Abstract
Ras proteins are synthesized as cytosolic precursors, but then undergo posttranslational lipid addition, membrane association, and subcellular targeting to the plasma membrane. Although the enzymes responsible for farnesyl and palmitoyl lipid addition have been described, the mechanism by which these modifications contribute to the subcellular localization of Ras is not known. Following addition of the farnesyl group, Ras associates with the endoplasmic reticulum (ER), where palmitoylation occurs in Saccharomyces cerevisiae. The subsequent translocation of Ras from the ER to the plasma membrane does not require the classical secretory pathway or a functional Golgi apparatus. Vesicular and nonvesicular transport pathways for Ras proteins have been proposed, but the pathway is not known. Here we describe a genetic screen designed to identify mutants defective in Ras trafficking in S. cerevisiae. The screen implicates, for the first time, the class C VPS complex in Ras trafficking. Vps proteins are best characterized for their role in endosome and vacuole membrane fusion. However, the role of the class C Vps complex in Ras trafficking is distinct from its role in endosome and vacuole vesicle fusion, as a mitochondrial involvement was uncovered. Disruption of class C VPS genes results in mitochondrial defects and an accumulation of Ras proteins on mitochondrial membranes. Ras also fractionates with mitochondria in wild-type cells, where it is detected on the outer mitochondrial membrane by virtue of its sensitivity to protease treatment. These results point to a previously uncharacterized role of mitochondria in the subcellular trafficking of Ras proteins.
Ras proteins are highly conserved small GTPases that cycle between an active GTP-bound and an inactive GDP-bound state. Depending on the cell type, activation results in cell proliferation, differentiation, and other cellular responses (11, 36). Constitutive activation of Ras contributes to the oncogenic and cellular hypertrophy phenotype of many types of cancer cells (29). Ras proteins localize to the cytoplasmic face of the plasma membrane, with individual Ras subtypes residing in distinct microdomains (45). H-Ras is localized to lipid rafts and caveolae in the GDP-bound state and is redistributed to the bulk membrane upon GTP loading. In contrast, K-Ras is located outside the lipid rafts in both the GDP- and GTP-bound states (12, 23, 45). Ras signaling on the plasma membrane is the most well studied, but Ras also engages effectors on endomembranes (15, 66). This emphasizes the importance of understanding the contribution of subcellular localization to Ras signaling.
Plasma membrane targeting of Ras proteins requires posttranslational modification of the C-terminal CaaX box (C is cysteine, a is any aliphatic residue, and X is the carboxy-terminal residue) and either palmitoylation or the presence of a polybasic domain (13, 18, 24, 58). Farnesylation of the CaaX box cysteine occurs in the cytoplasm and results in targeting of Ras to the cytosolic surface of the endoplasmic reticulum (ER), where the -aaX residues are removed and the C terminus is methylated by an ER protease and a methyltransferase, respectively (7, 18, 26, 31, 55, 57, 58). For most Ras proteins, a cysteine residue or residues adjacent to the CaaX box are palmitoylated, which serves as an additional targeting signal for translocation of Ras proteins from the ER to the plasma membrane (10, 33). Saccharomyces cerevisiae Ras proteins also require a hypervariable domain on their C termini for efficient translocation (19). In the case of K-Ras, the hypervariable domain (polybasic domain) and CaaX modifications are sufficient for plasma membrane targeting (24).
The subcellular trafficking of most membrane-associated proteins and secreted proteins occurs via the classical secretory pathway and involves the ER, the Golgi apparatus, secretory vesicles, and the plasma membrane (32). However, Ras trafficking appears to utilize a different mechanism. In mammalian cells, a fraction of both H-Ras and N-Ras is transported to and from the Golgi complex via a nonvesicular pathway (21, 51, 54). In some cell lines, the plasma membrane localization of H-Ras does not require a functional Golgi complex (74). K-Ras is also directed to the plasma membrane through a Golgi apparatus-independent pathway (2). Like K-Ras, yeast Ras proteins localize to the plasma membrane in the absence of a functional classical secretory pathway (19, 75).
To elucidate the Golgi complex-independent pathway in yeast, we performed a genetic screen designed to identify components involved in the Golgi complex-independent trafficking of Ras proteins. In this report, we describe the isolation of two class C VPS genes and an analysis of their effect on Ras trafficking. The class C Vps complex is composed of Vps11, Vps16, Vps18, and Vps33 and was first uncovered in genetic screens for mutants with altered vacuole morphology (47). Vps33 belongs to the Sec1/Munc family that conveys vesicle fusion specificity by interacting with syntaxins (43). The class C Vps complex also regulates vesicle fusion at the endosome and between the endosome and the vacuole (70). In addition, all of the class C vps deletion strains exhibit growth defects on nonfermentable carbon sources and have defects in the integrity of their mitochondria (68). Unexpectedly, it appears that the mitochondrial defect of class C Vps mutants is responsible for the Ras trafficking defect. This has led us to investigate the role of mitochondria in the subcellular localization of Ras proteins.
MATERIALS AND METHODS
Strains, media, and yeast techniques.
Media were prepared as described previously (61). The yeast strains used in this study are listed in Table 1. Cells were grown in synthetic complete (SC) medium or YPD (1% yeast extract, 2% peptone, and 2% glucose) medium (61). Induction of the GAL1 and -10 promoters was achieved by adding 4% galactose to SC medium. Yeast transformations were done by the lithium acetate procedure (28).
TABLE 1.
Yeast strains used for this study
| Straina | Genotype |
|---|---|
| LRB937 | MATα his3 leu2 ura3-52 sec23-ts |
| LRB938 | MATahis3 leu2 ura3-52 |
| RK511-6B-1 | MATaade2 ura3-52 his3-1 his6 leu2-3 trp1-1 sst1-2 ste18::URA3 |
| RJY1616 | MATaade2 ura3-52 his3-1 his6 leu2-3 trp1-1 sst1-2 ste18 gpa1::KANr |
| RJY1654 | MATahis3 leu2 ura3-52 vps33::KANr |
| RJY1672 | MATahis3 leu2 ura3-52 vps18::KANr |
| RJY1695 | MATahis3 leu2 ura3-52 vps33 end3::KANr |
| RJY1695 | MATahis3 leu2 ura3-52 pep5::KANr |
| RJY1702 | MATahis3 leu2 ura3-52 vps16::KANr |
| RJY1705 | MATahis3 leu2 ura3-52 pep12::KANr |
| RJY1706 | MATahis3 leu2 ura3-52 vam3::KANr |
| RJY1707 | MATahis3 leu2 ura3-52 pep12 vam3::KANr |
| RJY1722 | MATα his3 leu2 lys2 ura3 COP1 mRFP-KANr |
| W303 | MATaade2 his3 leu2 trp1 ura3 |
Strains designated LRB were obtained from L. Robinson (Louisiana State University Health Sciences Center). RK511-6B-1 was obtained from M. E. Linder (Washington University School of Medicine). RJY1722 was obtained from E. K. O'Shea (27). RJY1616 was obtained from RK511-6B-1 by two gene replacements. URA3 was replaced by a loxP-kanMX-loxP cassette. The KAN marker gene was then removed, and STE18 was replaced by another loxP-kanMX-loxP cassette (22). RJY1654, RJY1672, RJY1695, RJY1702, RJY1705, and RJY1706 were obtained from LRB938 by single-step gene replacement with vps33::KAN, vps18::KAN, pep5::KAN, vps16::KAN, pep12::KAN, and vam3::KAN, respectively. RJY1695 was obtained from RJY1654 by single gene replacement with an end3::KAN fragment after KAN marker removal. RJY1707 was obtained from RJY1705 by single gene replacement with a vam3::KAN fragment after KAN marker removal.
Plasmid construction.
YEp55-GFP-Ras2 (B828) was constructed as described previously (19). The GFP(K)CaaX plasmid was created by PCR amplification of the 54-bp fragment encoding the C-terminal 18 amino acids of K-Ras. The fragment was digested with EcoRI and ligated into pGPD-GFP-Ras2 (B701) (33) to create pGPD-GFP(K)CaaX (B1452). The 768-bp GFP(K)CaaX fragment was PCR amplified. It was then digested with BamHI and HindIII and ligated into YEp55c (B19) (53) to create the GFP(K)CaaX plasmid (B1454). The expression of GFP(K)CaaX is under the control of the GAL10 promoter.
To create the Ste18(K)CaaX plasmid, the 270-bp STE18 fragment encoding the N-terminal 70 amino acids of Ste18 was PCR amplified together with the upstream 1,053-bp region. The PCR fragment was digested with BamHI and EcoRI and ligated into YEplac112 (20) to create B1441 (Ste18ΔC). The 54-bp fragment encoding the C-terminal 18 amino acids of K-Ras was digested with EcoRI and ligated into Ste18ΔC digested with EcoRI to create Ste18(K)CaaX (B1452).
To create MET25-GFP-Ras2 (B1510), 380 bp of the MET25 promoter was inserted into the HindIII and XbaI sites of pRS315 (63) to generate pRS-MET25 (B803). RAS2, amplified from wild-type (LBR938) genomic DNA, was then inserted into the XbaI and SacI sites of pRS-MET25 to generate MET25-Ras2 (B805). The 720-bp GFP fragment was PCR amplified from pBS-3GFP-TRP1 (a generous gift from John Cooper, Washington University, St. Louis, MO) and inserted into the XbaI site of MET25-Ras2 to create MET25-GFP-Ras2 (B1510).
For colocalization studies, RFP was isolated by PCR amplification from the genomic DNA of Cop1-mRFP (27). The 940-bp PCR fragment contains sequences at both ends that are homologous to the sequences at both sides of the BamHI site in pRS316 (63). The PCR fragment and BamHI-digested pRS316 were cotransformed into LRB938. After transformation, the PCR fragment was integrated into pRS316 to form pRS316-RFP (B1467). The recombinant plasmid was isolated from yeast and transformed into Escherichia coli DH5α cells. Cox4-RFP was created by PCR amplification of COX4 with the upstream 600-bp region. The 1,168-bp PCR fragment was cotransformed with BamHI-digested pRS316-RFP (B1467) into LRB938. The recombinant plasmid Cox4-RFP (B1468) was isolated from yeast.
Genetic screen for mutants that affect the subcellular localization of Ras.
The screen for Ras trafficking mutants was based on a chimeric protein consisting of the plasma membrane-targeting sequences of Ras fused to the C terminus of the Gγ subunit of the heterotrimeric G protein complex that controls yeast mating. The G protein is composed of three subunits, namely, Gpa1 (Gα), Ste4 (Gβ), and Ste18 (Gγ) (35). Pheromone binding of the receptor results in GTP binding to Gpa1, dissociation of Gα (Gpa1) from Gβγ (Ste4/Ste18), and activation of the downstream mitogen-activated protein kinase cascade. This in turn causes cell cycle arrest and the induction of genes required for conjugation. In the absence of Gpa1, the pathway is constitutively activated, and cells are arrested in G1. This requires plasma membrane localization of the Gβγ (Ste4/Ste18) complex. Thus, plasma membrane localization of Gβγ can be monitored by growth arrest in the absence of Gpa1. This is the basis of the genetic screen for Ras trafficking mutants.
The Ste18(K)CaaX protein (B1452) functions the same as wild-type Ste18 but is targeted to the plasma membrane via the Ras localization sequence. The Ste18(K)CaaX plasmid and the GPA1 URA3 plasmid (pG1301) (39) were introduced into a ste18Δ gpa1Δ double deletion strain (RJY1616). Gpa1 encoded by the GPA1 URA3 plasmid negatively regulates Ste18(K)CaaX, preventing it from causing growth arrest (16), and the plasmid is required for cell growth when both Ste18(K)CaaX transport and signaling are intact. Under these conditions, the wild-type URA3 gene on the plasmid complements the ura3 mutation on the chromosome, and the cells are 5-fluoroorotic acid (5-FOA) sensitive (5). When the transport pathway for Ste18(K)CaaX is disrupted, plasma membrane localization of the protein is prevented, and the mating pathway is not activated. Under these conditions, the loss of Gpa1 does not cause growth arrest, and cells are 5-FOA resistant. Cells (RJY1616) harboring pG1301 (GPA1 URA3) and Ste18(K)CaaX were plated on SC−Trp−Ura (synthetic complete medium lacking tryptophan and uracil) plates and mutagenized with UV light to obtain approximately 90% killing. Plates were incubated at 30°C for 2 days and replicated on SC−Trp plates containing 5-FOA (1.0 g/liter) (5). Colonies growing on 5-FOA-containing plates are cells capable of losing the GPA1 URA3 plasmid and were selected. To test whether these mutants were trafficking mutants, they were transformed with the GFP(K)CaaX plasmid. Mutants in which GFP(K)CaaX and green fluorescent protein-Ras2 (GFP-Ras2) were mislocalized were characterized further.
The wild-type genes corresponding to the trafficking mutants were cloned by complementation, using a YSB32-based genomic library (ATCC 77162). To select for cells incapable of losing the GPA1 URA3 plasmid, transformants were replicated on 5-FOA-containing plates. Cells growing on SC−Trp−Ura but not on SC−Trp, 5-FOA-containing plates were selected. The plasmids that complemented the mutations were isolated and mapped by a combination of deletion analysis and subcloning strategies to identify the open reading frames of the genes.
Preparation of yeast extracts and immunoblot analysis.
Cells were grown in SC medium to exponential phase (A600, ∼0.5 to 1.0). Cells were then collected and lysed in sorbitol buffer (300 mM sorbitol, 100 mM NaCl, 5 mM MgCl2, 10 mM Tris-HCl, pH 7.5) with protease inhibitors (1 mM phenylmethylsulfonyl fluoride and 2.5 μg/ml [each] of chymostatin, leupeptin, aprotinin, and pepstatin) by vortexing with glass beads (425 to 600 μm; Sigma). Unbroken cells and debris were removed by centrifugation (500 × g for 10 min). The postnuclear supernatant was fractionated into a crude membrane pellet (P100) and a cytosolic fraction (S100) by centrifugation at 100,000 × g for 1 h in a TLA100.2 rotor (Beckman). The distribution of GFP-Ras2 in P100 and S100 was analyzed by immunoblotting with an anti-GFP antibody (Molecular Probes). The GFP-Ras2 expression level was analyzed with the Ras monoclonal antibody Y13-259 as previously described (5).
Fluorescence microscopy.
To visualize fluorescent proteins, cultures were grown to exponential phase in SC medium. Cells were collected by centrifugation and gently resuspended in residual medium. Aliquots (1 to 3 μl) were spotted onto microscopy slides, and images were visualized using a Nikon Eclipse 80i microscope equipped for epifluorescence. Images were captured using a photometric Cascade camera controlled by MetaMorph imaging software (Universal Imaging Corporation). The fluorescein isothiocyanate-Texas Red filter set used was obtained from Chroma Technology Corp.
Subcellular fractionation.
Sucrose gradient fractionation was performed as described previously (50). Briefly, NaN3 (10 mM) and KF (10 mM) were added to cell cultures grown to exponential phase. Cells were collected and washed in buffer containing 10 mM NaN3, 10 mM KF, and 5 mM Tris-HCl, pH 7.6, and resuspended in STE10 buffer (10% sucrose, 10 mM Tris-HCl, pH 7.6, 10 mM EDTA) with protease inhibitors (1 mM phenylmethylsulfonyl fluoride and 2.5 μg/ml [each] of chymostatin, leupeptin, antipain, and pepstatin). Cells were broken with glass beads (425 to 600 μm; Sigma). The cell lysate was centrifuged (500 × g, 10 min), and the postnuclear supernatant (700 μl) was loaded on a 20 to 60% linear sucrose gradient (in STE10 buffer). After centrifugation at 100,000 × g for 16 h in an SW41 rotor (Beckman), fractions were collected from the top, and the proteins were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, followed by immunoblotting. Ras was detected with the monoclonal antibody Y13-259 as previously described (5). Anti-GFP, anti-Dpm1 (ER), anti-Por1 (mitochondria), and anti-Pma1 (plasma membrane) were purchased from Molecular Probes. Anti-Cox2 (mitochondria) was a generous gift from Rosemary Stuart (Marquette University, Milwaukee, WI).
Azide treatment.
Cells harboring GFP-Ras2 (B828) were grown to exponential phase in minimal SC medium and induced with galactose (4%) and azide (2 mM), and the cultures were incubated for an additional 4 h. The localization of GFP-Ras2 was examined by fluorescence microscopy. For ATP repletion, azide-treated cells were collected and washed twice with TE (10 mM Tris, pH 8.0, 1 mM EDTA) buffer. The expression of GFP fusion proteins was stopped by the addition of glucose (2%). The cultures were incubated for another 2 h in SC-glucose medium. The localization of the GFP fusion proteins was examined by fluorescence microscopy.
Mitochondrion isolation.
Yeast spheroplasts and crude mitochondria were isolated as described previously (17). Additional purification of mitochondria was done by suspending crude mitochondria in 250 mM sucrose EM buffer (10 mM MOPS [morpholinepropanesulfonic acid], pH 7.2, 1 mM EDTA) and layering them on a three-step sucrose gradient (60%, 32%, 23%, 15%) in EM buffer (37). After centrifugation at 134,000 × g for 1 h in an SW41 rotor, mitochondria were harvested from the 60%-32% interface. The purity of the mitochondrion fraction was assessed by probing the fraction with antibodies for ER (Dpm1) and plasma membrane (Pma1) markers.
Proteinase K treatment.
One hundred micrograms of mitochondria was resuspended in 500 ml of SH buffer (0.6 M sorbitol, 20 mM HEPES, pH 7.2), HEPES buffer (20 mM HEPES, pH 7.2), or SH buffer with 0.08% Triton X-100. A protease sensitivity experiment was performed by adding proteinase K (20 μg/ml) and bovine serum albumin (1 mg/ml), followed by incubation at 4°C for 20 min. Proteolysis was halted by the addition of phenylmethylsulfonyl fluoride to a final concentration of 1 mM. Intact and hypotonically shocked mitochondria were pelleted and resuspended in 40 μl 1× SDS loading buffer. Proteins in Triton-solubilized mitochondria were precipitated with trichloroacetic acid and resuspended in 40 μl 1× SDS loading buffer. Twenty microliters of each final sample was loaded for immunoblot analysis with antibodies for Ras, the mitochondrial outer membrane protein Tom70, and the mitochondrial inner membrane protein Aac1. Anti-Tom70 and anti-Aac1 were generous gifts from Rosemary Stuart (Marquette University, Milwaukee, WI).
RESULTS
The C terminus of K-Ras targets GFP to the plasma membrane through a Golgi complex-independent pathway in yeast.
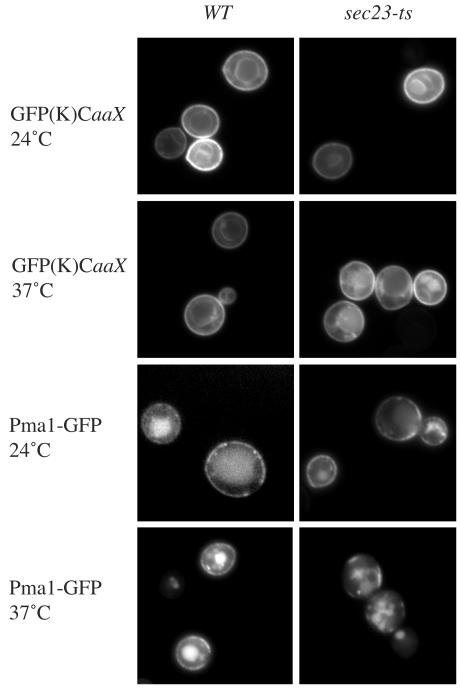
The C-terminal hypervariable domain and the CaaX box are necessary and sufficient for plasma membrane localization of Ras proteins (19, 75). In addition, plasma membrane localization of K-Ras in mammalian cells and Ras proteins in yeast does not require the classical secretory pathway or a functional Golgi apparatus (2). To determine if K-Ras also localizes to the plasma membrane in yeast, the hypervariable domain (18 residues) and the CaaX box of mammalian K-Ras were fused to the C terminus of GFP and expressed under the control of a galactose-inducible promoter in yeast. Like GFP-Ras2, GFP(K)CaaX was localized to the plasma membrane (Fig. 1). To examine if the classical secretory pathway is required, a temperature-sensitive sec23-ts strain was employed (19, 75). GFP(K)CaaX localized to the plasma membrane of the sec23-ts strain at the permissive (25°C) and the nonpermissive temperature (37°C). In contrast, Pma1-GFP, which requires the classical secretory pathway for plasma membrane localization (14), was found primarily on endomembranes when the sec23-ts strain was grown at the nonpermissive temperature (37°C) (Fig. 1). Thus, it appears that ER-to-plasma-membrane translocation of K-Ras, like that of yeast Ras1 and Ras2, does not require the classical secretory pathway in yeast.
FIG. 1.
The classical secretory pathway is not required for the plasma membrane localization of GFP(K)CaaX. GFP(K)CaaX and Pma1-GFP were expressed from the galactose-inducible promoter in wild-type (LRB938) and sec23-ts (LRB937) strains, and the cultures were grown at 25°C. GFP(K)CaaX and Pma1-GFP expression was induced by the addition of 4% galactose, and cells were grown at 25°C or 37°C for 4 h. Fluorescence images were collected using a Nikon Eclipse 80i microscope.
Isolation of mutants defective in nonclassical plasma membrane localization of Ras.
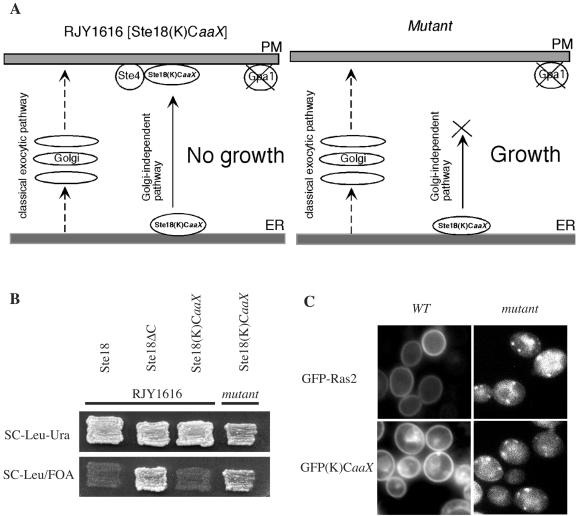
A genetic screen was designed to identify mutants defective in plasma membrane localization of Ras. The screen is based on the well-characterized yeast pheromone signal transduction pathway and is described in detail in Materials and Methods. Briefly, the loss of Gpa1 results in Gßγ-dependent growth arrest due to constitutive activation of the pheromone response pathway (35). This response requires plasma membrane localization of the Gßγ complex, which in turn requires posttranslational modification of Ste18 (Gγ) by farnesylation and palmitoylation (35). To establish a screen for Ras trafficking mutants, the C terminus of Ste18 was replaced by the hypervariable domain and CaaX box of K-Ras to create Ste18(K)CaaX (Fig. 2A). The expression of wild-type Ste18 or Ste18(K)CaaX resulted in growth arrest in a strain lacking Gpa1 (Fig. 2B). This growth arrest depends on the C-terminal targeting sequences and plasma membrane localization, because removal of the Ste18 C terminus restored cell growth (Fig. 2B). Mutants of interest blocked the plasma membrane localization of Ste18(K)CaaX and appeared as 5-FOA-resistant colonies. An example is shown in Fig. 2B. In addition to the desired trafficking mutants, the screen was expected to identify mutations in components of the signal transduction pathway downstream of Gßγ. However, these are readily distinguished from trafficking mutants by introducing a Ste18-expressing plasmid and testing for growth on 5-FOA-containing plates.
FIG. 2.
Genetic screen for isolation of mutants defective in Ras trafficking. (A) Schematic diagram of trafficking mutant screen based on the subcellular localization of Ste18(K)CaaX and the growth inhibition resulting from plasma membrane localization of Ste18(K)CaaX in yeast strain RJY1616 (see the text for a detailed description of the screen). (B) Patch assay demonstrating the growth defect resulting from the expression of Ste18 or Ste18(K)CaaX. Strains expressing a C-terminal deletion of the Ste18 CaaX box (Ste18ΔC) are able to grow under these conditions. One of the mutants isolated by the Ste18(K)CaaX-based trafficking screen is also shown. (C) GFP(K)CaaX and GFP-Ras2 exhibit plasma membrane localization (rim staining) in wild-type cells (RJY1616) but endomembrane localization in one of the mutants isolated by the trafficking screen (WTS92). Cells were examined by fluorescence microscopy (Nikon Eclipse 80i microscope).
Approximately 60,000 UV-mutagenized colonies were screened by 5-FOA selection, and 5-FOA-resistant colonies were identified at a frequency of 0.5% (approximately 300 colonies). The majority of these mutants were defective in the pheromone signaling pathway, but three appeared to be trafficking mutants by the criteria described above. Transformation of these three strains with GFP(K)CaaX or GFP-Ras2 revealed that all three mislocalized GFP-Ras fusion proteins. An example of one putative trafficking mutant is shown in Fig. 2C.
ER-to-plasma-membrane trafficking of Ras proteins is disrupted in class C vps mutants.
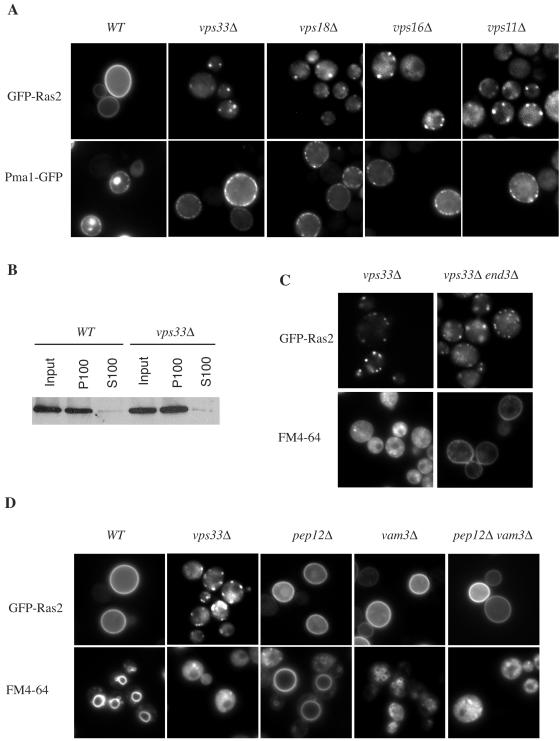
The wild-type genes corresponding to three putative Ras trafficking mutants were cloned by complementation of the growth arrest phenotype using a low-copy-number (YSB32) yeast genomic DNA library. Complementing plasmids were mapped by a combination of deletion analysis and subcloning strategies to identify the open reading frame responsible for the complementation. Two of the three mutants isolated in the screen were complemented by the VPS33 gene, whereas the third mutant was complemented by VPS11/PEP5. Vps33 and Vps11, together with Vps16 and Vps18, constitute the class C Vps complex that regulates vesicle docking and fusion of endosome and vacuole membranes (43, 56, 60, 67). To examine if the Ras trafficking defect extends to the other members of the class C Vps complex, strains harboring deletions of each of the four genes (vps33Δ, vps18Δ, vps16Δ, and vps11Δ) were constructed. Plasma membrane localization of newly synthesized GFP-Ras2 was defective in every case, whereas the localization of Pma1-GFP was not affected, indicating that the mutants did not cause a general defect in protein trafficking to the plasma membrane (Fig. 3A). Although GFP-Ras2 localized on endomembranes in the vps33Δ mutant strain, it remained membrane associated, as in wild-type cells (Fig. 3B).
FIG. 3.
Mislocalization of GFP-Ras2 in class C vps mutants. (A) GFP-Ras2 and Pmal-GFP were expressed in wild-type (LRB938) and vps33Δ (RJY1654), vps18Δ (RJY1672), vps16Δ (RJY1695), and vps11Δ (RJY1702) mutant cells by galactose induction and observed by fluorescence microscopy. (B) The membrane association of GFP-Ras2 in wild-type (LBR938) or vps33Δ mutant (RJY1654) cells was assessed by subcellular fractionation of total lysates (input) into cytosolic (S100) and crude membrane (P100) fractions, as described in Materials and Methods. GFP-Ras2 was detected by immunoblotting with an anti-GFP antibody, followed by chemiluminescence (Pierce). (C) Endocytosis does not play a role in endomembrane localization of GFP-Ras in vps33Δ mutant cells. GFP-Ras2 was expressed from a galactose-inducible promoter in the vps33Δ (RJY1654) or vps33Δ end3Δ (RJY1695) mutant strain. Endocytosis was monitored by staining the cells with FM4-64, a dye that is taken up in an End3-dependent manner. (D) Mutations in the class C VPS genes, but not in PEP12 or VAM3, affect the subcellular localization of Ras. GFP-Ras2 was examined in the wild-type (LBR938) and vps33Δ (RJY1654), pep12Δ (RJY1705), vam3Δ (RJY1706), and pep12Δ vam3Δ (RJY1707) mutant strains. FM4-64 was used to monitor endocytosis and to visualize vacuoles.
The class C Vps complex has been shown to be involved in the recycling step of endocytosis, raising the possibility that the intracellular accumulation of Ras2 results from a defect in membrane recycling via an endosomal compartment (9). If this is true, then GFP-Ras2 should remain on the plasma membrane when endocytosis is blocked. This was tested by deleting the END3 gene and examining GFP-Ras2 localization in the vps33Δ end3Δ mutant strain. END3 encodes a component of the endocytic complex which is essential for the internalization step of endocytosis (46). Deletion of END3 did not restore the plasma membrane localization of GFP-Ras2 in the vps33Δ mutant strain (Fig. 3C).
The class C Vps complex is also involved in endosome and vacuole vesicle trafficking. Vps33 interacts with the vacuolar syntaxin Vam3 and the endosomal syntaxin Pep12 to regulate SNARE pairing in these two membrane compartments (43). To determine if these steps are involved in Ras trafficking, VAM3 and/or PEP12 was deleted, and GFP-Ras2 localization was examined. In contrast to VPS33 deletion, the loss of VAM3 or PEP12 had no observable effect on Ras localization (Fig. 3D). To further test for an involvement of endosomes in Ras trafficking, the localization of GFP-Ras2 was compared to that of a red fluorescent protein (RFP) fusion to the endosome resident protein Vps32 (27). There was no overlap of the Vps32-RFP and GFP-Ras2 signals in the wild-type or vps33 mutant strain (data not shown). Taken together, these results suggest that translocation of Ras2 from the ER to the plasma membrane does not involve endosome-related functions of class C Vps proteins.
Class C vps mutants exhibit mitochondrial defects.
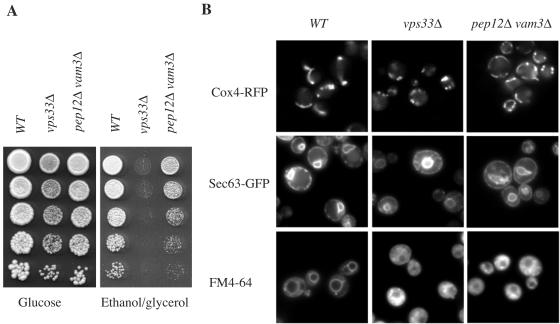
Other potential mechanisms of class C Vps involvement in Ras trafficking were investigated. The data shown in Fig. 4A confirm a report that class C vps deletion strains are unable to grow on nonfermentable carbon sources, indicating a mitochondrial defect (68). However, the nature of the Vps-related mitochondrial defect is not known. To begin to examine the mitochondrial defect in class C vps mutants, mitochondrial morphology was examined in wild-type and vps33 mutant strains by use of an RFP fusion with Cox4, a subunit of cytochrome oxidase found on the inner mitochondrial membrane (27). Mitochondria in wild-type cells and pep12Δ vam3Δ mutant cells exhibited a tubular structure, whereas mitochondria in the vps33Δ mutant strain appeared as punctate spots. This does not appear to be a general membrane defect, because the appearance of the ER, as indicated by Sec63-GFP, was unaffected by the deletion of VPS33. On the other hand, vacuolar morphology was affected by the deletion of VPS33, as expected (Fig. 4B). The vacuolar membrane defect, however, does not appear to be the cause of the mitochondrial defect. pep12Δ vam3Δ mutant cells exhibit the same vacuolar defect as vps33Δ mutant cells but have normal mitochondrial structure and Ras localization. It appears that the mitochondrial defect of the vps33Δ mutant strain may contribute to the Ras trafficking defect.
FIG. 4.
Class C vpsΔ strains exhibit mitochondrial defects. (A) Serial dilutions (1:5) of cultures (∼5 × 104) of wild-type (LBR938) and vps33Δ (RJY1654) and pep12Δ vam3Δ (RJY1707) mutant cells were spotted (5 μl) onto rich medium (yeast extract-peptone) containing 2% glucose or ethanol (2%)-glycerol (2%). The plates were incubated at 25°C for 3 days. (B) Localization of Cox4-RFP was used to assess mitochondrial morphology in the wild-type (LBR938) and vps33Δ (RJY1654) and pep12Δ vam3Δ (RJY1707) mutant strains (top panels). The ER marker Sec63-GFP was used to monitor ER morphology in the wild-type (LBR938) and vps33Δ (RJY1654) and pep12Δ vam3Δ (RJY1707) mutant strains (middle panels), and FM4-64 was used to monitor endocytosis and vacuole morphology (bottom panels).
GFP-Ras2 accumulates with mitochondria in class C vps mutants.
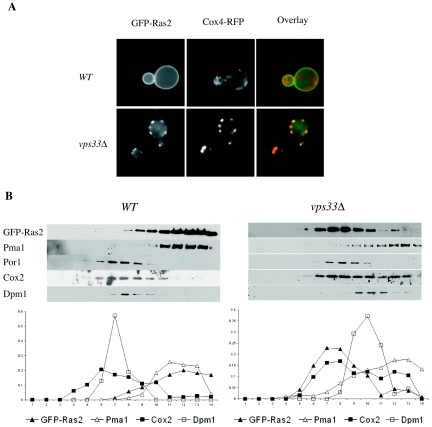
To further address the role of mitochondria in Ras trafficking, the localization of GFP-Ras2 was compared with that of Cox4-RFP, a mitochondrial marker (52). GFP-Ras2 and Cox4-RFP colocalized in vps33Δ mutant cells but not in wild-type cells (Fig. 5A). Wild-type and vps33 mutant cells expressing GFP-Ras2 were also lysed and fractionated by sucrose gradient centrifugation. GFP-Ras2 cofractionated primarily with the plasma membrane marker Pma1 in wild-type cells. In contrast, GFP-Ras2 cofractionated with two mitochondrial markers, Por1 and Cox2, in the vps33Δ mutant strain (Fig. 5B). The colocalization of GFP-Ras2 with mitochondrial markers in the trafficking mutants suggests a more direct involvement of mitochondria in the Golgi complex-independent trafficking of Ras proteins.
FIG. 5.
Newly synthesized GFP-Ras2 colocalizes with mitochondria in class C vps mutants. (A) GFP-Ras2 was expressed from the galactose-inducible promoter in wild-type (LBR938) and vps33Δ mutant (RJY1654) cells harboring Cox4-RFP. The localization of Cox4-RFP and GFP-Ras2 was examined by fluorescence microscopy, and representative images are shown. In each case, at least five fields and approximately 100 cells were examined. (B) The wild-type (LRB938) and vps33Δ mutant (RJY1654) strains expressing GFP-Ras2 were lysed, fractionated by a sucrose gradient as described in Materials and Methods, and immunoblotted with anti-GFP antibody. The plasma membrane fractions were identified by blotting with a polyclonal antibody to Pma1, the mitochondrial fractions were identified using anti-Por1 and anti-Cox2 antibodies, and an antibody to Dpm1 was used for the ER fractions. The immunoblots were quantified by densitometry and plotted as a function of fraction number, with fraction 1 being the top of the gradient.
Endogenous Ras2 colocalizes with mitochondria in wild-type yeast cells.
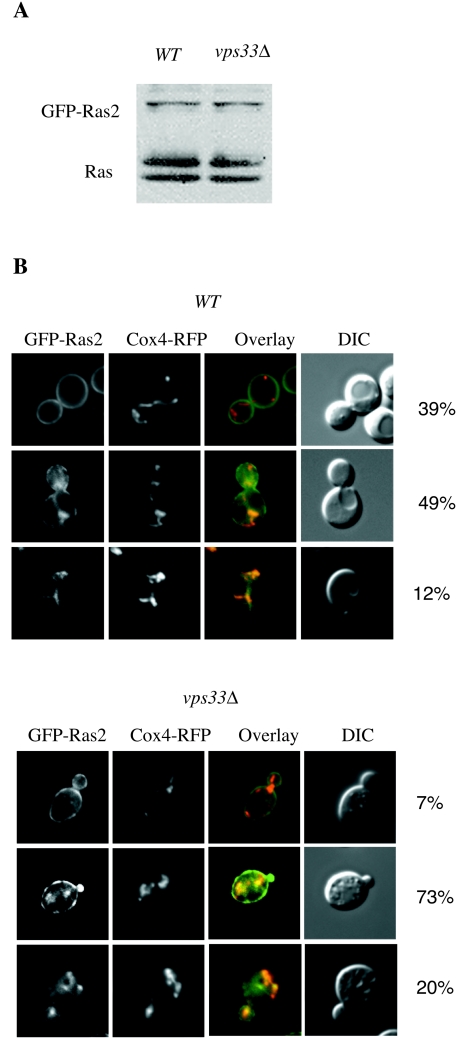
To rule out that the trafficking defects described above result from overexpression of Ras2, we next examined if GFP-Ras2 expressed at physiological levels associates with mitochondria. This was done by expressing GFP-Ras2 from a MET25 promoter (34). When cells were grown on medium supplemented with low levels of methionine (12 mM), GFP-Ras2 was expressed at approximately the same level as endogenous Ras2, as seen by immunoblotting with an anti-Ras antibody (Fig. 6A). Under these conditions, three groups of GFP-Ras2-expressing cells were observed. In the first group, GFP-Ras2 was found primarily on the plasma membrane; in the second, GFP-Ras2 was observed on the plasma membrane and endomembranes; and in the last group, GFP-Ras2 was localized primarily on endomembranes. In wild-type cells, 39% showed plasma membrane staining only, 49% exhibited plasma membrane and endomembrane staining, and only 12% showed endomembrane staining and no plasma membrane localization. However, in the absence of VPS33, the percentage of cells with plasma membrane staining was reduced to 7%, with the majority of cells exhibiting partial or complete endomembrane localization (93%) (Fig. 6B). In wild-type and vps33 mutant cells, the endomembrane-localized GFP-Ras2 overlapped with the mitochondrial marker Cox4-RFP.
FIG. 6.
Subcellular localization of GFP-Ras2 expressed at physiological levels. (A) GFP-Ras2 was expressed from a MET25 promoter in the wild-type (LRB938) and vps33Δ mutant (RJY1654) strains. Cells were grown to exponential phase in SC-glucose medium supplemented with 12 mM methionine. The immunoblot compares the expression of GFP-Ras2 with that of the endogenous Ras1 (upper band) and Ras2 (lower band) proteins, using the anti-Ras monoclonal antibody Y13-259. (B) Wild-type (LRB938) or vps33Δ mutant (RJY1654) cells harboring MET25-GFP-Ras2 and Cox4-RFP were grown to exponential phase in SC-glucose medium supplemented with 12 mM methionine. Cox4-RFP and GFP-Ras2 were visualized by fluorescence microscopy. The percentages of cells with predominantly plasma membrane-localized fluorescence (top), plasma membrane and endomembrane fluorescence (middle), and only endosomal fluorescence (bottom) were compared. For each sample, at least six fields and approximately 150 cells were examined. A representative example is shown.
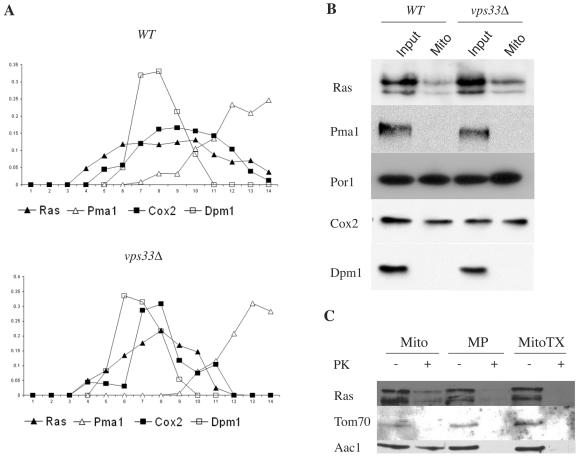
To further examine the role of mitochondria in Ras trafficking, endogenous Ras1 and Ras2 were examined by subcellular fractionation and sucrose density gradient centrifugation. In wild-type cells, Ras fractionates as a broad peak that includes the plasma membrane marker Pma1, but also the ER and mitochondrial markers Dpm1 and Cox2, respectively (Fig. 7A). Deletion of VPS33 resulted in a reduction of Ras in the plasma membrane fraction and an increase in the Cox2-enriched fractions. While these results are consistent with the colocalization of GFP-Ras2 with endomembrane and plasma membrane markers observed in vivo, the distribution of Ras with several compartments prevents the conclusion that endogenous Ras cofractionates with mitochondria. In fact, a small fraction of Ras is expected to fractionate with the ER, where posttranslational modifications occur.
FIG. 7.
Mitochondrial association of endogenous Ras proteins in wild-type and vps33Δ mutant strains. (A) Wild-type (LRB938) and vps33Δ mutant (RJY1654) cells were grown to exponential phase in yeast extract-peptone-dextrose medium. Cells were lysed and fractionated by a sucrose gradient as described in Materials and Methods. Endogenous Ras1 and Ras2 were visualized using the anti-Ras antibody Y13-259. The distributions of organelles were followed by immunoblotting with the following antibodies specific to organelles: anti-Pma1 (plasma membrane), anti-Por1 (mitochondria), anti-Cox2 (mitochondria), and anti-Dpm1 (ER). Relative levels of Ras and the organelle markers in each fraction were quantified by densitometry and plotted as a function of the fraction number (fraction 1 is the top of the gradient). (B) Cell lysates and purified mitochondria were prepared as described in Materials and Methods. Lysates (input) and mitochondria (Mito) from wild-type (LRB938) and vps33Δ mutant (RJY1654) cells were resolved by SDS-polyacrylamide gel electrophoresis (10%) and probed with antibodies for the plasma membrane (anti-Pma1), mitochondria (anti-Por1 and anti-Cox2), and the ER (anti-Dpm1). Ras1 and Ras2 were detected using the anti-Ras antibody Y13-259. (C) Mitochondria isolated from wild-type cells (W303) were treated as described in Materials and Methods to generate intact mitochondria (Mito), mitoplasts (MP), and Triton X-100-solubilized mitochondria (MitoTX). After treatment with proteinase K (PK) and processing as described in Materials and Methods, samples were separated in SDS gels and probed with an antibody for the mitochondrial outer membrane protein Tom70 and an antibody for the mitochondrial inner membrane protein Aac1. Ras1 and Ras2 were detected using the anti-Ras antibody Y13-259.
To determine directly whether there is a mitochondrial pool of Ras, mitochondria were purified from wild-type and vps33 mutant yeast cells. Mitochondria were purified by established procedures (17, 37) and found to be enriched for the mitochondrial markers Cox2 and Por1 but lacking detectable amounts of Pma1 (plasma membrane) and Dpm1 (ER). The Ras1 and Ras2 proteins cofractionated with purified mitochondria isolated from both the wild-type and vps33 mutant strains. Using Cox2 and Por1 to normalize the amount of mitochondrial protein loaded, there appeared to be more Ras associated with mitochondria isolated from vps33 mutant cells, consistent with the subcellular fractionation and in vivo localization results (Fig. 7B). Based on its protease sensitivity, Ras appears to be associated with the cytosolic surface of the outer mitochondrial membrane (Fig. 7C).
Azide treatment results in mitochondrial accumulation of Ras.
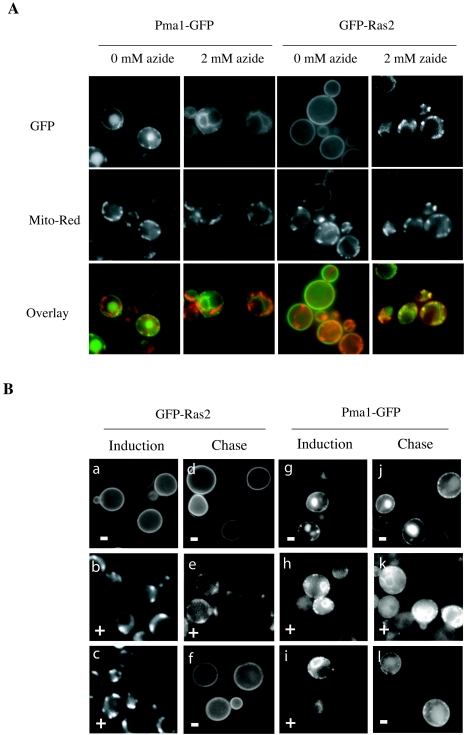
Azide has been used to study the energy dependence of lipid and protein transport (3, 40). For example, inhibition of the classical secretory pathway by azide has been taken as an indication of the ATP dependence of vesicle-mediated protein translocation. To assess the ATP dependence of Ras trafficking, cells were treated with 2 mM azide, and GFP-Ras2 or Pma1-GFP expression was induced. The addition of azide resulted in the mislocalization of GFP-Ras2 and Pma1-GFP. Pma1-GFP accumulated in a perinuclear compartment, presumably the ER. In contrast, GFP-Ras2 colocalized with mitochondria (Fig. 8A). The azide dependence of Ras trafficking indicates that it is an active, energy-dependent process rather than a diffusion-mediated mechanism. Furthermore, the azide-dependent mitochondrial localization of Ras is reversible. Upon removal of azide, intracellularly accumulated GFP-Ras2 was found at the plasma membrane (Fig. 8B). The reversible colocalization of GFP-Ras2, but not Pma1-GFP, with mitochondria in azide-treated cells is consistent with a role for mitochondria in the normal trafficking of Ras proteins.
FIG. 8.
Azide treatment causes reversible association of GFP-Ras2 with mitochondria. (A) Wild-type (LRB938) cells harboring galactose-inducible GFP-Ras2 or Pma1-GFP were grown and induced as described in the legend to Fig. 1, except that azide (2 mM) was added at the time of induction. The mitochondrion-specific dye MitoTracker red CMXRos (0.1 μg/ml) was added 4 h after the induction of GFP-Ras2 or Pma1-GFP expression, and the culture was incubated for another 30 min. Cells were collected and washed twice with TE buffer. GFP fluorescence (top) and MitoTracker red staining (middle) were examined by fluorescence microscopy. In the overlay images (bottom), pseudocolors were introduced, with green being GFP fluorescence and red being MitoTracker red fluorescence. (B) Cells (LRB938) harboring galactose-inducible GFP-Ras2 (a to f) or Pma1-GFP (g to l) were grown at 25°C to mid-exponential phase. Four hours after galactose (4%) induction in SC medium (−) (a and g) or SC medium supplemented with azide (2 mM; +) (b, c, h, and i), GFP images were collected. Cells were harvested and resuspended in SC-glucose (2%) medium (repressing conditions). After a 2-h chase with azide (+) (e and k) or without azide (−) (d, f, j, and l), the localization of GFP-Ras2 or Pma1-GFP was examined by fluorescence microscopy.
DISCUSSION
The classical secretory pathway is used by most proteins destined for the plasma membrane or secretion from the cell. However, there are examples of nonclassical membrane trafficking of some proteins. For example, the plasma membrane localization heterotrimeric G proteins, Gs and Gq, F3/contactin, caspr/paranodin, and flotillin-1/reggie-2 occur even in the presence of the Golgi complex-disrupting drug brefeldin A (4, 6, 41, 72). The present study focuses on the Golgi complex-independent trafficking of yeast Ras proteins as an example of proteins that associate with membranes via lipid anchors. The membrane association and subcellular trafficking of Ras proteins require that the CaaX box undergoes farnesylation, -aaX proteolysis, and methyl esterification (1, 7, 18, 26, 31, 55, 57, 58). However, these modifications are not sufficient for a stable membrane association or subsequent subcellular localization of Ras. Stable membrane association requires a second signal, which can be either a polybasic domain, in the case of K-Ras, or palmitoylation (S acylation) (19, 24). The functional significance of these modifications is still a matter of debate.
Modification of proteins or peptides with a farnesyl moiety results in a weak membrane association and rapid exchange between membrane compartments, whereas dual lipid modification (farnesyl and palmitoyl) results in a stable membrane association (64). This has led to the proposal of a kinetic trap model, in which farnesylation mediates a transient membrane association followed by palmitoylation and stable association with a target membrane. A palmitoylation-depalmitoylation cycle has been shown to regulate the trafficking of mammalian H-Ras and N-Ras between the late Golgi compartment and the plasma membrane (21, 51, 54). Palmitoylation of Ras on the Golgi apparatus results in vesicle-dependent delivery to the plasma membrane. Depalmitoylation of Ras on the plasma membrane allows the rapid exchange of Ras with endomembranes via a nonvesicular mechanism. A recently described Golgi complex-associated Ras palmitoyltransferase may be the palmitoylating enzyme predicted by the palmitoylation-depalmitoylation cycle model (71). The palmitoylthioesterase, on the other hand, has not been identified.
It is not clear whether the palmitoylation-depalmitoylation pathway described above operates during the earlier steps of Ras trafficking that begin in the ER. Yeast Ras is palmitoylated in the ER following CaaX box processing by the Erf2/Erf4 palmitoyltransferase (19, 33, 75). The model above predicts that the dual-lipid-modified form of Ras will undergo vesicle-mediated translocation from the ER. Consistent with this, we found that dual-lipid-modified Ras remains membrane associated. No cytosolic pool of Ras has been detected, and there has been no escort protein identified for Ras analogous to the Rab escort protein required for the cytosolic form of prenylated Rab proteins. In addition, we have shown in the present study that plasma membrane localization of Ras requires ATP (Fig. 6), arguing against a nonvesicular model. While these data point to a membrane-mediated trafficking pathway, it is clearly distinct from the classical secretory pathway. Blocking the classical secretory pathway with sec mutants or brefeldin A has no effect on plasma membrane localization of yeast Ras (19). Similar results have been seen with K-Ras in mammalian cells.
The genetic screen described in this report implicates the class C Vps complex in some aspect of Ras trafficking. The class C Vps complex is best characterized for its involvement in the biogenesis and function of vacuoles (47). It conveys vesicle fusion specificity by interacting with syntaxins on endosome and vacuole membranes (43, 56, 60). The identification of class C vps mutants defective in Ras trafficking points to an involvement of a vesicle fusion process. Our results, however, suggest that endosome and vacuole functions of the class C Vps complex are not responsible for the defects in Ras trafficking. Instead, a less well-characterized role of the class C Vps complex in mitochondrial function appears to be responsible. Mitochondrial function is compromised in class C vps mutant strains, resulting in a respiratory defect, as monitored by the inability to grow on nonfermentable carbon sources (68). Mitochondrial morphology was also altered in class C vps mutant strains (Fig. 4B). The mitochondrial localization of Ras proteins was also observed at a steady state in both the wild-type and vps33Δ mutant strains. Together, these results implicate mitochondria in the nonclassical pathway of Ras trafficking.
The unexpected mitochondrial dependence of Ras trafficking raises a number of interesting questions and suggests several new avenues of investigation. The class C Vps complex is required for vesicle fusion involving endosome and vacuole membranes. Is the class C Vps complex involved in mitochondrial vesicle fusion? Deletion of any one of the class C Vps genes leads to a respiration-deficient phenotype (68). However, the mitochondrial defect has not been characterized. It is tempting to speculate that the class C Vps complex plays a role in the cycles of fusion and fission that mitochondria undergo. Yeast mitochondria, like their metazoan counterparts, comprise a dynamic reticular network of membranes that undergo continuous remodeling in response to changes in growth and nutrient availability (69). Mitochondria also serve as a major cellular ATP source under respiring conditions. Plasma membrane localization of Ras requires ATP, raising the possibility that the mitochondrial requirement is simply to provide energy. This is unlikely to be the case, because the trafficking of Pma1 via the classical secretory pathway is also inhibited by azide but is not affected by the loss of class C Vps function (Fig. 6). Ras is also found associated with mitochondria, consistent with a more direct role of mitochondria in Ras trafficking.
How does Ras become associated with mitochondria following CaaX modification and palmitoylation on the ER membrane? It has been known for some time that the ER and mitochondria are physically associated through specialized regions called mitochondrion-associated ER membranes (MAMs). Lipids, ATP, and calcium have been shown to exchange between the ER and the mitochondria through MAMs (49, 65, 73). Mitochondria also play a key role in phospholipid synthesis and transport (73). MAM-mediated protein exchange has not been previously reported, but lipid-anchored proteins such as Ras have not been directly investigated. Given our results, this issue needs to be addressed. The lack of a protein marker of the MAM fraction has made these studies difficult. Recent progress on the mitochondrial proteome may be helpful in this regard (25, 59).
Other reports have found Ras associated with mitochondria. In the human kidney, H-Ras localizes with mitochondria in proximal and distal convoluted tubules (30). Bcl-2 and Ras proteins interact at mitochondria in the murine T-cell line TS1αß, and the mitochondrial association of the three Ras proteins is differentially regulated by interleukin-2 supplementation (48). Several downstream effectors of Ras proteins were also found at mitochondria. In mammalian cells, Grb10 is detected at mitochondria and interacts with the mitochondrion-associated Raf-1 pool (42). In yeast, the Ras GTPase activator Ira1 copurifies with yeast mitochondria (25, 38, 62), and a homolog of the yeast prenylcyteine methyltransferase, Ste14, has been reported to be part of the mitochondrial proteome of Neurospora crassa (59). Ras-cyclic AMP (Ras-cAMP) signaling has been linked to the release of reactive oxygen species and mitochondrial damage in yeast (8). Finally, Philips recently showed that phosphorylation results in translocation of K-Ras from the plasma membrane to the mitochondria, where it appears to play a role in apoptosis (44).
In summary, our results demonstrate the requirement of the class C Vps complex and functional mitochondria for the plasma membrane targeting of yeast Ras proteins through a Golgi complex-independent pathway. This has important implications for our understanding of Ras function. Mounting evidence points to Ras playing distinct signaling roles depending on its subcellular location. However, our understanding of Ras trafficking and targeting is still incomplete. Genetic approaches afforded by the yeast system have led to the identification of the Ras palmitoyltransferases as well as palmitoyltransferases for other acylated proteins. It is now known that palmitoylation plays a key role in Ras trafficking. Interfering with the palmitoylation of Ras by targeting the Ras palmitoyltransferase might be an effective strategy for blocking Ras activation. Given the prevalence of Ras mutations in human cancers, this is an approach that will be actively pursued.
Acknowledgments
We thank Won-Ki Huh, Erin K. O'Shea, Maurine E. Linder, Peter M. Pryciak, John Cooper, and Lucy Robinson for plasmids and yeast strains. We also thank Rosemary A. Stuart for antibodies and assistance with experiments demonstrating the association of Ras with yeast mitochondria.
This work was supported by an NIH grant (CA050211) to R.J.D.
REFERENCES
- 1.Achleitner, G., B. Gaigg, A. Krasser, E. Kainersdorfer, S. D. Kohlwein, A. Perktold, G. Zellnig, and G. Daum. 1999. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur. J. Biochem. 264:545-553. [DOI] [PubMed] [Google Scholar]
- 2.Apolloni, A., I. A. Prior, M. Lindsay, R. G. Parton, and J. F. Hancock. 2000. H-Ras but not K-Ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 20:2475-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balasubramanian, K., and C. M. Gupta. 1996. Transbilayer phosphatidylethanolamine movements in the yeast plasma membrane. Evidence for a protein-mediated, energy-dependent mechanism. Eur. J. Biochem. 240:798-806. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin, T. A., and H. L. Ostergaard. 2002. The protein-tyrosine phosphatase CD45 reaches the cell surface via Golgi-dependent and -independent pathways. J. Biol. Chem. 277:50333-50340. [DOI] [PubMed] [Google Scholar]
- 5.Bartels, D. J., D. A. Mitchell, X. Dong, and R. J. Deschenes. 1999. Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6775-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnon, C., L. Goutebroze, N. Denisenko-Nehrbass, J. A. Girault, and C. Faivre-Sarrailh. 2003. The paranodal complex of F3/contactin and caspr/paranodin traffics to the cell surface via a non-conventional pathway. J. Biol. Chem. 278:48339-48347. [DOI] [PubMed] [Google Scholar]
- 7.Boyartchuk, V. L., M. N. Ashby, and J. Rine. 1997. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science 275:1796-1800. [DOI] [PubMed] [Google Scholar]
- 8.Breitenbach, M., P. Laun, and M. Gimona. 2005. The actin cytoskeleton, RAS-cAMP signaling and mitochondrial ROS in yeast apoptosis. Trends Cell Biol. 15:637-639. [DOI] [PubMed] [Google Scholar]
- 9.Bugnicourt, A., M. Froissard, K. Sereti, H. D. Ulrich, R. Haguenauer-Tsapis, and J. M. Galan. 2004. Antagonistic roles of ESCRT and Vps class C/HOPS complexes in the recycling of yeast membrane proteins. Mol. Biol. Cell 15:4203-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buss, J. E., and B. M. Sefton. 1986. Direct identification of palmitic acid as the lipid attached to p21ras. Mol. Cell. Biol. 6:116-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell, S. L., R. Khosravi-Far, K. L. Rossman, G. J. Clark, and C. J. Der. 1998. Increasing complexity of Ras signaling. Oncogene 17:1395-1413. [DOI] [PubMed] [Google Scholar]
- 12.Carozzi, A. J., S. Roy, I. C. Morrow, A. Pol, B. Wyse, J. Clyde-Smith, I. A. Prior, S. J. Nixon, J. F. Hancock, and R. G. Parton. 2002. Inhibition of lipid raft-dependent signaling by a dystrophy-associated mutant of caveolin-3. J. Biol. Chem. 277:17944-17949. [DOI] [PubMed] [Google Scholar]
- 13.Casey, P. J., P. A. Solski, C. J. Der, and J. E. Buss. 1989. p21ras is modified by a farnesyl isoprenoid. Proc. Natl. Acad. Sci. USA 86:8323-8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, A., and C. W. Slayman. 1991. Maturation of the yeast plasma membrane [H+]ATPase involves phosphorylation during intracellular transport. J. Cell Biol. 115:289-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu, V. K., T. Bivona, A. Hach, J. B. Sajous, J. Silletti, H. Wiener, R. L. Johnson II, A. D. Cox, and M. R. Philips. 2002. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4:343-350. [DOI] [PubMed] [Google Scholar]
- 16.Clark, K. L., D. Dignard, D. Y. Thomas, and M. Whiteway. 1993. Interactions among the subunits of the G protein involved in Saccharomyces cerevisiae mating. Mol. Cell. Biol. 13:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daum, G., P. C. Bohni, and G. Schatz. 1982. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257:13028-13033. [PubMed] [Google Scholar]
- 18.Deschenes, R. J., J. B. Stimmel, S. Clarke, J. Stock, and J. R. Broach. 1989. RAS2 protein of Saccharomyces cerevisiae is methyl-esterified at its carboxyl terminus. J. Biol. Chem. 264:11865-11873. [PubMed] [Google Scholar]
- 19.Dong, X., D. A. Mitchell, S. Lobo, L. Zhao, D. J. Bartels, and R. J. Deschenes. 2003. Palmitoylation and plasma membrane localization of Ras2p by a nonclassical trafficking pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:6574-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 21.Goodwin, J. S., K. R. Drake, C. Rogers, L. Wright, J. Lippincott-Schwartz, M. R. Philips, and A. K. Kenworthy. 2005. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J. Cell Biol. 170:261-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hancock, J. F. 2003. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 4:373-384. [DOI] [PubMed] [Google Scholar]
- 24.Hancock, J. F., H. Paterson, and C. J. Marshall. 1990. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63:133-139. [DOI] [PubMed] [Google Scholar]
- 25.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 26.Hrycyna, C. A., S. K. Sapperstein, S. Clarke, and S. M. Michaelis. 1991. The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and RAS proteins. EMBO J. 10:1699-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 28.Ito, H., Y. Fukada, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joneson, T., and D. Bar-Sagi. 1997. Ras effectors and their role in mitogenesis and oncogenesis. J. Mol. Med. 75:587-593. [DOI] [PubMed] [Google Scholar]
- 30.Kocher, H. M., R. Senkus, M. Al-Nawab, and B. M. Hendry. 2005. Subcellular distribution of Ras GTPase isoforms in normal human kidney. Nephrol. Dial. Transplant. 20:886-891. [DOI] [PubMed] [Google Scholar]
- 31.Kohl, N. E., R. E. Diehl, M. D. Schaber, E. Rands, D. D. Soderman, B. He, S. L. Moores, D. L. Pompliano, S. Ferro-Novick, S. Powers, K. A. Thomas, and J. B. Gibbs. 1991. Structural homology among mammalian and Saccharomyces cerevisiae isoprenyl-protein transferases. J. Biol. Chem. 266:18884-18888. [PubMed] [Google Scholar]
- 32.Kuchler, K. 1993. Unusual routes of protein secretion: the easy way out. Trends Cell Biol. 3:421-426. [DOI] [PubMed] [Google Scholar]
- 33.Lobo, S., W. K. Greentree, M. E. Linder, and R. J. Deschenes. 2002. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277:41268-41273. [DOI] [PubMed] [Google Scholar]
- 34.Luo, W., and A. Chang. 2000. An endosome-to-plasma membrane pathway involved in trafficking of a mutant plasma membrane ATPase in yeast. Mol. Biol. Cell 11:579-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manahan, C. L., M. Patnana, K. J. Blumer, and M. E. Linder. 2000. Dual lipid modification motifs in G(alpha) and G(gamma) subunits are required for full activity of the pheromone response pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 11:957-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall, M. S. 1995. Ras target proteins in eukaryotic cells. FASEB J. 9:1311-1318. [DOI] [PubMed] [Google Scholar]
- 37.Meisinger, C., T. Sommer, and N. Pfanner. 2000. Purification of Saccharomyces cerevisiae mitochondria devoid of microsomal and cytosolic contaminations. Anal. Biochem. 287:339-342. [DOI] [PubMed] [Google Scholar]
- 38.Mitts, M. R., J. Bradshaw-Rouse, and W. Heideman. 1991. Interactions between adenylate cyclase and the yeast GTPase-activating protein IRA1. Mol. Cell. Biol. 11:4591-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyajima, I., K. Arai, and K. Matsumoto. 1989. GPA1Val-50 mutation in the mating-factor signaling pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:2289-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mondola, P., G. Ruggiero, R. Seru, S. Damiano, S. Grimaldi, C. Garbi, M. Monda, D. Greco, and M. Santillo. 2003. The Cu,Zn superoxide dismutase in neuroblastoma SK-N-BE cells is exported by a microvesicles dependent pathway. Brain Res. Mol. Brain Res. 110:45-51. [DOI] [PubMed] [Google Scholar]
- 41.Morrow, I. C., S. Rea, S. Martin, I. A. Prior, R. Prohaska, J. F. Hancock, D. E. James, and R. G. Parton. 2002. Flotillin-1/reggie-2 traffics to surface raft domains via a novel Golgi-independent pathway. Identification of a novel membrane targeting domain and a role for palmitoylation. J. Biol. Chem. 277:48834-48841. [DOI] [PubMed] [Google Scholar]
- 42.Nantel, A., M. Huber, and D. Y. Thomas. 1999. Localization of endogenous Grb10 to the mitochondria and its interaction with the mitochondrial-associated Raf-1 pool. J. Biol. Chem. 274:35719-35724. [DOI] [PubMed] [Google Scholar]
- 43.Peterson, M. R., and S. D. Emr. 2001. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic 2:476-486. [DOI] [PubMed] [Google Scholar]
- 44.Philips, M. R. 2005. Compartmentalized signalling of Ras. Biochem. Soc. Trans. 33:657. [DOI] [PubMed] [Google Scholar]
- 45.Prior, I. A., A. Harding, J. Yan, J. Sluimer, R. G. Parton, and J. F. Hancock. 2001. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat. Cell Biol. 3:368-375. [DOI] [PubMed] [Google Scholar]
- 46.Raths, S., J. Rohrer, F. Crausaz, and H. Riezman. 1993. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 120:55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raymond, C. K., I. Howald-Stevenson, C. A. Vater, and T. H. Stevens. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3:1389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rebollo, A., D. Perez-Sala, and A. C. Martinez. 1999. Bcl-2 differentially targets K-, N-, and H-Ras to mitochondria in IL-2 supplemented or deprived cells: implications in prevention of apoptosis. Oncogene 18:4930-4939. [DOI] [PubMed] [Google Scholar]
- 49.Rizzuto, R., M. R. Duchen, and T. Pozzan. 2004. Flirting in little space: the ER/mitochondria Ca2+ liaison. Sci. STKE 2004:re1. [DOI] [PubMed] [Google Scholar]
- 50.Roberg, K. J., N. Rowley, and C. A. Kaiser. 1997. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J. Cell Biol. 137:1469-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rocks, O., A. Peyker, M. Kahms, P. J. Verveer, C. Koerner, M. Lumbierres, J. Kuhlmann, H. Waldmann, A. Wittinghofer, and P. I. Bastiaens. 2005. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307:1746-1752. [DOI] [PubMed] [Google Scholar]
- 52.Roeder, A. D., G. J. Hermann, B. R. Keegan, S. A. Thatcher, and J. M. Shaw. 1998. Mitochondrial inheritance is delayed in Saccharomyces cerevisiae cells lacking the serine/threonine phosphatase PTC1. Mol. Biol. Cell 9:917-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rose, M. D., and J. R. Broach. 1990. Cloning genes by complementation in yeast. Methods Enzymol. 194:195-229. [DOI] [PubMed] [Google Scholar]
- 54.Roy, S., S. Plowman, B. Rotblat, I. A. Prior, C. Muncke, S. Grainger, R. G. Parton, Y. I. Henis, Y. Kloog, and J. F. Hancock. 2005. Individual palmitoyl residues serve distinct roles in H-Ras trafficking, microlocalization, and signaling. Mol. Cell. Biol. 25:6722-6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sapperstein, S., C. Berkower, and S. Michaelis. 1994. Nucleotide sequence of the yeast STE14 gene, which encodes farnesylcysteine carboxyl methyltransferase, and demonstration of its essential role in a-factor export. Mol. Cell. Biol. 14:1438-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato, T. K., P. Rehling, M. R. Peterson, and S. D. Emr. 2000. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell 6:661-671. [DOI] [PubMed] [Google Scholar]
- 57.Schaber, M. D., M. B. O'Hara, V. M. Garsky, S. D. Mosser, J. D. Bergstrom, S. L. Moores, M. S. Marshall, P. A. Friedman, R. A. F. Dixon, and J. B. Gibbs. 1990. Polyisoprenylation of Ras in vitro by a farnesyl-protein transferase. J. Biol. Chem. 265:14701-14704. [PubMed] [Google Scholar]
- 58.Schmidt, W. K., A. Tam, K. Fujimura-Kamada, and S. Michaelis. 1998. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc. Natl. Acad. Sci. USA 95:11175-11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmitt, S., H. Prokisch, T. Schlunck, D. G. Camp II, U. Ahting, T. Waizenegger, C. Scharfe, T. Meitinger, A. Imhof, W. Neupert, P. J. Oefner, and D. Rapaport. 2006. Proteome analysis of mitochondrial outer membrane from Neurospora crassa. Proteomics 6:72-80. [DOI] [PubMed] [Google Scholar]
- 60.Seals, D. F., G. Eitzen, N. Margolis, W. T. Wickner, and A. Price. 2000. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA 97:9402-9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual: methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 62.Sickmann, A., J. Reinders, Y. Wagner, C. Joppich, R. Zahedi, H. E. Meyer, B. Schonfisch, I. Perschil, A. Chacinska, B. Guiard, P. Rehling, N. Pfanner, and C. Meisinger. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA 100:13207-13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Silvius, J. R., P. Bhagatji, R. Leventis, and D. Terrone. 2005. K-ras4B and prenylated proteins lacking “second signals” associate dynamically with cellular membranes. Mol. Biol. Cell 17:192-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simmen, T., J. E. Aslan, A. D. Blagoveshchenskaya, L. Thomas, L. Wan, Y. Xiang, S. F. Feliciangeli, C. H. Hung, C. M. Crump, and G. Thomas. 2005. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 24:717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sobering, A. K., M. J. Romeo, H. A. Vay, and D. E. Levin. 2003. A novel Ras inhibitor, Eri1, engages yeast Ras at the endoplasmic reticulum. Mol. Cell. Biol. 23:4983-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Srivastava, A., C. A. Woolford, and E. W. Jones. 2000. Pep3p/Pep5p complex: a putative docking factor at multiple steps of vesicular transport to the vacuole of Saccharomyces cerevisiae. Genetics 156:105-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steinmetz, L. M., C. Scharfe, A. M. Deutschbauer, D. Mokranjac, Z. S. Herman, T. Jones, A. M. Chu, G. Giaever, H. Prokisch, P. J. Oefner, and R. W. Davis. 2002. Systematic screen for human disease genes in yeast. Nat. Genet. 31:400-404. [DOI] [PubMed] [Google Scholar]
- 69.Stevens, B. J., and J. G. White. 1979. Computer reconstruction of mitochondria from yeast. Methods Enzymol. 56:718-728. [DOI] [PubMed] [Google Scholar]
- 70.Subramanian, S., C. A. Woolford, and E. W. Jones. 2004. The Sec1/Munc18 protein, Vps33p, functions at the endosome and the vacuole of Saccharomyces cerevisiae. Mol. Biol. Cell 15:2593-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Swarthout, J. T., S. Lobo, L. Farh, M. R. Croke, W. K. Greentree, R. J. Deschenes, and M. E. Linder. 2005. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J. Biol. Chem. 280:31141-31148. [DOI] [PubMed] [Google Scholar]
- 72.Takida, S., and P. B. Wedegaertner. 2004. Exocytic pathway-independent plasma membrane targeting of heterotrimeric G proteins. FEBS Lett. 567:209-213. [DOI] [PubMed] [Google Scholar]
- 73.Voelker, D. R. 2003. New perspectives on the regulation of intermembrane glycerophospholipid traffic. J. Lipid Res. 44:441-449. [DOI] [PubMed] [Google Scholar]
- 74.Watson, R. T., M. Furukawa, S. H. Chiang, D. Boeglin, M. Kanzaki, A. R. Saltiel, and J. E. Pessin. 2003. The exocytotic trafficking of TC10 occurs through both classical and nonclassical secretory transport pathways in 3T3L1 adipocytes. Mol. Cell. Biol. 23:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao, L., S. Lobo, X. Dong, A. D. Ault, and R. J. Deschenes. 2002. Erf4p and Erf2p form an endoplasmic reticulum-associated complex involved in the plasma membrane localization of yeast Ras proteins. J. Biol. Chem. 277:49352-49359. [DOI] [PubMed] [Google Scholar]