Abstract
p21-activated kinase 5 (Pak5) is an effector for the small GTPase Cdc42, known to activate cell survival signaling pathways. Previously, we have shown that Pak5 localizes primarily to mitochondria. To study the relationship between Pak5 localization and its effects on apoptosis, we identified three N-terminal regions that regulate the localization of this kinase: a mitochondrial targeting sequence, a nuclear export sequence, and a nuclear localization sequence. When the first two sequences are deleted, Pak5 is retained in the nucleus and no longer protects cells from apoptosis. Moreover, blockade of nuclear export with leptomycin B causes endogenous Pak5 to accumulate in the nucleus. Additionally, the removal of the N-terminal nuclear localization sequence abolishes Pak5 translocation to the nucleus. Finally, we show that reduction of endogenous Pak5 expression in neuroblastoma and neural stem cells increases their sensitivity to apoptosis and that this effect is reversed upon reexpression of wild-type Pak5 but not of a mutant form of Pak5 that cannot localize to mitochondria. These results show that Pak5 shuttles from mitochondria to the nucleus and that the mitochondrial localization of Pak5 is vital to its effects on cell survival.
p21-activated kinases (Paks) are small GTPase effectors that have a wide range of effects on cell shape, movement, proliferation, and survival (1, 3, 24). Six members of the Pak family are known; these can be classified into two groups based on their sequence homology and regulatory properties. Group I comprises Paks 1 to 3, and group II comprises Paks 4 to 6 (11). Both group I and group II Paks are serine/threonine kinases that contain an N-terminal p21-binding domain (PBD) and a C-terminal protein kinase domain. The binding of Cdc42 and Rac to group I Paks such as Pak1 unfolds the kinase and leads to its activation (15, 19). Group II Paks are not strongly activated by GTPase binding, and the physiologic activation mechanism(s) for these kinases is not known. Functionally, the two groups are also distinct, as shown by the ability of group I, but not group II, Paks to complement the function of the Pak homolog STE20 in Saccharomyces cerevisiae (budding yeast) (7).
Of the three group II Paks, Pak5 is the least understood. Pak5 is expressed primarily in the brain and appears to be constitutively active (8, 18). Overexpression of Pak5 induces neurite extension in N1E-115 cells, whereas a dominant-negative Pak5 mutant inhibits neurite outgrowth (8). Unlike Pak4, overexpression of Pak5 has not been associated with cancers, and its deletion is not associated with an overt phenotype in mice (4, 16, 20). Overexpression of Pak5 activates the Jun N-terminal protein kinase pathway but not the p38 or ERK pathway (8, 18). Despite its ability to activate Jun N-terminal protein kinase, which is generally associated with apoptosis, we have previously shown that Pak5 induces resistance to apoptosis by phosphorylating BAD and that Pak5 is localized to mitochondria (7). While Paks 1, 2, and 4 have also been reported to phosphorylate BAD and can induce resistance to apoptosis (9, 12, 23), the function of these other Paks has not been related to mitochondrial localization. We therefore sought to determine how Pak5 localizes to this organelle and whether this localization is required for its effects on cell survival.
For this study, we identified sequence elements in Pak5 that are required for mitochondrial localization. In doing so, we also identified functional nuclear export and nuclear localization sequences. We found that endogenous Pak5 shuttles between mitochondria and the nucleus and that, when excluded from mitochondria, Pak5 does not efficiently phosphorylate BAD and no longer confers protection against apoptotic stimuli. In addition, growth factor treatment leads to a rapid relocalization of endogenous Pak5 to the nucleus. These findings suggest that Pak5 may be regulated by trafficking between mitochondria and the nucleus and that Pak5 has critical mitochondrial targets that mediate its antiapoptotic effects.
MATERIALS AND METHODS
Reagents and antibodies.
Camptothecin (CPT), leptomycin B (LMB), and staurosporine (STS) were purchased from Sigma, as were antiactin antibodies. Polyclonal anticalnexin and anti-Pak5 antibodies were obtained from EMD Biosciences. Mouse monoclonal anti-c-Myc (sc-40; clone 9E10), rabbit polyclonal anti-c-Myc (clone A14), anti-glutathione S-transferase (anti-GST), and anti-green fluorescent protein (anti-GFP) antibodies were purchased from Santa Cruz Biotechnology. Mouse monoclonal antihemagglutinin (anti-HA; 12CA5J) and anti-COX IV antibodies were purchased from Babco and Molecular Probes, respectively. Polyclonal antiphosphoserine and anti-BAD phosphoserine 112 antibodies were purchased from Zymed and Cell Signaling Technology, respectively.
Plasmids.
pCMV6-Pak5 and a plasmid encoding GST-tagged and HA-tagged Cdc42 L61 were constructed as described previously (7, 25). Pak5 truncations and internal deletions were obtained by PCR or site-directed mutagenesis (QuikChange kit; Stratagene). Wild-type and mutant forms of Pak5 were subcloned into the pEGFP-C1 (Clontech) and pCMV6 (25) vectors. pEBG-BAD, encoding GST-BAD, was purchased from Cell Signaling Technology, and a pET vector encoding His-tagged BAD was a gift from Michael Greenberg (Harvard Medical School, Boston, MA).
Cell culture, transfection, and treatment.
IMR32 and CHO cell lines were obtained from ATCC. C17.2 neural stem cells were obtained from Connie Cepko (Harvard Medical School, Boston, MA). Cell lines were grown at 37°C in 5% CO2 and cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS) and 5% horse serum for C17.2 cells, minimum essential medium plus 10% FBS for IMR32 neuroblastoma cells, and Dulbecco's modified Eagle's medium-F12 containing 10% FBS for CHO cells. Puromycin (2 μg/ml; Sigma) was added to the medium for stable CHO cell lines transfected with either empty vector (pCMV6; control cells) or wild-type (wt-Pak5) or mutant Pak5, and 1 mg/ml of G418 was added to cells transfected with GFP vectors containing the same cDNAs. Myc-Pak5-expressing stable cell lines were used for biochemistry studies, and GFP-Pak5-expressing vectors were used for immunofluorescence studies. For transient expression, cells were transfected using Lipofectamine (Gibco), and lysates were collected 48 h after transfection. When LMB was used, cells were treated for 12 h and then lysed for nuclear extraction or fixed for immunofluorescence (see below).
siRNA.
Smart-pool human and murine Pak5 small interfering RNA (siRNA) sets were purchased from Dharmacon Technologies. Cells plated onto coverslips precoated with poly-l-lysine in six-well plates were transfected with 150 pmol of siRNA and 10 μl of Lipofectamine 2000. Cells were processed 48 h after transfection for apoptosis and/or immunofluorescence.
GST pull-down assay.
To perform Cdc42 binding assays, 50 μg of purified GST fusion proteins coupled to glutathione-Sepharose beads (Amersham Pharmacia Biotech) were incubated with buffer A (40 mM HEPES, pH 7.4, 1% NP-40, 1 mM EDTA, 150 mM NaCl) supplemented with 10 mM EDTA for 15 min at room temperature to release any nucleotide, washed with buffer A, incubated with buffer B (buffer A supplemented with 1 mM GTP-γS and 10 mM MgCl2) for 30 min at room temperature, and then washed with buffer B. Next, the proteins were incubated with 250 μl of cell lysates for 3 h at 4°C. Precipitates were washed three times with lysis buffer (described below). Bound proteins were eluted in sodium dodecyl sulfate (SDS) sample buffer and subjected to immunoblotting with the anti-Myc monoclonal antibody 9E10 and a monoclonal antibody against GST.
Immunoblotting and immunoprecipitation.
Cell extracts were obtained in lysis buffer (50 mM Tris, pH 8, 100 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.5 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin), protein concentrations were assessed using bicinchoninic acid (Pierce), and equal amounts of proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (Immobilon P; Millipore Corp.). Blots were blocked for 1 h with Tris-buffered saline-0.1% Tween 20 supplemented with 5% nonfat milk and incubated with primary antibodies for an hour or overnight at 4°C. After a washing, blots were incubated with alkaline phosphatase-conjugated secondary antibodies (Jackson Laboratories), which were detected using the CPD Star reagent (New England Nuclear). For the detection of endogenous Pak5, 80 μg of protein was loaded, with 10 μg of lysates from stable CHO cell lines added as a control. The immunoprecipitation assay was carried out as follows. Whole-cell lysates (WCL) were incubated with polyclonal anti-Myc A14 antibodies overnight at 4°C and then for 1 h at 4°C with protein A-agarose beads (Pierce). Beads were collected by centrifugation and washed with the lysis buffer described above. Proteins were eluted by boiling in SDS sample buffer, separated by SDS-PAGE, immunoblotted, and probed with anti-HA (12CA5J) antibody.
Protein kinase assay.
CHO cells were transfected with appropriate expression vectors. Transfected cells were collected in lysis buffer 48 h after transfection, and equal amounts of the Myc-tagged proteins, as assayed by immunoblotting, were then immunoprecipitated with anti-Myc antibody A14 and protein A-agarose. After incubation, the immunoprecipitates were washed twice in lysis buffer and twice in kinase buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 1 mM MnCl2, 10 mM MgCl2, 1 mM dithiothreitol) and then incubated with 1.5 μg myelin basic protein in kinase buffer supplemented with 20 μM ATP and 5 μCi [γ-32P]ATP for 30 min at 30°C. Reactions were terminated with SDS-PAGE sample buffer, followed by SDS-PAGE and autoradiography.
For the in vitro BAD phosphorylation assay, immunoprecipitates were incubated with 5 μg of recombinant His-tagged BAD in kinase buffer supplemented with 20 μM ATP for 30 min at 30°C. Reactions were terminated with SDS-PAGE sample buffer, followed by SDS-PAGE and immunoblotting. Phosphorylation was revealed using antiphosphoserine antibodies.
In vivo BAD phosphorylation.
CHO cells were cotransfected with pCMV6-wt-Pak5, pCMV6-mut-Pak5, or empty vector and a mammalian expression vector for GST-BAD. Cells were lysed 24 h after transfection, and GST-BAD was pulled down using glutathione-Sepharose beads. After being washed, bound proteins were eluted in SDS sample buffer and subjected to immunoblotting with an anti-BAD phosphoserine 112 polyclonal antibody.
Apoptosis assays.
To estimate apoptosis in stably transfected cell lines, equal numbers of cells were seeded in six-well plates. CHO cells were treated for 12 h with CPT (10 μM) and collected for flow cytometry. Cells were collected by trypsinization and, after combination with the floating cells, labeled with annexin V-fluorescein isothiocyanate and propidium iodide according to the manufacturer's recommendations (Clontech) for fluorescence-activated cell sorting analysis (Becton Dickinson). IMR32 and C17.2 cells were seeded onto coverslips precoated with poly-l-lysine (Sigma). Cells were transfected with human Pak5 siRNA for IMR32 cells and with GFP, GFP-wt-Pak5, or GFP-mut-Pak5 and murine Pak5 siRNA for C17.2 cells. Cells were treated with 10 nM or 25 nM STS for 8 h. For the rescue experiment, C17.2 cells were cotransfected with murine Pak5 siRNA and GFP, GFP-wt-Pak5 (human), or GFP-mut-Pak5 (human). Cells were left untreated or were treated with 25 nM STS for 8 h. Cells were then fixed in 4% paraformaldehyde, stained for endogenous Pak5 expression, and counterstained with DAPI (4′,6′-diamidino-2-phenylindole). Two hundred fifty nuclei were counted, either in the whole population or only in the GFP-positive cells.
Cellular fractionation.
Cellular fractionation was performed as previously described (7). Briefly, cells were trypsinized and resuspended in isotonic mitochondrial buffer (MB; 210 mM mannitol, 70 mM sucrose, 1 mM EDTA, 10 mM HEPES, pH 7.5, supplemented with protease inhibitors). Cells were broken by passage through a 25-gauge needle fitted on a syringe, and the suspension was centrifuged at 2,000 × g at 4°C. The supernatant containing heavy membranes was subjected to centrifugation at 13,000 × g at 4°C, the supernatant corresponding to the cytosolic fraction was centrifuged at 100,000 × g for 30 min, and the pellet containing mitochondria was resuspended in MB buffer and subjected to a 500 × g centrifugation. The resultant supernatant was centrifuged for 10 min at 10,000 × g to collect the crude mitochondrial fraction. The crude mitochondrial fraction was resuspended in MB buffer and purified on a sucrose gradient as described previously (7). The S100 cytosolic fraction also generated the light microsomal pellet after centrifugation, which was resuspended in buffer H (20 mM Tris-HCl, 1 mM EDTA, 1 mM EGTA, 6 mM β-mercaptoethanol, pH 7.5, 1% SDS).
Nuclear extracts were isolated by first incubating cells for 15 min on ice in buffer A (10 mM HEPES, 2.5 mM MgCl2, 10 mM KCl, 0.5% NP-40) containing protease inhibitors. After swelling, nuclei were pelleted for 5 min at 12,000 × g. The supernatant corresponding to the cytosolic fraction was collected, while the nuclei were resuspended in buffer B (20 mM HEPES, 2.5 mM MgCl2, 20 mM EDTA, 420 mM NaCl, 25% glycerol) supplemented with protease inhibitors. After a 20-min incubation on ice with mixing of lysates every 5 min, the soluble nuclear fraction was collected by centrifugation for 15 min at 15,000 × g. The protein concentration in each fraction was assessed using bicinchoninic protein assay reagent (Pierce), and equal amounts of proteins were loaded and subjected to SDS-PAGE.
Immunofluorescence.
For colocalization with mitochondria, stable or transfected CHO cell lines expressing GFP vectors containing different constructs or IMR32 and C17.2 cells were stained for 15 min at 37°C with 5 ng/ml of the mitochondrion-specific dye MitoTracker red CMXRos (Molecular Probes Inc.) 24 h after transfection and then fixed in methanol for 5 min at −20°C. For detection of endogenous Pak5 or Myc-Pak5 and apoptosis assays, cells were fixed in 4% paraformaldehyde and permeabilized in phosphate-buffered saline-0.1% NP-40, followed by blocking for 15 min with 10% bovine serum albumin. Cells were stained with polyclonal anti-Pak5 or monoclonal anti-Myc antibodies and incubated with anti-rabbit immunoglobulin G conjugated with rhodamine X (Jackson Laboratories) or Alexa 488-conjugated anti-rabbit or anti-mouse serum (Molecular Probes). Cells were then washed and mounted, and photomicrographs were obtained using a Bio-Rad Radiant 2000 laser scanning confocal microscope with a 60× 11.20 objective (Nikon E800 Eclipse) or a Nikon TE300 microscope equipped with a Spot RT monochrome digital camera (Diagnostics Instruments) running MetaVue (Universal Imaging/Molecular Devices) software. Photographs were taken with a 60× Oil Plan Apo objective.
RESULTS
Identification of Pak5 mitochondrial targeting sequences.
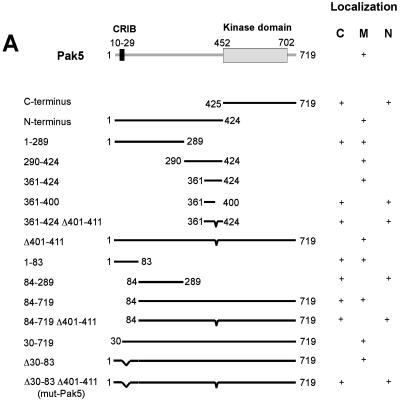
We have shown previously that transgenic Pak5 localizes predominantly to mitochondria in CHO cells (7). To identify the sequence(s) that targets Pak5 to this organelle, we generated deletion mutants by linking GFP to different regions of Pak5 (Fig. 1A). The expression of all the constructs was checked by immunoblotting (data not shown), and the localization of these mutants was assessed by subcellular fractionation and immunofluorescence (Fig. 1B).
FIG.1.
Mapping of a minimal region in Pak5 for localization to mitochondria. (A) Schematic representation of different Pak5 deletion constructs. (B) Confocal microscopy. CHO cells were transfected with pEGFP-C1 vector or the indicated constructs. Cells were stained with MitoTracker and fixed 24 h after transfection, as described in Materials and Methods. The fields shown were analyzed independently by confocal fluorescence microscopy at the appropriate wavelengths for GFP (Pak5) and MitoTracker red CMXRos (Mito), and the two images were overlaid (overlay). Bar = 20 μm. In parallel with microscopy, cellular fractionation was performed for each construct 48 h after transfections, as described in Materials and Methods, to assess the presence of Pak5 in the cellular compartments. Equal amounts of proteins from the cytosolic (S100), light microsome (L), and mitochondrial (M) fractions were loaded onto a gel, and immunoblotting was performed using an anti-GFP antibody. Anti-COX IV (for mitochondria) and anticalnexin (for light membranes) antibodies were used as controls. (C) CHO cells were transfected with empty vector (pCMV6) or with a vector carrying Myc-wt-Pak5 or Myc-mut-Pak5 cDNA. Nuclear extraction was performed as described in Materials and Methods. Equal amounts of lysates were loaded onto a gel and subjected to immunoblot analysis using anti-Myc antibodies. (D) CHO cells were transiently transfected with pCMV6 Myc-wt-Pak5 or pCMV6 Myc-mut-Pak5. Cells were processed for immunofluorescence using MitoTracker anti-Myc antibodies as described in Materials and Methods. Bar = 20 μm. (E) CHO cells were transfected with a GFP vector encoding GFP fused to the mitochondrial targeting sequence, corresponding to amino acids 30 to 83 of the Pak5 sequence. For the first three panels, cells were stained with MitoTracker red CMXRos (Mito) and then fixed in methanol 24 h after transfection, and confocal microscopy was used to visualize colocalization. For the right panel, cellular fractionation was performed to assess the presence of Pak5 in the different cellular fractions. Equal amounts of proteins from the cytosolic S100, light microsome (L), and mitochondrial (M) fractions were loaded on a gel, and immunoblotting was performed using anti-GFP antibodies. Anti-COX IV (for mitochondria) and anticalnexin (for light membranes) antibodies were used as controls. Bar = 20 μm.
Pak5 comprises at least two recognizable domains: the N-terminal regulatory domain and the C-terminal kinase domain (Fig. 1A). As shown in Fig. 1A and B (panels c and d), the N terminus is required for the mitochondrial localization of Pak5, as its removal resulted in a form of the protein that was cytosolic and nuclear but not mitochondrial. We therefore focused more closely on the N terminus to better define the mitochondrial localization motif(s).
As summarized in Fig. 1A, we determined the minimal sequences within the N terminus responsible for the mitochondrial localization of Pak5. We subdivided the N-terminal half of Pak5 into two segments, amino acids 1 to 289 and 290 to 424 (Fig. 1A). Interestingly, both fragments displayed mitochondrial localization (although the former was also found in the cytosol), indicating that the N-terminal half of the protein contained more than one mitochondrial localization motif (Fig. 1B, panels e and f). Analysis of the segment containing amino acids 290 to 424 revealed that the smallest GFP-Pak5 fragment that localized to mitochondria comprised amino acids 361 to 424. Interestingly, this region of Pak5 is not well conserved in the other group II Paks. Deletion of amino acids 401 to 411 within this region abolished mitochondrial localization and resulted in accumulation of the protein in the cytosol and the nucleus. However, when introduced in the context of the full-length molecule, this small deletion did not diminish the localization of Pak5 to mitochondria (Fig. 1A and data not shown). These results are consistent with the notion that Pak5 contains more than one motif that affects mitochondrial localization. For this reason, we proceeded to examine Pak5 for additional mitochondrial targeting regions, concentrating on the region encompassing residues 1 to 289. Deletion of the PBD, which resides within this region (amino acids 10 to 29), did not affect Pak5 localization (data not shown), suggesting that mitochondrial localization of Pak5 does not require direct binding to Cdc42 or related GTPases. However, we observed that the fragment comprising residues 1 to 83 localized to mitochondria, whereas the fragment comprising residues 84 to 289 did not. Therefore, we reasoned that a second mitochondrial targeting sequence must be present within residues 1 to 83. As expected, deletion of amino acids 1 to 83 did not affect the localization of longer Pak5 constructs that included amino acids 401 to 411; however, combining this deletion with the deletion of amino acids 401 to 411 (Δ1-83, Δ401-411) caused Pak5 to relocalize to the nucleus. To minimize the disruption of Pak5 structure and function, we constructed another version of this protein that retained the PBD (Pak5 Δ30-83). A combined deletion of the two minimal regions that affect mitochondrial localization (Δ30-83, Δ401-411; henceforth termed mut-Pak5) resulted in a nearly full-length form of Pak5 that was nonmitochondrial. Rather, this mutant protein localized primarily to the nucleus, with small amounts localized to the cytosol (Fig. 1A and B, panel h). Attempts to further reduce the size of the N-terminal Δ30-83 deletion resulted in a partial retention of Pak5 in mitochondria (data not shown).
To confirm the localization of these mutants, subcellular fractionation and nuclear extraction were performed, along with immunostaining. wt-Pak5 was present in the mitochondrial fraction (Fig. 1B, panel b), whereas mut-Pak5, containing the double deletion, was undetectable in the mitochondrial fraction and only slightly present in the light-membrane fraction (the nucleus is not represented in this fractionation) (Fig. 1B, panel h). To further assess the presence of Pak5 in the nucleus, we performed nuclear extraction (Fig. 1C). mut-Pak5, but not wt-Pak5, was found in nuclear extracts, consistent with the results from immunofluorescence studies. To ensure that the observed localization was not related to the presence of the GFP tag, we also performed immunofluorescence with cells expressing Myc-tagged Pak5 constructs. As shown in Fig. 1D, Myc-tagged mut-Pak5, like the GFP fusion, was found primarily in the nucleus. Finally, the fusion of GFP to Pak5 amino acids 30 to 83 resulted in a predominantly mitochondrial localization of the fusion protein, suggesting that this region of Pak5 is sufficient to confer targeting to mitochondria (Fig. 1E).
Biochemical characterization.
Since mut-Pak5 lacks a substantial portion of the N terminus, we compared it to wt-Pak5 in terms of GTPase binding and protein kinase activities. Pak5 is known to interact preferentially with Cdc42 (7, 8, 18). To study the interaction of the Pak5 mutant with this GTPase, we performed pull-down and coimmunoprecipitation assays. Figures 2A and B show that the nonmitochondrial form of Pak5, like the wild-type protein, bound to Cdc42 in both assays. Furthermore, the wild-type and mutant forms were both catalytically active, although mut-Pak5 appeared to be slightly more active than the wild-type enzyme in autophosphorylation assays (Fig. 2C). These results suggest that removal of the Pak5 regions that are required for mitochondrial localization does not grossly affect the structure or enzymatic properties of this kinase. To further investigate the properties of mut-Pak5, we assessed its activity toward a known substrate, BAD, in vitro. We found that wt-Pak5 and mut-Pak5 had similar kinase activities towards BAD (Fig. 2D). Thus, while mut-Pak5 had slightly increased autophosphorylation relative to wt-Pak5, their activities towards a bona fide exogenous substrate were indistinguishable.
FIG. 2.
Biochemical characterization of Pak5. (A) CHO cells were cotransfected with equal amounts of expression vectors encoding HA-tagged Cdc42 L61 and Myc-tagged wt-Pak5 or mut-Pak5 or with an empty vector. After transient expression, Myc-wt-Pak5 and Myc-mut-Pak5 were immunoprecipitated from total lysates with anti-Myc antibodies and protein A-agarose. Immunoblotting was then performed, and the complexes were revealed with anti-HA antibodies. The bottom panel shows the expression of Pak5, revealed with anti-Myc antibodies, and Cdc42, revealed with anti-HA antibodies, from extracts of WCL. (B) CHO cells were transfected with equal amounts of expression vectors encoding Myc-tagged wt-Pak5 and mut-Pak5 or with an empty vector. After transient expression, Myc-Pak5 proteins were pulled down from WCL with GST-Cdc42 loaded with GTP. Immunoblotting was then performed using anti-Myc and anti-GST antibodies. The expression level of Pak5 in the lysates used for the pull-down assay is shown in the bottom panel (expression was detected by immunoblotting using anti-Myc antibodies). (C) CHO cells were transfected with expression vectors for Myc-tagged wt- or mut-Pak5. Proteins were immunoprecipitated with an anti-Myc antibody, and a kinase assay was performed as described in Materials and Methods, using [γ-32P]ATP and myelin basic protein (MBP) as a substrate. Reaction products were separated by SDS-PAGE and subjected to autoradiography. The bottom panel shows the expression levels of proteins in the WCL prior to immunoprecipitation. (D) CHO cells were transfected with expression vectors for Myc-tagged wt- or mut-Pak5. Proteins were immunoprecipitated with an anti-Myc antibody, and a kinase assay was performed as described in Materials and Methods, using His-tagged recombinant BAD as a substrate. Reaction products were separated by SDS-PAGE and subjected to immunoblotting using antiphosphoserine antibodies. The bottom panels show the expression levels of proteins in the WCL prior to immunoprecipitation and the amount of His-BAD added to each reaction.
Pak5 shuttles to the nucleus.
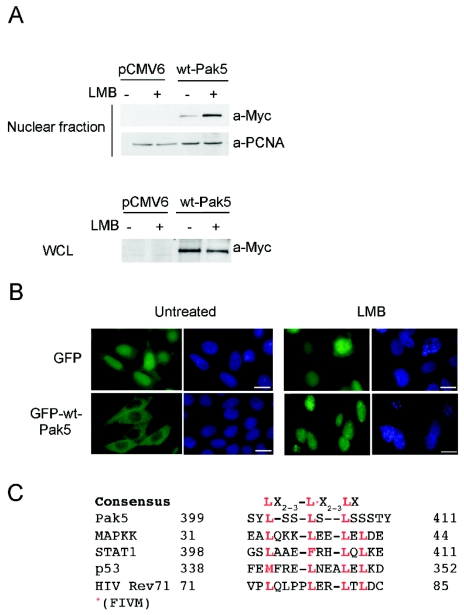
Since mut-Pak5 localized mainly to the nucleus, we wondered whether wt-Pak5 could shuttle to and from this location. To answer this question, we established stable CHO cell lines expressing GFP and GFP-wt-Pak5 for immunofluorescence (Fig. 3) and their pCMV6 equivalents for apoptosis (Fig. 4). We then treated these cells with LMB, an inhibitor of nuclear export, and performed nuclear extraction. Figure 3A shows that wt-Pak5 accumulated in the nucleus upon LMB treatment.
FIG. 3.
Nucleocytoplasmic shuttling of Pak5. (A) CHO control cells and stable cell lines expressing Myc-tagged Pak5 were left untreated or were treated for 12 h with LMB. Nuclear extraction was performed, and equal amounts of lysates were subjected to SDS-PAGE and revealed with anti-Myc antibodies. Anti-PCNA antibodies were used as a qualitative and quantitative control. (B) Stable CHO cell lines expressing GFP or GFP-Pak5 were treated for 12 h with LMB, fixed, and analyzed by fluorescence microscopy. Bar = 20 μm. (C) Sequence alignment of the functional Pak5 NES, as defined by deletion studies, and known NES elements in representative proteins.
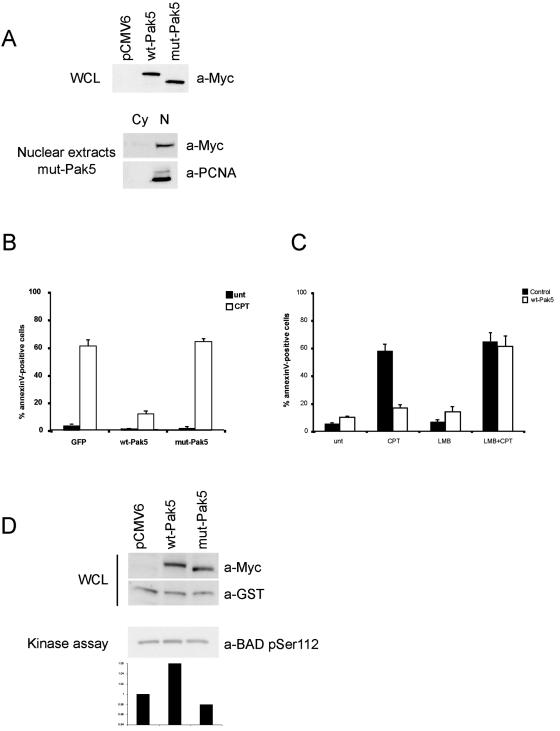
FIG. 4.
Nuclear Pak5 does not protect cells from apoptosis. (A) Expression of Pak5 in CHO cell lines. (Top panel) Equal amounts of lysates from stable CHO cells containing empty pCMV6 vector, Myc-wt-Pak5, or Myc-mut-Pak5 were analyzed for the expression of Pak5 by immunoblotting using anti-Myc antibodies. (Bottom panel) Nuclear extraction was performed for mut-Pak5 as previously described. Cy, cytosolic fraction; N, nuclear fraction. (B) Stable cell lines expressing Myc-tagged proteins were treated with CPT (10 μM) for 12 h, collected, and stained with annexin V and propidium iodide. Apoptosis was assessed by flow cytometry. The percentages of total cells positive for annexin V are shown in the graph and represent the averages of three experiments. (C) Stable cell lines expressing Myc-wt-Pak5 were pretreated for 1 hour with LMB and then treated with CPT (10 μM) for 12 h. Cells were then collected and stained with annexin V and propidium iodide. Apoptosis was assessed by flow cytometry. The percentages of total cells positive for annexin V are shown in the graph and represent the averages of three experiments. (D) CHO cells cotransfected with wt-Pak5, mut-Pak5, or empty vector and pEBG-BAD and lysed 24 h after transfection. Equal amounts of cell extracts were analyzed by immunoblotting to assess Myc-tagged proteins and BAD expression levels using anti-GST and anti-Myc antibodies. A pull-down assay of GST-BAD was performed, using antibodies directed against phospho-Ser 112 to assess the BAD phosphorylation status, which was evaluated by densitometry.
We also assessed the presence of wt-Pak5 in the nucleus by immunofluorescence. As shown in Fig. 3B, wt-Pak5 was present in the nucleus after LMB treatment, whereas it was rarely seen in the nuclei of untreated cells. LMB did not affect the localization of GFP. The sum of these results suggests that wt-Pak5 contains a functional nuclear exclusion sequence (NES) and that this sequence is absent from mut-Pak5. Interestingly, the amino acid sequence from residues 401 to 411, which is deleted in mut-Pak5, nearly conforms to the NES consensus sequence, as shown by multiple amino acid sequence alignments of these residues with known NES motifs (Fig. 3C). To confirm that this sequence does indeed represent a functional NES, we mutated three conserved leucine residues within this region (Fig. 3C; see the underlined residues in Fig. S1 in the supplemental material) and studied the effect on the cellular localization of Pak5 in LMB-treated cells. Cells were transfected with either wt-Pak5, mut-Pak5, or mut-Pak5-L→A (see Fig. S1 in the supplemental material). We found that in untreated cells, both mut-Pak5 (in which residues 400 to 411, encoding the putative NES, are deleted) and mut-Pak5-L→A were nuclear. These results confirm that the 400-411 region in Pak5 functions as an NES.
Nuclear Pak5 does not protect against apoptosis.
To examine the relationship between the localization of Pak5 and its effects on apoptosis, we asked if mut-Pak5 could confer protection to cells treated with a DNA-damaging agent. Control cells and stable cell lines expressing wt-Pak5 or mut-Pak5 expressed similar levels of proteins (Fig. 4A). These proteins localized to the mitochondria (data not shown) and the nucleus, respectively (Fig. 4A, bottom panel). These cells were treated with vehicle or CPT. In control cells, CPT rapidly induced apoptosis in a large percentage of cells (Fig. 4B). As we reported previously (7), wt-Pak5 blocked this effect. Notably, mut-Pak5 did not protect cells against CPT-induced apoptosis.
We then asked if retaining wt-Pak5 in the nucleus by inhibiting its export would affect its protective effects. We pretreated control CHO cells or CHO cells expressing wt-Pak5 with LMB and then induced apoptosis with CPT. LMB treatment alone had a minor effect on cell survival (Fig. 4C). When cells expressing wt-Pak5 were treated with LMB and then challenged with CPT, they underwent apoptosis to the same extent as control cells (Fig. 4C). These results are consistent with the data obtained with mut-Pak5 and suggest that the cellular localization of Pak5 is vital for its effects on apoptotic signaling.
One reason that nuclear Pak5 might exert lesser protective effects on cell survival is that this mutant kinase cannot access substrates such as BAD. To test this hypothesis, we determined the phosphorylation status of BAD in cells expressing wt-Pak5 or mut-Pak5. CHO cells were cotransfected with vectors expressing GST-BAD and wt-Pak5 or mut-Pak5 (Fig. 4D). We found that, consistent with our earlier report (7), BAD was phosphorylated on serine-112 in cells expressing wt-Pak5. This level of phosphorylation was substantially decreased in cells expressing mut-Pak5.
Pak5 contains a functional NLS.
The presence of Pak5 in the nucleus suggested that this protein contains a nuclear localization signal (NLS). A polylysine (KKKK) sequence is present at amino acids 5 to 10 that might represent a noncanonical NLS (Fig. 5A). To determine if this sequence is functional, we deleted this sequence in the context of wt-Pak5 and mut-Pak5 and assessed the localization of these proteins by immunofluorescence (Fig. 5B), subcellular fractionation (Fig. 5C), and nuclear extraction (Fig. 5D). Deletion of these residues from wt-Pak5 did not overtly affect localization, which remained largely mitochondrial (Fig. 5B and C). Cellular fractionation showed that mut-Pak5 Δ5-10 was present in the cytosolic and light-membrane fractions, as opposed to wt-Pak5 Δ5-10, which remained mitochondrial (Fig. 5C). Nuclear extraction showed the absence of both mut-Pak5 Δ5-10 and wt-Pak5 Δ5-10 in the nuclear fraction (Fig. 5D). These data showed that the lysine-rich motif at the N terminus of Pak5 is required for the nuclear localization of this protein. Immunofluorescence analysis of cells expressing Myc-tagged Pak5 mutants showed a similar profile (data not shown). To emphasize the importance of each of these sequence elements, we transfected cells with wt-Pak5 or the different mutants (mut-Pak5, wt-Pak5 Δ5-10, and mut-Pak5 Δ5-10) and assessed the effect of LMB on Pak5 localization (see Fig. S2 in the supplemental material). Mutants lacking the NLS did not shuttle to the nucleus after LMB treatment. Instead, the Pak5 Δ5-10 protein remained localized in the mitochondria, and the mut-Pak5 Δ5-10 protein remained in the cytosol. These findings support the view that the NLS is functional and is required for Pak5 entry into the nucleus.
FIG.5.
Pak5 contains a functional NLS. (A) Schematic representation of primary structure of Pak5 showing the various protein domains, in particular the NLS. (B) CHO cells were transfected with GFP-wt-Pak5 Δ5-10, GFP-mut-Pak5 Δ5-10, or GFP-mut-Pak5 as a control. Cells were stained with MitoTracker, and cellular localization was visualized by confocal microscopy. Bar = 20 μm. (C) Stable cell lines expressing Myc-tagged wt-Pak5 Δ5-10 or mut-Pak5 Δ5-10 were established. (Left panel) Immunoblot showing expression of both proteins. (Right panels) Cellular fractionation was performed 48 h after transfections, as described in Materials and Methods, to assess Pak5 distribution to various cellular compartments. Equal amounts of proteins from the cytosolic (S100), light microsome (L), and mitochondrial (M) fractions were loaded on a gel, and immunoblotting was performed using anti-Myc antibodies. Anti-COX IV (for mitochondria) and anticalnexin (for light membranes) antibodies were used as controls. (D) Nuclear extraction was performed as described in Materials and Methods on the stable cell lines described above. Anti-PCNA antibodies were used as a loading control. (E) Stable CHO cells expressing Myc-wt-Pak5 Δ5-10 or Myc-mut-Pak5 Δ5-10 were treated for 12 h with CPT (10 μM). Cells were then collected and stained with annexin V and propidium iodide. Apoptosis was assessed by flow cytometry. The percentages of total cells positive for annexin V are shown in the graph and represent the averages of three experiments.
We then tested whether cytosolic, but nonmitochondrial, Pak5 retained its antiapoptotic properties. To test this idea, we used cells that stably expressed mut-Pak5 Δ5-10 or wt-Pak5 Δ5-10. As shown in Fig. 5D, the former localized to the cytosol but not to the nucleus (because it lacks an NLS) or mitochondria (because it lacks the mitochondrial targeting motifs). These cells were treated with CPT, and the degree of cell death was assessed. We found that this form of Pak5 did not confer protection against apoptosis (Fig. 5E). These results are consistent with the notion that Pak5 must reside in the mitochondria to exert its prosurvival effects.
Prosurvival function of endogenous Pak5 in neuroblastoma cells and neural stem cells.
Pak5 is highly expressed in neurons (8, 16, 18). We found that Pak5 is expressed in the neuroblastoma cell line IMR32 (6) as well as in C17.2 neural stem cells (21) (Fig. 6A). We had previously shown that transgenic Pak5 localized to mitochondria in CHO and HMN1 cells (7) (Fig. 1B, panel b), but the localization of endogenous Pak5 had not previously been reported. We therefore determined the cellular localization of endogenous Pak5 by immunofluorescence (Fig. 6B) and cellular fractionation (Fig. 6C) in IMR32 and C17.2 cells. We found that endogenous Pak5 localized primarily to mitochondria in both cell types (Fig. 6B). Not all Pak5 was present on mitochondria; a small fraction was also detected in the cytosol. The mitochondrial localization was confirmed by cellular fractionation, which showed that the mitochondrial fraction was enriched in Pak5 compared to the whole-cell lysate (Fig. 6C). These findings support our previous observations using transgenic Pak5.
FIG. 6.
Expression/characterization of endogenous Pak5 in IMR32 and C17.2 cells. (A) Immunoblot using anti-Pak5 antibodies. CHO and stably Myc-Pak5-expressing CHO cells were used as negative and positive controls, respectively. (B) Immunofluorescent staining of Pak5 in IMR32 and C17.2 cells showing Pak5 colocalization with mitochondria. Cells were stained with anti-Pak5 antibodies followed by Alexa 488-conjugated anti-rabbit secondary antibodies. Mitochondria were stained with MitoTracker red, and colocalization was analyzed by confocal microscopy. Bar = 20 μm. (C) Cellular fractionation was performed as described in Materials and Methods. WCL and mitochondrial (m) fractions were loaded on a 7.5% gel, and blots were stained with anti-Pak5 antibodies. Mitochondria were loaded on a separate 12% gel to assess the presence of COX IV in the fraction, as indicated. Myc-Pak5 was used as a positive control.
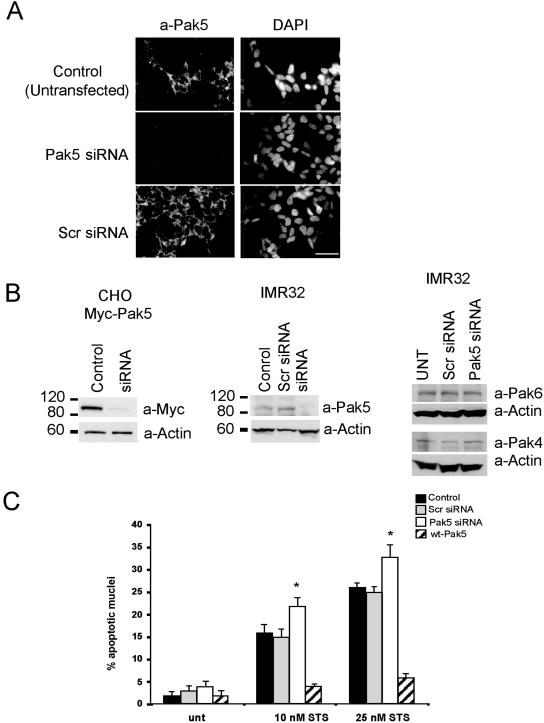
To determine if the endogenous kinase contributes to cell survival, we knocked down Pak5 expression in IMR32 cells and examined their ability to survive when treated with STS. We transfected IMR32 cells with either scrambled siRNA or Pak5-specific siRNA. As shown in Fig. 7A, cells transfected with Pak5-specific siRNA showed greatly reduced Pak5 staining relative to control cells, indicating a successful knockdown of Pak5. The knockdown was also documented by immunoblotting (Fig. 7B). CHO cells stably transfected with Myc-Pak5 were used as a control and showed approximately 90% inhibition of Pak5 expression when transfected with Pak5-specific siRNA (Fig. 7B, left panel). Similar results were obtained with IMR32 cells (Fig. 7B, middle panel). The knockdown of Pak5 with a specific siRNA did not affect the expression of the other group II Paks, Pak4 and Pak6 (Fig. 7B, right panel). As shown in Fig. 7C, Pak5 knockdown in IMR32 cells increased STS-induced apoptosis ∼10% (P < 0.001), whether cells were treated with 10 nM or 25 nM of STS, whereas control siRNA had no effect on the sensitivity to STS. The opposite trends were observed in cells overexpressing wt-Pak5, which conferred resistance to STS, similar to the results obtained with CHO cells (Fig. 5E).
FIG. 7.
Role of Pak5 in apoptosis in neuroblastoma cells. (A) IMR32 cells were grown on coverslips and transfected with scrambled siRNA or Pak5 siRNA or left untransfected. Pak5 expression was assessed by fluorescence microscopy. The same settings (exposure time and gain) were used to take photographs of controls and Pak5 siRNA-transfected cells. Bar = 20 μm. (B) Pak5 immunoblot of CHO cells expressing Pak5 (left panel) or IMR32 cells (middle panel) transfected with Pak5-specific siRNA or scrambled siRNA (Scr). The right panel shows the expression of the group II Paks Pak4 and Pak6 in IMR32 cells transfected with Pak5-specific siRNA or scrambled siRNA (Scr). Actin was used as a loading control. (C) IMR32 cells were transfected with GFP-wt-Pak5 or scrambled or Pak5-specific siRNA or left untransfected and then treated for 8 h with STS (10 nM or 25 nM) 48 h after transfection. Cells were then fixed and counterstained with DAPI. Two hundred fifty nuclei were counted in the whole population for the control cells (untransfected [unt], Scr) and Pak5 siRNA-treated cells, and only GFP-positive cells were counted for cells expressing GFP-wt-Pak5. Data are means plus standard deviations (n = 5). *, P < 0.001.
We also examined the role of Pak5 in apoptosis in C17.2 cells (Fig. 8). Because these cells are difficult to transfect (∼3% transfection efficiency for a GFP plasmid and ∼50% transfection efficiency for fluorescein isothiocyanate-labeled oligonucleotides; data not shown), we cotransfected cells with Pak5-specific siRNA and GFP and assessed apoptosis in GFP-positive cells. The knockdown of Pak5 was assessed by immunofluorescence, as shown in Fig. 8A. The expression of GFP did not affect Pak5 levels, but cotransfection of the GFP plasmid and Pak5-specific siRNAs resulted in a decrease in Pak5 staining in GFP-positive cells (Fig. 8A). The knockdown of Pak5 reproducibly increased the sensitivity of C17.2 cells to STS-induced apoptosis, by about 15% compared to control cells, using two different concentrations of STS (Fig. 8B). Overexpression of Pak5 dramatically reduced the percentage of apoptotic cells. These results confirm the observations made with IMR32 cells and support the idea that Pak5 plays a role in neuronal cell survival.
FIG. 8.
Role of Pak5 in apoptosis in C17.2 neural stem cells. (A) C17.2 cells were transfected with a GFP-expressing plasmid alone (control) or a GFP-expressing plasmid plus Pak5-specific siRNA. Immunofluorescence microscopy was used to assess Pak5 expression. Note that GFP-positive cells (arrowheads) showed reduced Pak5 expression. Bar = 20 μm. (B) Cells were treated with STS (10 nM or 25 nM, 8 h) 48 h after transfection. Cells were fixed and counterstained with DAPI. Two hundred fifty nuclei from the GFP-positive cells only were counted. Data are means plus standard deviations (n = 3). *, P < 0.001. (C) Cells were transfected with GFP-, GFP-wt-Pak5-, or GFP-mut-Pak5-expressing plasmids plus Pak5-specific siRNA. Immunofluorescence microscopy was used to assess Pak5 expression. Forty-eight hours after transfection, cells were left untreated (UNT) or were treated with 25 nM STS for 8 h. Cells were then fixed and counterstained with DAPI. Five hundred nuclei from the GFP-positive cells only were counted. The percentages of apoptotic nuclei are shown in the graph and represent the averages of three experiments.
We then determined if reintroducing wt-Pak5 could rescue cells in which endogenous Pak5 had been knocked down (Fig. 8C). Murine C17.2 cells were cotransfected with murine Pak5 siRNAs to knock down endogenous Pak5 and with an empty GFP vector or a GFP vector encoding siRNA-resistant human wt-Pak5 or human mut-Pak5. The expression of Pak5 in GFP-positive cells was assessed by immunofluorescence using anti-Pak5 antibodies. Cells transfected with GFP-wt-Pak5 displayed a low percentage of apoptotic cells compared to control cells (transfected with GFP alone) or cells transfected with mut-Pak5. These results show that the expression of human wt-Pak5 can efficiently rescue murine cells from Pak5 siRNA-induced apoptotic sensitivity. In contrast, the nonmitochondrial mutant of human Pak5 (mut-Pak5) failed to reverse this defect.
Endogenous Pak5 shuttles to the nuclei of neuronal cells.
Having demonstrated that endogenous Pak5 has a role in neuronal cell survival and is a strong inhibitor of STS-induced apoptosis when overexpressed, we determined if endogenous Pak5 could shuttle to the nuclei of neuronal cells, as observed in Pak5-transgenic CHO cells. We treated IMR32 cells with LMB and stained the cells with anti-Pak5 antibodies. As shown in Fig. 9A, while the vast majority of Pak5 resided in mitochondria in untreated cells, in LMB-treated cells, Pak5 was concentrated in the nucleus in approximately 70% of the cells. Similar results were obtained with C17.2 cells (data not shown).
FIG.9.
Endogenous Pak5 shuttles to the nucleus in neuronal cells. (A) IMR32 cells were treated or not with LMB for 12 h. Cells were fixed and stained with anti-Pak5 antibody followed by Alexa 488-conjugated anti-rabbit antibodies. Pak5 localization was analyzed by fluorescence microscopy. Bar = 20 μm. Arrowheads indicate cells displaying nuclear Pak5. (B) C17.2 cells were transfected with GFP-wt-Pak5 or GFP-mut-Pak5, fixed 24 h later, and counterstained with DAPI. Pak5 localization was analyzed by fluorescence microscopy. (C) C17.2 cells transfected with GFP-wt-Pak5 or GFP-mut-Pak5 were treated 48 h later with 25 nM STS for 8 h. Cells were fixed and counterstained with DAPI. Only GFP-positive cells were analyzed for apoptosis measurement. Two hundred fifty nuclei were counted. Representative data from three experiments are shown. (D) Asynchronous C17.2 and IMR32 cells showing the presence of Pak5 in the nucleus. Cells were grown on coverslips, fixed, and stained with Pak5 antibodies. Random fields were photographed and show nuclear Pak5, as indicated by arrowheads. The graph on the right shows the percentages of cells containing nuclear Pak5 in asynchronous populations of C17.2, IMR32, and CHO stable cell lines expressing exogenous Myc-Pak5 or GFP-Pak5. Five hundred cells were counted. (E) C17.2 cells were serum starved for 36 h and then treated with 100 ng/ml EGF, 100 ng/ml FGF, or 20% FBS for 1 h. Cells were fixed and immunostained with anti-Pak5 antibodies. The upper panel shows fluorescence microscopy. Five hundred cells were counted. The graph in the lower panel represents the percentages of cells containing nuclear endogenous Pak5.
We next determined if the nonmitochondrial mutant of Pak5 (mut-Pak5) localizes to the nucleus in these cells and if the localization of Pak5 in the nucleus has an effect on apoptosis. Figure 9B shows that GFP-mut-Pak5 localized to the nucleus, whereas GFP-wt-Pak5 localized to the cytosol, consistent with our previous observations in CHO cells (Fig. 1C). To assess the effect of this mutant on apoptosis, C17.2 cells were grown on coverslips, transfected with GFP-wt-Pak5 or GFP-mut-Pak5, and then treated with STS. The nuclear morphology of GFP-positive cells was assessed for evidence of apoptosis. As in CHO cells (Fig. 4B), wt-Pak5, but not mut-Pak5, conferred resistance to apoptosis (Fig. 9C). Thus, the relationship between mitochondrial localization and resistance to apoptosis is not restricted to CHO cells but also occurs in a cell type (neuronal cells) in which Pak5 is normally expressed.
To determine if Pak5 physiologically shuttles to the nucleus in neuronal cell lines, we studied asynchronous populations and found that endogenous Pak5 is present in the nucleus in approximately 5% of the cells (Fig. 9D). CHO cell lines stably expressing Myc-wt-Pak5 or GFP-wt-Pak5 were used as controls, and these cells displayed the same percentage of nuclear Pak5 as IMR32 and C17.2 cells (data not shown).
C17.2 cells have been shown to proliferate when treated with epidermal growth factor (EGF) or fibroblast growth factor (FGF) (13). We therefore sought to determine if these two growth factors triggered Pak5 shuttling to the nucleus. Cells were starved for 36 h and then treated with epidermal growth factor, fibroblast growth factor, or fetal bovine serum. We found that these growth factors caused up to 50% of the Pak5 to accumulate in the nucleus (Fig. 9E). These data clearly demonstrate that Pak5 relocalizes to the nucleus in response to various external stimuli.
DISCUSSION
Prior to this study, there has been very limited information about the structural elements that regulate Pak5 activity or localization. The group II Paks are more restrictive in their binding to small GTPases (Cdc42 ≫ Rac) and, unlike group I Paks, are not strongly activated upon binding Cdc42. By truncation analysis, Ching et al. have identified a putative autoinhibitory domain within the N terminus of Pak5 (5), but it is not clear how or if this domain normally regulates the activity of this kinase. In this report, we present evidence that Pak5 contains functional elements that regulate its intracellular localization, that this localization is critical to the biological functions of this kinase, and that localization is regulated both by internal programs and by external stimuli.
We had previously established that transgenic forms of Pak5 localize primarily to mitochondria; here we show that this is also true for the endogenous protein in two different neuronal cell types—IMR32 neuroblastoma cells and C17.2 neural stem cells—and that such localization requires an N-terminal segment located between residues 30 and 83 and the presence of an intact NES. Deletion of this NES or treatment of cells with a nuclear export inhibitor leads to an accumulation of Pak5 in the nucleus. Conversely, deletion of an NLS that we identified in the extreme N terminus (residues 5 to 10) prevents the accumulation of Pak5 in the nucleus, even in mutants lacking the NES. This NLS is functional and required for import of the nuclear mutant of Pak5 but is less dominant than the NES, since the flow of Pak5 appears to be from the nucleus to the cytosol by reexport via an LMB-sensitive CRM1-dependent pathway. The mutant Pak5 84-719 Δ400-411, which lacks all three identified localization sequences (mitochondrial targeting sequence, NES, and NLS), is still present in the nucleus, suggesting that Pak5 contains a second, nonconsensus, cryptic NLS. This sequence has weaker effects than the one already identified (amino acids 5 to 10) because the LMB treatment of cells expressing mutants lacking the consensus NLS does not induce translocation to the nuclei of these mutant cells. These findings also suggest that Pak5 shuttles between various cellular compartments and that it might therefore have several distinct targets and functions that depend on its location. For example, mutations that prevent Pak5 from localizing to mitochondria also prevent Pak5 from conferring antiapoptotic properties. This loss of function is not due to impaired kinase activity or impaired binding to Cdc42 (Fig. 2). Instead, we suggest that nonmitochondrial forms of Pak5 do not have access to the requisite substrates that affect cell survival. One of these might be BAD, which we previously showed to be a potential target of Pak5 (Fig. 4D) (7), but there are likely also others that affect cell survival and other functions. For example, Matenia et al. recently reported that exogenous Pak5 binds to the kinase MARK2 (also called Par-1) and partially colocalizes with AP-1/2-containing endosomes (17). While MARK2 itself does not appear to be a substrate for Pak5, it is nevertheless inhibited by its interactions with this kinase.
We have noted that in a small proportion of cells, Pak5 resides in the nucleus, and that this fraction can be substantially increased by blockade of nuclear export induced by chemical agents or by removal of the NES. Conversely, we showed that removal of the NLS from Pak5 prevents its accumulation in the nucleus. Neither the kinase activity of Pak5 nor its ability to bind Cdc42 is required for its nuclear localization, as kinase-dead and PBD mutants of Pak5 were also retained in the nucleus after treatment of cells with LMB (data not shown). These results suggest that Pak5 normally shuttles to and from the nucleus and that this kinase has one or more nuclear functions. What might these nuclear functions be? The related enzyme Pak6 was discovered as a binding partner and inhibitor of the androgen receptor (AR) and the estrogen receptor, and therefore Pak5 might affect the transcriptional activities of these factors (14, 27). Active Pak6 has been reported to cotranslocate with the AR into the nucleus and to inhibit AR-mediated transcription (27). However, another group has reported that active Pak6 inhibits the nuclear translocation of the stimulated AR, suggesting that Pak6 acts in the cytosol (22) or, alternatively, that it acts in the nucleus to promote the export of the AR. Whatever the mechanism underlying the nuclear location and functions of Pak6, Pak5 has not been reported to bind to or act upon steroid receptors, and its restricted pattern of expression suggests that its functions do not overlap substantially with those of Pak6.
Recently, Pak1 (a group I Pak) was found to translocate to the nucleus upon growth factor stimulation (26). This enzyme associates with particular enhancer and promoter elements and appears to regulate the expression of a number of genes, in particular NFAT and the muscle form of phosphofructokinase. In addition, Pak1 has been shown to bind to and phosphorylate a transcriptional corepressor, CtBP (2). By analogy, it is possible that Pak5, which translocates to the nucleus upon growth factor stimulation (Fig. 9E), also plays a role in regulating gene expression.
Mice lacking the Pak5 gene develop normally (16). Like Pak5, several other members of the Pak family are highly expressed in the brain, and it is possible that these possess certain redundant functions. However, it is clear that these functions cannot be completely overlapping, as both over- and underexpression of the various Pak isoforms leads to distinct phenotypes (10). Pak5 possesses unique features, as exemplified by its mitochondrial localization and the requirement of this localization for its effects on cell survival. Since the knockdown of endogenous Pak5 in neuronal cell types renders these cells more sensitive to apoptosis (Fig. 7 and 8), it is possible that this kinase plays a role in the survival of neurons under stress. Such a function might not be easily revealed during the development and maintenance of mice under standard laboratory conditions but might be elicited only by specific stresses that affect neuronal survival in vivo. We are currently examining these possibilities.
Supplementary Material
Acknowledgments
We thank Connie Cepko for the C17.2 cell line, Sandy Jablonski (Cell Imaging Core Facility) for assistance, and Peter Adams and Maureen Murphy for reviewing the manuscript.
This work was supported by grants from the National Institutes of Health and the American Cancer Society, by a National Cancer Institute CORE grant to the Fox Chase Cancer Center, and by an appropriation from the Commonwealth of Pennsylvania.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bagrodia, S., and R. A. Cerione. 1999. PAK to the future. Trends Cell Biol. 9:350-355. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, C. J., R. K. Vadlamudi, S. K. Mishra, R. H. Jacobson, F. Li, and R. Kumar. 2003. Functional inactivation of a transcriptional corepressor by a signaling kinase. Nat. Struct. Biol. 10:622-628. [DOI] [PubMed] [Google Scholar]
- 3.Bokoch, G. M. 2003. Biology of the p21-activated kinases. Annu. Rev. Biochem. 72:743-781. [DOI] [PubMed] [Google Scholar]
- 4.Callow, M. G., F. Clairvoyant, S. Zhu, B. Schryver, D. B. Whyte, J. R. Bischoff, B. Jallal, and T. Smeal. 2002. Requirement for PAK4 in the anchorage-independent growth of human cancer cell lines. J. Biol. Chem. 277:550-558. [DOI] [PubMed] [Google Scholar]
- 5.Ching, Y. P., V. Y. Leong, C. M. Wong, and H. F. Kung. 2003. Identification of an autoinhibitory domain of p21-activated protein kinase 5. J. Biol. Chem. 238:33621-33624. [DOI] [PubMed] [Google Scholar]
- 6.Clementi, F., D. Cabrini, C. Gotti, and E. Sher. 1986. Pharmacological characterization of cholinergic receptors in a human neuroblastoma cell line. J. Neurochem. 47:291-297. [DOI] [PubMed] [Google Scholar]
- 7.Cotteret, S., Z. M. Jaffer, A. Beeser, and J. Chernoff. 2003. p21-activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol. Cell. Biol. 23:5526-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dan, C., N. Nath, M. Liberto, and A. Minden. 2002. PAK5, a new brain-specific kinase, promotes neurite outgrowth in N1E-115 cells. Mol. Cell. Biol. 22:567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gnesutta, N., J. Qu, and A. Minden. 2001. The serine/threonine kinase PAK4 prevents caspase activation and protects cells from apoptosis. J. Biol. Chem. 276:14414-14419. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann, C., M. Shepelev, and J. Chernoff. 2004. The genetics of Pak. J. Cell Sci. 117:4343-4354. [DOI] [PubMed] [Google Scholar]
- 11.Jaffer, Z. M., and J. Chernoff. 2002. p21-activated kinases: three more join the Pak. Int. J. Biochem. Cell. Biol. 34:713-717. [DOI] [PubMed] [Google Scholar]
- 12.Jakobi, R., E. Moertl, and M. A. Koeppel. 2001. p21-activated protein kinase gamma-PAK suppresses programmed cell death of BALB3T3 fibroblasts. J. Biol. Chem. 276:16624-16634. [DOI] [PubMed] [Google Scholar]
- 13.Kitchens, D. L., E. Y. Snyder, and D. I. Gottlieb. 1994. FGF and EGF are mitogens for immortalized neural progenitors. J. Neurobiol. 25:797-807. [DOI] [PubMed] [Google Scholar]
- 14.Lee, S. R., S. M. Ramos, A. Ko, D. Masiello, K. D. Swanson, M. L. Lu, and S. P. Balk. 2002. AR and ER interaction with a p21-activated kinase (PAK6). Mol. Endocrinol. 16:85-99. [DOI] [PubMed] [Google Scholar]
- 15.Lei, M., W. Lu, W. Meng, M.-C. Parrini, M. J. Eck, B. J. Mayer, and S. C. Harrison. 2000. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell 102:387-397. [DOI] [PubMed] [Google Scholar]
- 16.Li, X., and A. Minden. 2003. Targeted disruption of the gene for the PAK5 kinase in mice. Mol. Cell. Biol. 23:7134-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matenia, D., B. Griesshaber, X. Y. Li, A. Thiessen, C. Johne, J. Jiao, E. Mandelkow, and E. M. Mandelkow. 2005. PAK5 kinase is an inhibitor of MARK/Par-1, which leads to stable microtubules and dynamic actin. Mol. Biol. Cell 16:4410-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey, A., I. Dan, T. Z. Kristiansen, N. M. Watanabe, J. Voldby, E. Kajikawa, R. Khosravi-Far, B. Blagoev, and M. Mann. 2002. Cloning and characterization of PAK5, a novel member of mammalian p21-activated kinase-II subfamily that is predominantly expressed in brain. Oncogene 21:3939-3948. [DOI] [PubMed] [Google Scholar]
- 19.Parrini, M. C., M. Lei, S. C. Harrison, and B. J. Mayer. 2002. Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol. Cell 9:73-83. [DOI] [PubMed] [Google Scholar]
- 20.Qu, J., M. S. Cammarano, Q. Shi, K. C. Ha, P. deLanerolle, and A. Minden. 2001. Activated PAK4 regulates cell adhesion and anchorage-independent growth. Mol. Cell. Biol. 21:3523-3533.11313478 [Google Scholar]
- 21.Ryder, E. F., E. Y. Snyder, and C. L. Cepko. 1990. Establishment and characterization of multipotent neural cell lines using retrovirus vector-mediated oncogene transfer. J. Neurobiol. 21:356-375. [DOI] [PubMed] [Google Scholar]
- 22.Schrantz, N., J. da Silva Correia, B. Fowler, Q. Ge, Z. Sun, and G. M. Bokoch. 2004. Mechanism of p21-activated kinase 6-mediated inhibition of androgen receptor signaling. J. Biol. Chem. 279:1922-1931. [DOI] [PubMed] [Google Scholar]
- 23.Schurmann, A., A. F. Mooney, L. C. Sanders, M. A. Sells, H.-G. Wang, J. C. Reed, and G. M. Bokoch. 2000. p21-activated kinase 1 (PAK1) phosphorylates the death agonist Bad and protects cells from apoptosis. Mol. Cell. Biol. 20:453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sells, M. A., and J. Chernoff. 1997. Emerging from the Pak: the p21-activated protein kinase family. Trends Cell Biol. 7:162-167. [DOI] [PubMed] [Google Scholar]
- 25.Sells, M. A., U. G. Knaus, S. Bagrodia, D. M. Ambrose, G. M. Bokoch, and J. Chernoff. 1997. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr. Biol. 7:202-210. [DOI] [PubMed] [Google Scholar]
- 26.Singh, R., C. Song, Z. Yang, and R. Kumar. 2005. Nuclear localization and chromatin targets of p21-activated kinase. J. Biol. Chem. 280:18130-18137. [DOI] [PubMed] [Google Scholar]
- 27.Yang, F., X. Li, M. Sharma, M. Zarnegar, and Z. Lim. 2001. Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J. Biol. Chem. 276:15345-15353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.