ testes could reflect a requirement for PA200 in the repair of DNA DSBs generated in these meiotic spermatocytes. However, any such requirement would not be generally applicable to all meiotic cells, as PA200Δ/Δ females did not exhibit fertility defects. In addition, Dmc1- and Msh5-deficient male mice have a complete block at the spermatocyte stage (13, 31). In contrast, spermatogenesis proceeds beyond this stage in PA200Δ/Δ mice with defects in spermatid maturation that cannot be easily attributed to defects in meiotic DNA DSB repair. Thus, the defects in spermatogenesis observed in PA200Δ/Δ males cannot be due solely to a requirement for PA200 in the repair of DNA DSBs generated in meiotic cells.
How then might PA200 function during spermatogenesis? The requirement for PA200 could be intrinsic and/or extrinsic to developing spermatocytes. Extrinsic requirements could reflect, for example, a role for PA200 in the normal function of cells that support spermatogenesis, such as Sertoli cells. In this regard, the accumulation of residual bodies in PA200-deficient testes are consistent with a Sertoli cell dysfunction, as these cells normally clear discarded residual bodies. However, Sertoli cell function can also be compromised by the release of toxic compounds, such as protamine, from dying spermatids which are present at increased levels in PA200Δ/Δ testes (28).
The defects in spermiation in PA200Δ/Δ mice demonstrate that PA200 is required for the development of postmeiotic spermatids. In this regard, developing spermatids undergo dramatic morphological and biochemical changes marked notably by a change in the composition of DNA-associated proteins throughout maturation. DNA-associated histone proteins are initially removed and replaced with the transitional proteins, TP1 and TP2, as spermatocytes become haploid spermatids (3). These transition proteins must then be removed and replaced with protamine, permitting packaging of the DNA in the confined space of the acrosomal head. Notably, disturbances in transition protein expression and metabolism lead to abnormal spermatogenesis and diminished fertility (30, 38). Thus, the defect in spermatid maturation in PA200Δ/Δ mice could, among other things, reflect a requirement for PA200 in the metabolism of DNA-associated proteins.
Several lines of evidence implicate the ubiquitin-proteasome system in the metabolism of DNA-associated proteins during spermatogenesis. Ubiquitin is found in high levels in the testes, and during mouse spermatogenesis H2A, a major histone protein, is highly ubiquitinylated in spermatocytes just prior to its replacement with transition proteins (4, 24). Moreover, expression of the HR6B ubiquitin-conjugating enzyme is upregulated in spermatocytes (22). Interestingly, HR6B-deficient mice have an isolated defect in male fertility similar to that observed in PA200-deficient mice (34). Furthermore, the histological findings with HR6B-deficient testes are reminiscent of those observed in PA200-deficient males (34). In this regard, HR6B-deficient mice have a block in spermatogenesis with marked defects in the maturation of haploid spermatids coupled with the production of reduced numbers and grossly abnormal spermatozoa (34). Notably, the requirement for PA200 in spermatogenesis is not shared by the PA28α, -β, or -γ proteasome activators, as mice deficient in these proteins do not have reported fertility defects (5, 25, 26, 32). Given this and the isolated defects in spermatogenesis noted in both PA200- and HR6B-deficient mice, it is tempting to speculate that spermatogenesis poses unique protein metabolic challenges that are met by specific nonredundant proteolytic pathways.
Acknowledgments
We thank Daniel Finley and Marion Schmidt for critical review of the manuscript.
This work is supported in part by National Institutes of Health grants AI47829 and AI47829 (B.P.S.) and American Cancer Society grant RSG-05-070-01-LIB (B.P.S.). C.H. is supported by a postdoctoral training grant from the NIH. Mice were produced by a transgenic core facility supported by the Rheumatic Diseases Core Center at Washington University (NIH P30-AR48335) and housed in a facility supported by NCRR grant RR012466.
REFERENCES
- 1.Ahn, J. Y., N. Tanahashi, K. Akiyama, H. Hisamatsu, C. Noda, K. Tanaka, C. H. Chung, N. Shibmara, P. J. Willy, J. D. Mott, et al. 1995. Primary structures of two homologous subunits of PA28, a gamma-interferon-inducible protein activator of the 20S proteasome. FEBS Lett. 366:37-42. [DOI] [PubMed] [Google Scholar]
- 2.Aifantis, I., C. Borowski, F. Gounari, H. D. Lacorazza, J. Nikolich-Zugich, and H. von Boehmer. 2002. A critical role for the cytoplasmic tail of pTα in T lymphocyte development. Nat. Immunol. 3:483-488. [DOI] [PubMed] [Google Scholar]
- 3.Alfonso, P. J., and W. S. Kistler. 1993. Immunohistochemical localization of spermatid nuclear transition protein 2 in the testes of rats and mice. Biol. Reprod. 48:522-529. [DOI] [PubMed] [Google Scholar]
- 4.Baarends, W. M., J. W. Hoogerbrugge, H. P. Roest, M. Ooms, J. Vreeburg, J. H. Hoeijmakers, and J. A. Grootegoed. 1999. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 207:322-333. [DOI] [PubMed] [Google Scholar]
- 5.Barton, L. F., H. A. Runnels, T. D. Schell, Y. Cho, R. Gibbons, S. S. Tevethia, G. S. Deepe, Jr., and J. J. Monaco. 2004. Immune defects in 28-kDa proteasome activator gamma-deficient mice. J. Immunol. 172:3948-3954. [DOI] [PubMed] [Google Scholar]
- 6.Bassing, C. H., W. Swat, and F. W. Alt. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109(Suppl.):S45-S55. [DOI] [PubMed] [Google Scholar]
- 7.Baudat, F., K. Manova, J. P. Yuen, M. Jasin, and S. Keeney. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6:989-998. [DOI] [PubMed] [Google Scholar]
- 8.Baumeister, W., J. Walz, F. Zuhl, and E. Seemuller. 1998. The proteasome: paradigm of a self-compartmentalizing protease. Cell 92:367-380. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhuri, J., and F. W. Alt. 2004. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4:541-552. [DOI] [PubMed] [Google Scholar]
- 10.Cogne, M., R. Lansford, A. Bottaro, J. Zhang, J. Gorman, F. Young, H. L. Cheng, and F. W. Alt. 1994. A class switch control region at the 3′ end of the immunoglobulin heavy chain locus. Cell 77:737-747. [DOI] [PubMed] [Google Scholar]
- 11.de Rooij, D. G. 1998. Stem cells in the testis. Int. J. Exp. Pathol. 79:67-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelmann, W., P. E. Cohen, B. Kneitz, N. Winand, M. Lia, J. Heyer, R. Kolodner, J. W. Pollard, and R. Kucherlapati. 1999. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21:123-127. [DOI] [PubMed] [Google Scholar]
- 13.Edelmann, W., K. Yang, A. Umar, J. Heyer, K. Lau, K. Fan, W. Liedtke, P. E. Cohen, M. F. Kane, J. R. Lipford, N. Yu, G. F. Crouse, J. W. Pollard, T. Kunkel, M. Lipkin, R. Kolodner, and R. Kucherlapati. 1997. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell 91:467-477. [DOI] [PubMed] [Google Scholar]
- 14.Fugmann, S. D., A. I. Lee, P. E. Shockett, I. J. Villey, and D. G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495-527. [DOI] [PubMed] [Google Scholar]
- 15.Gao, Y., Y. Sun, K. M. Frank, P. Dikkes, Y. Fujiwara, K. J. Seidl, J. M. Sekiguchi, G. A. Rathbun, W. Swat, J. Wang, R. T. Bronson, B. A. Malynn, M. Bryans, C. Zhu, J. Chaudhuri, L. Davidson, R. Ferrini, T. Stamato, S. H. Orkin, M. E. Greenberg, and F. W. Alt. 1998. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 95:891-902. [DOI] [PubMed] [Google Scholar]
- 16.Hendil, K. B., S. Khan, and K. Tanaka. 1998. Simultaneous binding of PA28 and PA700 activators to 20S proteasomes. Biochem. J. 332:749-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, C. Y., B. P. Sleckman, and O. Kanagawa. 2005. Revision of T cell receptor α chain genes is required for normal T lymphocyte development. Proc. Natl. Acad. Sci. USA 102:14356-14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacks, T., L. Remington, B. O. Williams, E. M. Schmitt, S. Halachmi, R. T. Bronson, and R. A. Weinberg. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 19.Kim, J. M., J. M. White, A. S. Shaw, and B. P. Sleckman. 2005. MAPK p38 alpha is dispensable for lymphocyte development and proliferation. J. Immunol. 174:1239-1244. [DOI] [PubMed] [Google Scholar]
- 20.Kloetzel, P. M. 2004. Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat. Immunol. 5:661-669. [DOI] [PubMed] [Google Scholar]
- 21.Kohda, K., T. Ishibashi, N. Shimbara, K. Tanaka, Y. Matsuda, and M. Kasahara. 1998. Characterization of the mouse PA28 activator complex gene family: complete organizations of the three member genes and a physical map of the approximately 150-kb region containing the alpha- and beta-subunit genes. J. Immunol. 160:4923-4935. [PubMed] [Google Scholar]
- 22.Koken, M. H., J. W. Hoogerbrugge, I. Jasper-Dekker, J. de Wit, R. Willemsen, H. P. Roest, J. A. Grootegoed, and J. H. Hoeijmakers. 1996. Expression of the ubiquitin-conjugating DNA repair enzymes HHR6A and B suggests a role in spermatogenesis and chromatin modification. Dev. Biol. 173:119-132.8575614 [Google Scholar]
- 23.Krogan, N. J., M. H. Lam, J. Fillingham, M. C. Keogh, M. Gebbia, J. Li, N. Datta, G. Cagney, S. Buratowski, A. Emili, and J. F. Greenblatt. 2004. Proteasome involvement in the repair of DNA double-strand breaks. Mol. Cell 16:1027-1034. [DOI] [PubMed] [Google Scholar]
- 24.Lanneau, M., and M. Loir. 1982. An electrophoretic investigation of mammalian spermatid-specific nuclear proteins. J. Reprod. Fertil. 65:163-170. [DOI] [PubMed] [Google Scholar]
- 25.Murata, S., H. Kawahara, S. Tohma, K. Yamamoto, M. Kasahara, Y. Nabeshima, K. Tanaka, and T. Chiba. 1999. Growth retardation in mice lacking the proteasome activator PA28γ. J. Biol. Chem. 274:38211-38215. [DOI] [PubMed] [Google Scholar]
- 26.Murata, S., H. Udono, N. Tanahashi, N. Hamada, K. Watanabe, K. Adachi, T. Yamano, K. Yui, N. Kobayashi, M. Kasahara, K. Tanaka, and T. Chiba. 2001. Immunoproteasome assembly and antigen presentation in mice lacking both PA28α and PA28β. EMBO J. 20:5898-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortega, J., J. B. Heymann, A. V. Kajava, V. Ustrell, M. Rechsteiner, and A. C. Steven. 2005. The axial channel of the 20S proteasome opens upon binding of the PA200 activator. J. Mol. Biol. 346:1221-1227. [DOI] [PubMed] [Google Scholar]
- 28.Peterson, M. W., and D. Gruenhaupt. 1992. Protamine interaction with the epithelial cell surface. J. Appl. Physiol. 72:236-241. [DOI] [PubMed] [Google Scholar]
- 29.Pickart, C. M., and R. E. Cohen. 2004. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5:177-187. [DOI] [PubMed] [Google Scholar]
- 30.Pina-Guzman, B., M. J. Solis-Heredia, and B. Quintanilla-Vega. 2005. Diazinon alters sperm chromatin structure in mice by phosphorylating nuclear protamines. Toxicol. Appl. Pharmacol. 202:189-198. [DOI] [PubMed] [Google Scholar]
- 31.Pittman, D. L., J. Cobb, K. J. Schimenti, L. A. Wilson, D. M. Cooper, E. Brignull, M. A. Handel, and J. C. Schimenti. 1998. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1:697-705. [DOI] [PubMed] [Google Scholar]
- 32.Preckel, T., W. P. Fung-Leung, Z. Cai, A. Vitiello, L. Salter-Cid, O. Winqvist, T. G. Wolfe, M. Von Herrath, A. Angulo, P. Ghazal, J. D. Lee, A. M. Fourie, Y. Wu, J. Pang, K. Ngo, P. A. Peterson, K. Fruh, and Y. Yang. 1999. Impaired immunoproteasome assembly and immune responses in PA28−/− mice. Science 286:2162-2165. [DOI] [PubMed] [Google Scholar]
- 33.Rechsteiner, M., and C. P. Hill. 2005. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 15:27-33. [DOI] [PubMed] [Google Scholar]
- 34.Roest, H. P., J. van Klaveren, J. de Wit, C. G. van Gurp, M. H. Koken, M. Vermey, J. H. van Roijen, J. W. Hoogerbrugge, J. T. Vreeburg, W. M. Baarends, D. Bootsma, J. A. Grootegoed, and J. H. Hoeijmakers. 1996. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell 86:799-810. [DOI] [PubMed] [Google Scholar]
- 35.Rooney, S., F. W. Alt, D. Lombard, S. Whitlow, M. Eckersdorff, J. Fleming, S. Fugmann, D. O. Ferguson, D. G. Schatz, and J. Sekiguchi. 2003. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 197:553-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt, M., W. Haas, B. Crosas, P. G. Santamaria, S. P. Gygi, T. Walz, and D. Finley. 2005. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat. Struct. Mol. Biol. 12:294-303. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt, M., A. N. Lupas, and D. Finley. 1999. Structure and mechanism of ATP-dependent proteases. Curr. Opin. Chem. Biol. 3:584-591. [DOI] [PubMed] [Google Scholar]
- 38.Shirley, C. R., S. Hayashi, S. Mounsey, R. Yanagimachi, and M. L. Meistrich. 2004. Abnormalities and reduced reproductive potential of sperm from Tnp1- and Tnp2-null double mutant mice. Biol. Reprod. 71:1220-1229. [DOI] [PubMed] [Google Scholar]
- 39.Song, X., J. D. Mott, J. von Kampen, B. Pramanik, K. Tanaka, C. A. Slaughter, and G. N. DeMartino. 1996. A model for the quaternary structure of the proteasome activator PA28. J. Biol. Chem. 271:26410-26417. [DOI] [PubMed] [Google Scholar]
- 40.Soza, A., C. Knuehl, M. Groettrup, P. Henklein, K. Tanaka, and P. M. Kloetzel. 1997. Expression and subcellular localization of mouse 20S proteasome activator complex PA28. FEBS Lett. 413:27-34. [DOI] [PubMed] [Google Scholar]
- 41.Tanahashi, N., Y. Murakami, Y. Minami, N. Shimbara, K. B. Hendil, and K. Tanaka. 2000. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J. Biol. Chem. 275:14336-14345. [DOI] [PubMed] [Google Scholar]
- 42.Tanahashi, N., K. Yokota, J. Y. Ahn, C. H. Chung, T. Fujiwara, E. Takahashi, G. N. DeMartino, C. A. Slaughter, T. Toyonaga, K. Yamamura, N. Shimbara, and K. Tanaka. 1997. Molecular properties of the proteasome activator PA28 family proteins and gamma-interferon regulation. Genes Cells 2:195-211. [DOI] [PubMed] [Google Scholar]
- 43.Ustrell, V., L. Hoffman, G. Pratt, and M. Rechsteiner. 2002. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 21:3516-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wojcik, C., K. Tanaka, N. Paweletz, U. Naab, and S. Wilk. 1998. Proteasome activator (PA28) subunits, alpha, beta and gamma (Ki antigen) in NT2 neuronal precursor cells and HeLa S3 cells. Eur. J. Cell Biol. 77:151-160. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Z., A. Krutchinsky, S. Endicott, C. Realini, M. Rechsteiner, and K. G. Standing. 1999. Proteasome activator 11S REG or PA28: recombinant REG alpha/REG beta hetero-oligomers are heptamers. Biochemistry 38:5651-5658.10220354 [Google Scholar]
- 46.Zheng, B., M. Sage, E. A. Sheppeard, V. Jurecic, and A. Bradley. 2000. Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol. Cell. Biol. 20:648-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, C., K. D. Mills, D. O. Ferguson, C. Lee, J. Manis, J. Fleming, Y. Gao, C. C. Morton, and F. W. Alt. 2002. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 109:811-821. [DOI] [PubMed] [Google Scholar]