Abstract
Aberrant gene silencing accompanied by DNA methylation is associated with neoplastic progression in many tumors that also show global loss of DNA methylation. Using conditional inactivation of de novo methyltransferase Dnmt3b in ApcMin/+ mice, we demonstrate that the loss of Dnmt3b has no impact on microadenoma formation, which is considered the earliest stage of intestinal tumor formation. Nevertheless, we observed a significant decrease in the formation of macroscopic colonic adenomas. Interestingly, many large adenomas showed regions with Dnmt3b inactivation, indicating that Dnmt3b is required for initial outgrowth of macroscopic adenomas but is not required for their maintenance. These results support a role for Dnmt3b in the transition stage between microadenoma formation and macroscopic colonic tumor growth and further suggest that Dnmt3b, and by extension de novo methylation, is not required for maintaining tumor growth after this transition stage has occurred.
Altered DNA methylation in the form of global hypomethylation and regional hypermethylation is one of the most consistent epigenetic changes in cancer (18). Global hypomethylation, which is frequently observed at early stages of tumorigenesis in human cancer (10, 11), promotes tumor development in several mouse models and causes chromosomal instability in cultured fibroblasts (9, 12). After the initial observation of DNA hypermethylation within the retinoblastoma tumor suppressor gene (14), dozens of genes have been shown to be hypermethylated and transcriptionally silenced in tumors (2, 20). Although the consequences of global hypomethylation and gene-specific hypermethylation have been mechanistically connected to chromosome instability and transcriptional silencing, respectively, the causes of aberrant DNA methylation patterns are currently unknown.
DNA methylation is catalyzed by a family of three DNA methyltransferases: Dnmt1, Dnmt3a, and Dnmt3b. Although the three Dnmts partially cooperate to establish and maintain genomic methylation patterns (21), they also have distinctive functions. Dnmt1 has a preference for hemimethylated DNA (1, 15, 38), and indeed a hypomorphic allele of Dnmt1 has been shown to cause global DNA hypomethylation (12). Dnmt1 is therefore considered the major maintenance methyltransferase. Dnmt3a and -3b probably function as de novo DNA methyltransferases because these enzymes were shown to have equal preferences in vitro for unmethylated and hemimethylated DNA (25, 26). Furthermore, de novo methylation of a subset of the CpG sites on stable episomes is detected in human cells overexpressing the murine Dnmt3a or Dnmt3b1 protein (17). Consistent with these notions, inactivation of both Dnmt3a and Dnmt3b by gene targeting blocks de novo DNA methylation in embryonic stem (ES) cells and early embryos, as well as de novo methylation of imprinted genes in germ cells (25, 26). These findings support the view that Dnmt3a and Dnmt3b function primarily as de novo methyltransferases during normal development. Nevertheless, the role of the de novo Dnmts in cancer is unresolved.
Evidence from humans and mouse models has shown that Dnmt3b is important for maintaining methylation of pericentromeric repetitive elements (5, 25) and of single copy genes in cooperation with Dnmt1 in a human colon cancer cell line (29, 37). Mouse embryonic fibroblasts expressing large T antigen and Ras form soft agar colonies and large tumors, but similarly treated fibroblasts from Dnmt3b−/− mice do not grow in soft agar and are much less tumorigenic in vivo (34). These findings suggest that deletion of Dnmt3b inhibits cell transformation. In addition, Dnmt3b deficiency promotes the chromosomal instability of mouse embryonic fibroblasts, which in turn promotes spontaneous immortalization or premature senescence (7). Furthermore, deletion of Dnmt3b was shown to induce apoptosis in a human cancer cell line (3). Taken together, these observations implicate a role for Dnmt3b in oncogenesis.
Despite these in vitro studies, there is no direct evidence in vivo to clarify the role of Dnmt3b in tumorigenesis. Here we show that Dnmt3b expression is elevated in colonic adenomas derived from the ApcMin/+ mouse model. We therefore chose this in vivo tumor model to analyze the role of Dnmt3b in tumor initiation and progression by using Cre-Lox mediated conditional Dnmt3b gene deletion in the intestinal mucosa. We found that the removal of Dnmt3b does not affect tumor initiation in the form of microadenomas but does inhibit the formation of macroscopic tumors at an early stage of tumor development.
MATERIALS AND METHODS
Construction of the Dnmt3b conditional knockout targeting vector.
A 10-kb XhoI-EcoRV genomic DNA fragment containing the catalytic domain and surrounding regions from the Dnmt3b locus was isolated from a 129/SvEvTac female mouse bacterial artificial chromosome library (Roswell Park Cancer Institute) by hybridization to a Dnmt3b cDNA probe. The fragment was subsequently cloned into the pBluescript II SK vector (Stratagene), generating plasmid pBS3bXEV, which was used to generate the conditional knockout construct.
A loxP site was inserted into the intron just 3′ of the exon 19, which encodes DNA methyltransferase motif VI. In addition, a drug selection marker in the form of a neomycin phosphotransferase gene under the control of the phosphoglycerate kinase 1 gene promoter (pgk-neo cassette flanked by Frt sites) (28) was inserted into the intron between exons 15 and 16 of the Dnmt3b locus, which are 5′ to the catalytic domain. The pgk-neo cassette was flanked by frt sites to facilitate removal by flp recombinase. A loxP site was also included just 5′ to the pgk-neo cassette.
In the final targeting construct, pBS3blfN, the loxP sites flank exons 16 to 19, which encode the catalytic domains. Arms of genomic DNA from the Dnmt3b locus—a 1.9-kb HindII-EcoRV fragment on the 5′ side and a 4.5-kb HindII fragment on the 3′ side—surround the loxP-flanked region. The orientation of pieces in the targeting construct was confirmed by restriction digest analysis and sequencing.
Generation of ES cells and mice with conditional knockout alleles of Dnmt3b.
Plasmid pBS3blfN was linearized with SacII and electroporated into F1 (129SvJae × C57BL/6; v6.5 line) mouse ES cells and selected for drug resistance against neomycin as described previously. Neomycin-resistant ES colonies were analyzed by Southern blotting. In ca. 45% of the picked colonies, the targeting construct had integrated properly into the Dnmt3b locus.
In order to remove the pgk-neo cassette, ES cells from a targeted clone were transiently transfected with enhanced flp (Flp-e) recombinase-encoding plasmids (a gift from Francis Stewart) (4) by lipofection with Cytofectene (Bio-Rad). Removal of the pgk-neo cassette was confirmed by PCR, Southern blotting, and by death in G418-containing medium (350 μg/ml). Sixteen of twenty-four subclones in this experiment no longer contained the neomycin resistance gene.
For injection into blastocysts, we used ES cells from one properly targeted subclone in which the neomycin resistance cassette was removed. The resulting chimeras were crossed with C57BL/6 mice to generate the founder colony.
Mice and polyp analysis.
Mice were maintained in the facilities of Whitehead Institute for Biomedical Research. The conditional Dnmt3b allele was backcrossed to the C57BL/6 strain prior to crossing with ApcMin/+ mice. All tumor bearing mice analyzed in this experiment were backcrossed at least six generations into C57BL/6 mice. For tumor analysis, the entire intestine was excised immediately after sacrifice and subjected to a systematic microscopic screen for tumor formation along the entire length of the intestine. The investigator counting the adenomas was blind with regard to mouse genotype.
Genotyping.
DNA was isolated from tail tips and amplified by PCR for the Dnmt3b locus as follows: Dnmt3bF (5′-GCTGAATTATACCCGCCCCAAGGAGGGCG-3′; located just 5′ of the 3′loxP site) and Dnmt3bR (5′-CCCTTCAAAGGTGCATCGTGGCCAGTGTGC-3′; located in the 3′ arm of the conditional construct). PCR products are 410 bp for the wild-type allele and 550 bp for the 2lox allele. For the Dnmt3b 1lox allele, Dnmt3bR was used in conjunction with Dnmt3b1F (5′-CCTGAGAGACTGGCCGGCTTTTCCTCAGAC-3′; located in the 5′ arm of the conditional construct), generating a roughly 500-bp product from the Dnmt3b 1lox allele. C57BL/6 ApcMin/+ mice were purchased from Jackson Laboratories, Bar Harbor, Maine. Oligonucleotides RM21a (5′-TTCTGAGAAAGACAGAAGTTA-3′), RM20 (5′-TTCCACTTTGGCATAAGGC-3′), and RM19 (5′-GCCATCCCTTCACGTTAG-3′ were used for genotyping. The Fabp-Cre mice (a gift from Jeffrey I. Gordon, Washington University, St. Louis, MO) were backcrossed to the C57BL/6 strain for more than 10 generations. Cre-positive mice were determined by PCR with the oligonucleotides CreF (5′-TGGGCGGCATGGTGCAAGTT-3′) and CreR (5′-CGGTGCTAACCAGCGTTTTC-3′).
Southern blotting, Western blotting, and histological analysis.
Southern blotting, Western blotting, and histological analysis were performed as described previously (16). Total DNA was isolated from ES cells, intestinal mucosa, and tumor tissue and digested by BamHI for Southern blotting. The probe is a genomic fragment of Dnmt3b located downstream of the 3′ end of the Dnmt3b targeting construct. Dnmt3b antibody (1:200 dilution for Western blotting; 1:100 for immunohistochemical analysis, IMGENEX), anti-β-catenin antibody (1:1,000 dilution; BD Transduction Laboratories), and anti-Ki-67 antibody (1:250; Dako) were used in these analyses. The fluorescein in situ cell death detection kit, (Roche) was used to detect apoptosis.
RESULTS
Dnmt3b is activated in the intestinal tumors of ApcMin/+ mice.
Elevated expression of Dnmt3b has been observed in many human tumors (13, 23, 24, 30, 32). We therefore sought to identify a mouse tumor model that recapitulated Dnmt3b overexpression for in vivo analysis of the functional importance of de novo methyltransferase upregulation. Western blot analysis of intestinal macroscopic tumors from ApcMin/+ mice revealed that expression of Dnmt3b is substantially higher in tumors compared to normal epithelium (Fig. 1A). Consistent with this result, immunohistochemical staining of sections through polyps identified strong and specific staining with an anti-Dnmt3b antibody, in contrast to the adjacent normal epithelium, which showed weak staining (Fig. 1B). These results demonstrate that Dnmt3b expression is elevated in the intestinal tumors of ApcMin/+ mice.
FIG. 1.
Expression level of Dnmt3b in intestinal tumors of ApcMin/+ mice. (A) Western blot showing expression levels of Dnmt3b in intestinal macroadenomas and adjacent normal mucosa from ApcMin/+ mice. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) serves as the loading control. (B) Immunohistochemical analysis with an anti-Dnmt3b antibody of a macroscopic colon tumor and adjacent normal crypts.
Deletion of Dnmt3b in the intestine.
To investigate whether Dnmt3b overexpression plays a causal role in intestinal carcinogenesis, we generated a conditional Dnmt3b allele that would allow intestine-specific deletion of this gene, since Dnmt3b-null mutant mice do not survive past midgestation (25). Deletion of the exons encoding highly conserved PC and ENV DNA methyltransferase catalytic motifs (motifs IV and VI) of Dnmt3b has previously been shown to result in a null mutation in this gene (25). We therefore created an analogous conditional mutation in Dnmt3b, by placing loxP sites flanking exons 16 through 19 using homologous recombination in V6.5 ES cells (Fig. 2A and see also Materials and Methods). Successfully targeted ES cells bearing a conditional knockout (“2lox”) allele were analyzed by Western blotting with a Dnmt3b antibody and shown to express normal levels of Dnmt3b protein, confirming that the Dnmt3b2lox allele is a silent mutation (Fig. 2B). Western analysis of Dnmt3b1lox/1lox ES cells, similarly confirmed that the deleted Dnmt3b1lox allele is null (Fig. 2B).
FIG. 2.
Targeting vector of conditional knockout and the frequency of Fabp-Cre mediated the recombination of conditional Dnmt3b allele in the intestine. (A) Restriction map of the Dnmt3b conditional knockout targeting vector. Restriction sites: B, BamHI; H, HindIII; RV, EcoRV; X, XhoI. (B) Western blot demonstrating the protein level of Dnmt3b in Dnmt3b 2lox/wild-type ES cells and Dnmt3b 1lox/1lox ES cells. The Dnmt3b knockout ES cell is a negative control (a gift from E. Li). (C) Southern blot demonstrating Fabp-Cre-mediated recombination of the conditional Dnmt3b allele in the colon and small intestinal mucosa harvested from Dnmt3b2lox/2lox Fabp-Cre-positive and -negative mice (14 kb for the 2lox allele and 11 kb for the 1lox allele). (D and E) Longitudinal section and cross section, respectively, of the colonic mucosa from a Dnmt3b2lox/2lox Fabp-Cre-positive and -negative mouse stained with an anti-Dnmt3b antibody, demonstrating the Fabp-Cre-mediated recombination of the conditional Dnmt3b allele.
Dnmt3b2lox/+ ES cells were injected into blastocysts to generate chimeric mice, which were backcrossed to the C57BL/6 strain for subsequent crosses with ApcMin/+ mice. An intercross between Dnmt3b2lox/+ mice generated 13 2lox/2lox, 30 2lox/+, and 17 +/+ mice (total n = 60), a finding consistent with expected Mendelian ratios (P < 0.05, chi-square analysis) and, in contrast to the embryonic lethal phenotype observed for the Dnmt3b-null mutants (25), Dnmt3b2lox/2lox mice are healthy and fertile with no phenotype observed through 1 year of age.
To achieve intestine-specific deletion, a Cre transgene under the control of transcriptional regulatory elements from the fatty acid-binding protein gene (Fabp-Cre) (31) was used to generate Dnmt3b2lox/2lox, Fabp-Cre mice. Cre-mediated deletion of Dnmt3b was assessed by Southern blotting, which revealed ca. 50% recombination in the colon, but no recombination was detected in the small intestine from the same mice (Fig. 2C). To assess further the efficiency of Cre-mediated deletion within individual crypts, sections of the colon and small intestine of 8- and 21-week-old animals were stained with an anti-Dnmt3b antibody. Immunostaining showed ca. 50% of colon crypts stained positive for Dnmt3b in animals of both ages (Fig. 2D and 2E). Similar analysis of small intestinal crypts showed uniform expression of Dnmt3b in Dnmt3b2lox/2lox Fabp-Cre mice (data not shown), a finding consistent with the lack of recombination observed by Southern blotting in this tissue (Fig. 2C). Within the colon, immunostaining demonstrated that all cells of a given colon crypt were either Dnmt3b positive or negative, indicating that deletion of Dnmt3b likely occurred in crypt forming stem cells. Histological analysis did not reveal any significant phenotypic differences between Dnmt3b-positive and -negative crypts (Fig. 2D and E and data not shown).
Deletion of Dnmt3b decreases the number of colon tumors but not the formation of microadenomas.
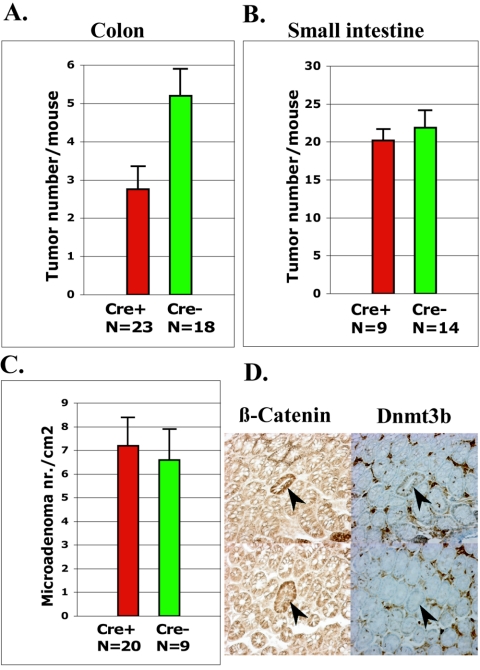
To study the effects of Dnmt3b deletion on intestinal tumorigenesis in ApcMin/+ mice, cohorts of 23 ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ and 18 ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre− mice were sacrificed at 150 days of age to quantify intestinal tumors. Since Fabp-Cre has different recombination efficiencies in different regions of the intestinal tract (Fig. 2C), tumor numbers in the small intestine and the colon were scored separately. Conditional deletion of Dnmt3b reduced the total number of colonic tumors by ca. 40% (P = 0.0012; Fig. 3A) as shown by the average colonic tumor burden of 2.75 ± 0.6 polyps in ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ mice, whereas ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre− mice had a mean macroscopic tumor load of 5.2 ± 0.7 colonic polyps. Based on the low recombination rate detected in the small intestine of Fabp-Cre mice, we anticipated little difference in tumor load in the small intestine of Fabp-Cre-positive and -negative cohorts. No significant difference was observed (P = 0.89, Fig. 3B), which shows the specificity of the result for Dnmt3b deletion in colon and supports the notion that Dnmt3b deficiency inhibits tumor formation in ApcMin/+ mice.
FIG. 3.
Effect of Dnmt3b deletion on the number of macroscopic tumors and microadenomas in the colon and small intestine of Apcmin/+mice. (A and B) Number of macroscopic tumors in the colons and small intestines of Apcmin/+ Dnmt3b2lox/2lox mice that either carried (ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+) or did not carry (ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre−) the Fabp-Cre transgene. (C) The number of colonic microadenomas in ApcMin/+ Dnmt3b2lox/2lox mice that either carried or did not carry the Fabp-Cre transgene was determined by immunohistochemical analysis for β-catenin. (D) Dnmt3b-positive and Dnmt3b-negative microadenomas in the colonic mucosa of ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ mice. The left panel illustrates our method of microadenoma detection using an anti-β-catenin antibody. The right panel illustrates immunohistochemical analysis of microadenomas with an anti-Dnmt3b antibody. The right upper picture shows a Dnmt3b-positive microadenoma, and the right lower picture shows a Dnmt3b-negative microadenoma.
Tumor formation in ApcMin/+ mice occurs in two distinct stages, which are referred to as microadenoma and macroscopic adenoma. Microadenomas are considered to be the first stage of tumorigenesis and are detectable as histologically abnormal crypts that have mostly undergone loss of heterozygosity (LOH) at the Apc locus and therefore stain positive for β-catenin (39). To discern whether Dnmt3b affects the initial stage of tumor formation, we compared the number of microadenomas in the colons of ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre-positive and -negative mice. Microadenomas were scored on en face sections of the colonic mucosa by staining with anti-β-catenin antibody. ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ mice had a mean microadenoma load of 7.2 ± 1.2/cm2 in the colon and ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre− mice had a mean microadenoma load of 6.5 ± 1.3/cm2 (Fig. 3C). The marginal decrease in microadenomas observed was not statistically significant (P = 0.08). These results suggest that Dnmt3b deletion has no impact on the formation of microadenomas. To support this notion further, microadenomas of ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ mice were stained with anti-Dnmt3b antibody, revealing ca. 50% Dnmt3b-positive and 50% Dnmt3b-negative microadenomas (Fig. 3D). Since this ratio reflects the recombination frequency in the colonic mucosa (Fig. 2C), these results are consistent with the conclusion that Dnmt3b deficiency has no effect on microadenoma formation. Taken together, these data suggest that the inhibition of macroscopic tumor formation in ApcMin/+ mice by Dnmt3b conditional mutants occurs later during either initiation or growth of macroscopic tumors.
Dnmt3b is not essential for tumor growth.
The consistency in the frequency of Cre-mediated recombination within the colon (50%, Fig. 2C to E), along with the nearly twofold reduction in colonic polyps in the Cre-expressing cohort (Fig. 3A), suggested that deletion of Dnmt3b might provide a strong block to macroscopic tumor formation. To assess whether loss of Dnmt3b was compatible with macroscopic tumor growth, immunohistochemical staining of tumor sections using a Dnmt3b antibody was performed to determine the number of Dnmt3b-positive and -negative tumors. This analysis revealed that polyps from ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ mice were predominantly composed of both Dnmt3b-positive and -negative sectors, with a few sections showing homogeneous wild-type or mutant staining patterns (Fig. 4A and Table 1). To quantitate the extent of recombination throughout the tumors, we developed a competitive PCR assay to measure the ratio of 1lox to 2lox Dnmt3b alleles in DNA isolated from individual whole colon tumors (Fig. 4B). DNA samples from 2lox/2lox and 1lox/1lox ES cells were mixed in defined ratios to standardize the PCR conditions. This analysis revealed that most macroscopic tumors contained 2lox as well as 1lox Dnmt3b alleles, a few contained only the nonrecombined 2lox allele, and almost no tumors were observed with greater than 50% contribution of 1lox alleles.
FIG. 4.
Most colon tumors from ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ mice are both Dnmt3b positive and negative. (A) Immunohistochemical analysis with an anti-Dnmt3b antibody illustrating expression of Dnmt3b in colon tumors of ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ mice. The left picture illustrates a purely Dnmt3b-positive section, the middle picture shows a mixed Dnmt3b-positive and -negative section, and the right picture shows a pure Dnmt3b-negative section. (B) PCR analysis shows that most colon tumors from ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ mice contain both 1lox and 2lox alleles of Dnmt3b. The top panel shows a reference PCR using different ratios of 2lox to 1lox Dnmt3b genomic DNA. The top right and lower panels show the PCR analysis of colon tumors from ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ mice.
TABLE 1.
Loss of Dnmt3b observed in early-stage microadenomas is selected against in later-stage colonic tumors
| Tissue analyzed | n | % Dnmt3b expression (n)a
|
||
|---|---|---|---|---|
| Positive | Mixed | Negative | ||
| Colon crypts | 98 | 48 (49) | 50 (51) | |
| Microadenomas | 23 | 11 (48) | 12 (52) | |
| Tumors | 45b | 14 (31) | 26 (58) | 5 (11) |
Dnmt3b expression was determined by immunohistochemistry on sections of Dnmt3b conditional mutant colonic tissue. Microadenomas were identified by beta-catenin staining of adjacent sections.
These data include 14 tumors analyzed by PCR, using a genotyping assay for recombination at the Dnmt3b locus shown in Fig. 4B, and 31 tumors analyzed by immunostaining tumor sections.
The immunostaining and PCR analysis together demonstrated a partial contribution of Dnmt3b mutant cells in a majority of the macroscopic colon tumors in ApcMin/+ Dnmt3b conditional mutant animals (as summarized in Table 1). Since Cre-mediated recombination is irreversible and the majority of tumors in ApcMin/+ mice are considered to be monoclonal (22, 27, 36), this suggests that Dnmt3b deletion occurred after tumor initiation during tumor growth. Two observations are consistent with this idea. First, we observed Cre expression in macroscopic tumors by reverse transcription-PCR (data not shown). Second, the areas lacking Dnmt3b expression were always seen as coherent patches (Fig. 4A, middle panel), indicating that the Dnmt3b-negative tumor sections were clonally derived from a tumor cell that had recombined Dnmt3b 2lox alleles during tumor growth.
The presence of clonal sectors of Dnmt3b mutant cells within the macroscopic tumor lesions suggested that Dnmt3b function is dispensable in an already-formed colon tumor. To assess whether Dnmt3b deletion affects the proliferation rate of tumors, histological sections of colon polyps were stained with a cellular replication marker (Ki67 antibody). Consistent with the notion that Dnmt3b is not required in adenomas, no significant difference in Ki67 staining was observed between Dnmt3b-positive (45.7% ± 1.4% positive cells per crypt) and -negative (42.8% ± 1.45% positive cells per crypt) tumor sectors (Fig. 5A, P = 0.185). In addition, TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) staining revealed 0.90% ± 0.2% and 0.82% ± 0.3% TUNEL-positive cells/crypt in Dnmt3b-positive and -negative areas, respectively (P = 0.66), indicating there was no effect of the Dnmt3b deletion on tumor cell apoptosis (Fig. 5B). Furthermore, we calculated the tumor volume from ApcMin/+ Dnmt3b2lox/2lox mice that either carried or did not carry the Fabp-Cre transgene. Figure 5C shows that the average volume of colon tumors from ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ mice (11.84 ± 1.5 mm3) was not significantly different from the average tumor volume in ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre− mice (11.04 ± 1.4 mm3; P = 0.68). We therefore conclude that Dnmt3b deficiency impairs the initiation of macroscopic tumors but has no noticeable effect on later stages of tumor growth (Fig. 6).
FIG. 5.
Effect of Dnmt3b deletion on tumor growth and apoptosis. (A) Percentage of Ki67-positive cells per crypt in either the Dnmt3b-positive or -negative region of the mixed tumor. (B) Percentage of TUNEL-positive cells per crypt in either the Dnmt3b-positive or the Dnmt3b-negative region of the mixed tumor. (C) Average volume (in cubic millimeters) of colon macroscopic tumors from ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ and Fabp-Cre− mice.
FIG. 6.
Model depicting the effects of Dnmt1 and Dnmt3b on various stages of colon tumorigenesis in ApcMin mice.
DISCUSSION
In this study we investigated the role of Dnmt3b in intestinal tumorigenesis of ApcMin/+ mice. Colon tumorigenesis in ApcMin/+ mice occurs in two distinct stages, with microadenomas representing the first stage, followed by the development of macroscopic adenomas. Western blot and immunohistochemical analyses indicated that Dnmt3b was overexpressed in adenomas of ApcMin/+ mice compared to normal intestinal mucosa. To assess whether Dnmt3b was causally involved in tumorigenesis, we generated a conditional allele of Dnmt3b gene and used a Fabp-Cre transgene to delete this enzyme in the intestinal epithelium. The frequency of Cre-mediated Dnmt3b deletion was 50% in the colon crypts, with little or no deletion in the small intestine. When the number of microadenomas in the colons of ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ and Fabp-Cre− mice were compared, we found no differences, indicating that Dnmt3b deficiency had no effect on the formation of microadenomas. Consistent with this conclusion was the finding that 50% of all microadenomas in the colon of ApcMin/+ Dnmt3b2lox/2lox Fabp-Cre+ were positive and 50% were negative for Dnmt3b, a ratio that corresponds to the Fabp-Cre-mediated recombination frequency in normal crypts. In contrast to microadenoma formation, Dnmt3b deficiency reduced the load of macroscopic adenomas in the colon by ca. 40%. No changes in tumor multiplicity were observed in the small intestine that could be attributed to Dnmt3b inactivation, because of the low to undetectable recombination frequency in this region. Together, these results support the notion that Dnmt3b deletion selectively inhibits the formation of macroscopic colonic tumors but does not affect the formation of colonic microadenomas.
Two findings indicate that Dnmt3b activity and, by extension, de novo methylation are required specifically at the transition from microadenoma to macroscopic tumor outgrowth. First, as summarized in Table 1, roughly 50% of microadenomas lacked Dnmt3b expression, which was equated to the recombination frequency in the colon. However, only 10% of macroscopic tumors developed without Dnmt3b, suggesting that the outgrowth of the Dnmt3b deficient microadenomas was selected against. Second, the subsequent maintenance of a tumor outgrowth was not dependent upon Dnmt3b since both DNA and immunohistochemical analyses indicated that most tumors in Fabp-Cre+ mice were mixed, being composed of cells carrying the unrecombined 2lox Dnmt3b allele as well as the deleted Dnmt3b 1lox allele. Together, these two results strongly suggest that Dnmt3b deletion selectively inhibits the transition from microadenoma to macroscopic adenoma and that the majority of macroscopic adenomas in these mice were initiated in cells with a nonrecombined Dnmt3b conditional allele (2lox). Persistent or stochastic expression of Cre recombinase within an already formed tumor may have allowed recombination to occur at some later point after adenoma outgrowth, which resulted in clonal expansion of Dnmt3b-negative sectors in a given adenoma. This interpretation is supported by immunostaining, which showed that the region of the tumor lacking Dnmt3b expression always appeared as coherent patches of variable size. Although immunohistochemical analyses indicated that ca. 10% of the adenomas lacked Dnmt3b expression, these were analyzed on representative sections and may even be an overestimate of the number of tumors that formed entirely lacking Dnmt3b (Fig. 4A). In a less biased analysis, DNA isolated from whole tumor samples failed to confirm the presence of a tumor with greater than 50% contribution by Dnmt3b mutant cells (Fig. 4B), and in fact most of the mixed tumors had a contribution of less than 20% based on the competitive PCR analysis. Together, these results suggest that Dnmt3b deficiency resulted in a more than three- to fourfold reduction in macroscopic tumor incidence, owing to a selective requirement for de novo methylation at the transition from micro- to macroadenoma (Fig. 6).
It has been suggested, based on the analysis of chimeric ApcMin/+ mice, that tumors in the small intestine arise by active interactions between crypts promoting loss of Apc and thus resulting in the formation of polyclonal adenomas (36). In that study, however, only 22% of all tumors were identified as polyclonal. A similar experiment analyzing chimeric ApcMin/+ mice, using lacZ as a reporter, yielded only 9% mixed tumors (22). Also, in our experimental system, an average of only three adenomas were observed in the colon, in contrast to an average of 17 tumors in the small intestine of the chimeric mice (36). Because the same frequency of recombination in crypts and microadenomas was observed in our study and the microadenomas were homogeneous in Dnmt3b expression (Table 1), our data indicate the initiating event of LOH at the APC locus must precede the local recruitment of a similarly mutated crypt to produce the mixed tumor class according to the collision model. The low incidence of lesions in our system virtually excludes the possibility that the random collision model accounts for a substantial fraction of the majority class of mixed tumors observed in the ApcMin/+ Dnmt3b conditional mutants. Consistent with this conclusion are other studies that also demonstrated a monoclonal origin of colon tumors in mice (27). We think it more likely that persistent and stochastic expression of Cre recombinase within colonic tumors, which we observed by reverse transcription-PCR, explains the formation of the mixed class of tumors in these animals.
It has been shown previously that global DNA hypomethylation can protect as well as enhance tumor formation (8, 12, 19). For example, reduced Dnmt1 activity was demonstrated to inhibit adenoma formation in ApcMin/+ mice but to promote formation of lymphomas in the thymus and of sarcomas in soft tissues (9, 12). Enhanced thymoma and sarcoma formation was due to increased genetic instability and elevated LOH frequencies of tumor suppressor genes. Consistent with this finding is our recent observation that reduced levels of Dnmt1 expression increased the number of microadenomas in ApcMin/+ mice (39) by enhancing the rate of Apc LOH (Fig. 6). In contrast to reduced Dnmt1 activity, the somatic deletion of Dnmt3b would be predicted to have no effect on global methylation levels and thus would not be expected to affect APC LOH. In agreement with this prediction is the finding that microadenoma formation is unaffected by loss of Dnmt3b (Fig. 6).
Protection against macroscopic tumors by reduced Dnmt1 expression was suggested to be due to an epigenetic mechanism such as the impaired maintenance of methylation marks that are acquired during tumorigenesis. For example, de novo methylation of tumor suppressor genes of the H19 DMR has been shown to promote intestinal tumor load by loss of imprinting of Igf2 (6, 33). Because the maintenance of methylation marks depends on Dnmt1 activity, a reduced Dnmt1 level may lead to loss of DMR methylation causing reduced Igf2 expression and thus protect against intestinal tumors. Our results indicate that somatic deletion of Dnmt3b also protects against polyp formation, suggesting that this de novo methyltransferase is causally involved in tumor formation, possibly by promoting de novo methylation that mediates silencing of tumor suppressor genes (35) or loss of imprinting of genes such as Igf2 (6, 33). This predicts that, once tumor growth is initiated, continuous expression of Dnmt3b would not be required for further tumor growth (Fig. 6). Indeed, we found that loss of Dnmt3b has no noticeable effect on the growth of established tumors as the proliferation and apoptosis rate of cells was similar in Dnmt3b-positive and -negative sectors of a given tumor. Also, we failed to detect any significant difference in the average tumor volume when comparing Dnmt3b2lox/2lox ApcMin/+ mice with or without the Fabp-Cre transgene, which is consistent with tumor growth being unaffected by Dnmt3b. These studies are consistent with the results obtained by targeted disruption of the human DNMT3B gene in the human colon cancer cell line HCT116, which resulted in little to no change in the global DNA methylation or growth of these cells. Although the disruption of both DNMT1 and DNMT3B caused DNA hypomethylation in the same colorectal cancer cell line, our results suggests that Dnmt1 alone is sufficient to support the proliferation and maintenance of tumors in ApcMin mice (29, 37). Thus, although Dnmt3b is required during the initial outgrowth phase of macroscopic colonic adenomas in these mice, it has no effect on subsequent tumor growth and maintenance (Fig. 6).
Our previous observation that inhibition of Dnmt1 protected against intestinal tumorigenesis (19) sparked efforts to develop inhibitors of Dnmt1 that could be administered prophylactically to FAP patients carrying an Apc mutation. However, the later recognition that Dnmt1 inhibition can impair genomic stability and enhance tumor formation in other tissues such as the thymus or in soft tissues (9, 12) cautioned against long-term treatment with Dnmt1 inhibitors. Since Dnmt3b inhibition has no effect on microadenoma formation and, by implication, on genomic stability but strongly inhibits macroscopic tumor initiation, drug-mediated inhibition of this enzyme may be effective as a prophylactic agent to protect against tumors without the potential side effects of Dnmt1 inhibitors.
Acknowledgments
We thank David Pellman, Konrad Hochedlinger, Caroline Beard, Qiang Chang, Marius Wernig, and Taiping Chen for helpful discussions. We are indebted to all members of the Jaenisch lab for critical comments and in particular to Jessica Dausman, Ruth Flannery, and Dongdong Fu for help with the mouse colony and histological analysis.
H.L. was supported by a fellowship from the Canadian Institutes of Health Research. R.J. received support from the NIH (R37-CA84198, RO1-HD0445022, and RO1-CA87869).
REFERENCES
- 1.Bacolla, A., S. Pradhan, J. E. Larson, R. J. Roberts, and R. D. Wells. 2001. Recombinant human DNA (cytosine-5) methyltransferase. III. Allosteric control, reaction order, and influence of plasmid topology and triplet repeat length on methylation of the fragile X CGG.CCG sequence. J. Biol. Chem. 276:18605-18613. [DOI] [PubMed] [Google Scholar]
- 2.Baylin, S. B., M. Esteller, M. R. Rountree, K. E. Bachman, K. Schuebel, and J. G. Herman. 2001. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 10:687-692. [DOI] [PubMed] [Google Scholar]
- 3.Beaulieu, N., S. Morin, I. C. Chute, M. F. Robert, H. Nguyen, and A. R. MacLeod. 2002. An essential role for DNA methyltransferase DNMT3B in cancer cell survival. J. Biol. Chem. 277:28176-28181. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz, F., P. O. Angrand, and A. F. Stewart. 1998. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 16:657-662. [DOI] [PubMed] [Google Scholar]
- 5.Chen, T., N. Tsujimoto, and E. Li. 2004. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 24:9048-9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui, H., M. Cruz-Correa, F. M. Giardiello, D. F. Hutcheon, D. R. Kafonek, S. Brandenburg, Y. Wu, X. He, N. R. Powe, and A. P. Feinberg. 2003. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science 299:1753-1755. [DOI] [PubMed] [Google Scholar]
- 7.Dodge, J. E., M. Okano, F. Dick, N. Tsujimoto, T. Chen, S. Wang, Y. Ueda, N. Dyson, and E. Li. 2005. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J. Biol. Chem. 280:17986-17991. [DOI] [PubMed] [Google Scholar]
- 8.Eads, C. A., A. E. Nickel, and P. W. Laird. 2002. Complete genetic suppression of polyp formation and reduction of CpG-island hypermethylation in ApcMin/+ Dnmt1-hypomorphic mice. Cancer Res. 62:1296-1299. [PubMed] [Google Scholar]
- 9.Eden, A., F. Gaudet, A. Waghmare, and R. Jaenisch. 2003. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300:455. [DOI] [PubMed] [Google Scholar]
- 10.Feinberg, A. P., C. W. Gehrke, K. C. Kuo, and M. Ehrlich. 1988. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 48:1159-1161. [PubMed] [Google Scholar]
- 11.Feinberg, A. P., and B. Vogelstein. 1983. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301:89-92. [DOI] [PubMed] [Google Scholar]
- 12.Gaudet, F., J. G. Hodgson, A. Eden, L. Jackson-Grusby, J. Dausman, J. W. Gray, H. Leonhardt, and R. Jaenisch. 2003. Induction of tumors in mice by genomic hypomethylation. Science 300:489-492. [DOI] [PubMed] [Google Scholar]
- 13.Girault, I., S. Tozlu, R. Lidereau, and I. Bieche. 2003. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin. Cancer Res. 9:4415-4422. [PubMed] [Google Scholar]
- 14.Greger, V., E. Passarge, W. Hopping, E. Messmer, and B. Horsthemke. 1989. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum. Genet. 83:155-158. [DOI] [PubMed] [Google Scholar]
- 15.Gruenbaum, Y., H. Cedar, and A. Razin. 1982. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature 295:620-622. [DOI] [PubMed] [Google Scholar]
- 16.Hochedlinger, K., Y. Yamada, C. Beard, and R. Jaenisch. 2005. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121:465-477. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh, C. L. 1999. In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell. Biol. 19:8211-8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, P. A., and S. B. Baylin. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3:415-428. [DOI] [PubMed] [Google Scholar]
- 19.Laird, P. W., L. Jackson-Grusby, A. Fazeli, S. L. Dickinson, W. E. Jung, E. Li, R. A. Weinberg, and R. Jaenisch. 1995. Suppression of intestinal neoplasia by DNA hypomethylation. Cell 81:197-205. [DOI] [PubMed] [Google Scholar]
- 20.Levine, J. J., K. M. Stimson-Crider, and P. M. Vertino. 2003. Effects of methylation on expression of TMS1/ASC in human breast cancer cells. Oncogene 22:3475-3488. [DOI] [PubMed] [Google Scholar]
- 21.Liang, G., M. F. Chan, Y. Tomigahara, Y. C. Tsai, F. A. Gonzales, E. Li, P. W. Laird, and P. A. Jones. 2002. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22:480-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merritt, A. J., K. A. Gould, and W. F. Dove. 1997. Polyclonal structure of intestinal adenomas in ApcMin/+ mice with concomitant loss of Apc+ from all tumor lineages. Proc. Natl. Acad. Sci. USA 94:13927-13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuno, S., T. Chijiwa, T. Okamura, K. Akashi, Y. Fukumaki, Y. Niho, and H. Sasaki. 2001. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood 97:1172-1179. [DOI] [PubMed] [Google Scholar]
- 24.Nagai, M., A. Nakamura, R. Makino, and K. Mitamura. 2003. Expression of DNA (5-cytosin)-methyltransferases (DNMTs) in hepatocellular carcinomas. Hepatol. Res. 26:186-191. [DOI] [PubMed] [Google Scholar]
- 25.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 26.Okano, M., S. Xie, and E. Li. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19:219-220. [DOI] [PubMed] [Google Scholar]
- 27.Ponder, B. A., and M. M. Wilkinson. 1986. Direct examination of the clonality of carcinogen-induced colonic epithelial dysplasia in chimeric mice. J. Natl. Cancer Inst. 77:967-976. [PubMed] [Google Scholar]
- 28.Possemato, R., K. Eggan, B. J. Moeller, R. Jaenisch, and L. Jackson-Grusby. 2002. Flp recombinase regulated lacZ expression at the ROSA26 locus. Genesis 32:184-186. [DOI] [PubMed] [Google Scholar]
- 29.Rhee, I., K. E. Bachman, B. H. Park, K. W. Jair, R. W. Yen, K. E. Schuebel, H. Cui, A. P. Feinberg, C. Lengauer, K. W. Kinzler, S. B. Baylin, and B. Vogelstein. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416:552-556. [DOI] [PubMed] [Google Scholar]
- 30.Robertson, K. D., E. Uzvolgyi, G. Liang, C. Talmadge, J. Sumegi, F. A. Gonzales, and P. A. Jones. 1999. The human DNA methyltransferases (DNMTs) 1, 3a, and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 27:2291-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saam, J. R., and J. I. Gordon. 1999. Inducible gene knockouts in the small intestinal and colonic epithelium. J. Biol. Chem. 274:38071-38082. [DOI] [PubMed] [Google Scholar]
- 32.Saito, Y., Y. Kanai, M. Sakamoto, H. Saito, H. Ishii, and S. Hirohashi. 2002. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc. Natl. Acad. Sci. USA 99:10060-10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakatani, T., A. Kaneda, C. A. Iacobuzio-Donahue, M. G. Carter, S. de Boom Witzel, H. Okano, M. S. Ko, R. Ohlsson, D. L. Longo, and A. P. Feinberg. 2005. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science 307:1976-1978. [DOI] [PubMed] [Google Scholar]
- 34.Soejima, K., W. Fang, and B. J. Rollins. 2003. DNA methyltransferase 3b contributes to oncogenic transformation induced by SV40T antigen and activated Ras. Oncogene 22:4723-4733. [DOI] [PubMed] [Google Scholar]
- 35.Takada, J., D. J. Baylink, and K. H. Lau. 1995. Pretreatment with low doses of norethindrone potentiates the osteogenic effects of fluoride on human osteosarcoma cells. J. Bone Miner. Res. 10:1512-1522. [DOI] [PubMed] [Google Scholar]
- 36.Thliveris, A. T., R. B. Halberg, L. Clipson, W. F. Dove, R. Sullivan, M. K. Washington, S. Stanhope, and M. A. Newton. 2005. Polyclonality of familial murine adenomas: analyses of mouse chimeras with low tumor multiplicity suggest short-range interactions. Proc. Natl. Acad. Sci. USA 102:6960-6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ting, A. H., K. W. Jair, H. Suzuki, R. W. Yen, S. B. Baylin, and K. E. Schuebel. 2004. CpG island hypermethylation is maintained in human colorectal cancer cells after RNAi-mediated depletion of DNMT1. Nat. Genet. 36:582-584. [DOI] [PubMed] [Google Scholar]
- 38.Vilkaitis, G., I. Suetake, S. Klimasauskas, and S. Tajima. 2005. Processive methylation of hemimethylated CpG sites by mouse Dnmt1 DNA methyltransferase. J. Biol. Chem. 280:64-72. [DOI] [PubMed] [Google Scholar]
- 39.Yamada, Y., L. Jackson-Grusby, H. Linhart, A. Meissner, A. Eden, H. Lin, and R. Jaenisch. 2005. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc. Natl. Acad. Sci. USA 102:13580-13585. [DOI] [PMC free article] [PubMed] [Google Scholar]