Abstract
p63, a p53 family member, is essential for the development of various stratified epithelia and is one of the earliest markers of many ectodermal structures, including the epidermis, oral mucosa, apical ectodermal ridge, and mammary gland. Genetic regulatory mechanisms controlling p63 spatial expression during development have not yet been defined. Using a genomic approach, we identified an evolutionarily conserved cis-regulatory element, located 160 kb downstream of the first p63 exon, which functions as a keratinocyte-specific enhancer and is sufficient to recapitulate expression of the endogenous gene during mouse embryogenesis. Dissection of the p63 enhancer activity revealed a positive autoregulatory loop in which the p63 proteins directly bind to and are essential regulators of the enhancer. Accordingly, transactivating p63 isoforms induce endogenous p63 expression in cells that do not normally express this gene, whereas dominant negative isoforms suppress p63 expression in keratinocytes. In addition the transcription factor AP-2 also binds to the enhancer and cooperates with p63 to induce its activity. These results demonstrate that a long-range autoregulatory loop is involved in the regulation of p63 expression during embryonic development and in adult cells.
p63 is highly expressed in embryonic ectoderm as well as in the nuclei of basal regenerative cells of many epithelial tissues in the adult (19, 26, 41). In contrast to its family members p53 and p73, p63 plays an essential role in the formation and maintenance of various stratified epithelia, including skin and its appendages, in both humans and mice. In humans, heterozygous mutations of the gene cause a number of malformation syndromes whose phenotypic features include skin defects, mammary gland hypoplasia, and limb and craniofacial abnormalities (36). Mice lacking the p63 gene are devoid of all squamous epithelia and their derivatives (skin, hair, and teeth, as well as mammary, lachrymal, and salivary glands), and they die from dehydration shortly after birth (26, 41). In addition, limb formation is severely impaired, probably due to defects in the apical ectodermal ridge, a specialized epithelium required for limb outgrowth.
The p63 gene encodes a tetrameric transcription factor that is expressed as at least six isoforms, driven by two independent promoters and with widely different transactivation potentials (2, 29, 40). The first promoter, located upstream from exon 1, drives the expression of transcripts encoding proteins that have an amino-terminal transactivation (TA) domain similar to the TA domain of p53. The other promoter, located more than 30 kb downstream from the first starting site, gives rise to N-terminally truncated (ΔN) products that lack the TA domain. Both TA and ΔN isoforms contain a common transactivation domain, rendering the ΔN isoforms also capable of some transactivation activity (8, 10, 39).
Alternative splicing gives rise to three different carboxyl termini designated α, β, and γ. Whereas the p63γ isoforms are transcriptionally the most active, at least in vitro, the p63α isoforms contain a longer carboxy-terminal region that includes a protein-protein interaction domain (SAM domain), which is mutated in the Hay-Wells syndrome (24), and a transcription inhibitory domain (TID) that is responsible for dominant negative function toward transactivation by p53 and p63 (32, 40). ΔNp63α is the most abundant isoform class in mature proliferating epithelia such as the skin (17, 30, 40).
Given its selective expression and crucial role in stratified epithelia, understanding of the molecular mechanisms underlying the specific expression of p63 is crucial to unravel the molecular pathways controlling the specification and maintenance of these tissues. A 10-kb human genomic fragment upstream from the ΔNp63 transcription start site is insufficient to mimic endogenous ΔNp63 expression in mammary epithelial cells (11), suggesting that epithelial cell-specific elements may reside elsewhere in the genome.
With the completion of genomic sequences from several vertebrate species, a number of strongly conserved genomic regions can be identified as candidate cis-acting regulatory elements (6). To identify such elements, we performed a functional analysis of evolutionarily conserved sequences located in the p63 genomic region. We isolated a robust keratinocyte-specific enhancer located in intron 5 that recapitulates p63 expression during embryonic development in transgenic mice. The enhancer sequence is evolutionarily conserved in vertebrates from humans to fish and contains a highly conserved binding site for the p63 protein that is essential for its function. p63 associates with the enhancer both in primary keratinocytes and in skin. Reduced p63 expression in primary keratinocytes results in suppression of the enhancer activity, suggesting that p63 regulates its own expression. In addition, we provide strong evidence that a complex balance among transactivating and dominant negative p63 isoforms controls endogenous p63 expression, thus establishing an autoregulatory feedback mechanism. Lastly, we found that the transcription factor AP-2 binds to the enhancer and cooperates with p63 to stimulate enhancer activity.
MATERIALS AND METHODS
Cell cultures, transfection, and reporter assays.
Mouse primary keratinocytes and dermal fibroblasts were isolated from 2-day-old Swiss CD1 mice and cultured as previously described (7, 12). Human primary keratinocytes were grown in supplemented keratinocyte-SFM medium (Invitrogen). NIH 3T3, HT1080, HeLa, HEK293, and HepG2 cells were cultured in Dulbecco's modified Eagle's medium-10% fetal bovine serum. C2C12 cells were grown in Dulbecco's modified Eagle's medium-20% fetal bovine serum. All cell types were transfected using Lipofectamine 2000 (Invitrogen), except for fibroblasts, which were transfected by the calcium phosphate method. Cells were transfected with the thymidine kinase (TK) minimal promoter construct pGL3 containing the various conserved regions (28); effector plasmids (0.5 to 1 μg) for Myc-TAp63γ (40), FLAG-ΔNp63α, FLAG-ΔNp63γ, and FLAG-ΔNp63R304Q, under the control of the cytomegalovirus promoter; shp63 and shp53 (5); and pCMV-Renilla or TK-Renilla (20 ng; Promega). Primary keratinocytes were transfected with stealth small interfering RNA (siRNA) (Invitrogen) using Lipofectamine 2000, following the manufacturer's protocol. Luciferase activity was determined 48 h after transfection with the dual-luciferase reporter assay kit (Promega). Renilla luciferase activity was used to normalize transfection efficiency.
Identification and cloning of conserved noncoding elements.
Identification of conserved noncoding elements was performed with the Multi LAGAN program (http://lagan.stanford.edu) using NCBI Build 33 for the mouse genome, NCBI Build 35 for the human genome, the February 2004 chicken draft assembly produced by the Genome Sequencing Center at Washington University (St. Louis, Mo.), and the whole-genome shotgun assembly for Fugu rubripes (v.3.0; August 2002). The mouse trp63 genome region from kb −100 to +10 was the reference sequence. The parameters used selected contiguous segments of 100 bp that have a minimum of 75% identity between the mouse and human genomes. Genomic regions had a PhastCons score equal to or above 60, taking into account the lowest score among the exons (exon 9 had a score of 64). Each conserved element was amplified from mouse genomic DNA using oligonucleotide primers designed just outside the conserved region (see Table S1 in the supplemental material), and the sequence was verified and cloned in the TK minimal promoter-Luc vector (28).
Generation of transgenic mice and β-galactosidase staining.
Four copies of the C40 enhancer were subcloned in tandem in the modified β-globin-lacZ vector p1229 (42). The resulting construct was tested for β-galactosidase activity by transient transfection in mouse primary keratinocytes. To generate transgenic mice, the construct was linearized, purified, and injected into fertilized oocytes of DBA × C57BL/6 mice at the CBRC Transgenic Facility (Massachusetts General Hospital, Boston, MA). Integration of the transgene was confirmed by PCR as previously described (21). Founders were backcrossed with C57BL/6 mice to establish lines. Mice were housed and treated according to the guidelines of the local Institutional Animal Care and Use Committee. β-Galactosidase staining was performed as described elsewhere (21).
Constructs and analysis of transcription factor binding site.
The ΔNp63α and -γ expression vectors were obtained by amplifying the coding sequences that lacked the ATG from keratinocyte cDNA and cloning in frame in pCMVFLAG2 (Sigma). The constructs were sequence verified and tested for their expression and transactivating properties. The ΔNp63γ R304Q mutant was obtained by site-directed mutagenesis (see the supplemental material). The ΔNp63 promoter (4.4 kb from the start codon) was amplified from genomic DNA and cloned in pGL3-basic Luc. A 2.2-kb promoter region was obtained by deletion using the restriction enzyme SacI. Expression vector short hairpin RNAs (shRNAs) for p63 were generated by insertion in the pSUPER vector (5) of double-stranded oligonucleotides containing the specific target sequence (see the supplemental material for oligonucleotide sequences). Analysis of transcription factor binding sites was performed using a combination of MatInspector (GenomatixSuite 3.1.0) (31), TRANSFAC (TRANSFAC Professional 8.2) (16), and the binding factor identification algorithm (A. Ambesi and D. Di Bernardo, unpublished results), a novel algorithm that takes account of conservation among species.
Immunoblotting analysis.
For induction of the endogenous p63 gene, HeLa cells were transfected with 2 μg expression vectors carrying p53, ΔNp63α, ΔNp63γ, ΔNp63γR304Q, or an empty vector as the control, in the presence of Lipofectamine 2000 (Invitrogen). For RNA interference experiments, HeLa cells were cotransfected with 0.2 μg of pCMV-ΔNp63α or pcDNA3myc-TAp63γ and 2 μg of empty pSUPER vector, pSUPER p53, or various pSUPER p63 constructs; 20 μg of protein lysates was run on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gel and transferred on an Immobilon-P transfer membrane (Millipore). The membrane was probed with anti-p63 monoclonal antibodies (4A4; Santa Cruz Biotechnology) and with anti-ERK antibodies (Santa Cruz Biotechnology) or with antitubulin antibodies (Santa Cruz Biotechnology) as loading controls.
Real-time RT-PCR.
Forty-eight hours after transfection, RNA was extracted using TRIzol reagent (Invitrogen) and treated with RNase-free DNase I (Promega). cDNA was synthesized using Superscript II (Invitrogen) and random primers. Two-step real-time reverse transcription (RT)-PCR was performed using the SYBR Green PCR core kit (Applied Biosystems). Expression of the exogenous and endogenous p63 transcripts, as well as glyceraldehyde-3-phosphate dehydrogenase and green fluorescent protein, was quantified. pCMV-GFP (0.1 μg) was cotransfected to normalize for transfection efficiency. Oligonucleotide sequences are given in the supplemental material.
Chromatin immunoprecipitation.
Approximately 3 × 106 mouse keratinocytes or dermal fibroblasts were fixed with 1% formaldehyde in growth medium at 37°C for 10 min. For chromatin immunoprecipitation in vivo, total skin was isolated from 1-day-old CD1 mice and fixed immediately with 1% formaldehyde for 20 min at room temperature. Tissue disaggregation was obtained by chopping tissue into small pieces and by using a Dounce homogenizer. Extracts were extensively sonicated on ice to obtain DNA fragments ranging from 400 to 800 bp in length. Chromatin was immunoprecipitated as in the Upstate protocol (http://www.upstate.com). Immunoprecipitation was performed using anti-p63 (H-137; Santa Cruz Biotechnology), anti-p53 (Ab-7; Oncogene Research Products), and anti-ERK-1 (K23; Santa Cruz Biotechnology) antibodies. Real-time PCR was performed using the SYBR Green PCR master mix in an ABI PRISM 7000 (Applied Biosystems). Oligonucleotide sequences are given in the supplemental material.
Electrophoretic mobility shift assays.
Total protein extracts were obtained from HEK293 cells 48 h after transfection with pcDNA3.1 AP-2 γ (25) or with an unrelated control, by centrifugation in 40 mM Tris (pH 7.5), 1 mM EDTA, and 150 mM NaCl. The cell pellets were then lysed in a buffer containing 10 mM HEPES (pH 7.9), 400 mM NaCl, 0.1 mM EGTA (pH 7.8), 5% glycerol, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride and centrifuged for 30 min at 4°C (12,000 × g). The protein concentration was determined using the Bio-Rad DC protein assay (Bio-Rad). The double-stranded oligonucleotide (CCCCATGGCCTGCAGCGTTTACGTAGAAATTGGGGATG) was labeled with γ-32P-labeled ATP and T4 polynucleotide kinase and used as the probe. The binding reactions were carried out in 10 mM HEPES (pH 7.9), 10% glycerol, 0.1 mM EDTA, 8 mM MgCl2, 1 mM dithiothreitol, and 0.15 μg/ml of poly(dI-dC) for 30 min at room temperature. DNA-protein complexes were resolved on a 6% nondenaturing polyacrylamide gel and visualized by autoradiography. For DNA competition experiments, unlabeled double-stranded oligonucleotide was added 10 min before the probe. As unlabeled competitor, the following oligonucleotide primer was used: CCCCATGCGTTGCAGCGTTTACGTAGAAATTGGGGATG.
RESULTS
Identification of evolutionarily conserved genomic sequences that act as keratinocyte-specific enhancers in the p63 genomic locus.
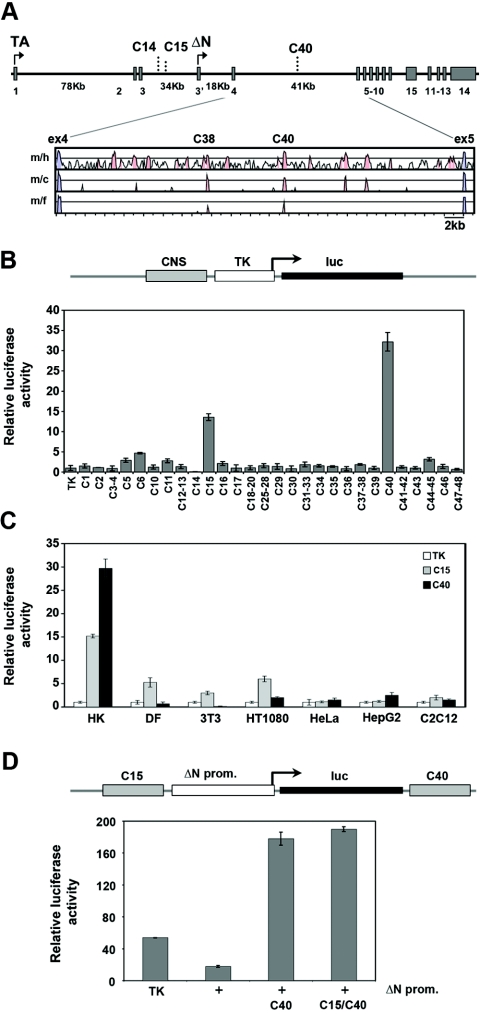
The mouse p63 gene (Trp63) spans 205 kb of genomic DNA on chromosome 16, and contains 16 exons, four large introns, and two independent promoters that drive the expression of the TA and the ΔN isoforms (Fig. 1A). Neither promoters nor nearby conserved sequences (Table 1) were sufficient to confer keratinocyte-specific expression on a luciferase reporter gene (data not shown).
FIG. 1.
Identification of candidate enhancer regions in the p63 locus. (A) Diagram of the p63 genome organization. Exons and functional noncoding elements (C14, C15, and C40) are indicated. TAp63 transcripts start from exon 1, whereas ΔNp63 transcripts start from the alternative exon 3′ (arrows). The sizes of large introns are indicated in kb. A VISTA plot of the alignment of p63 intron 5 in different species is shown. Exons are indicated in blue, and the conserved noncoding regions are in pink. The mouse genomic sequence (m) was compared to the human (h), chicken (c), and Fugu (f) sequences by MultiLagan and is represented on the x axis. The y axis represents percent identity, with a scale between 50% and 100%. (B) Enhancer/silencer activity of the conserved noncoding elements in the p63 genomic locus. Each conserved noncoding sequence (CNS) was cloned in front of a TK minimal promoter that drives the expression of the luciferase reporter. The transactivation activity of each element was assayed in mouse primary keratinocytes by transient-transfection and luciferase assays. The transfection value of the TK promoter was set at 1. (C) The C15 and C40 enhancers were tested in human primary keratinocytes (HK), mouse dermal fibroblasts (DF), and NIH 3T3, HT1080, HeLa, HepG2, and C2C12 cells. (D) The activity of the ΔNp63 promoter (2.2 kb upstream of the start codon; +) was tested in mouse primary keratinocytes in the presence or in the absence of the C40 element alone or the C40 and C15 elements and compared with that of the TK minimal promoter (TK). The pGL3 promoterless construct value was set at 1. All transfection data are expressed as relative luciferase activity corrected for transfection efficiency, using the cytomegalovirus-Renilla reporter as the internal control, and are representative of at least three independent experiments.
TABLE 1.
Evolutionarily conserved genomic elements in the p63 locus
| Conserved sequence block | Position in mouse sequence | Length in mice (bp) | Identity (%) with human sequence | Identity with chicken gene | Identity with fugu gene | Conserved sequence block | Position in mouse sequence | Length in mice (bp) | Identity (%) with human sequence | Identity with chicken gene | Identity with fugu gene | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 | −81276 | 301 | 79.1 | Ab | A | C25 | 117813 | 334 | 75.1 | A | A | |
| C2 | −69112 | 323 | 82.0 | A | A | C26 | 118155 | 102 | 75.5 | A | A | |
| C3 | −61994 | 436 | 78.0 | P | A | C27 | 118612 | 148 | 75.7 | P | A | |
| C4 | −61549 | 101 | 75.2 | A | A | C28 | 118880 | 252 | 79.0 | A | A | |
| C5 | −23704 | 382 | 80.9 | P | A | C29 | 125167 | 470 | 82.1 | P | A | |
| C6 | −3829 | 257 | 85.6 | A | A | C30 | 127699 | 338 | 75.7 | A | A | |
| C7a | −1717 | 437 | 78.5 | P | A | C31 | 133418 | 100 | 75.8 | A | A | |
| C8a | −1280 | 200 | 75.0 | P | A | C32 | 133585 | 278 | 79.5 | P | A | |
| C9a | −666 | 689 | 79.4 | P | A | C33 | 133968 | 201 | 78.1 | P | A | |
| C10 | 2462 | 165 | 83.0 | A | A | C34 | 141708 | 409 | 79.7 | A | A | |
| C11 | 23985 | 316 | 82.0 | A | A | C35 | 143848 | 350 | 77.1 | P | A | |
| C12 | 62281 | 248 | 78.2 | A | A | C36 | 145277 | 283 | 78.1 | A | A | |
| C13 | 62615 | 336 | 81.5 | P | A | C37 | 151138 | 179 | 75.1 | A | A | |
| C14 | 90480 | 631 | 82.3 | P | A | C38 | 151534 | 238 | 79.4 | P | P | |
| C15 | 95830 | 187 | 75.3 | P | A | C39 | 153501 | 290 | 76.6 | A | A | |
| C16 | 100054 | 204 | 76.0 | A | A | C40 | 159458 | 264 | 88.8 | P | P | |
| C17 | 105908 | 203 | 77.8 | A | A | C41 | 165358 | 139 | 75.1 | A | A | |
| C18 | 108206 | 200 | 81.5 | A | A | C42 | 165691 | 342 | 80.1 | P | A | |
| C19 | 108441 | 126 | 79.4 | A | A | C43 | 167904 | 378 | 77.5 | P | A | |
| C20 | 108606 | 107 | 79.4 | A | A | C44 | 189508 | 204 | 75.5 | A | A | |
| C21a | 113833 | 100 | 75.8 | A | A | C45 | 189753 | 257 | 76.7 | A | A | |
| C22a | 115080 | 127 | 75.2 | A | A | C46 | 214848 | 227 | 75.3 | A | A | |
| C23a | 115431 | 108 | 77.8 | A | A | C47 | 216140 | 341 | 80.4 | A | A | |
| C24a | 117300 | 473 | 82.5 | P | A | C48 | 216486 | 100 | 75.0 | A | A |
These conserved noncoding regions were cloned as part of either the TA (C7 to C9) or ΔN (C21 to C24) promoter.
A, absent; P, present.
To locate conserved noncoding sequences that might regulate p63 transcription, a 315-kb region of the mouse genome encompassing the p63 locus was compared to the corresponding genomic sequence of humans, rats, dogs, chickens, and pufferfish (Fugu rubripes) using a combination of LAGAN alignment (4) and the PhastCons program (33) (see Table S1 in the supplemental material). Forty-one highly conserved elements were cloned into a reporter vector upstream from the thymidine kinase minimal promoter. Their ability to induce or repress promoter activity was tested by transient transfection and luciferase assays in mouse primary keratinocytes, which express high levels of p63. Most of the conserved regions did not display any activity. In contrast, elements C15 (187 bp long; 75.3% identity with human sequence) and C40 (264 bp; 88.3%) strongly induced transcriptional activity of the minimal TK promoter, while a single element, C14 (631 bp; 82.3%), acted as a repressor (Fig. 1B).
To assess whether the transcription-modulatory activity of these elements was cell type specific, we tested them in several cell types. Human primary keratinocytes, which, like their murine counterparts, express high levels of endogenous p63, were compared to cells that express p63 at very low to undetectable levels (data not shown). C40 exerted a strong enhancer activity in human primary keratinocytes but was inactive in all other cell types, including primary dermal fibroblasts (Fig. 1C). In contrast, the C15 enhancer, whose activity was less robust than that of C40 in human and mouse primary keratinocytes, was also active in fibroblasts and fibrosarcoma cells, albeit less so than in keratinocytes (Fig. 1C). As in mouse primary keratinocytes, C14 acted as a strong repressor in all cell types tested (data not shown), and thus its activity was not characterized further.
C40 enhancer was also assayed in the context of the ΔNp63 promoter, as ΔNp63 is highly expressed in keratinocytes. Promoter activity was strongly induced by C40 independently of its orientation (Fig. 1D and data not shown). In contrast, addition of the C15 element did not further increase the promoter activity in the presence of C40. For this reason further studies were focused on the C40 element.
C40 enhancer recapitulates p63 expression in transgenic mice.
We next asked whether the C40 element is active in vivo and whether its activity mimics the pattern of p63 expression occurring during embryogenesis, i.e., in the ectodermal surfaces of the limb buds, branchial arches, and epidermal appendages (26, 41).
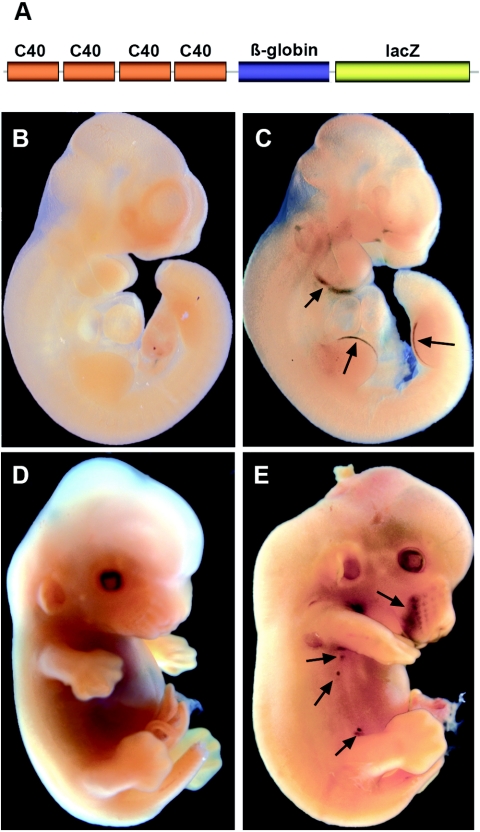
Towards this aim, we engineered transgenic mice harboring four copies of the C40 sequence coupled to the heterologous β-globin minimal promoter to drive a lacZ reporter gene (Fig. 2A). The construct was first tested for its enhancer activity in cell cultures and was found to possess the same specificity as the single-copy element in keratinocytes and fibroblasts (data not shown). Seven independent lines expressed the transgene in a similar pattern and showed remarkable tissue-specific lacZ expression, mostly recapitulating the expression of endogenous p63. lacZ expression was already detectable at embryonic day 8.5 (E8.5) in the branchial arches (data not shown). At E10.5, β-galactosidase staining was restricted to the first and second branchial arches and to the apical ectodermal ridge of both the fore- and hind limbs (Fig. 2C). At E13.5, β-galactosidase staining was still present at the tip of the developing limbs and was also detected in the mammary buds and developing whiskers (Fig. 2E), consistent with the p63 expression pattern previously reported for the endogenous gene. At E15.5, the lacZ reporter was expressed in the developing digits, in the epithelial cells of the nasal cavity, the whiskers, the external genitalia, and to a lesser extent in the skin and hair follicles throughout the body (Fig. 3B and C).
FIG. 2.
Activity of the C40 enhancer in the developing embryo. (A) Schematic structure of the C40 transgenic construct containing four copies of the C40 enhancer in front of the minimal β-globin promoter driving expression of the lacZ reporter gene. Embryos were processed for β-galactosidase activity. At E10.5 lacZ was expressed only in the branchial arches and the apical ectodermal ridge of the fore- and hind limb bud (C; arrows), whereas no expression was detected in a negative littermate (B). At E13.5, lacZ expression was also present in the mammary buds, in the skin around the mouth, and in the lower vibrissae (E, arrows). Residual staining is visible in the distal limb bud and in the neck. There was no staining in a wild-type littermate (D).
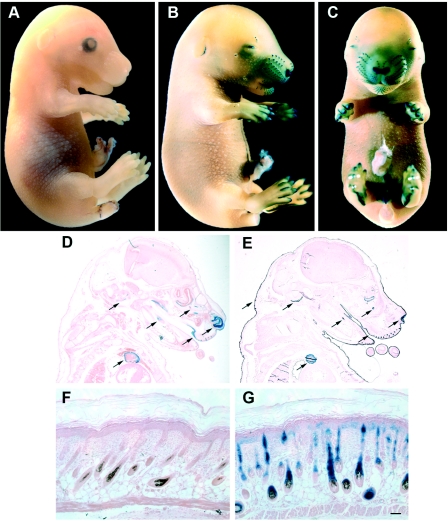
FIG. 3.
Activity of the C40 enhancer in developing tissues. At E15.5, enhancer activity was present in the developing digits, whiskers, and snout skin, as well as in the external genitalia (B and C); no expression was detected in the wild-type counterpart (A). (D) Histological sections were obtained after β-galactosidase staining and were subsequently paraffin embedded and counterstained with eosin. Staining was present in the olfactory and oral epithelium, in the tooth bud and in the atrium. (E) An adjacent section from the same embryo was immunostained using anti-p63 antibodies, after β-galactosidase staining. Arrows indicate p63-expressing tissues and/or the C40 enhancer activity. In newborn transgenic mouse skin the enhancer activity was exclusively localized in the epithelial component, with intense β-galactosidase staining in the hair follicle, and weaker staining in the epidermis (G), whereas no staining was detected in wild-type littermates (F). Scale bar, 50 μm.
Sectioning revealed strong lacZ expression in the olfactory epithelium, as well as in the oral cavity, in the tooth primordial, and in the developing heart (Fig. 3D). p63 expression in these tissues, including the heart, was confirmed by immunohistochemistry with anti-p63 antibodies (Fig. 3E). In embryonic skin, lacZ expression was strongest in the follicles of the developing vibrissae and was present to a lesser extent in the epidermis and in the hair buds throughout the body. These data were confirmed by immunofluorescence using a β-galactosidase-specific antibody (data not shown). Overall, the enhancer activity was highly tissue specific and was not seen in surrounding mesenchyme, cartilage, or other nonepithelial cell types. Similarly, in newborn skin β-galactosidase staining was strong in the hair follicle, less intense in the epidermis, and absent in the dermis (Fig. 3G).
Thus, C40 contained the information required for tissue- and differentiation-specific gene expression in a pattern that markedly resembled that of the endogenous p63 gene.
p63 is required for C40 enhancer activity.
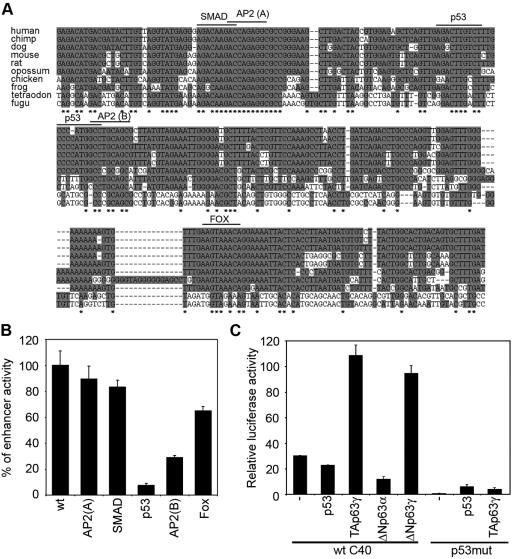
Multiple alignment of the mouse C40 enhancer sequence with the DNA sequences from several vertebrate species, including Gallus gallus, Xenopus laevis, Tetraodon nigroviridis, and Fugu rubripes, revealed a remarkably high sequence identity (Fig. 4A), which further supports its role as a cis-acting element. Extensive bioinformatics analyses of the C40 enhancer revealed a number of putative sequence motifs for known transcription factors, including SMAD, AP-2 (two sites), p53, and the transcription factors of the forkhead Fox family (Fig. 4A). To evaluate whether any of these sequence motifs is involved in the enhancer activity, we mutated three base pairs of each binding core by site-directed mutagenesis and tested the mutant C40 elements versus the wild type in keratinocytes (Fig. 4B). Mutations in the p53-binding site inhibited transcription by 90%, whereas mutations in the AP-2(B) binding site partially impaired the enhancer activity.
FIG. 4.
Evolutionary conservation of the C40 enhancer and its functional response to p63. (A) Multiple sequence alignment of the C40 enhancer showing a high level of conservation among several vertebrate sequences. Conserved nucleotides are shown in gray, and nucleotides that are identical in all species are indicated by stars. The conserved binding sites for SMAD, AP-2(A) and AP-2(B), p53, and Fox are indicated. (B) Mutations in the conserved binding sites were tested by transient transfection in mouse primary keratinocytes for their ability to affect C40 enhancer activity. The C40-TK-Luc construct was mutated in the indicated binding site. Data are presented as percentages of enhancer activity compared to that with the wild-type control (wt). (C) p63 controls the C40 enhancer activity in mouse primary keratinocytes. Wild-type C40-TK-Luc (wt C40) was transiently transfected into keratinocytes in the absence (−) or in the presence of expression constructs for p53, TAp63γ, ΔNp63γ, and ΔNp63α. As a control, the C40 enhancer carrying a mutation in the p53 binding site (p53mut) was transfected alone or with TAp63γ.
Given the strong effect obtained by mutating the putative p53-binding site, we then asked whether p53 was sufficient and/or required for C40 enhancer activity. In fibroblasts, in which the enhancer is inactive, p53 expression caused only a modest increase of enhancer activity (fivefold induction; data not shown). Moreover, in p53-null primary keratinocytes C40 was as active as in wild-type keratinocytes (see Fig. S1 in the supplemental material), suggesting that p53 is not a crucial regulator of C40 activity.
Since p63 can bind to the p53 binding site (3, 13, 29, 40), we evaluated whether p63 rather than p53 regulates C40 enhancer activity. In fibroblasts, expression of TAp63γ induced the C40-TK-Luc construct more efficiently than p53 (30-fold) (data not shown). Importantly, while p53 expression did not induce C40 activity in keratinocytes, exogenous TAp63γ and ΔNp63γ isoforms strongly induced C40 activity, whereas ΔNp63α suppressed it (Fig. 4C), consistent with their previously described roles as transactivator and dominant negative factors, respectively. Mutation of the putative p53 binding site resulted in complete suppression of TAp63γ-mediated activation (Fig. 4C), demonstrating that the identified p53 binding site is essential for p63 activity.
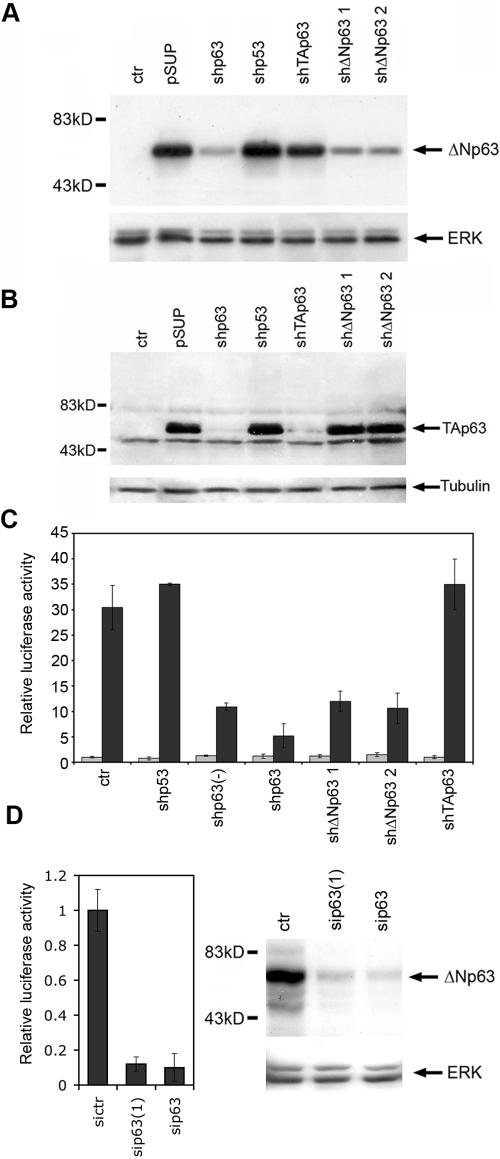
We next investigated whether p63 is required for C40 activity in keratinocytes. Because primary keratinocytes cannot be efficiently isolated from p63 null mice, we engineered pSUPER constructs expressing p63-specific short hairpin RNAs either targeted to a region in common to all isoforms (shp63) or designed to interfere specifically with TAp63 (shTAp63) or ΔNp63 (shΔNp63) (Fig. 5A and B). In primary keratinocytes, shp63, but not shp53, markedly inhibited C40 enhancer activity in a dose-dependent manner, but did not affect the TK minimal promoter (Fig. 5C), demonstrating that p63 is required for C40 enhancer activity. A similar strong inhibition of C40 enhancer activity was obtained by transfecting two independent p63-specific stealth siRNAs, which were able to efficiently suppress the endogenous p63 expression at the RNA and protein levels (Fig. 5D and data not shown).
FIG. 5.
p63 is required for C40 enhancer activity. (A) Downregulation of the ΔNp63 protein by specific shRNAs. HeLa cells were either untransfected (ctr) or transfected with constructs expressing ΔNp63 in the presence of the pSUPER empty vector (pSUP) or of the pSUPER vector carrying a short hairpin RNA for p63 (shp63), TAp63, and ΔNp63 (shΔNp63 1 and shΔNp63 2). Immunoblotting of total cell extracts was performed using anti-p63 antibodies. Membranes were reprobed with anti ERK-1 antibodies as a loading control. (B) Downregulation of the TAp63 protein by specific shRNAs. HeLa cells were either untransfected (ctr) or transfected with a construct expressing TAp63γ in the presence of the various pSUPER constructs. Immunoblotting of total cell extracts was performed using anti-p63 antibodies. The membrane was reprobed with antitubulin antibodies as a loading control. (C) shp63 RNA suppresses C40 enhancer activity. TK-Luc (gray bars) or C40-TK-Luc (black bars) constructs were transfected into mouse primary keratinocytes in the absence (ctr) or in the presence of shp53, shp63, a half-dose of shp63 (−), shΔNp63 1 and 2, and shTAp63. The value for the TK promoter transfection in the control was set at 1. (D) Two independent p63-specific siRNAs, sip63(1) and sip63, strongly inhibit C40 enhancer activity (left panel) in parallel with suppression of endogenous p63 protein expression in keratinocytes (right panel). In contrast to plasmid DNA, stealth siRNAs are efficiently transfected in all cells, allowing simultaneous assessment of sip63 efficiently in downregulating endogenous p63. sip63 recognizes the same sequence as sip63, whereas sip63(1) recognizes an independent sequence common to all p63 isoforms. A medium-length GC-rich stealth siRNA was used as a negative control (sictr). The C40 enhancer activity of cells transfected with sictr was set to 1. Immunoblotting was performed as described for panel A.
Given that TAp63γ is a strong transactivator, it could be responsible for C40 activity, even if its expression is very low in keratinocytes. However, a TAp63-specific shRNA did not inhibit the C40 enhancer, whereas two ΔNp63-specific shRNAs severely impaired enhancer activity (Fig. 5C). Taken together, these data suggest that in keratinocytes the C40 enhancer is controlled by p63 and, more specifically, is regulated by ΔNp63 isoforms.
p63 associates in vivo with the C40 enhancer region.
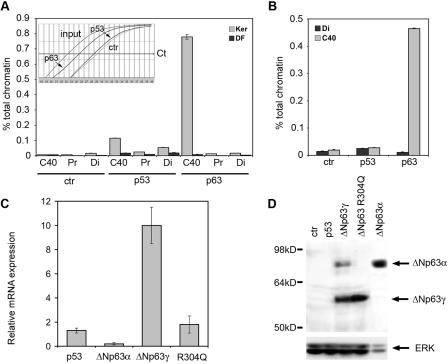
To assess whether p63 plays a significant role in regulating the C40 enhancer, we tested whether p63 binds to the enhancer in intact primary keratinocytes by chromatin immunoprecipitation assays. The protein-DNA complexes were immunoprecipitated with control antibodies and antibodies specific for p63 and p53. The DNA content of immunoprecipitates was then analyzed by real-time PCR. The C40 enhancer region was more enriched in immunoprecipitates obtained with anti-p63 antibodies than with control and anti-p53 antibodies (Fig. 6A). In contrast, in dermal fibroblasts immunoprecipitation with anti-p63 antibodies resulted in no enrichment of the C40 enhancer, consistent with the fact that p63 is not expressed in these cells.
FIG. 6.
p63 binds to the C40 enhancer in the chromatin context and can induce its own transcription. (A) p63 specifically associates with C40 in keratinocyte nuclei. Chromatin extracted from cross-linked mouse primary keratinocytes (Ker) or dermal fibroblasts (DF) was immunoprecipitated in parallel using anti-p63, anti-p53, or anti-ERK (ctr) antibodies. Precipitated DNA samples were used as a template for real-time PCR with oligonucleotide primers recognizing the C40 enhancer, a flanking genomic region 1.8 kb from C40 (Pr), and a distal genomic region (Di: C44 to C45 conserved region). The amount of precipitated DNA was calculated relative to the total input chromatin and expressed as percentage of the total according to the following formula: % total = 2ΔCt × 5, where ΔCt = Ct (input) − Ct (immunoprecipitation), and Ct is cycle threshold, as previously described (9). (Inset) Representative PCR amplification curves. PCR cycles are represented on the x axis, whereas the y axis represents log mean fluorescence. (B) p63 specifically associates with C40 in intact skin. Chromatin extracted from cross-linked newborn mouse skin was immunoprecipitated in parallel using anti-p63, anti-p53, or anti-ERK (ctr) antibodies. The C40 enhancer and the distal genomic region were amplified. (C) p63 proteins affect expression of the endogenous p63 gene. HeLa cells were transiently transfected with expression vectors for the indicated proteins. The mRNA levels of the endogenous p63 transcripts were quantified by real-time RT-PCR using oligonucleotides directed to the 5′ untranslated region of the human ΔNp63 mRNA. Values are expressed as induction (n-fold) of the control transfection. Transfection efficiency was normalized by cotransfecting an expression vector for green fluorescent protein (GFP). (D) Exogenous expression of ΔNp63γ in HeLa cells turns on expression of the endogenous ΔNp63α protein. HeLa cells were transiently transfected as described for panel C. Immunoblotting of total cell extracts was performed using anti-p63 antibodies. As a control, an extract of HeLa cells transfected with ΔNp63α was also run at the same time. Due to the higher stability of ΔNp63α than of ΔNp63γ, approximately one-fifth as much protein extract was loaded. Membranes were reprobed with anti-ERK antibodies as a loading control.
To determine whether p63 directly associates with the endogenous enhancer region in intact skin, we performed quantitative chromatin immunoprecipitation assays on newborn mouse skin. Importantly, we found that p63 efficiently binds the endogenous C40 enhancer in vivo, whereas p53 does not (Fig. 6B). These data demonstrate that p63 associates with the enhancer in the chromatin context both in isolated keratinocytes and in skin.
Selective regulation of p63 gene expression by different p63 protein isoforms.
Regulation of the C40 enhancer by p63 suggests that p63 may be able to regulate its own expression. To directly test this possibility, we expressed exogenous p53 and the various p63 isoforms in HeLa cells and measured expression of the endogenous human ΔNp63 gene using oligonucleotides primers specific for the 5′ untranslated region. HeLa cells expressed low levels of ΔNp63 transcripts as measured by real-time RT-PCR. ΔNp63γ protein expression resulted in robust induction of the endogenous ΔNp63 gene, whereas p53 expression was unable to significantly affect its expression (Fig. 6C), consistent with the ability of p63 proteins, but not of p53, to directly bind and control the C40 enhancer. A missense mutation affecting the DNA-binding domain of ΔNp63γ (R304Q) (35) completely abolished the ability to induce the p63 transcript. ΔNp63α protein repressed endogenous ΔNp63 gene expression (Fig. 6C), in agreement with the dominant negative function exerted by this protein on the C40 enhancer (Fig. 4C).
At the protein level, p63 was undetectable in HeLa cells, and expression of p53 was unable to induce p63 expression. In contrast, exogenous expression of the ΔNp63γ isoform turned on expression of the endogenous ΔNp63α isoform, whereas the mutation in the DNA-binding domain impaired this function (Fig. 6D). Consistent with these observations, in keratinocytes overexpression of ΔNp63α, as achieved by adenovirus infection, significantly impaired endogenous gene expression (data not shown).
Taken together these data indicate that the various p63 isoforms differentially regulate expression of their own transcripts, and this autoregulatory loop is likely to occur through direct p63 binding to the C40 enhancer.
AP-2 cooperates with p63 to modulate C40 enhancer activity.
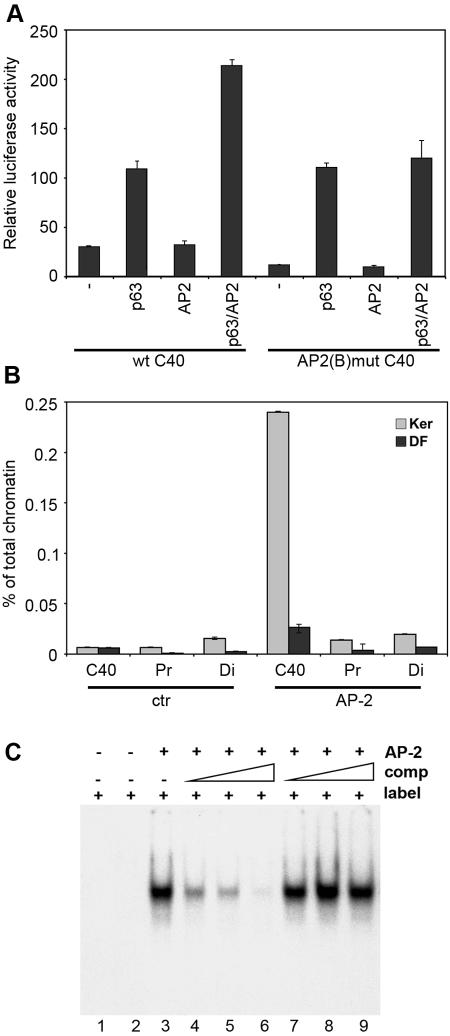
We next investigated the contribution of AP-2 to the C40 enhancer activity, since mutations in the AP-2(B) binding site affect its activity (Fig. 4B). Many epidermal promoters contain functional binding sites for the AP-2 family of transcription factors, which however are expressed not only in skin but also in a number of other cells (20, 22, 27). Overexpression of two members of the AP-2 family, α and γ, did not significantly induce C40 activity in either fibroblasts or keratinocytes (Fig. 7A and data not shown). Interestingly however, AP-2α and -γ expression resulted in induction of C40 activity in the presence of either TAp63γ or ΔNp63γ, suggesting that AP-2 cooperates with p63 to induce enhancer activity (Fig. 7A and data not shown).
FIG. 7.
AP-2 directly binds the C40 enhancer and cooperates with p63 to induce its activity. (A) C40 enhancer activity in the presence of AP-2. The wild-type C40-TK-Luc construct (wt C40) was transiently transfected in keratinocytes in the absence (−) or in the presence of expression constructs for TAp63γ, AP-2γ, or both. As a control, the activity of a C40 enhancer carrying a mutation in the AP-2(B) binding site, AP-2(B)mut C40, was measured. The results are presented as relative luciferase activity, with the TK promoter transfection value set at 1 (TK). Similar results were obtained using AP-2α. (B) AP-2 specifically associates with C40 in keratinocyte nuclei. Chromatin extracted from cross-linked mouse primary keratinocytes (Ker) or dermal fibroblasts (DF) was immunoprecipitated in parallel using anti-AP-2 or anti-ERK (ctr) antibodies and AP-2 binding to the various genomic regions was evaluated as described for Fig. 6A. (C) AP-2 binds to the AP-2(B) motif within the C40 enhancer. The electrophoretic mobility shift assay was performed by incubating radiolabeled AP-2(B) oligonucleotide (label) either by itself (lane 1) or with 6 μg of total cell extract from HEK 293 cells transiently transfected with the AP-2γ expression vector (lanes 3 to 9). Labeled oligonucleotide was challenged with unlabeled wild-type (lanes 4 to 6) or mutated (lanes 7 to 9) oligonucleotide competitors (comp; 15-fold, 20-fold, and 30-fold excesses of each), as indicated by the triangle over each lane.
We used quantitative chromatin immunoprecipitation to investigate AP-2 binding to the C40 element. The C40 enhancer region was enriched in immunoprecipitates obtained with AP-2 antibodies over those with control antibodies, although binding was weaker than for p63, possibly due to less efficient immunoprecipitation (Fig. 7B). We then evaluated whether AP-2 factors could bind directly to the C40 element in vitro. Electrophoretic mobility shift assays revealed that AP-2 binds directly to the portion of the enhancer sequence containing the AP-2(B) binding site (Fig. 7C). Mutations in the AP-2 binding core abolished this interaction.
Thus, AP-2 binds to the enhancer both in vitro and in vivo and cooperates with p63 to induce its activity in keratinocytes.
DISCUSSION
In contrast to its homologue p53, p63 is an essential gene in humans and mice as mutations or deletion of it cause a severe phenotype affecting limbs and/or several stratified epithelia and their derivatives. Thus, an understanding of the regulation of p63 expression and function is of crucial relevance. We describe here the identification and characterization of a long-range enhancer that regulates p63 expression through an autoregulatory mechanism. We took advantage of the completion of genome sequences for several vertebrate species to identify evolutionarily conserved genomic regions in the p63 locus that may be functionally relevant. A functional analysis of these conserved sequences identified three potential cis-regulatory sequences. The C40 enhancer, located 160 kb and 42 kb downstream from the TA and ΔN transcriptional start sites, respectively, provides the first example of a long-range cis-regulatory element specific for keratinocytes. Its functional significance is supported by its remarkable conservation even in lower vertebrates such as chickens, Xenopus, Fugu, and Tetraodon spp.
It has recently been recognized that long-range enhancers can control the transcription of genes located as far as 1 Mb from the enhancer (18). Here we report compelling evidence that the C40 enhancer specifically controls p63 expression, as none of the other genes located within 2.2 Mb from the p63 gene in the mouse has the complicated expression pattern of p63. Remarkably, in transgenic mice during embryonic development, lacZ expression under the control of the C40 enhancer almost entirely overlaps the expression of endogenous p63 in most cells and tissues. lacZ expression was also detected in the embryonic heart, in parallel with the finding that TAp63 is expressed at the RNA level in the adult heart (40). Interestingly, in skin, even though the enhancer activity is confined to the epithelial components, C40 directs expression primarily to the hair follicle and to a lesser extent the epidermis, suggesting that other as-yet-unknown regulatory elements may contribute to the epidermis-specific expression of p63. These unknown elements may reside either in less-conserved genomic regions or in genomic regions outside of the genomic portion taken into consideration. In addition, functional elements that may be essential in vivo may be inactive in transient-transfection assays where the chromatin context is altered. These hypotheses will be the subject of future studies aimed at fully characterizing the transcriptional regulation of the p63 gene.
In addition to the specific pattern of expression elicited by the C40 enhancer during embryogenesis, we present strong evidence of a direct feedback regulation of p63 on its own gene expression through the enhancer. p63 is required for C40 enhancer activity, is sufficient to stimulate the enhancer activity in p63-expressing and -nonexpressing cell types, and binds to it in isolated keratinocytes as well as in skin. In addition, in cells that express very low levels of p63, expression of the ΔNp63γ isoform, but not of ΔNp63α, results in induction of ΔNp63 endogenous gene expression. Binding to the DNA is required for ΔNp63γ-mediated induction of the endogenous p63 genes, as its ability is impaired by a binding-defective ΔNp63γ mutant.
Consistent with its previously described function as dominant negative and with its activity on the enhancer, overexpression of ΔNp63α results in inhibition of the endogenous p63 gene. Using isoform-specific shRNAs, however, we showed that ΔNp63 positively controls the enhancer activity, suggesting either that endogenous ΔNp63α may be required for the enhancer function or that even small amounts of ΔNp63γ in keratinocytes are sufficient to positively regulate the enhancer activity. In other contexts, such as during embryonic development, when the TAp63 transcripts are readily detectable (19; our unpublished observations), the TA isoforms may also play an important role in regulating the enhancer activity. p63 activity and function are complex, as TAp63γ and, to a lesser extent, ΔNp63γ are extremely active transactivators that, similar to p53, induce apoptosis when overexpressed (40). In contrast, the highly abundant ΔNp63α is a stable nontoxic protein that is endowed with both transcription-activating (8, 39), and -repressing (3, 38, 40) functions.
Taken together, these data suggest a model in which a fine balance between the α and γ isoforms exists in keratinocytes in controlling p63 expression, with the γ isoform inducing the C40 enhancer and activating endogenous gene expression, thus bursting the α isoform. In turn, the α isoform may negatively control p63 expression as shown by downmodulation of the p63 gene in response to ΔNp63α overexpression in primary keratinocytes, which is consistent with its ability to inhibit the C40 enhancer.
Two recent studies have pointed to the presence of a conserved p63-binding site in the ΔNp63 promoter sequence (11, 37). Interestingly, we find that TAp63γ expression exerts a modest inhibitory effect on the ΔNp63 promoter, whereas ΔNp63α has a modest stimulatory effect (D. Antonini and C. Missero, unpublished observations), possibly partially counteracting the effect of the enhancer on ΔNp63 expression, but not on TAp63.
As p63 binds to its own enhancer and autoregulation cannot initiate expression of a gene, the initiation of p63 expression must be triggered by other means. p63 expression in zebra fish is mediated by bone morphogenetic protein signaling (1). In the mouse C40 enhancer, mutation of the binding site for SMAD does not affect the enhancer activity in keratinocytes, which may be due to the fact that in isolated cells p63 expression has already been initiated and that it is by itself sufficient to maintain the loop. These data raise the interesting possibility that bone morphogenetic protein signaling may also be involved in initial transcription activation of p63 in the mouse, possibly through the highly conserved SMAD site that we identified. After the first induction, p63 is required in cooperation with AP-2 to maintain its own expression by a positive autoregulatory feedback.
AP-2 participates in the control of the C40 enhancer, as a mutation in the AP-2 binding sites affects enhancer activity. In addition, AP-2 directly binds the C40 enhancer in keratinocytes and cooperates with p63 to induce enhancer activity. The relevance of AP-2 in directing skin-specific expression is widely documented, as many epidermis-specific upstream regulatory regions contain functional binding sites for AP-2 (14, 15, 20, 23, 34). Our data indicate that AP-2 may fulfill its function in skin at least in part through regulation of p63 expression.
In conclusion, identification of the mechanisms that regulate this novel keratinocyte-specific enhancer shed light on the complex regulation of p63 expression during embryonic development and in keratinocytes.
Supplementary Material
Acknowledgments
We thank Andrea Ballabio, Graciana Diez-Roux, and Domenico Salvatore for their critical reading of the manuscript and Stefano Piccolo for helpful discussion. We are grateful to Ronald J. Weigel for CMV AP-2α and -γ constructs. We are indebted to Jean Gilder (Scientific Communication) for text editing.
T.D.P. is the recipient of an AIRC (Associazione Italiana per la Ricerca sul Cancro) fellowship. This work was supported by grants from the Italian Telethon Foundation (TCMP14TELB), from the Ministry of Instruction, University and Research (MIUR FIRB), from the European Union (LSHG-CT-2004-511990) to C.M., from AIRC to M.Z., and from the NIH (AR45284) to J.L.B. J.L.B. was also funded by the Cutaneous Biology Research Center through the Massachusetts General Hospital/Shiseido Co. Ltd. agreement.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bakkers, J., M. Hild, C. Kramer, M. Furutani-Seiki, and M. Hammerschmidt. 2002. Zebrafish deltaNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev. Cell 2:617-627. [DOI] [PubMed] [Google Scholar]
- 2.Bamberger, C., and H. Schmale. 2001. Identification and tissue distribution of novel KET/p63 splice variants. FEBS Lett. 501:121-126. [DOI] [PubMed] [Google Scholar]
- 3.Barbieri, C. E., C. A. Perez, K. N. Johnson, K. A. Ely, D. Billheimer, and J. A. Pietenpol. 2005. IGFBP-3 is a direct target of transcriptional regulation by deltaNp63alpha in squamous epithelium. Cancer Res. 65:2314-2320. [DOI] [PubMed] [Google Scholar]
- 4.Brudno, M., C. B. Do, G. M. Cooper, M. F. Kim, E. Davydov, E. D. Green, A. Sidow, and S. Batzoglou. 2003. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13:721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 6.Dermitzakis, E. T., A. Reymond, and S. E. Antonarakis. 2005. Conserved non-genic sequences—an unexpected feature of mammalian genomes. Nat. Rev. Genet. 6:151-157. [DOI] [PubMed] [Google Scholar]
- 7.Dotto, G. P., R. A. Weinberg, and A. Ariza. 1988. Malignant transformation of mouse primary keratinocytes by Harvey sarcoma virus and its modulation by surrounding normal cells. Proc. Natl. Acad. Sci. USA 85:6389-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellisen, L. W., K. D. Ramsayer, C. M. Johannessen, A. Yang, H. Beppu, K. Minda, J. D. Oliner, F. McKeon, and D. A. Haber. 2002. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol. Cell 10:995-1005. [DOI] [PubMed] [Google Scholar]
- 9.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghioni, P., F. Bolognese, P. H. Duijf, H. Van Bokhoven, R. Mantovani, and L. Guerrini. 2002. Complex transcriptional effects of p63 isoforms: identification of novel activation and repression domains. Mol. Cell. Biol. 22:8659-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmes, D. C., E. Bresnick, E. A. Lubin, J. K. Watson, K. E. Heim, J. C. Curtin, A. M. Suskind, J. Lamb, and J. DiRenzo. 2003. Positive and negative regulation of deltaN-p63 promoter activity by p53 and deltaN-p63-alpha contributes to differential regulation of p53 target genes. Oncogene 22:7607-7616. [DOI] [PubMed] [Google Scholar]
- 12.Hennings, H., D. Michael, C. Cheng, P. Steinert, K. Holbrook, and S. H. Yuspa. 1980. Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell 19:245-254. [DOI] [PubMed] [Google Scholar]
- 13.Ihrie, R. A., M. R. Marques, B. T. Nguyen, J. S. Horner, C. Papazoglu, R. T. Bronson, A. A. Mills, and L. D. Attardi. 2005. Perp is a p63-regulated gene essential for epithelial integrity. Cell 120:843-856. [DOI] [PubMed] [Google Scholar]
- 14.Jang, S. I., and P. M. Steinert. 2002. Loricrin expression in cultured human keratinocytes is controlled by a complex interplay between transcription factors of the Sp1, CREB, AP1, and AP2 families. J. Biol. Chem. 277:42268-42279. [DOI] [PubMed] [Google Scholar]
- 15.Kaufman, C. K., S. Sinha, D. Bolotin, J. Fan, and E. Fuchs. 2002. Dissection of a complex enhancer element: maintenance of keratinocyte specificity but loss of differentiation specificity. Mol. Cell. Biol. 22:4293-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kel, A. E., E. Gossling, I. Reuter, E. Cheremushkin, O. V. Kel-Margoulis, and E. Wingender. 2003. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 31:3576-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King, K. E., R. M. Ponnamperuma, T. Yamashita, T. Tokino, L. A. Lee, M. F. Young, and W. C. Weinberg. 2003. DeltaNp63alpha functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene 22:3635-3644. [DOI] [PubMed] [Google Scholar]
- 18.Kleinjan, D. A., and V. van Heyningen. 2005. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76:8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koster, M. I., S. Kim, A. A. Mills, F. J. DeMayo, and D. R. Roop. 2004. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 18:126-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leask, A., C. Byrne, and E. Fuchs. 1991. Transcription factor AP2 and its role in epidermal-specific gene expression. Proc. Natl. Acad. Sci. USA 88:7948-7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, D., D. M. Prowse, and J. L. Brissette. 1999. Association between mouse nude gene expression and the initiation of epithelial terminal differentiation. Dev. Biol. 208:362-374. [DOI] [PubMed] [Google Scholar]
- 22.Luo, T., M. Matsuo-Takasaki, M. L. Thomas, D. L. Weeks, and T. D. Sargent. 2002. Transcription factor AP-2 is an essential and direct regulator of epidermal development in Xenopus. Dev. Biol. 245:136-144. [DOI] [PubMed] [Google Scholar]
- 23.Maytin, E. V., J. C. Lin, R. Krishnamurthy, N. Batchvarova, D. Ron, P. J. Mitchell, and J. F. Habener. 1999. Keratin 10 gene expression during differentiation of mouse epidermis requires transcription factors C/EBP and AP-2. Dev. Biol. 216:164-181. [DOI] [PubMed] [Google Scholar]
- 24.McGrath, J. A., P. H. Duijf, V. Doetsch, A. D. Irvine, R. de Waal, K. R. Vanmolkot, V. Wessagowit, A. Kelly, D. J. Atherton, W. A. Griffiths, S. J. Orlow, A. van Haeringen, M. G. Ausems, A. Yang, F. McKeon, M. A. Bamshad, H. G. Brunner, B. C. Hamel, and H. van Bokhoven. 2001. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum. Mol. Genet. 10:221-229. [DOI] [PubMed] [Google Scholar]
- 25.McPherson, L. A., and R. J. Weigel. 1999. AP2alpha and AP2gamma: a comparison of binding site specificity and trans-activation of the estrogen receptor promoter and single site promoter constructs. Nucleic Acids Res. 27:4040-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills, A. A., B. Zheng, X. J. Wang, H. Vogel, D. R. Roop, and A. Bradley. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398:708-713. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell, P. J., P. M. Timmons, J. M. Hebert, P. W. Rigby, and R. Tjian. 1991. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 5:105-119. [DOI] [PubMed] [Google Scholar]
- 28.Ohno, M., M. Zannini, O. Levy, N. Carrasco, and R. di Lauro. 1999. The paired-domain transcription factor Pax8 binds to the upstream enhancer of the rat sodium/iodide symporter gene and participates in both thyroid-specific and cyclic-AMP-dependent transcription. Mol. Cell. Biol. 19:2051-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osada, M., M. Ohba, C. Kawahara, C. Ishioka, R. Kanamaru, I. Katoh, Y. Ikawa, Y. Nimura, A. Nakagawara, M. Obinata, and S. Ikawa. 1998. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat. Med. 4:839-843. [DOI] [PubMed] [Google Scholar]
- 30.Parsa, R., A. Yang, F. McKeon, and H. Green. 1999. Association of p63 with proliferative potential in normal and neoplastic human keratinocytes. J. Investig. Dermatol. 113:1099-1105. [DOI] [PubMed] [Google Scholar]
- 31.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serber, Z., H. C. Lai, A. Yang, H. D. Ou, M. S. Sigal, A. E. Kelly, B. D. Darimont, P. H. Duijf, H. Van Bokhoven, F. McKeon, and V. Dotsch. 2002. A C-terminal inhibitory domain controls the activity of p63 by an intramolecular mechanism. Mol. Cell. Biol. 22:8601-8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siepel, A., and D. Haussler. 2004. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol. Biol. Evol. 21:468-488. [DOI] [PubMed] [Google Scholar]
- 34.Sinha, S., L. Degenstein, C. Copenhaver, and E. Fuchs. 2000. Defining the regulatory factors required for epidermal gene expression. Mol. Cell. Biol. 20:2543-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Bokhoven, H., B. C. Hamel, M. Bamshad, E. Sangiorgi, F. Gurrieri, P. H. Duijf, K. R. Vanmolkot, E. van Beusekom, S. E. van Beersum, J. Celli, G. F. Merkx, R. Tenconi, J. P. Fryns, A. Verloes, R. A. Newbury-Ecob, A. Raas-Rotschild, F. Majewski, F. A. Beemer, A. Janecke, D. Chitayat, G. Crisponi, H. Kayserili, J. R. Yates, G. Neri, and H. G. Brunner. 2001. p63 gene mutations in EEC syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am. J. Hum. Genet. 69:481-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Bokhoven, H., and F. McKeon. 2002. Mutations in the p53 homolog p63: allele-specific developmental syndromes in humans. Trends Mol. Med. 8:133-139. [DOI] [PubMed] [Google Scholar]
- 37.Waltermann, A., N. N. Kartasheva, and M. Dobbelstein. 2003. Differential regulation of p63 and p73 expression. Oncogene 22:5686-5693. [DOI] [PubMed] [Google Scholar]
- 38.Westfall, M. D., D. J. Mays, J. C. Sniezek, and J. A. Pietenpol. 2003. The ΔNp63α phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol. Cell. Biol. 23:2264-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, G., S. Nomoto, M. O. Hoque, T. Dracheva, M. Osada, C. C. Lee, S. M. Dong, Z. Guo, N. Benoit, Y. Cohen, P. Rechthand, J. Califano, C. S. Moon, E. Ratovitski, J. Jen, D. Sidransky, and B. Trink. 2003. DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 63:2351-2357. [PubMed] [Google Scholar]
- 40.Yang, A., M. Kaghad, Y. Wang, E. Gillett, M. D. Fleming, V. Dotsch, N. C. Andrews, D. Caput, and F. McKeon. 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2:305-316. [DOI] [PubMed] [Google Scholar]
- 41.Yang, A., R. Schweitzer, D. Sun, M. Kaghad, N. Walker, R. T. Bronson, C. Tabin, A. Sharpe, D. Caput, C. Crum, and F. McKeon. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398:714-718. [DOI] [PubMed] [Google Scholar]
- 42.Yee, S. P., and P. W. Rigby. 1993. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 7:1277-1289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.