Abstract
Translation initiation in eukaryotic cells is known to be a complex multistep process which involves numerous protein factors. Here we demonstrate that leaderless mRNAs with initiator Met-tRNA can bind directly to 80S mammalian ribosomes in the absence of initiation factors and that the complexes thus formed are fully competent for the subsequent steps of polypeptide synthesis. We show that the canonical 48S pathway of eukaryotic translation initiation has no obvious advantage over the 80S pathway of translation initiation on leaderless mRNAs and suggest that, in the presence of competing mRNAs containing a leader, the latter mechanism will be preferred. The direct binding of the leaderless mRNA to the 80S ribosome was precluded when such an mRNA was supplied with a 5′ leader, irrespective of whether it was in a totally single-stranded conformation or was prone to base pairing. The striking similarity between the mechanisms of binding of leaderless mRNAs with mammalian 80S or bacterial 70S ribosomes gives support to the idea that the alternative mode of translation initiation used by leaderless mRNAs represents a relic from early steps in the evolution of the translation apparatus.
More than a decade ago, we showed using the toeprint assay that the prokaryotic leaderless mRNA coding for the cI repressor of lambda phage assisted by initiator Met-tRNA was able to bind directly to undissociated 70S ribosomes in the absence of initiation factor 1 (IF1), IF2, and IF3 (1). Under the same conditions, no initiation complex was observed with the 30S ribosomal subunit. In the presence of initiation factors, the 30S subunit invariably bound to internal sites on the mRNA immediately downstream of the closest Shine-Dalgarno (SD) sequence, whereas undissociated 70S ribosome strongly preferred the initiation triplet as close as possible to the 5′ end of the mRNA. These studies were substantially extended and confirmed in other laboratories both in vitro and in vivo (12, 13, 15, 28) and resulted in similar conclusions. Natural leaderless mRNAs have been identified in all kingdoms of life (see references 10 and 11). This class of mRNAs is well represented in archaea and mammalian mitochondria. Although they have not yet been found among cellular mammalian mRNAs, leaderless monocistronic N and dicistronic N-P RNAs of measles virus were found to be associated with polysomes in infected mammalian cells (4). In addition, it is unlikely that a systematic search for such mRNAs in mammalian cells has ever been undertaken. There is one well-documented example of a translatable leaderless mRNA in unicellular eukaryotes. It encodes a variant specific surface protein of Giardia lamblia and is certainly translated in these cells (14). Leaderless mRNAs are efficiently translated in mammalian cell-free systems (see reference 12). However, it is not known whether they use a similar (80S) pathway for translation initiation in eukaryotes, given the remarkable quantitative and qualitative differences in the organization of eukaryotic and prokaryotic translation initiation machinery.
Unlike bacteria and lower eukaryotes, mammalian cells are poorly amenable to genetic approaches to dissect molecular mechanisms of gene expression. To study pathways of recruitment of leaderless mRNAs onto mammalian 80S ribosomes, we have decided to use our system of assembly of mammalian translation complexes from totally purified components combined with toeprinting (16). This approach has proven to be very fruitful in elucidating initiation factor requirements for several viral internal ribosome entry site elements (for a review, see reference 9) and standard cap-dependent mRNAs (6, 17). Here, using this technique, we show for the first time that leaderless mRNAs with initiator Met-tRNA can bind directly to 80S mammalian ribosomes in the absence of initiation factors and that the complexes thus formed are fully competent for the subsequent steps of polypeptide synthesis. As with 70S ribosomes, the direct binding of a leaderless mRNA to the 80S ribosome was precluded when such an mRNA was supplied with a 5′ leader. We present interpretations of these effects which are based on the current model of organization of the mRNA-binding site in the ribosome.
MATERIALS AND METHODS
Plasmid constructs.
The initial construct pcIlacZ, containing the T7 promoter, one extra G for more efficient T7 transcription, the first 13 triplets of cI, and most of the lacZ coding sequence, was created using the backbone of vector pUC18. The cI fragment was produced by PCR from the cI plasmid previously described (1), using a forward primer containing the EcoRI restriction site and the T7 promoter and a reverse primer containing the site for AvrII. The lacZ fragment was also prepared by PCR from a lacZ-containing plasmid using a forward primer containing the AvrII restriction site and a reverse primer containing the termination codon for lacZ and the BamHI site. After cleavage with the corresponding restriction endonucleases, the fragments were ligated and inserted between the EcoRI-BamHI sites of pUC18.
The construct pCAA-cIlacZ was obtained using plasmid pCAA-GUS (27) by replacement of the GUS-coding sequence with the coding sequence from pcIlacZ. For the construction of pbAct-cIlacZ, the coding sequence of plasmid pAbG (6) was replaced by that from pcIlacZ in the same way as that for the construct pCAA-cIlacZ. By PCR amplification of the construct pcIlacZ using oligonucleotides containing the T7 promoter, corresponding mutations, and a part of the downstream coding sequence of the cI mRNA as direct primers and a common reverse primer containing the AvrII restriction site (the same primer as that which was used to create the initial plasmid pcIlacZ), followed by ligation of these PCR products and insertion into pUC18 at the SmaI site, the following constructs were prepared: (i) constructs pcI(GTG) and pcI(TAA), where the ATG initiation codon was replaced by GTG and TAA, respectively; (ii) construct pcIA4G2, where the leader GAAAAG was inserted between the T7 promoter and the cI AUG initiation codon; (iii) construct pcIG4C2, which contained the 5′-untranslated region (5′-UTR) GGGCCG followed by the cI coding sequence starting with its AUG initiation codon; and (iv) construct pcI(TAA-7), where the seventh triplet of the cI coding sequence was replaced by the termination codon TAA.
In vitro transcription and translation.
The initial plasmid pcIlacZ and all its derivatives were linearized prior to T7 transcription by digestion with AvrII or HindIII for toeprinting or in vitro translation, respectively. For sucrose gradient analysis, the RNA was synthesized in the presence of [α-32P]UTP as described previously (16). To obtain a double-stranded RNA (dsRNA) for activation of the protein kinase R (PKR) activity in rabbit reticulocyte lysate (RRL), the long polylinker of pBS DNA was transcribed in opposite directions from the T7 or T3 promoter. The complementary RNAs thus obtained were annealed, and the resulting dsRNA (∼60 bp) was digested with nuclease S1 to remove unpaired nucleotide sequences followed by phenol purification. The RRL translation samples were supplied with the indicated amounts (see Fig. 4C) of the dsRNA and incubated for 5 min at 30°C, followed by addition of the mRNAs. In vitro translation in RRL was performed in the presence of [35S]methionine (Amersham Biosciences) essentially as recommended by the manufacturer (Promega). To observe the dsRNA-induced phosphorylation of translation initiation factor 2α (eIF2α), 20 μl of the RRL translation mixture was supplied with an additional amount of eIF2 (4.5 μg), 200 ng of dsRNA, and 6 μCi of [γ-32P]ATP. The mixture was incubated for 30 min at 30°C, followed by addition of 20 μl of goat eIF2 antiserum (kindly provided by W. C. Merrick) and 460 μl of binding buffer (BB; 50 mM Tris-HCl, pH 8.0, 120 mM NaCl, 1 mM EDTA, and 0.5% Tween 20). After a gentle shaking for 3 h at 0°C, the mixture was combined with 40 μl of 50% suspension of A-protein Sepharose (Amersham Biosciences), and the incubation was continued for 1 h more at 0°C. The Sepharose was then carefully washed with BB, the adsorbed proteins were eluted by boiling in the sample buffer for sodium dodecyl sulfate (SDS)-electrophoresis, and the proteins were resolved by SDS-12% polyacrylamide gel electrophoresis (PAGE). The protein bands were stained with Coomassie blue, and their radioactivity was visualized with a PhosphorImager (Molecular Dynamics).
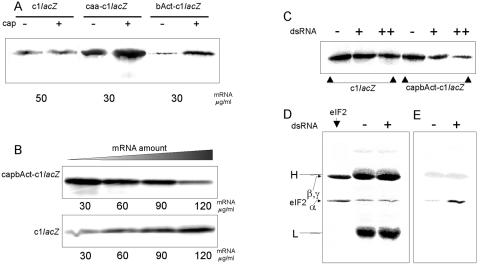
FIG. 4.
Translation of the leaderless cIlacZ mRNA and leadered mRNAs possessing the same coding sequence in RRL. (A) Effect of capping of the leaderless and leadered transcripts on their translational efficiency. Constructs caa-cIlacZ and bAct-cIlacZ contained the (CAA)19 5′ leader and the 5′-UTR from the beta-actin mRNA, respectively. (B) Effect of mRNA concentration in the cell-free system on the translation efficiency of cIlacZ mRNA and a capped mRNA where the 5′UTR of beta-actin mRNA was fused to the cIlacZ coding sequence (capbAct-cIlacZ). (C) Effect of the PKR activity induced in RRL by incubation with the dsRNA on the translation of the cIlacZ mRNA and the capbAct-cIalcZ mRNA. Signs “+” and “++” correspond to the additions of 10 and 100 ng of dsRNA per 10 μl of the translation mixture, respectively. (D and E) dsRNA-induced phosphorylation of eIF2α. (D) Electrophoresis of proteins eluted from anti-eIF2 adsorbents. The gel was stained with Coomassie blue. “H” and “L” denote the heavy and light chains of immunoglobulin G. (E) The central and right lanes of the gel shown in panel D but developed with PhosphorImager. For other experimental details, see Materials and Methods.
Preparation of 40S ribosomal subunits, 80S ribosomes, initiation factors, and Met-tRNA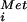 .
.
The 40S and 60S ribosomal subunits and mammalian translation initiation factors were purified from cytoplasmic extracts of Krebs-2 ascites cells as indicated previously (16, 17). Elongation factor eEF1H was purified to homogeneity as described previously (21). The homogeneous eEF2 was kindly provided by L. Ovchinnikov.
To prepare 80S ribosomes, 10 A260 units of 40S subunits and 20 A260 units of 60S subunits were reassociated in buffer [20 mM Tris-HCl, pH 7.5, 150 mM KCl, 6 mM Mg(Ac)2, 1 mM dithiothreitol (DTT)], incubated for 10 min at 30°C, layered onto a 5 to 20% sucrose gradient prepared with the same buffer, and centrifuged at 16,000 rpm for 13 h at 4°C. The 80S fractions of the gradients were combined, and the reassociated 80S ribosomes were pelleted and dissolved in 20 mM Tris-HCl, pH 7.5, 0.25 M sucrose, 50 mM KCl, 1 mM Mg(Ac)2, 1 mM EDTA, and 1 mM DTT. The individual 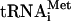 from Escherichia coli was kindly provided by V. Makhno and Y. Semenkov. It was aminoacylated using recombinant MetRSase (24) according to the protocol described previously for the total aminoacyl tRNA synthetase (ARSase) pool from E. coli cells (16). The total aminoacylated tRNA was prepared from calf liver tRNA (Novagen) using a protocol similar to that which was employed for the aminoacylation of
from Escherichia coli was kindly provided by V. Makhno and Y. Semenkov. It was aminoacylated using recombinant MetRSase (24) according to the protocol described previously for the total aminoacyl tRNA synthetase (ARSase) pool from E. coli cells (16). The total aminoacylated tRNA was prepared from calf liver tRNA (Novagen) using a protocol similar to that which was employed for the aminoacylation of 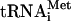 . The S100 supernatant from Krebs-2 ascites cells, freed from nucleic acids by passing through DEAE-cellulose with 0.25 M KCl and ammonium sulfate precipitation, was used to provide a source of mammalian ARSases.
. The S100 supernatant from Krebs-2 ascites cells, freed from nucleic acids by passing through DEAE-cellulose with 0.25 M KCl and ammonium sulfate precipitation, was used to provide a source of mammalian ARSases.
Assembly and analysis of 48S and 80S complexes.
The reconstitution of 48S complexes from purified components and their analysis by toeprinting were performed as described previously (5, 6, 16, 17). For the assembly and toeprinting analysis of 80S translation initiation complexes, the reaction mixture (20 μl) contained 80S reassociated ribosomes (2.5 pmol), an mRNA (0.5 pmol), Met-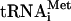 (5 pmol), and, where indicated, the initiation factors at the same molar ratio as for the assembly and analysis of 48S translation initiation complexes (5). The mixture was incubated for 5 min at 30°C and then analyzed by toeprinting or sucrose gradient sedimentation. To perform the step of polypeptide elongation from preformed 80S initiation complexes, the 80S ribosomes were first incubated in 10 μl with cI(TAA-7) mRNA and Met-
(5 pmol), and, where indicated, the initiation factors at the same molar ratio as for the assembly and analysis of 48S translation initiation complexes (5). The mixture was incubated for 5 min at 30°C and then analyzed by toeprinting or sucrose gradient sedimentation. To perform the step of polypeptide elongation from preformed 80S initiation complexes, the 80S ribosomes were first incubated in 10 μl with cI(TAA-7) mRNA and Met-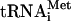 for 5 min at 30°C, followed by adding elongation factors eEF1H (4 μg) and eEF2 (1 μg), total aminoacylated tRNA (2 μg), and 0.2 mM GTP or GMPPNP, the nonhydrolyzable analog of GTP, and a further incubation for 10 min at 30°C. Primer extension analysis was performed with the oligonucleotide 5′-CCAGGGTTTTCCCAGTCACG-3′ that is complementary to nucleotides 63 to 83 from the 5′ end of cIlacZ mRNA. Analysis of the resulting cDNA was performed using denaturing 6% PAGE as described before (5). Radioactive bands were visualized with a PhosphorImager (Molecular Dynamics). The yield of 48S and 80S complexes was determined as the ratio of the radioactivity in the toeprint bands compared to the total radioactivity in the corresponding lanes, including the toeprint bands, using ImageQuant 5.0 software.
for 5 min at 30°C, followed by adding elongation factors eEF1H (4 μg) and eEF2 (1 μg), total aminoacylated tRNA (2 μg), and 0.2 mM GTP or GMPPNP, the nonhydrolyzable analog of GTP, and a further incubation for 10 min at 30°C. Primer extension analysis was performed with the oligonucleotide 5′-CCAGGGTTTTCCCAGTCACG-3′ that is complementary to nucleotides 63 to 83 from the 5′ end of cIlacZ mRNA. Analysis of the resulting cDNA was performed using denaturing 6% PAGE as described before (5). Radioactive bands were visualized with a PhosphorImager (Molecular Dynamics). The yield of 48S and 80S complexes was determined as the ratio of the radioactivity in the toeprint bands compared to the total radioactivity in the corresponding lanes, including the toeprint bands, using ImageQuant 5.0 software.
RESULTS
The leaderless λ phage cI mRNA forms an 80S-mRNA-Met-tRNAiMet complex in the absence of eukaryotic translation initiation factors.
To analyze binding properties of leaderless mRNAs with the 40S and 80S ribosomes, we deliberately selected as a model an mRNA that starts with the 5′-terminal sequence of the leaderless λ phage cI mRNA, i.e., the same sequence which was used in similar studies with E. coli 70S ribosomes (1). The cIlacZ mRNA used in this work lacks all translation initiation regulatory elements inherent to eukaryotic or prokaryotic mRNAs and hence is an ideal model to reveal parallels between translation initiation on eukaryotic and prokaryotic ribosomes. To exclude any contamination of 80S ribosomes with translation initiation factors, 80S ribosomes reassociated from KCl-washed and purified 40S and 60S ribosomes followed by their separation from free ribosomal subunits by sucrose gradient sedimentation were used throughout these studies. The complexes of 80S ribosomes with the cIlacZ mRNA were assembled in the presence of prokaryotic 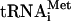 , which fully substituted for the mammalian
, which fully substituted for the mammalian 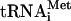 in the formation of 40S and 80S translation initiation complexes (see below). As seen from Fig. 1A and B, lane 4, the cIlacZ mRNA formed a stable ternary complex with the 80S ribosome and Met-
in the formation of 40S and 80S translation initiation complexes (see below). As seen from Fig. 1A and B, lane 4, the cIlacZ mRNA formed a stable ternary complex with the 80S ribosome and Met-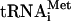 , whereas no such complex was observed for the 40S subunit (Fig. 1B, lane 2). The replacement of the 5′-terminal AUG codon by GUG resulted in a dramatic decrease of the intensity of the toeprint signal (Fig. 1B, lane 5) which only slightly exceeded the background intensity observed for the RNA with the AUG→UAA replacement (Fig. 1B, lane 6). This indicates that the 5′-terminal AUG triplet plays a critical role in the stability of the 80S-mRNA-Met-
, whereas no such complex was observed for the 40S subunit (Fig. 1B, lane 2). The replacement of the 5′-terminal AUG codon by GUG resulted in a dramatic decrease of the intensity of the toeprint signal (Fig. 1B, lane 5) which only slightly exceeded the background intensity observed for the RNA with the AUG→UAA replacement (Fig. 1B, lane 6). This indicates that the 5′-terminal AUG triplet plays a critical role in the stability of the 80S-mRNA-Met-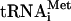 ternary complex.
ternary complex.
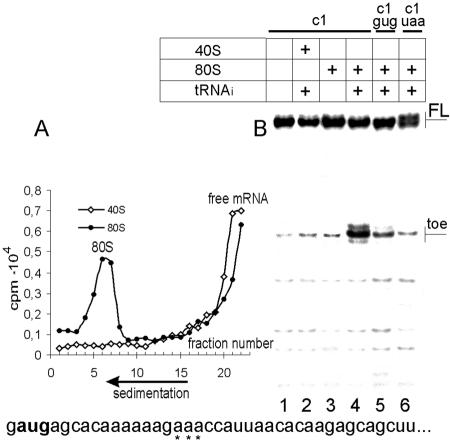
FIG. 1.
Formation of the ternary complex 80S ribosome-cIlacZ mRNA-Met-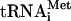 . (A) Sucrose gradient sedimentation of the incubation mixtures of the cI mRNA and Met-
. (A) Sucrose gradient sedimentation of the incubation mixtures of the cI mRNA and Met-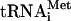 with mammalian 40S ribosomal subunits or 80S ribosomes. (B) The toeprint analysis of the same mixtures. Lanes 5 and 6 show toeprinting for mutant cIlacZ mRNAs where the initiation AUG codon was replaced by the GUG and UAA triplets, respectively. The 5′-terminal sequence of cIlacZ mRNA is shown at the bottom of the figure. Positions of toeprint bands are indicated by asterisks.
with mammalian 40S ribosomal subunits or 80S ribosomes. (B) The toeprint analysis of the same mixtures. Lanes 5 and 6 show toeprinting for mutant cIlacZ mRNAs where the initiation AUG codon was replaced by the GUG and UAA triplets, respectively. The 5′-terminal sequence of cIlacZ mRNA is shown at the bottom of the figure. Positions of toeprint bands are indicated by asterisks.
The complex 80S-cI mRNA-Met-tRNAiMet formed without translation initiation factors is totally competent for the elongation step of polypeptide synthesis.
It may be argued that the 80S-cIlacZ mRNA-Met-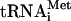 complex formed in the absence of translation initiation factors may be nonfunctional or required an additional initiation factor to be converted into a form competent for the subsequent elongation of the polypeptide. To check this possibility, the seventh triplet of the cI coding sequence was mutated into the nonsense UAA codon. Such an mRNA was used in the assay where the preformed 80S ternary complexes were supplied with elongation factors eEF1H and eEF2, and the total aminoacylated mammalian tRNA, incubated for an additional 10 min prior to analysis by toeprinting. If the 80S-cI mRNA-Met-
complex formed in the absence of translation initiation factors may be nonfunctional or required an additional initiation factor to be converted into a form competent for the subsequent elongation of the polypeptide. To check this possibility, the seventh triplet of the cI coding sequence was mutated into the nonsense UAA codon. Such an mRNA was used in the assay where the preformed 80S ternary complexes were supplied with elongation factors eEF1H and eEF2, and the total aminoacylated mammalian tRNA, incubated for an additional 10 min prior to analysis by toeprinting. If the 80S-cI mRNA-Met-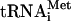 is able to be engaged in the synthesis of polypeptide, then the ribosomes having attained the termination codon must stop at the sixth triplet of the cIlacZ mRNA in the P site of the ribosome. In this case, the resulting toeprint bands should be observed at 15 nucleotides downstream from those originating from the initiation AUG codon. As seen from Fig. 2, this is exactly what was observed in the experiment: addition of eEF1H and eEF2 produced a toeprint at the expected position (Fig. 2, lane 4). No toeprint at this position was seen when either of the elongation factors was omitted from the incubation mixture or when GTP was substituted for its nonhydrolyzable analogue GMPPNP. Thus, the 80S-cIlacZ mRNA-Met-
is able to be engaged in the synthesis of polypeptide, then the ribosomes having attained the termination codon must stop at the sixth triplet of the cIlacZ mRNA in the P site of the ribosome. In this case, the resulting toeprint bands should be observed at 15 nucleotides downstream from those originating from the initiation AUG codon. As seen from Fig. 2, this is exactly what was observed in the experiment: addition of eEF1H and eEF2 produced a toeprint at the expected position (Fig. 2, lane 4). No toeprint at this position was seen when either of the elongation factors was omitted from the incubation mixture or when GTP was substituted for its nonhydrolyzable analogue GMPPNP. Thus, the 80S-cIlacZ mRNA-Met-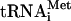 complex formed without translation initiation factors was totally competent for the elongation step of polypeptide synthesis.
complex formed without translation initiation factors was totally competent for the elongation step of polypeptide synthesis.
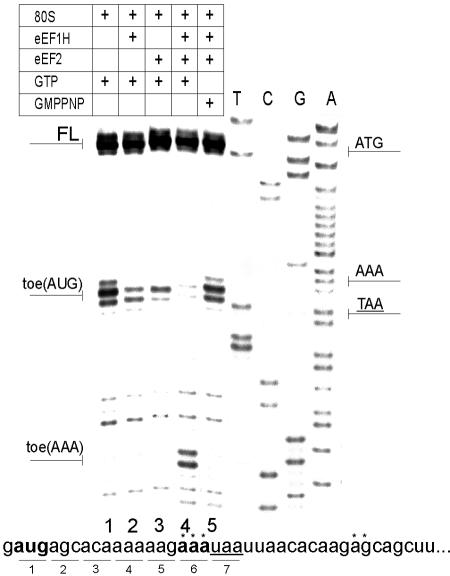
FIG. 2.
Elongation of the polypeptide by the 80S ribosome-cI mRNA-Met-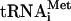 complex assembled without translation initiation factors. Positions of the toeprint bands originated from the AUG initiation codon and the triplet (AAA) preceding the termination codon are shown on the left of the gel. A dideoxynucleotide sequence generated with the same primer was run in parallel (shown on the right of the gel). The full-length product of primer extension is denoted “FL.” The sequence of the mutant cI(UAA-7) mRNA is presented below the autograph. The termination nucleotide triplet is underlined, and the initiation codon and the codon preceding the termination one are shown in boldface. Asterisks above the sequence show positions of the toeprint bands.
complex assembled without translation initiation factors. Positions of the toeprint bands originated from the AUG initiation codon and the triplet (AAA) preceding the termination codon are shown on the left of the gel. A dideoxynucleotide sequence generated with the same primer was run in parallel (shown on the right of the gel). The full-length product of primer extension is denoted “FL.” The sequence of the mutant cI(UAA-7) mRNA is presented below the autograph. The termination nucleotide triplet is underlined, and the initiation codon and the codon preceding the termination one are shown in boldface. Asterisks above the sequence show positions of the toeprint bands.
Formation of 48S translation initiation complexes on the cIlacZ mRNA.
Since under natural conditions 40S subunits are present in the system, it was important to see whether and how efficiently the cIlacZ mRNA formed complexes with 40S subunits in the presence of translation initiation factors. As seen from Fig. 3 (lanes 1 to 4), such complexes did form, and in the presence of initiation factors 1A, 2, 3, 4A, and 4F (eIF4B was not used in the assembly, since it is required only for mRNAs with structured 5′-UTRs [6]), their assembly occurred more efficiently than with 80S ternary complexes (compare lanes 2 and 8). The stimulatory effect of eIF4F (in the absence of eIF1) on the binding of uncapped, leaderless, and single-stranded cI mRNAs with the 40S ribosomal subunit was surprising. This intriguing issue is now under investigation. However, when the incubation mixture was complemented with the missing factor eIF1, the yield of the 48S complex dramatically dropped and became comparable to that for the 80S complex assembled in the absence of initiation factors (compare lanes 1 and 8). Thus, eIF1 should counteract leaderless mRNAs from using the standard 40S pathway of eukaryotic translation initiation.
FIG. 3.
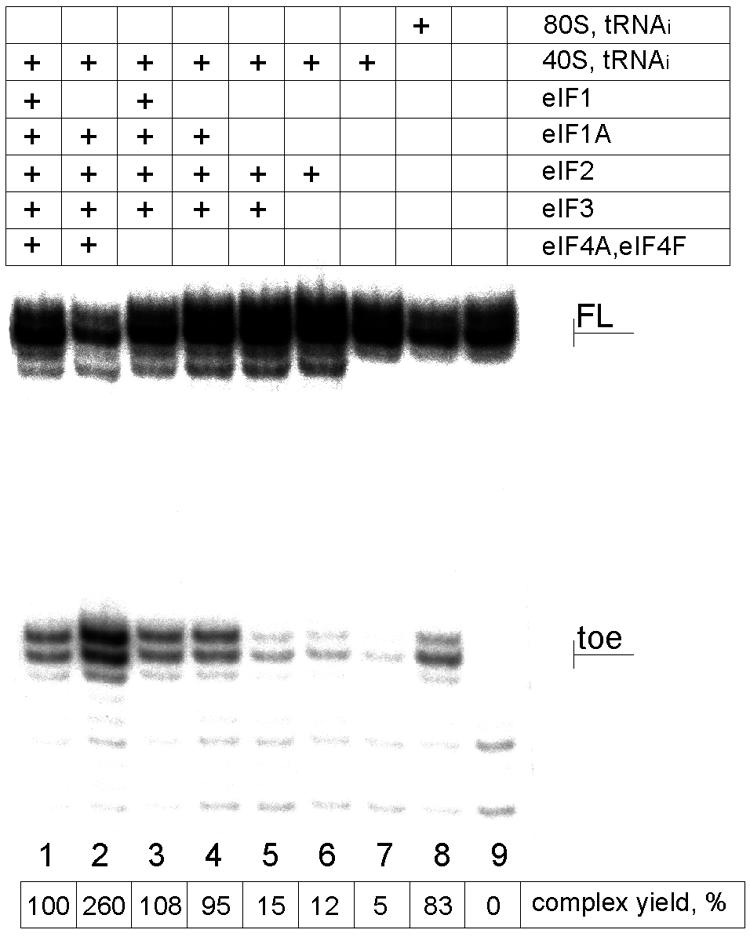
Effect of translation initiation factors on the formation of 48S translation initiation complexes. The molar concentrations of 40S ribosomes and cIlacZ mRNA used in these assays were exactly the same as those in the experiments on formation of 80S ribosome-cI mRNA-Met-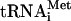 complexes. The yield of 48S initiation complexes was estimated as described in Materials and Methods and is presented below the toeprints. The values represent the average from three independent assembly experiments. For other designations, see the legends to Fig. 1 and 2.
complexes. The yield of 48S initiation complexes was estimated as described in Materials and Methods and is presented below the toeprints. The values represent the average from three independent assembly experiments. For other designations, see the legends to Fig. 1 and 2.
Translational properties of cIlacZ mRNA in RRL confirm its relaxed dependence on mRNA-recruiting initiation factors.
The translational behavior of cIlacZ mRNA in RRL was compared with that for leadered control mRNAs. All these mRNAs carried the same coding sequence. One of these control mRNAs had the 5′-UTR of beta-actin mRNA, whereas another one carried a totally unstructured (CAA)19 5′ leader (27). The cIlacZ mRNA was translated with similar efficiency, whether it was capped or uncapped. In contrast, the capping strongly stimulated the translation of both beta-actin and (CAA)19 mRNAs, suggesting that eIF4E was not essential for the translational initiation on the leaderless cIlacZ mRNA (Fig. 4A). As seen in Fig. 4B, the translation of the mRNA with the beta-actin 5′-UTR was inversely proportional to the amount of RNA added to the RRL system, whereas, in contrast, the translation of cIlacZ mRNA progressively increased in parallel with the concentration of the leaderless mRNA. As has been recently demonstrated in our laboratory (7), the inhibition of translation of mRNAs in RRL at their elevated concentrations is primarily accounted for by a gradual sequestration of initiation factor eIF2 at spurious sites on an mRNA, since replenishment of the system with eIF2 fully restored the translation. Clearly, the leaderless cI mRNA easily tolerated sequestering of eIF2 and probably of other mRNA binding initiation factors.
To confirm this conclusion, we induced the PKR activity in our cell-free system by treatment of the RRL with a dsRNA. PKR is known to phosphorylate the α subunit of eIF2, thereby sequestering this initiation factor in an inactive complex with initiation factor eIF2B. As evident from Fig. 4C, the translation of the leadered mRNA was strongly inhibited by the dsRNA, whereas only a weak effect was observed for the leaderless cIlacZ mRNA. The data which confirm the dsRNA-induced phosphorylation of eIF2α in our system are presented in Fig. 4D and E.
The 5′-terminal position of the initiation codon and the downstream sequence are essential elements for the ability of leaderless mRNAs to program 80S ribosomes.
Addition of 5′ leaders to the cI coding sequence, irrespective of whether the 5′-UTR was derived from a natural mRNA (beta-actin mRNA) or was artificial and possessed a 100% single-stranded conformation (CAA19 leader), completely abrogated the ability of such mRNAs to bind 80S ribosomes (Fig. 5, lanes 1 and 2). This inhibitory effect of 5′-UTRs was even observed for much shorter leaders, such as GAAAAGAUG and GGGCCGAUG (Fig. 5, lanes 3, and 4), being significantly higher for the GC-rich leader. This is probably accounted for by a higher potential of G residues compared to A residues for base pairing, thereby resulting in a more drastic change of the initial (competent for the direct binding with 80S ribosomes) conformation of the cI 5′-terminal segment. Thus, the very 5′-terminal position of the initiation triplet is one of the essential features of the cI mRNA which allows it to use the 80S pathway of translation initiation. Another essential feature may be the structure or nature of the sequence immediately downstream from the AUG codon. The single-stranded nature of the 5′ terminus of cI mRNA has been shown experimentally (see reference 1). In contrast, a leaderless mRNA derived from the beta-actin mRNA did not show any toeprint with 80S ribosomes. The computer-predicted folding of the coding sequence of the beta-actin mRNA gave several alternative structures. All of them showed extensive base pairing within the part of the beta-globin coding sequence which should be covered by the ribosome in 80S-mRNA-Met-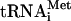 complexes (data not shown).
complexes (data not shown).
FIG. 5.
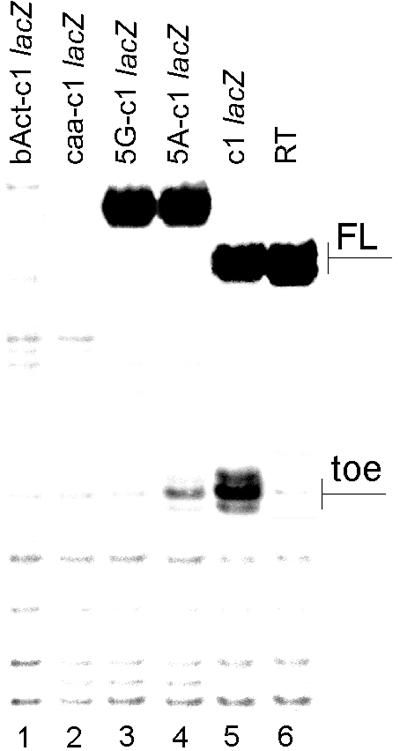
Effect of 5′-UTR additions and a nucleotide context of the AUG codon of cIlacZ mRNA on its ability to directly program 80S ribosomes. Abbreviated names of RNA constructs are shown above the lanes. The same cIlacZ coding sequence was linked to the beta-actin 5′-UTR (bAct-cIlacZ), unstructured (CAA)19 leader (caa-cIlacZ), and short leaders GGGCCGAUG (G4C2-cIlacZ) or GAAAAGAUG (A4G2-cIlacZ). The full-length products for the first two constructs are situated in the gel much above those for other transcripts. For space limitations, they are not shown in the figure.
DISCUSSION
As demonstrated in this paper, the binding of the leaderless cIlacZ mRNA to 80S mammalian ribosomes can occur without participation of any translation initiation factors, and the 80S initiation complex thus formed is fully competent for the subsequent elongation steps of polypeptide synthesis. In the purified in vitro system, the cIlacZ mRNA can also employ the classical mechanism of initiation, i.e., that mediated by the 40S ribosomal subunit loaded with translation initiation factors. However, in the presence of all translation initiation factors (including eIF1), the 40S pathway has no obvious advantage over the 80S pathway for the leaderless cIlacZ mRNA, at least in our experimental system. Therefore, it is quite reasonable to suggest that under less “ideal” conditions, when a leaderless mRNA is forced to compete for translation initiation components with the bulk of mRNAs containing a 5′ leader sequence, the 80S pathway will be preferred. The translation characteristics of the cIlacZ mRNA in RRL are quite distinct from those for 5′-UTR-containing mRNAs: (i) its translation continues to rise at concentrations when a control mRNA, containing a 5′ leader and the same coding sequence, already shows a dramatic decline in the yield of the encoded polypeptide; (ii) it does not depend on the cap; (iii) it easily tolerates strong inhibition of eIF2 activity. These facts offer additional arguments, albeit indirect, to support our conclusion that leaderless mRNAs have their own niche to initiate the polypeptide synthesis. It may be that this alternative pathway for forming the 80S initiation complex would allow leaderless mRNAs to avoid competition with standard mRNAs, thereby maintaining a minimum level of the proteins they encode under conditions of unfavorable cell growth.
A strong destabilizing effect of initiation factor eIF1 on the formation of 48S complexes at 5′-terminal initiation codons has been recently demonstrated (20) and confirmed in this paper. eIF1 triggers the process of scanning of 5′ leaders of eukaryotic mRNAs in search of an appropriate initiation codon in a good nucleotide context at both sides of the codon and is indispensable for standard mRNAs, whereas in the case of a leaderless mRNA the ribosome should remain at the 5′-terminal AUG triplet. The functional analog of eIF1, prokaryotic IF3, certainly plays a similar mechanistic role (1, 26).
The inhibitory effect of extra nucleotides added to the 5′-terminal initiation triplet reveals a further parallel with the analogous bacterial system. Addition of even short sequences to the 5′ terminus of cI mRNA causes a strong deleterious effect on the formation of 80S initiation complexes in the absence of initiation factors. It should be noted, however, that GC-poor and very short 5′-UTRs still allow mRNAs to bind 80S ribosomes, though with a low efficiency (see Fig. 5). One may not rule out that such mRNAs use to some extent the 80S pathway along with the canonical 48S pathway of the translation initiation. The efficiency of the direct 80S binding is also sensitive to the nucleotide composition of the sequence downstream from the 5′-terminal initiation codon. Our interpretation of these data is based on the current three-dimensional structures of 70S (80S) ribosomes derived from crystallographic (30) and cryoelectron microscopy studies (25) and our hypothesis that the mRNA forms a U-turn as it passes along the mRNA binding channel of the small ribosomal subunit (8, 23). This hypothesis has found support in recent crystallographic studies of the ternary complex 70S ribosome -tRNA-mRNA (31). One may conclude from these data that the 5′-untranslated region should be confined between the ribosomal subunits, whereas the sequence downstream from the AUG codon is open to the solvent. Presumably, it is difficult for an mRNA with a 5′ leader to diffuse between the subunits without dissociation of 80S (or 70S) ribosomes and assistance of scanning initiation factors. Base pairing within the 5′-UTRs or between them and the coding parts of an mRNA present additional topological and energy barriers to the direct mRNA accommodation into the mRNA-binding channel of 80S (70S) ribosomes, since the 80S (70S) ribosome by itself is unable to unwind the secondary structure of the mRNA. In contrast, a leaderless mRNA need not penetrate between the subunits of the 80S ribosome. The efficiency of its binding relies on the stability of the codon-anticodon interaction in the ribosomal P site, on a sufficiently low energy of the interaction of the coding sequence with the corresponding part of the mRNA-binding channel of the small ribosomal subunit, and on the interaction of the CCA end of initiator tRNA with the large ribosomal subunit. A low energy of interaction of the AUG downstream sequence with the 40S (30S) subunit is presumably provided by a single-stranded conformation of the coding sequence rather than by existence of a specific downstream box within this sequence, which, for instance, could form SD-like complementary interactions with rRNA from small ribosomal subunits. Indeed, no evidence for ribosomal recruitment signals downstream of the 5′-terminal AUG has been obtained (for a review, see reference 12) to explain the fairly efficient binding of the cI mRNA with the 70S ribosome.
Until recently, translation initiation mechanisms in prokaryotic and eukaryotic cells have been regarded as fundamentally dissimilar. However, several reports published in the last few years suggest that some basic translation initiation steps in bacteria (E. coli) and eukaryotes have more similarities than has been thought before. Initiation factors eIF1, eIF1A, and eIF5B have been proposed to be functional analogs of bacterial factors IF3, IF1, and IF2, respectively (2, 19, 20, 22). Internal ribosome entry site elements of RNAs from some mammalian viruses (hepatitis C virus and some pestiviruses) and insect viruses bind ribosomes directly at or close to the start codons of these RNAs and require neither scanning nor RNA-unwinding translation initiation factors (18, 29). On the other hand, the recruitment of the mRNA coding for the E. coli ribosomal protein S1 onto the 30S ribosomal subunit is directed by a complex secondary-tertiary structure which includes the start codon and is based on the RNA-protein recognition rather than on the SD interaction (3). In the case of leaderless mRNAs which lack specific features of eukaryotic or prokaryotic mRNAs, we see a full analogy in their molecular mechanisms of recruitment onto bacterial 70S and mammalian 80S ribosomes. One may think that the 80S (70S) pathway of translation initiation represents a relic from early steps in the evolution of the translation apparatus.
Acknowledgments
We acknowledge a great contribution of E. Skripkin to initiate this project many years ago. We thank I. Boni for prompting us to undertake these studies and for critical reading of the manuscript, G. Belsham for valuable comments, Y. Semenkov and V. Makhno for a generous gift of individual 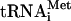 , L. Ovchinnikov for the elongation factor eEF2, and W. Merrick for polyclonal antibodies to eIF2.
, L. Ovchinnikov for the elongation factor eEF2, and W. Merrick for polyclonal antibodies to eIF2.
This work was supported by grant 02-04-48798 from the Russian Foundation for Basic Research to I.N.S.
REFERENCES
- 1.Balakin, A. G., E. A. Skripkin, I. N. Shatsky, and A. A. Bogdanov. 1992. Unusual ribosome binding properties of mRNA encoding bacteriophage lambda repressor. Nucleic Acids Res. 20:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battiste, J. L., T. V. Pestova, C. U. Hellen, and G. Wagner. 2000. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell 5:109-119. [DOI] [PubMed] [Google Scholar]
- 3.Boni, I. V., V. S. Artamonova, N. V. Tzareva, and M. Dreyfus. 2001. Non-canonical mechanism for translational control in bacteria: synthesis of ribosomal protein S1. EMBO J. 20:4222-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castaneda, S. J., and T. C. Wong. 1990. Leader sequence distinguishes between translatable and encapsidated measles virus RNAs. J. Virol. 64:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dmitriev, S. E., A. V. Pisarev, M. P. Rubtsova, Y. E. Dunaevsky, and I. N. Shatsky. 2003. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 533:99-104. [DOI] [PubMed] [Google Scholar]
- 6.Dmitriev, S. E., Y. M. Terenin, Y. E. Dunaevsky, W. C. Merrick, and I. N. Shatsky. 2003. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′ untranslated regions. Mol. Cell. Biol. 23:8925-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dmitriev, S. E., I. M. Terenin, M. P. Rubtsova, and I. N. Shatsky. 2003c. Minor secondary-structure variation in the 5′-untranslated region of the beta-globin mRNA changes the concentration requirements for eIF2. Mol. Biol. (Moscow) 7:494-503. [PubMed] [Google Scholar]
- 8.Evstafieva, A. G., I. N. Shatsky, A. A. Bogdanov, Y. P. Semenkov, and V. D. Vasiliev. 1983. Localization of 5′ and 3′ ends of the ribosome-bound segment of template polynucleotides by immune electron microscopy. EMBO J. 2:799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellen, C. U. T., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 10.Janssen, G. R. 1993. Eubacterial, archaebacterial and eukaryotic genes that encode leaderless mRNA, p. 59-67. In R. H. Baltz, G. D. Hegeman, and P. L. Scatrud (ed.), Industrial microorganisms: basic and applied molecular genetics. ASM Press, Washington, D.C.
- 11.Londei, P. 2004. Evolution of translational initiation: new insights from the archaea. FEMS Microbiol. Rev. 29:185-200. [DOI] [PubMed] [Google Scholar]
- 12.Moll, I., S. Grill, C. O. Gualerzi, and U. Blasi. 2002. Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control. Mol. Microbiol. 43:239-246. [DOI] [PubMed] [Google Scholar]
- 13.Moll, I., G. Hirokawa, M. C. Kiel, and U. Blasi. 2004. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res. 32:3354-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mowatt, M. R., A. Aggarwal, and T. E. Nash. 1991. Carboxy-terminal sequence conservation among variant-specific surface proteins of Giardia lamblia. Mol. Biochem. Parasitol. 49:215-227. [DOI] [PubMed] [Google Scholar]
- 15.O'Donnell, S. M., and G. R. Janssen. 2002. Leaderless mRNAs bind 70S ribosomes more strongly than 30S ribosomal subunits in Escherichia coli. J. Bacteriol. 184:6730-6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pestova, T. V., C. U. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pestova, T. V., S. I. Borukhov, and C. U. T. Hellen. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394:854-859. [DOI] [PubMed] [Google Scholar]
- 18.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pestova, T. V., I. B. Lomakin, J. H. Lee, S. K. Choi, T. E. Dever, and C. U. Hellen. 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403:332-335. [DOI] [PubMed] [Google Scholar]
- 20.Pestova, T. V., and V. G. Kolupaeva. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16:2906-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schreier, M. H., B. Erni, and T. Staehelin. 1977. Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors J. Mol. Biol. 116:727-753. [DOI] [PubMed] [Google Scholar]
- 22.Sette, M., P. van Tilborg, R. Spurio, R. Kaptein, M. Paci, C. O. Gualerzi, and R. Boelens. 1997. The structure of the translational initiation factor IF1 from E. coli contains an oligomer-binding motif. EMBO J. 16:1426-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shatsky, I. N., A. V. Bakin, A. A. Bogdanov, and V. D. Vasiliev. 1991. How does the mRNA pass through the ribosome? Biochimie 73:937-945. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu, Y., A. Inoue, Y. Tomari, T. Suzuki, T. Yokogawa, K. Nishikawa, and T. Ueda. 2001. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19:751-755. [DOI] [PubMed] [Google Scholar]
- 25.Spahn, C. M., R. Beckmann, N. Eswar, P. A. Penczek, A. Sali, G. Blobel, and J. Frank. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae-tRNA-ribosome and subunit-subunit interactions. Cell 107:373-386. [DOI] [PubMed] [Google Scholar]
- 26.Tedin, K., I. Moll, S. Grill, A. Resch, A. Graschopf, C. O. Gualerzi, and U. Blasi. 1999. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol. Microbiol. 31:67-77. [DOI] [PubMed] [Google Scholar]
- 27.Tzareva, N. V., V. I. Makhno, and I. V. Boni. 1994. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 337:189-194. [DOI] [PubMed] [Google Scholar]
- 28.Udagawa, T., Y. Shimizu, and T. Ueda. 2004. Evidence for the translation initiation of leaderless mRNAs by the intact 70 S ribosome without its dissociation into subunits in eubacteria. J. Biol. Chem. 279:8539-8546. [DOI] [PubMed] [Google Scholar]
- 29.Wilson, J. E., T. V. Pestova, C. U. Hellen, and P. Sarnow. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102:511-520. [DOI] [PubMed] [Google Scholar]
- 30.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 A resolution. Science 292:883-896. [DOI] [PubMed] [Google Scholar]
- 31.Yusupova, G. Z., M. M. Yusupov, J. H. Cate, and H. F. Noller. 2001. The path of messenger RNA through the ribosome. Cell 106:233-241. [DOI] [PubMed] [Google Scholar]