Abstract
Mediator is a key RNA polymerase II (Pol II) cofactor in the regulation of eukaryotic gene expression. It is believed to function as a coactivator linking gene-specific activators to the basal Pol II initiation machinery. In support of this model, we provide evidence that Mediator serves in vivo as a coactivator for the yeast activator Met4, which controls the gene network responsible for the biosynthesis of sulfur-containing amino acids and S-adenosylmethionine. In addition, we show that SAGA (Spt-Ada-Gcn5-acetyltransferase) is also recruited to Met4 target promoters, where it participates in the recruitment of Pol II by a mechanism involving histone acetylation. Interestingly, we find that SAGA is not required for Mediator recruitment by Met4 and vice versa. Our results provide a novel example of functional interplay between Mediator and coactivators involved in histone modification.
Synthesis of eukaryotic mRNA requires the assembly at the promoter of a basal transcription machinery comprising RNA polymerase II (Pol II) and a defined set of general transcription factors (GTFs), including TATA-binding protein (TBP), TFIIB, IIE, IIF, and IIH. The regulation of this process in response to environmental signals involves a number of additional factors termed coactivators, which are recruited to enhancers or upstream activating sequences (UAS) by gene-specific activators and operate by several distinct mechanisms involving alteration of the structure of chromatin and direct interaction with Pol II and GTFs. Mediator has emerged in recent years as a prominent coactivator linking activators with the basal transcription machinery from yeast to humans (for reviews, see references 7, 11, 28, 29, and 46).
The Saccharomyces cerevisiae core Mediator complex comprises 21 subunits and is found in free form and as a holoenzyme in association with Pol II (20, 27, 44, 59). Electron microscopy and image processing revealed that S. cerevisiae Mediator presents an extended conformation divided in three distinct submodules (termed head, middle, and tail domains) in association with Pol II (1, 15). The head and middle domains establish multiple contacts with Pol II, whereas the tail domain extends away from Pol II (14). The tail domain contains subunits involved in interactions with several activators, including Gal4 and Gcn4 (42, 48). In addition to supporting activated transcription in vitro, Mediator has the capacity to stimulate basal transcription, as well as TFIIH-dependent phosphorylation of the Pol II carboxy-terminal domain (CTD) (27).
The general requirement of Mediator for Pol II transcription in vivo was shown in genome-wide transcription analysis with a yeast temperature-sensitive mutant of Med17 (Srb4) (23). This analysis revealed that genome-wide transcription is as dependent on Med17 (Srb4) as it is on the largest Pol II subunit, Rpb1; however, this analysis did not distinguish whether Med17 (Srb4) was required for basal transcription or interaction between activators and the basal transcription machinery. In fact, only a limited number of activators have been shown to recruit Mediator in vivo (6, 9, 17, 51, 56), and the question whether Mediator is generally required as a coactivator remains unanswered.
The S. cerevisiae SAGA complex is an example of a coactivator that targets the chromatin. SAGA is a multisubunit complex that contains Gcn5, a histone acetyltransferase (HAT) protein, which preferentially acetylates nucleosomal histone H3 (19). Histone acetylation is thought to destabilize chromatin higher-order structure and, as a result, improve accessibility to DNA for transcription factors. Alternatively, histone acetylation may provide binding sites for bromodomain-containing proteins, such as the Swi2/Snf2 subunit of the chromatin remodeling complex Swi/Snf (22). At certain promoters, SAGA has been proposed to operate independently of its HAT activity by directly interacting with TBP through its Spt3 and Spt8 subunits (3, 4, 16, 38).
Met4 is the transcriptional activator which controls the gene network responsible for the biosynthesis of the sulfur-containing amino acids methionine and cysteine (58). Met4 is also involved in the regulation of the genes needed for the biosynthesis of S-adenosylmethionine (34), a sulfur-containing compound widely used as a methyl donor in methylation of proteins, nucleic acids, and lipids. Met4 activity is tightly regulated by the intracellular concentration of methionine, cysteine, and S-adenosylmethionine (58). Under conditions of excess, Met4 is inactivated by ubiquitination via the SCFMet30 ubiquitin ligase (49, 53). The consequence of Met4 ubiquitination depends on environmental growth conditions. When cells are grown in minimal medium, ubiquitination targets Met4 for degradation by the 26S proteasome (53). In contrast, when cells are grown in rich medium, ubiquitination does not affect Met4 stability but impairs its recruitment to DNA (26, 34). Under conditions of limitation in sulfur-containing amino acids, Met4 is tethered to its target genes through interaction with the DNA-binding factors Cbf1 and Met31/32, which bind two distinct DNA elements found, individually or in combination, upstream of all Met4-responsive genes (8, 31). In addition to its role in the recruitment of Met4, Cbf1 is also required for the function of centromeres (2, 10). Because Cbf1 is partly dispensable for MET gene regulation, Met31/32 provides the main platform for the recruitment of Met4 to DNA (8, 33, 37). Cbf1 and Met31/32 possess no intrinsic capacity to activate transcription, and they are thought to be mainly dedicated to the recruitment of the activator Met4 (8, 33, 57).
In this study, we present evidence that Met4 uses Mediator as a coactivator, reinforcing the notion that Mediator is a prevalent interface between enhancer-bound activators and Pol II transcription machinery in yeast. In addition, we find that SAGA is also recruited to Met4 target genes and catalyzes acetylation of histone H3 through its Gcn5 HAT subunit. Interestingly, the recruitment of Mediator by Met4 does not require SAGA, and vice versa. These results provide a novel instance of interplay between Mediator and SAGA.
MATERIALS AND METHODS
Yeast strains, plasmids, and media.
Yeast strains used in this study are listed in Table 1. Tandem affinity purification (TAP)-tagged Mediator subunits and hemagglutinin (HA)-tagged TBP were generated as previously described (32). Gene deletions and HA-tagged strains were generated using PCR-based, one-step integration strategies (45). Plasmids pGAL1-oplexA-lacZ, plexA-MET4, and plexA-MET4Δ12 were described previously (36). B minimal medium is a synthetic medium lacking organic and inorganic sulfur sources (37). YPD medium contains 1% yeast extract, 2% Bacto peptone, and 2% glucose. YNB medium contains 0.7% yeast nitrogen base, 0.5% ammonium sulfate, and 2% glucose.
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| W303-1A | matahis3 leu2 ura3 ade2 trp1 | R. Rothstein |
| Y62 | matapep4::HIS3 prb1::LEU2 prc1::hisG SPT15::HA3-SPT15-URA3 MED18::MED18-TAP-kl TRP1 | This study |
| Y69 | matapep4::HIS3 prb1::LEU2 prc1::hisG SPT15::HA3-SPT15 MED5::MED5-TAP-kl TRP1 | 32 |
| Y70 | matapep4::HIS3 prb1::LEU2 prc1::hisG SPT15::HA3-SPT15 MED14::MED14-TAP-kl TRP1 | 32 |
| Y84 | matα his3 leu2 ade2 ura3 trp1 SPT15::HA3-SPT15 MED14::MED14-TAP-kl TRP1 | This study |
| Y86 | matα his3 leu2 ade2 ura3 trp1 SPT15::HA3-SPT15 MED14::MED14-TAP-kl TRP | This study |
| CL7-1 | matahis3 leu2 ura3 ade2 trp1 can1-100 med3Δ::HIS3 MX6 | This study |
| CL8-1 | matahis3 leu2 ura3 ade2 trp1 can1-100 med1Δ::HIS3 MX6 | This study |
| CL9-1 | matahis3 leu2 ura3 ade2 trp1 can1-100 med9Δ::HIS3 MX6 | This study |
| CL10-1 | matahis3 leu2 ura3 ade2 trp1 can1-100 med5Δ::HIS3 MX6 | This study |
| CL11-1 | matahis3 leu2 ura3 ade2 trp1 can1-100 med18Δ::HIS3 MX6 | This study |
| CL12-1 | matahis3 leu2 ura3 ade2 trp1 can1-100 med20Δ::HIS3 MX6 | This study |
| CL18-1 | matahis3 leu2 ura3 ade2 trp1 med2Δ::HIS3 MX6 | This study |
| DY3168 | matahis3 leu2 ura3 ade2 trp1 can1-100 lys2 med14-100 | D. Stillman |
| DY1700 | matahis3 leu2 ura3 ade2 trp1 can1-100 lys2 med16Δ::LEU2 | D. Stillman |
| CC939-1A | his3 leu2 ura3 ade2 trp1 can1-100 lys2 med15Δ::LEU2 | This study |
| YCL10 | matα his3-Δ200 leu2-Δ1 ade2-101 ura3-52 lys2-801 trp1-Δ63 GAL med6::LEU2 [pRS313-MED6 HIS3] | 40 |
| YCL8 | matahis3-Δ200 leu2-Δ1 ade2-101 ura3-52 lys2-801 trp1-Δ63 GAL med6::LEU2 [pRS313-med6ts2 HIS3] | 40 |
| YSJ7 | matα his3-Δ200 leu2-Δ1 ade2-101 ura3-52 lys2-801 trp1-Δ63 GAL med11::TRP1 [pRS313-MED11 URA3] | 21 |
| YSJ8-4 | matahis3-Δ200 leu2-Δ1 ade2-101 ura3-52 lys2-801 trp1-Δ63 GAL med11::TRP1 [pRS313-med11ts URA3] | 21 |
| Z579 | matahis3 leu2 ura3 med17Δ2::HIS3 [MED17 LEU2 CEN] | 23 |
| Z628 | matahis3 leu2 ura3 med17Δ2::HIS3 [med17-138 LEU2 CEN] | 23 |
| Y217 | matahis3 leu2 ura3 ade2 trp1 SPT15-3HA::SPT15 MED6::MED6-TAP-kl TRP1 | This study |
| Y221 | matα his3 leu2 ura3 ade2 trp1 SPT15-3HA::SPT15 MED19::MED19-TAP-kl TRP1 | This study |
| Y225 | matα his3 leu2 ura3 ade2 trp1 SPT15-3HA::SPT15 MED2::MED2-TAP-kl TRP1 | This study |
| Y229 | matα his3 leu2 ura3 ade2 trp1 SPT15-3HA::SPT15 MED21::MED21-TAP-kl TRP1 | This study |
| Y233 | matα his3 leu2 ura3 ade2 trp1 SPT15-3HA::SPT15 MED10::MED10-TAP-kl TRP1 | This study |
| CL26-A | MED14::MED14-TAP-kl TRP1 GCN5::GCN5-3HA KAN MX6 med2Δ::HIS3 MX6 | This study |
| CL27-j | MED14::MED14-TAP-kl TRP1 GCN5::GCN5-3HA KAN MX6 med3Δ::HIS3 MX6 | This study |
| CL15-1 | matahis3 leu2 ura3 ade2 trp1 can1-100 spt3Δ::HIS3 MX6 | This study |
| CL16-1 | his3 leu2 ura3 ade2 trp1 spt20Δ::HIS3 MX6 | This study |
| CL17-1 | matα his3 leu2 ura3 ade2 trp1 gcn5Δ::HIS3 MX6 | This study |
| CL29-25 | mataMED14::MED14-TAP-kl TRP1 GCN5::GCN5-3HA KAN MX6 | This study |
| Y404 | matahis3 leu2 ura3 ade2 trp1 GCN5::GCN5-3HA-HIS3 MX6 | This study |
| Y421 | matahis3 leu2 ura3 ade2 trp1 SPT20::SPT20-3HA-HIS3 MX6 | This study |
| Y282 | matahis3 leu2 ura3 ade2 trp1 SPT3::SPT3-3HA-HIS3 MX6 MED14::MED14-TAP-kl TRP1 | This study |
| Y431 | matahis3 leu2 ura3 ade2 trp1 SPT3::SPT3-3HA-HIS3 MX6 MED14::MED14-TAP-kl TRP1 med3Δ::KAN MX6 | This study |
| Y432 | matahis3 leu2 ura3 ade2 trp1 SPT3::SPT3-3HA-KAN MX6 MED14::MED14-TAP-kl TRP1 med2Δ::HIS3 MX6 | This study |
| CL19-2 | mataSPT15::HA3-SPT15 MED14::MED14-TAP-kl TRP1 spt3Δ::HIS3 MX6 | This study |
| CL21-B | matα SPT15::HA3-SPT15 MED14::MED14-TAP-kl TRP1 gcn5Δ::HIS3 MX6 | This study |
| yJS6 | matahis3 leu2 ura3 trp1 taf1Δ gcn5Δ::KanR psW104-TAF1 pJW215-GCN5 | 24 |
| yJS7 | matahis3 leu2 ura3 trp1 taf1Δ gcn5Δ::KanR pSW104-TAF1 pKQL4-gcn5hat | 24 |
| CL33-4 | mataSPT15::HA3-SPT15 MED14::MED14-TAP-kl TRP1 spt20Δ::HIS3 MX6 | This study |
| Y364 | matahis3 leu2 ura3 ade2 trp1 med2Δ::HIS3 MX6 gcn5Δ::HIS3 MX6 | This study |
| CC849-8A | matα his3 leu2 ura3 trp1 met4Δ::TRP1 | 53 |
| CL37-B | matα met4Δ::TRP1 MED14::MED14-TAP-kl TRP1 GCN5::GCN5-3HA KAN MX6 | This study |
| CL23-2 | matα met4Δ::TRP1 ura3 lexAop lacZ::URA3 SPT15::HA3-SPT15 MED14::MED14-TAP-kl TRP1 | This study |
ChIP method.
Chromatin immunoprecipitation (ChIP) experiments were carried out essentially as previously described (32, 35). Cells (culture of 100 ml at a density of 1 × 107 to 2 × 107 cells/ml) were fixed with 1.4% formaldehyde for 15 min at 28°C. Fixation was stopped by the addition of glycine to a final concentration of 0.4 M. Cells were disrupted in FA lysis buffer (50 mM HEPES-KOH [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholic acid sodium salt, 0.1% sodium dodecyl sulfate) containing protease inhibitors and with a FastPrep instrument (Qbiogene). The cross-linked chromatin was collected by centrifugation for 15 min at 16,100 × g at 4°C in a microcentrifuge, resuspended in 2 ml FA lysis buffer containing protease inhibitors, and subjected to sonication yielding DNA fragments in a size range between 100 to 1,000 bp with an average of 400 bp. The insoluble debris was eliminated by centrifugation for 10 min at 6,100 × g at 4°C.
TAP-tagged proteins were immunoprecipitated by incubating chromatin from 2 × 108 to 3 × 108 cells with 15 μl of rabbit immunoglobulin G (IgG)-agarose (Sigma) or human IgG-Sepharose (Amersham Pharmacia) for 60 to 90 min at room temperature. Immune complexes were washed twice in FA lysis buffer containing 1 M NaCl, once in 10 mM Tris-HCl (pH 8.0)-0.25 M LiCl-1 mM EDTA-0.5% NP-40-0.5% Na deoxycholate, once in 10 mM Tris-HCl (pH 8.0)-1 mM EDTA-0.150 M urea, and once in 10 mM Tris-HCl (pH 8.0)-1 mM EDTA. Other proteins were immunoprecipitated by incubation for 60 to 90 min at room temperature chromatin from 2 × 108 to 3 × 108 cells with specific antibodies prebound to 15 μl of protein A-Sepharose (Amersham Pharmacia). The antibodies used in this study include monoclonal antibodies to HA (F7, Santa Cruz Biotechnology; 20 μl per IP) and Pol II CTD (8WG16, Covance; 5 μl per IP) and polyclonal antibodies to Met4 (kindly provided by Mike Tyers, Samuel Lunenfeld Research Institute; 10 μl per IP), Gal4 DNA-binding domain (DBD) (Santa Cruz Biotechnology; 20 μl per IP), H3 carboxy terminus (Abcam; 2 μl per IP), and acetyl K9 and acetyl K14 (both from Abcam or Upstate Biotechnology; 2 μl per IP). Immune complexes were washed three times in FA lysis buffer containing 0.5 M NaCl, once in 10 mM Tris-HCl (pH 8.0)-0.25 M LiCl-1 mM EDTA-0.5% NP-40-0.5% Na deoxycholate and once in 10 mM Tris-HCl (pH 8.0)-1 mM EDTA. IPs were eluted from the beads by being heated for 20 min at 65°C in 125 μl of 25 mM Tris-HCl (pH 7.5)-5 mM EDTA (0.5%). Formaldehyde cross-links were reversed by heating the eluates overnight at 70°C in the presence of 1-mg/ml Pronase (Roche). Aliquots of total chromatin input were processed in parallel for subsequent normalization. DNA was purified with the QIAquick DNA cleanup system (QIAGEN). A final elution was performed with 100 μl of TE (10 mM Tris-HCl [pH 8.0]-1 mM EDTA). Amounts of specific DNA targets present in input and IP samples were measured by real-time PCR using the LightCycler instrument and the FastStart DNA Master SYBR Green I mixture (Roche). Measures were calculated using two separate aliquots or two independent dilutions of the sample with a standard deviation of <15%. Primers used are listed in Table 2. A standard curve was generated for each run with a dilution series of input sample. This standard curve was used to assess the PCR efficiency and determine the relative concentration of target DNA in other samples. The specificity of the PCR products was assessed by performing a melting curve analysis. Data were analyzed with LightCycler software, version 3. The IP/Tot ratio corresponds to the concentration of target DNA in the IP sample relative to that in the corresponding input sample. For each immunoprecipitation, the highest value obtained was set at 100, and other values were expressed relative to this maximum.
TABLE 2.
Sequence and position of primers used in this study
| Gene | Position | Sequence | Expt |
|---|---|---|---|
| MET2-promoter fwd | −395 | ATTTCTTGCTATTGTTAGTGGCTCC | ChIP |
| MET2-promoter rev | −167 | CAACGAAGCGGAAGCTCATCTATT | ChIP |
| MET2-UAS (A) fwd | −395 | ATTTCTTGCTATTGTTAGTGGCTCC | ChIP |
| MET2-UAS (A) rev | −242 | GGTGTGTGCCAAATCCAAACGATTA | ChIP |
| MET2-core (B) fwd | −250 | GCACACACCCACAAATATACACATTAC | ChIP |
| MET2-core (B) rev | −51 | AAACTTTAGACGGACCCTGTGACT | ChIP |
| MET2-ORF fwd | +205 | GTAATTTGTCATGCCTTGACTGGGTC | ChIP/RT-PCR |
| MET2-ORF rev | +379 | ATCTAACGCCCGTCTCCTCATTTAT | ChIP/RT-PCR |
| MET3-promoter fwd | −402 | ACGGATTGCTGACAGAAAAAAAGG | ChIP |
| MET3-promoter rev | −191 | AGAAAGAGCCTCTATTTCTCATTGGT | ChIP |
| MET3-ORF fwd | +220 | TTAGCAGACGGCACATTGTGG | RT-PCR |
| MET3-ORF rev | +424 | TGGCTGGATGTTCTGGGTCA | RT-PCR |
| MET17-promoter fwd | −329 | AGGTCACATGATCGCAAAATGG | ChIP |
| MET17-promoter rev | −73 | GAAAAGACAAGAGAGCAAGAAAAAGG | ChIP |
| MET17-ORF fwd | +59 | ATGCTCACAGATCCAGAGCT | ChIP/RT-PCR |
| MET17-ORE rev | +309 | TGTCACCAGTGTGTGCCAAA | ChIP/RT-PCR |
| MET30-promoter fwd | −231 | GTGTTGGCGTGTGTGGTACAATGT | ChIP |
| MET30-promoter rev | +30 | ACTCATCATCCTTTGCCTCTCTCT | ChIP |
| MET30-ORF fwd | +819 | TTGGAAAGTCATCTACAGAGAACGGT | RT-PCR |
| MET30-ORF rev | +996 | CCCCGTGAATAAGTCCCATATACCTA | RT-PCR |
| CYS3-promoter fwd | −300 | GACCCCATACCACTTCTTTTTGTT | ChIP |
| CYS3-promoter rev | −36 | AGGTGCAAATGTCTATGTGTATAGGC | ChIP |
| CYS3-ORF fwd | +512 | GCCAAGACGTGATCTTGGTTGTCG | RT-PCR |
| CYS3-ORF rev | +697 | TTTGTAAGAACTGCAGACGCTCGTA | RT-PCR |
| GAL1-UAS fwd | −295 | AGAGGAAAAATTGGCAGTAACCTGG | ChIP |
| GAL1-UAS rev | −156 | TAGATCAAAAATCATCGCTTCGCTG | ChIP |
| GAL1-core fwd | −144 | ATAAATGGAAAAGCTGCATAACCAC | ChIP |
| GAL1-core rev | −9 | TTTCTCCTTGACGTTAAAGTATAGAGG | ChIP |
| IME2-ORF fwd | +1173 | ATCCCAAGTAGACGCAAGAGGCAAT | ChIP |
| IME2-ORF rev | +1377 | TTCTTGATTTAATGTTGGTGAGCACA | ChIP |
| POL1-ORF fwd | +2499 | TGCACCAGTTAATTCTAAAAAGGCA | ChIP |
| POL1-ORF rev 2 | +2717 | AAAACACCCTGATCCACCTCTGAA | ChIP |
| U4 fwd | +1 | ATCCTTATGCACGGGAAA | RT-PCR |
| U4 rev | +110 | CACCGAATTGACCATGAG | RT-PCR |
| 25S fwd | +3875 | GGTTATATGCCGCCCGTCTTGA | RT-PCR |
| 25S rev | +4051 | CCCAACAGCTATGCTCTTACTC | RT-PCR |
Analysis of mRNA levels.
Total RNA was isolated by extraction with hot acidic phenol (25). cDNA was generated by SuperScript II Reverse Transcriptase (Invitrogen) using p(dN)6 random hexamers (Roche) and anchored oligo(dT)23 primers (Sigma). Oligonucleotides specific for U4 small nuclear RNA or 25S rRNA (Table 2) were incorporated in the reaction to serve as internal reference. Individual cDNAs were quantified by real-time PCR using the LightCycler instrument and the FastStart DNA Master SYBR Green I reaction mixture (Roche). Data were analyzed with the LightCycler data analysis software.
RESULTS
Mediator is required for transcription of sulfur-containing amino acid biosynthesis genes.
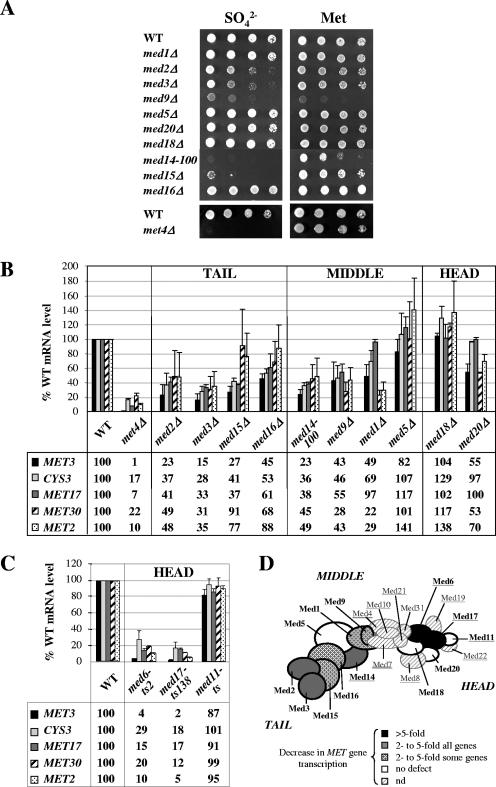
To determine whether Mediator is required for the regulation of the genes responsible for cysteine and methionine biosynthesis (referred to as MET genes), we first tested the ability of yeast strains containing null alleles of all nonessential subunit of core Mediator to metabolize inorganic sulfur sources. We tested in parallel a strain containing a viable mutation in the essential subunit Med14 (Rgr1) (med14-100) (13). As shown in Fig. 1A, mutation of Med2, Med3, Med14 (Rgr1), and Med15 (Gal11) led to a severe loss of viability on medium containing sulfate compared to methionine. Remarkably, the phenotypes of the med14-100 (rgr1-100) and med15Δ (gal11Δ) strains were very similar to that of the met4Δ strain, which lacks the activator controlling the MET genes, consistent with the possibility that Mediator plays a central role in expression of Met4 target genes.
FIG. 1.
Role of Mediator in transcription of the methionine biosynthesis genes. (A) Phenotypic analyses. Wild-type, med1Δ, med2Δ, med3Δ, med9Δ, med5Δ (nut1Δ), med20Δ (srb2Δ), med18Δ (srb5Δ), med14-100 (rgr1-100), med15Δ (gal11Δ), med16Δ (sin4Δ), and met4Δ strains were grown to exponential phase, collected, washed, and suspended in water. A serial fivefold dilution was spotted on B medium containing 0.05 mM ammonium sulfate (SO42−) or 0.05 mM l-methionine (Met). The most concentrated spot corresponds to a dilution at a density of 0.2 × 107 cells/ml. Plates were incubated at 28°C for 3 days. (B) Analysis of mRNA levels for selected Met4 target genes in strains used in the experiment shown in panel A. Cells were grown in YPD medium, collected, washed, and incubated for 60 min into B medium. RNA levels were quantified by reverse transcription, followed by real-time PCR, and normalized with U4 snRNA. Values represent the average of two independent experiments, and error bars indicate standard deviations. (C) Analysis of mRNA levels in cells containing temperature-sensitive mutations in Med6, Med17 (Srb4), and Med11. Strains containing wild-type or temperature-sensitive alleles of MED6 (40), MED17 (SRB4) (23), and MED11 (21) were grown in YPD medium at a permissive temperature (25°C), shifted at a restrictive temperature (38°C) for 90 min, collected, washed, and incubated in B medium at 38°C for 30 min. RNA levels were quantified as described in panel B, except that 25S rRNA was used for normalization. Values represent the average of two independent experiments. Error bars indicate standard deviations. (D) Summary of the effects of Mediator mutations on MET gene transcription. The model of topological organization of Mediator is adapted from reference 20. The Mediator subunits analyzed in this analysis are marked in boldface type. Subunits essential for viability are underlined.
To further define this role, we examined the effect of mutations in Mediator on MET gene transcription (Fig. 1B). The results showed that mutation of the tail domain subunit Med2 or Med3 and the middle subunit Med14 (Rgr1) or Med9 caused a marked decrease (two- to sevenfold) in mRNA levels of all five MET genes examined. Interestingly, deletion of the tail subunit Med15 (Gal11) caused a decrease by a factor of 2 or more in mRNA levels of MET3, MET17, and CYS3 but not MET2 and MET30. Conversely, deletion of the middle subunit Med1 caused a decrease by a factor of 2 or more in mRNA levels of MET2 and MET30 but not MET3, MET17, and CYS3. In contrast to other tail and middle subunit mutants, the med16Δ (sin4Δ) and med5Δ (nut1Δ) mutants showed only a weak or no defect in transcription of MET genes. Deletion of the two nonessential subunits of the head domain Med18 (Srb5) and Med20 (Srb2) had no major effect, either. To extend this analysis to essential subunits of the head domain, we measured mRNA levels in yeast strains bearing temperature-sensitive alleles of Med6, Med11, and Med17 (Srb4) (Fig. 1C). The results showed that inactivation of Med11 upon incubation at nonpermissive temperatures had no effect, whereas inactivation of Med6 and Med17 (Srb4) caused a major decrease (5 to 50 fold) in mRNA levels of all MET genes examined. These results, summarized in the schematic Fig. 1D, demonstrate as a whole that Mediator is required for the full activation of MET genes. In addition, individual subunits in each domain are differentially required, and some subunits seem to be required at only a subset of MET genes.
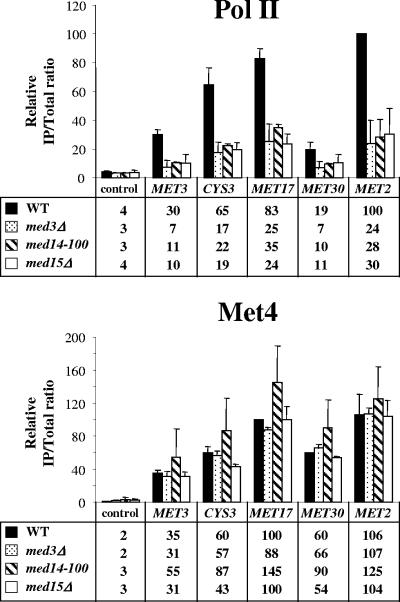
Transcription activation is a complex process with several successive steps, including activator binding, Pol II recruitment, transcription initiation, elongation, and termination. Defects in each step can have an impact on final mRNA levels. To clarify the function of Mediator in Met4-mediated activation, we performed ChIP experiments to measure Pol II and Met4 recruitment to MET promoters in cells containing mutations in the subunits Med3, Med14 (Rgr1), and Med15 (Gal11) (Fig. 2). The results showed that Pol II association with MET2, MET3, MET17, MET30, and CYS3 promoters was decreased by a factor of 2 to 5 in the mutants compared to the wild type. In contrast, Met4 association did not show any decrease. These results strongly suggest that Mediator operates in the first place during the recruitment of Pol II to Met4 target promoters, although they do not exclude additional roles in subsequent steps.
FIG. 2.
Association of Pol II and Met4 with MET genes in Mediator mutants. ChIP was performed on wild-type, med3Δ, med14-100 (rgr1-100), and med15Δ (gal11Δ) using antibodies against Pol II CTD or Met4 and PCR primers for the indicated Met4 target promoters and the IME2 open reading frame (ORF) as a control. Cells were grown as described in the legend to Fig. 1B. Values represent the average of two independent experiments. Error bars indicate standard deviations.
Mediator associates with MET genes under activating conditions.
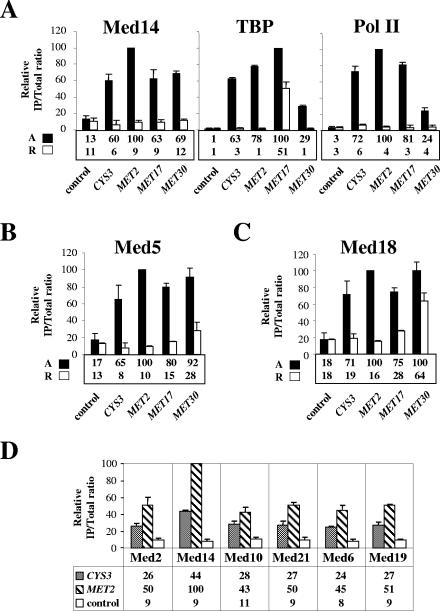
We next asked whether Mediator is physically recruited to Met4 target genes and whether its recruitment correlates with transcriptional activity. To address these questions, we first performed ChIP analysis with a strain expressing TAP-tagged Med14 (Rgr1) and HA-tagged TBP. Analysis of the immunoprecipitates showed association of Med14 (Rgr1) with MET promoters under inducing conditions but not under repressing conditions (Fig. 3A). To assess transcriptional activity, we measured TBP and RNA Pol II association in parallel. Comparison of the results for Med14 (Rgr1), TBP, and RNA Pol II revealed a strong correlation between Med14 (Rgr1) recruitment and transcriptional activity (note that the apparent high level of TBP occupancy at MET17 under repressing conditions was due to the presence of a tRNA gene just upstream of the MET17 promoter). Similar results were obtained with strains expressing TAP-tagged derivatives of the middle subunit Med5 (Nut1) and the head subunit Med18 (Srb5) (Fig. 3B and C). Additional ChIP experiments also showed association with MET genes of the tail subunit Med2, the middle subunits Med10 and Med21 (Srb7), and the head subunits Med6 and Med19 (Rox3) (Fig. 3D). We conclude that the entire Mediator associates with transcriptionally active but not inactive Met4 target promoters.
FIG. 3.
Association of Mediator with selected Met4-activated genes. (A) Med14 (Rgr1), TBP, and Pol II association. ChIP was performed on a strain expressing TAP-tagged Med14 and HA3TBP using IgG to immunoprecipitate Med14-TAP and antibodies to HA and Pol II CTD. Cells were grown in B medium containing 0.05 mM l-methionine until early exponential phase, filtered, washed, and transferred into B medium. After 1-h incubation (activating conditions [A]), half of the culture was cross-linked with formaldehyde and the other half was supplemented with 1 mM l-methionine and incubated an additional 40 min (repressing conditions [R]) prior to formaldehyde addition. IPs were quantified with PCR primers for the indicated MET promoters and the IME2 or POL1 ORF as a control. Values represent the average of two independent experiments. Error bars indicate standard deviations. (B and C) Med5 (Nut1) and Med18 (Srb5) association, respectively. ChIP was performed on strains expressing TAP-tagged Med5 or Med18 and HA3TBP as described in panel A. The results for TBP and Pol II were essentially the same as those shown in panel A (data not shown). (D) Med2, Med14 (Rgr1), Med10, Med21 (Srb7), Med6, and Med19 (Rox3) association. ChIP was performed on strains expressing TAP-tagged Mediator subunits and HA3TBP as described in the legend to panel A, except that the entire culture was cross-linked after a 1-h incubation in B medium. IPs were quantified using PCR primers for the CYS3 and MET2 promoters and the POL1 ORF as a control. Values represent the average of two independent experiments. Error bars indicate standard deviations.
The Met4 activator recruits Mediator through its activation domain.
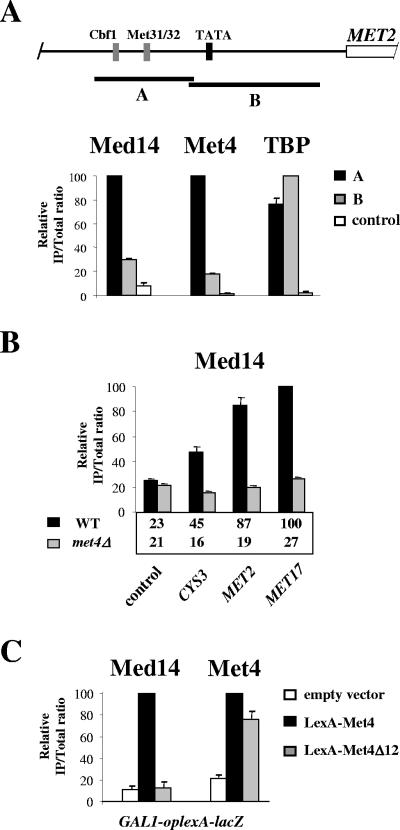
We next sought to establish Mediator localization along the prototype MET2 promoter. Like most Met4 target genes, the MET2 upstream regulatory region contains binding sites for the DNA-binding factors Cbf1 and Met31/32, which serve as anchorage platforms for Met4 (Fig. 4A). Med14 (Rgr1) and TBP immunoprecipitates from cells incubated under inducing conditions were analyzed with PCR primer pairs flanking Cbf1- and Met31/32-binding sites (fragment A) or flanking the TATA box (fragment B). Comparison of the results showed that fragment A was significantly more abundant than fragment B in Med14 (Rgr1) and Met4 immunoprecipitates. By contrast, fragments A and B were equally represented in the TBP immunoprecipitate (note that the TATA box was at the 5′ end of fragment B and very close to fragment A). Similar results were obtained with cells expressing TAP-tagged versions of the middle subunit Med5 (Nut1) and the tail subunit Med18 (Srb5) (data not shown). Therefore, Mediator shows preferential association with the upstream regulatory region and not to the core promoter at MET2. This observation is consistent with the idea that Mediator is targeted to MET genes primarily through interaction with Met4 and its cofactors.
FIG. 4.
Mediator is recruited through Met4 activation domain. (A) Localization of Med14 (Rgr1), Met4, and TBP along the MET2 promoter. (Top) Schematic of MET2 representing Cbf1- and Met31/32-binding sites (gray boxes), TATA element (black box), and open reading frame (open rectangle). (Bottom) ChIP on a strain expressing TAP-tagged Med14 and HA3TBP using IgG to immunoprecipitate Med14 and antibodies to Met4 and HA. Cells were grown as described in the legend to Fig. 1B. IPs were quantified using PCR primers for fragment A, fragment B, and the POL1 ORF as a control. Values represent the average of two independent experiments, and error bars indicate standard deviations. (B) Association of Med14 (Rgr1) with selected Met4 target genes in wild-type (WT) and met4Δ strains. ChIP was performed as described in the legend to panel A on WT and met4Δ strains expressing TAP-tagged Med14. IPs were quantified using PCR primers for the indicated MET promoters and the POL1 ORF as a control. Values represent the average of two independent experiments, and error bars indicate standard deviations. (C) A met4Δ strain expressing TAP-tagged Med14 (Rgr1) and bearing a GAL1-lexAop-lacZ chimeric gene (which contains four LexA operators in place of GAL1 UAS) integrated at the URA3 locus was transformed with a plasmid expressing the LexA DNA-binding domain fused to either wild-type Met4 or a derivative lacking the activation domain (Met4Δ12, deleted for residues 79 to 180). As a control, the strain was also transformed with an empty plasmid (pRS313). Cells were grown in YNB medium, collected, washed, incubated for 60 min into B medium, and subjected to ChIP with IgG to immunoprecipitate Med14 and antibodies against Met4. IPs were quantified using PCR primers for the GAL1-lexAop-lacZ promoter. Values represent the average of two independent experiments, and error bars indicate standard deviations.
To assess the respective contributions of Met4 and its cofactors Cbf1 and Met31/32 in recruitment of Mediator, we performed ChIP analysis of a met4Δ mutant (Fig. 4B). The results showed that association of Med14 (Rgr1) with MET2, MET17, and CYS3 was completely abolished in the met4Δ mutant, indicating that Met4 is essential for recruitment of Mediator to MET genes, whereas Met4 cofactors are not able to recruit Mediator on their own. We next asked whether Met4 would be able to recruit Mediator independently of its cofactors. To answer this question, ChIP analysis was performed on a strain expressing Met4 fused to the DNA-binding domain of LexA and containing a lacZ reporter gene driven by LexA operators integrated at the chromosome (Fig. 4C). The results showed that Med14 (Rgr1) was efficiently recruited to the reporter gene in cells expressing LexA-Met4, compared to cells expressing no LexA-Met4. Therefore, Met4 can recruit Mediator in the absence of its cofactors Cbf1 and Met31/32. To assess the requirement for Met4 activation domain, we performed ChIP analysis of a strain expressing a derivative of LexA-Met4 lacking the region spanning the activation domain. The results showed that deletion of Met4 activation domain completely abolished the recruitment of Med14 (Rgr1) without affecting the binding of LexA-Met4.
We conclude that Met4 has the intrinsic ability to target Mediator to DNA. Moreover, recruitment of Mediator by Met4 requires a functional activation domain.
SAGA serves as a coactivator for transcriptional activation by Met4.
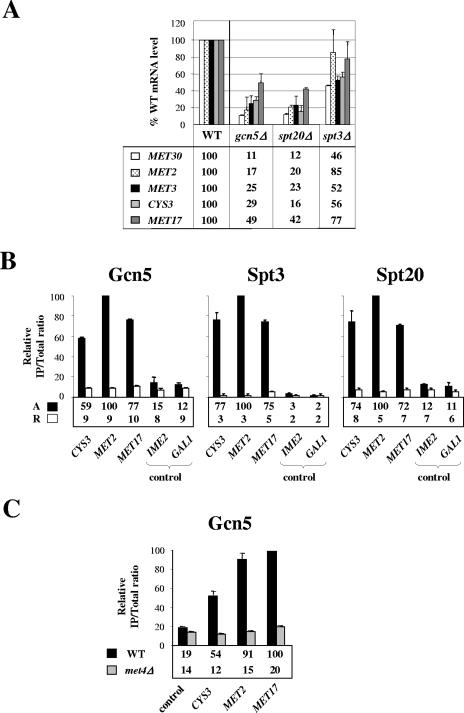
During the course of this work, Bhaumik et al. (5) found that recruitment of Mediator by Gal4 required SAGA, leading to the proposal that SAGA might function at some promoters as an “adaptor” that enables DNA-bound activators to recruit Mediator. To determine whether this model would apply to the MET gene network, we examined whether SAGA was involved in Met4-activated transcription. To this end, we first performed quantitative reverse transcription-PCR on mutant strains lacking Gcn5, Spt3, and Spt20, three distinct subunits of SAGA involved in histone acetylation, TBP recruitment, and structural integrity, respectively (3, 19, 38, 54). As shown in Fig. 5A, mRNA levels for MET genes were reduced by as much as 10 fold in the gcn5Δ and spt20Δ mutants compared to the wild-type cells. By contrast, MET gene transcription was only mildly affected in the spt3Δ mutant. These results demonstrate that some functions of SAGA, but not all, are required for transcriptional activation by Met4. We next performed ChIP analysis of strains expressing HA-tagged forms of Gcn5, Spt3, and Spt20 (Fig. 5B). The results showed association of the three subunits of SAGA with MET2, MET17, and CYS3 promoters under inducing conditions but not under repressing conditions. Additional ChIP experiments revealed that Gcn5 association with MET genes was reduced to background level in a met4Δ strain (Fig. 5C). Altogether, our results suggest that SAGA serves as a coactivator for transcriptional activation by Met4.
FIG. 5.
Role of SAGA in transcription of Met4-activated genes. (A) Analysis of mRNA levels in cells lacking Gcn5, Spt3, and Spt20. Wild-type, gcn5Δ, spt3Δ, and spt20Δ strains were grown in YPD medium, collected, washed, and incubated for 60 min into B medium. RNA levels were quantified by reverse transcription-PCR and normalized using 25S rRNA. Values represent the average of two independent experiments, and error bars indicate standard deviations. (B) Gcn5, Spt3, and Spt20 association with Met4 target promoters under activating and repressing conditions. ChIP was performed on strains expressing HA-tagged Gcn5, Spt3, or Spt20 as described in the legend to Fig. 3A. IPs were quantified using PCR primers for the indicated MET promoters and the IME2 ORF, as well as the GAL1 promoter as a control. Values represent the average of two independent experiments, and error bars indicate standard deviations. (C) Gcn5 association with Met4 target promoters in WT and met4Δ strains. ChIP was carried out on WT and met4Δ strains expressing HA-tagged Gcn5 using antibodies to HA and PCR primers for the indicated Met4 target promoters and the POL1 ORF as a control. Cells were grown in YPD medium, collected, washed, and incubated for 60 min into B medium prior to cross-linking. Values represent the average of two independent experiments, and error bars indicate standard deviations.
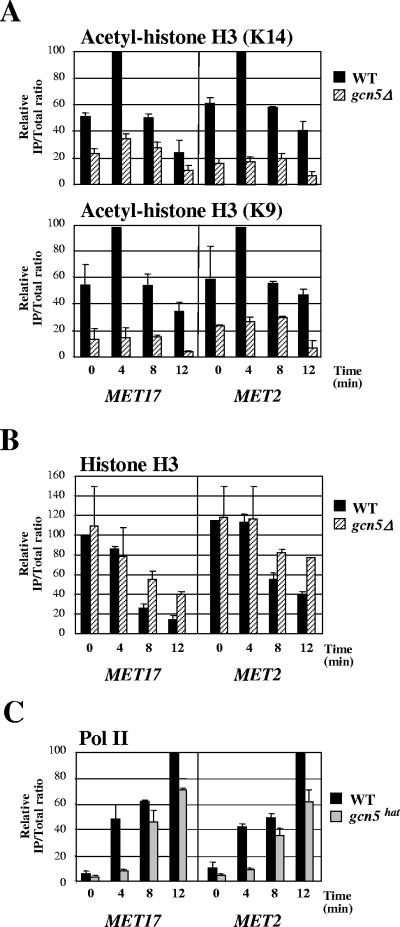
Comparison of transcription levels among the different SAGA mutants (Fig. 5A) showed that elimination of Gcn5 had a similar effect on MET mRNA levels as elimination of Spt20, which is required for structural integrity, suggesting that the function of SAGA in Met4-activated transcription might involve Gcn5-mediated histone acetylation. To test this possibility, we examined the levels of histone acetylation at MET genes at different times upon activation in parallel in the wild-type and gcn5Δ strains (Fig. 6A). The results showed higher levels of H3 acetylation at MET17 and MET2 in the wild type than in the gcn5Δ mutant. Moreover, following the induction, H3 acetylation was increased in the wild type but not in the gcn5Δ mutant. These results argue for a Gcn5-dependent H3 acetylation at Met4 target genes upon transcriptional induction. Note that the difference in acetylation levels between the wild-type and gcn5Δ strains at the zero time point was certainly due to the fact that Met4 was not completely absent from MET17 and MET2 in complete medium and was still able to weakly activate transcription (34). This difference might also reflect Gcn5 role in global, untargeted histone acetylation (30, 60). To directly assess the importance of Gcn5 HAT activity in Met4-dependent activation, we next examined the recruitment of Pol II at MET genes in a strain containing substitution mutations in Gcn5 that impairs its HAT activity (Fig. 6C). The results showed a defect in Pol II recruitment to MET17 and MET2 in the catalytic mutant compared to the wild type, supporting the hypothesis that function of SAGA in Met4-mediated activation depends, at least in part, on the HAT activity of its subunit Gcn5.
FIG. 6.
Levels of histone acetylation at Met4-activated genes. (A) Effect of Gcn5 inactivation on histone H3 acetylation at MET17 and MET2. ChIP of wild-type and gcn5Δ strains using antibodies specific for histone H3 acetylated at K9 or K14 and PCR primers for the 5′ end of MET17 and MET2 ORF was carried out. Cells were grown in YPD medium, filtered, and suspended into B medium. Aliquots were cross-linked at different times. Values represent the average of two independent experiments, and error bars indicate standard deviations. (B) Status of histone H3 at MET genes upon induction. Conditions were the same as described in the legend to panel A, except that an antibody specific for the carboxy-terminal end of histone H3 was used. (C) Recruitment of Pol II to MET17 and MET2 in a Gcn5 HAT mutant. ChIP of strains containing wild-type GCN5 or gcn5hat possessing the KQL mutation (24) using antibodies for Pol II CTD and PCR primers for MET17 and MET2 promoters was performed. Cells were grown and cross-linked as described in the legend to panel A. Values represent the average of two independent experiments, and error bars indicate standard deviations.
A remarkable feature of the results shown in Fig. 6A was that H3 acetylation in the wild-type strain peaked very rapidly following induction (within 4 min) and then diminished. To discriminate between removal of acetyl groups by deacetylases and loss of histone as a whole, the cross-linked chromatin was immunoprecipitated with an antibody specific for the carboxy-terminal end of histone H3. The results in Fig. 6B showed a substantial decrease over time in histone H3 at MET17 and MET2 in the wild type. Therefore, the apparent diminution in acetylation subsequent to the initial burst is certainly due to the loss of H3 and not to deacetylation. The results with the gcn5Δ mutant also showed a decrease in H3 association with MET17 and, to a lesser extent, MET2. These results indicate that eviction of histone H3 form MET17 and MET2 promoters can occur in the absence of Gcn5 HAT activity.
Altogether, our results suggest that SAGA is recruited to MET promoters through interaction with Met4 and then acetylates histone H3 through its Gcn5 HAT subunit.
Independent recruitments of Mediator and SAGA by Met4.
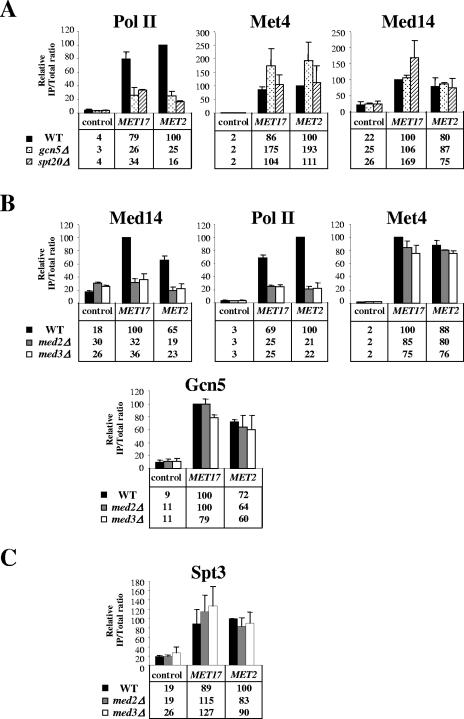
To characterize the functional relationship and respective mechanisms of action of Mediator and SAGA at MET genes, we performed ChIP analysis of mutant cells containing null alleles of SAGA and Mediator subunits. The results with the gcn5Δ and spt20Δ mutants showed a decrease in Pol II association with MET17 and MET2 in the mutants compared to the wild-type (by a factor of 2.4 to 6.7), but no significant decrease in Met4 association (Fig. 7A). Thus, SAGA facilitates the assembly of Pol II basal transcription machinery at MET genes at a postactivator-binding step. Moreover, inactivation of Gcn5 and Spt20 did not cause any decrease in Med14 (Rgr1) association either, strongly suggesting that the recruitment of Mediator by Met4 is independent of SAGA. The results with the med2Δ and med3Δ mutants showed a decrease in Med14 (Rgr1) and Pol II association with MET17 and MET2 in the mutants compared to the wild type (by a factor of 2.8 to 4.8) but no decrease in Met4 association (Fig. 7B). Thus, inactivation of the tail subunits Med2 and Med3 has no significant effect on the binding of the Met4 but impairs the recruitment of Mediator and prevents formation of Pol II basal transcription machinery. Moreover, inactivation of Med2 and Med3 did not affect Gcn5 and Spt3 association with MET17 and MET2 (Fig. 7B and C), suggesting that the recruitment of SAGA by Met4 is independent of Mediator.
FIG. 7.
Functional relationship between Mediator and SAGA in Met4-activated transcription. (A) Med14 (Rgr1), Pol II, and Met4 association with MET promoters in cells lacking Gcn5 and Spt20. ChIP of wild-type, gcn5Δ, and spt20Δ strains expressing Med14-TAP using IgG to immunoprecipitate Med14-TAP and antibodies to Pol II CTD and Met4 was carried out. Cells were grown in YPD medium, collected, washed, and incubated for 60 min into B medium prior to cross-linking. IPs were quantified using PCR primers for MET2 and MET17 promoters and the POL1 ORF as a control. Values represent the average of two independent experiments, and error bars indicate standard deviations. (B) Gcn5, Med14 (Rgr1), Pol II, and Met4 association with MET promoters in cells lacking Med2 and Med3. ChIP of wild-type, med2Δ, and med3Δ cells expressing Med14-TAP and Gcn5-HA3 using IgG to immunoprecipitate Med14-TAP, antibodies to HA to immunoprecipitate Gcn5-HA3, and antibodies to Pol II CTD and Met4 was carried out. Cells were grown in YPD medium, collected, washed, and incubated for 60 min into B medium prior to cross-linking. IPs were quantified using PCR primers for MET2 and MET17 promoters and the POL1 ORF as a control. Values represent the average of two independent experiments, and error bars indicate standard deviations. (C) Spt3 association with MET promoters in cells lacking Med2 and Med3. ChIP of wild-type, med2Δ, and med3Δ cells expressing Spt3-HA3 using antibodies to HA was carried out. Cells were grown in YPD medium, collected, washed, and incubated for 60 min into B medium prior to cross-linking. IPs were quantified using PCR primers for MET2 and MET17 promoters and the POL1 ORF as a control. Values represent the average of two independent experiments, and error bars indicate standard deviations.
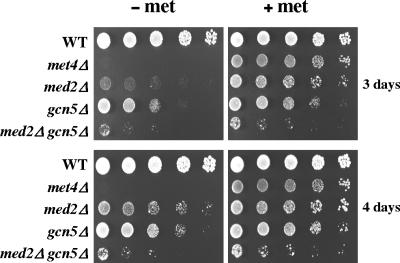
To further explore the functional interdependence between SAGA and Mediator in Met4-activated transcription, we compared the growth of med2Δ, gcn5Δ, and med2Δ gcn5Δ mutants on minimal medium containing either sulfate alone as a sulfur source or additional methionine (Fig. 8). The results showed that inactivation of Med2 and Gcn5 together had a more severe effect on growth in the absence of methionine than inactivation of Med2 and Gcn5 separately, suggesting that Mediator and SAGA can operate independently from each other and perform additive functions in the regulation of the MET gene network. Moreover, the growth of the med2Δ mutant was more severely affected than that of the gcn5Δ mutant, suggesting that the set of MET genes regulated by SAGA does not completely overlap with that regulated by Mediator. It is also possible that transcription of a MET gene particularly important for methionine biosynthesis is more affected in the med2Δ mutant than in the gcn5Δ mutant.
FIG. 8.
Growth phenotype on plates. Wild-type, met4Δ, med2Δ, gcn5Δ, and med2Δ gcn5Δ strains were grown to exponential phase, collected, washed, and suspended in water. Serial fivefold dilution was spotted on YNB minimal medium with or without 0.05 mM l-methionine. The most concentrated spot corresponds to a dilution at a density of 0.2 × 107 cells/ml. Plates were incubated at 28°C for 3 and 4 days.
DISCUSSION
In this report, we provide direct evidence that Mediator serves in vivo as a coactivator for transcriptional activation by the yeast activator Met4. First, mutations in several Mediator subunits impair transcription of the gene network regulated by Met4, leading ultimately to a defect in biosynthesis of methionine and cysteine. Second, Mediator associates with Met4 target promoters in vivo. This association maps to the Met4 binding region and happens only under inducing conditions. Third, artificial binding of Met4 to an ectopic site leads to recruitment of Mediator. Thus, Met4 provides an additional example of yeast activator that directs Mediator recruitment to target promoters during transcriptional activation, along with Gal4 (51), Gcn4 (56), Swi5 and Swi4/Swi6 (6, 12), and Pdr1 (17).
Differential requirement of Mediator subunits for Met4-activated transcription.
Our results provide new insights into the role of individual Mediator subunits in transcription activation. As already observed with Gal4 and Gcn4, elimination of Mediator subunits has various consequences on transcription activation by Met4, ranging from a severe decrease in mRNA to no visible defect. Remarkably, among the nonessential subunits, those which are the most critically required for Met4-activated transcription belong to the tail domain, which includes Med2, Med3, Med15 (Gal11), and Med16 (Sin4). The fact that elimination of Med2 and Med3 leads to the loss of Med14 (Rgr1) from MET2 and MET17 promoters (Fig. 7) suggests that the tail domain is required for Mediator recruitment to Met4 target genes. This supports the proposal that the tail domain serves as a binding surface for recruitment by gene-specific activators (48). The similarities in the transcriptional defects of the med2Δ, med3Δ, and, at some point, med15Δ (gal11Δ) mutants is probably due to the fact that elimination of one of the subunits causes dissociation of the others (43, 47, 61). Alternatively, it is also possible that Med2, Med3, and Med15 (Gal11) do not work individually but make concerted contributions to Mediator recruitment by Met4.
Contrary to Med2, Med3, and Med15 (Gal11), inactivation of Med16 (Sin4) has no significant effect on MET gene transcription. This is surprising, considering that Mediator isolated from med16Δ (sin4Δ) cells lacks the three other subunits (15, 43, 47). One explanation may be that elimination of Med16 (Sin4) destabilizes the tail domain but does not lead to its complete dissociation from the rest of Mediator within the cells. Consistently, Med2 and Med3 were observed to comigrate with other Mediator components during the first purification steps from med16Δ (sin4Δ) cells (47). Alternatively, Med2, Med3, and Med15 (Gal11) might be required for Mediator recruitment by Met4 only in the presence of Med16 (Sin4) but not in its absence. This hypothesis implies the existence of other binding sites for Met4 and suggests, furthermore, that the Mediator tail domain could be a regulatory module rather than an activator-binding surface. Finally, similar to what was observed in the case of some Gcn4-regulated genes (61), it is also possible that, in the med16Δ (sin4Δ) mutant, Med2/Med3/Med15 (Gal11) might be recruited to MET genes as a distinct functional entity able to function as a coactivator independently of the rest of Mediator. Further investigation will be necessary to decide between these possibilities.
There is strong evidence that the two yeast activators Gal4 and Gcn4 also recruit Mediator to their target genes in vivo (9, 32, 56). A comparison of the requirement in Mediator subunits for Gal4, Gcn4, and Met4 clearly indicates that the three activators function through overlapping but distinct sets of subunits. Indeed, inactivation of Med2, Med3, and Med15 (Gal11) impairs transcription activation by all three activators (Fig. 1C) (47, 50, 55, 56, 61). By contrast, inactivation of Med6 severely affects activation by Gal4 and Met4 but not Gcn4 (Fig. 1C) (41). On the other hand, inactivation of Med18 (Srb5) affects activation by Gcn4 (56) but not Gal4 and Met4 (Fig. 1B and data not shown). Finally, inactivation of the middle subunit Med9 impairs activation by Met4 (Fig. 1B) but not Gal4 and Gcn4 (21). Thus, Met4, Gal4, and Gcn4 activate transcription through Mediator by different mechanisms. The exact role of Med6, Med9, or Med18 (Srb5) is still unknown. It cannot be excluded that these subunits serve as binding sites for specific activators in concert with the tail subunits; however, we favor the hypothesis that they function at a postrecruitment stage. Additional experiments are in progress to address this point.
Contrary to other Mediator subunits, deletion of Med15 (Gal11) and Med1 has differential effects on MET gene transcription. Interestingly, the requirement for these two subunits seems to vary in an opposite direction: MET17 and CYS3 transcription is impaired in the med15Δ (gal11Δ) mutant but not in the med1Δ mutant, whereas MET2 and MET30 transcription is impaired in the med1Δ mutant but not in the med15Δ (gal11Δ) mutant (Fig. 1B). These results suggest that the requirement for individual Mediator subunits depends primarily on the activator but can also be influenced by the intrinsic structural characteristics of the promoter bound by the activator. It is tempting to speculate that some Mediator subunits may be essentially dedicated to accommodate the differences that may exist among promoters coregulated by a same activator.
Interplay between Mediator and SAGA at Met4-activated genes.
We found that SAGA is physically present at promoters of MET genes and is required for full transcriptional activity, demonstrating that MET genes are SAGA dependent (Fig. 5). Moreover, SAGA is required for Pol II but not Met4 association with MET promoters, suggesting a coactivator role for SAGA during transcription activation by Met4. Our results support a model in which SAGA would function during activation by Met4, primarily by modifying the structure of nucleosomes at MET promoters to promote formation of the Pol II basal transcription machinery. Indeed, elimination of SAGA HAT subunit causes similar defects in MET gene transcription as disruption of the whole complex (Fig. 5). Moreover, transcription activation by Met4 is accompanied by a Gcn5-dependent increase in acetylation of histone H3 at the promoter and 5′ end of the coding region of MET genes; in addition, mutation of Gcn5 HAT activity impairs recruitment of Pol II at the promoter (Fig. 6 and data not shown). The weak transcriptional defect resulting from Spt3 inactivation (Fig. 5A) makes unlikely a model where the function of SAGA at Met4 target promoters would involve, as in the case of Gal4-activated promoters (16, 38, 54), recruitment of TBP via interaction with Spt3 and Spt8.
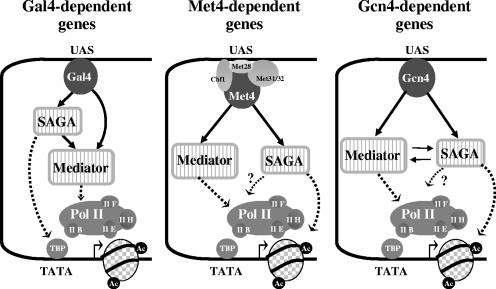
Our result that disruption of SAGA substantially impairs Pol II association with MET promoters but has no effect on Med14 (Rgr1) association with MET regulatory regions (Fig. 7A) has several important mechanistic implications. First, Mediator can associate with MET promoters independently of Pol II. Analyses of Mediator association with HO, GAL1,10, and ARG1 promoters have led to similar conclusions, supporting the notion that Mediator and Pol II are recruited separately and not as a preassembled holoenzyme complex in yeast (6, 9, 12, 32, 52). Secondly, the presence of Mediator at MET regulatory regions is not sufficient by itself to achieve high levels of Pol II association. The intervention of SAGA, and more particularly Gcn5, is certainly required to help removing nucleosomes likely to obstruct the access of Pol II and GTFs to the promoter. Finally, these results dismiss a role for SAGA in the recruitment of Mediator by Met4. In this respect, Met4 differentiates itself from Gal4 and Gcn4, which are also known to recruit both Mediator and SAGA (Fig. 9) (4, 9, 38, 51, 56). Indeed, recruitment of Mediator by Gal4 and Gcn4 is, at least in part, dependent on SAGA (5, 39, 52). Note, however, that there are conflicting reports regarding the actual contribution of SAGA in Mediator recruitment by Gal4. Monitoring Med15 (Gal11) and Med17 (Srb4) in a spt20Δ mutant, Bryant and Ptashne (9) reported that SAGA and Mediator were recruited to GAL1 promoter independently from each other; however, no direct comparison of the amount of bound Mediator in wild-type and spt20Δ strains was presented in the report, making it difficult to appreciate whether Mediator recruitment is optimal or not. On the other hand, Bhaumik et al. (5) reported a sharp diminution of Med17 (Srb4) association with GAL1 UAS in several mutants of SAGA, including an spt20Δ mutant, leading to the proposal that SAGA might serve as an “adaptor” that recruits Mediator to Gal4 activation domain. A possible explanation to account for this discrepancy might be differences in strain backgrounds and/or experimental conditions. We found under our own experimental conditions that Mediator association with GAL1 UAS was substantially impaired in the absence of SAGA (see Fig. S1 in the supplemental material), supporting the model that SAGA would indeed play a role in the recruitment of Mediator to Gal4-activated genes. Similarly, recruitment of Mediator was shown to involve an important contribution of SAGA at the Gcn4 target genes ARG4 and SNZ1 (52). However, at ARG1, Mediator recruitment by Gcn4 is largely independent of SAGA, possibly because this promoter possesses different structural features or binds additional transcription factors that render SAGA dispensable (18, 52).
FIG. 9.
Model for Mediator and SAGA interplay in transcriptional activation. Like Met4, Gal4 and Gcn4 are able to direct Mediator and SAGA recruitment to their target promoters (see the text for references). Recruitments of SAGA and Mediator are independent at Met4 target promoters and interdependent at Gcn4 target promoters. In contrast, at Gal4 target genes, optimal Mediator recruitment requires SAGA but SAGA recruitment does not require Mediator (see the text for details and references). The function of SAGA involves acetylation of histone H3 at Gcn4 (30) and Met4 target promoters and recruitment of TBP via interactions with Spt3/Spt8 at Gal4 target promoters (16, 38, 54). Mediator functions primarily by promoting recruitment of Pol II to the promoter, but roles at subsequent steps are not excluded.
As a whole, our results indicate, in combination with results from other laboratories, that the interplay between SAGA and Mediator can vary greatly, depending on the gene network and the nature of the activator (Fig. 8). As mentioned above, SAGA is required for optimal recruitment of Mediator at genes activated by Gal4 and Gcn4 but not at genes activated by Met4. In the other way, Mediator is required for optimal recruitment of SAGA at the Gcn4-activated genes ARG1, ARG4, and SNZ1 (52) but not at genes activated by Gal4 (5) and Met4. The molecular bases which underlie these mechanistic differences remain to be elucidated.
Altogether, our results emphasize the variety of mechanisms of action of coactivators and underline the necessity to diversify the model systems studied.
Supplementary Material
Acknowledgments
We are grateful to Dominique Thomas for constant support and critical reading of the manuscript. We thank Rick Young, Young Chul Lee, and Franklin Pugh for strains.
This work was supported by funds from the Centre National de la Recherche Scientifique and grants to L.K. from the Ministère de la Recherche (ACI Jeune Chercheur 2001) and the Association pour la Recherche sur le Cancer (subvention ARC no. 3578). C.L. was supported in part by a fellowship from the Ligue Nationale contre le Cancer.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Asturias, F. J., Y. W. Jiang, L. C. Myers, C. M. Gustafsson, and R. D. Kornberg. 1999. Conserved structures of mediator and RNA polymerase II holoenzyme. Science 283:985-987. [DOI] [PubMed] [Google Scholar]
- 2.Baker, R. E., and D. C. Masison. 1990. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol. Cell. Biol. 10:2458-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbaric, S., H. Reinke, and W. Horz. 2003. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell. Biol. 23:3468-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhaumik, S. R., T. Raha, D. P. Aiello, and M. R. Green. 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhoite, L. T., Y. Yu, and D. J. Stillman. 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 15:2457-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorklund, S., and C. M. Gustafsson. 2005. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 30:240-244. [DOI] [PubMed] [Google Scholar]
- 8.Blaiseau, P. L., and D. Thomas. 1998. Multiple transcriptional activation complexes tether the yeast activator Met4 to DNA. EMBO J. 17:6327-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant, G. O., and M. Ptashne. 2003. Independent recruitment in vivo by gal4 of two complexes required for transcription. Mol. Cell 11:1301-1309. [DOI] [PubMed] [Google Scholar]
- 10.Cai, M., and R. W. Davis. 1990. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell 61:437-446. [DOI] [PubMed] [Google Scholar]
- 11.Conaway, R. C., S. Sato, C. Tomomori-Sato, T. Yao, and J. W. Conaway. 2005. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30:250-255. [DOI] [PubMed] [Google Scholar]
- 12.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 7:1213-1220. [DOI] [PubMed] [Google Scholar]
- 13.Covitz, P. A., W. Song, and A. P. Mitchell. 1994. Requirement for RGR1 and SIN4 in RME1-dependent repression in Saccharomyces cerevisiae. Genetics 138:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, J. A., Y. Takagi, R. D. Kornberg, and F. A. Asturias. 2002. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell 10:409-415. [DOI] [PubMed] [Google Scholar]
- 15.Dotson, M. R., C. X. Yuan, R. G. Roeder, L. C. Myers, C. M. Gustafsson, Y. W. Jiang, Y. Li, R. D. Kornberg, and F. J. Asturias. 2000. Structural organization of yeast and mammalian mediator complexes. Proc. Natl. Acad. Sci. USA 97:14307-14310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, C., L. Wang, E. Milgrom, and W. C. Shen. 2004. On the mechanism of constitutive Pdr1 activator-mediated PDR5 transcription in Saccharomyces cerevisiae: evidence for enhanced recruitment of coactivators and altered nucleosome structures. J. Biol. Chem. 279:42677-42686. [DOI] [PubMed] [Google Scholar]
- 18.Govind, C. K., S. Yoon, H. Qiu, S. Govind, and A. G. Hinnebusch. 2005. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol. Cell. Biol. 25:5626-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 20.Guglielmi, B., N. L. van Berkum, B. Klapholz, T. Bijma, M. Boube, C. Boschiero, H. M. Bourbon, F. C. Holstege, and M. Werner. 2004. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 32:5379-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han, S. J., Y. C. Lee, B. S. Gim, G. H. Ryu, S. J. Park, W. S. Lane, and Y. J. Kim. 1999. Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation. Mol. Cell. Biol. 19:979-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 23.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 24.Huisinga, K. L., and B. F. Pugh. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13:573-585. [DOI] [PubMed] [Google Scholar]
- 25.Iyer, V., and K. Struhl. 1996. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5208-5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser, P., K. Flick, C. Wittenberg, and S. I. Reed. 2000. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell 102:303-314. [DOI] [PubMed] [Google Scholar]
- 27.Kim, Y. J., S. Bjorklund, Y. Li, M. H. Sayre, and R. D. Kornberg. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599-608. [DOI] [PubMed] [Google Scholar]
- 28.Kim, Y. J., and J. T. Lis. 2005. Interactions between subunits of Drosophila Mediator and activator proteins. Trends Biochem. Sci. 30:245-249. [DOI] [PubMed] [Google Scholar]
- 29.Kornberg, R. D. 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30:235-239. [DOI] [PubMed] [Google Scholar]
- 30.Kuo, M. H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 31.Kuras, L., R. Barbey, and D. Thomas. 1997. Assembly of a bZIP-bHLH transcription activation complex: formation of the yeast Cbf1-Met4-Met28 complex is regulated through Met28 stimulation of Cbf1 DNA binding. EMBO J. 16:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuras, L., T. Borggrefe, and R. D. Kornberg. 2003. Association of the Mediator complex with enhancers of active genes. Proc. Natl. Acad. Sci. USA 100:13887-13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuras, L., H. Cherest, Y. Surdin-Kerjan, and D. Thomas. 1996. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 15:2519-2529. [PMC free article] [PubMed] [Google Scholar]
- 34.Kuras, L., A. Rouillon, T. Lee, R. Barbey, M. Tyers, and D. Thomas. 2002. Dual regulation of the met4 transcription factor by ubiquitin-dependent degradation and inhibition of promoter recruitment. Mol. Cell 10:69-80. [DOI] [PubMed] [Google Scholar]
- 35.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 36.Kuras, L., and D. Thomas. 1995. Functional analysis of Met4, a yeast transcriptional activator responsive to S-adenosylmethionine. Mol. Cell. Biol. 15:208-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuras, L., and D. Thomas. 1995. Identification of the yeast methionine biosynthetic genes that require the centromere binding factor 1 for their transcriptional activation. FEBS Lett. 367:15-18. [DOI] [PubMed] [Google Scholar]
- 38.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larschan, E., and F. Winston. 2005. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol. Cell. Biol. 25:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, Y. C., and Y. J. Kim. 1998. Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol. Cell. Biol. 18:5364-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, Y. C., S. Min, B. S. Gim, and Y. J. Kim. 1997. A transcriptional mediator protein that is required for activation of many RNA polymerase II promoters and is conserved from yeast to humans. Mol. Cell. Biol. 17:4622-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, Y. C., J. M. Park, S. Min, S. J. Han, and Y. J. Kim. 1999. An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 19:2967-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, Y., S. Bjorklund, Y. W. Jiang, Y. J. Kim, W. S. Lane, D. J. Stillman, and R. D. Kornberg. 1995. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 92:10864-10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linder, T., and C. M. Gustafsson. 2004. The Soh1/MED31 protein is an ancient component of Schizosaccharomyces pombe and Saccharomyces cerevisiae Mediator. J. Biol. Chem. 279:49455-49459. [DOI] [PubMed] [Google Scholar]
- 45.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 46.Malik, S., and R. G. Roeder. 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30:256-263. [DOI] [PubMed] [Google Scholar]
- 47.Myers, L. C., C. M. Gustafsson, K. C. Hayashibara, P. O. Brown, and R. D. Kornberg. 1999. Mediator protein mutations that selectively abolish activated transcription. Proc. Natl. Acad. Sci. USA 96:67-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park, J. M., H. S. Kim, S. J. Han, M. S. Hwang, Y. C. Lee, and Y. J. Kim. 2000. In vivo requirement of activator-specific binding targets of mediator. Mol. Cell. Biol. 20:8709-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patton, E. E., A. R. Willems, D. Sa, L. Kuras, D. Thomas, K. L. Craig, and M. Tyers. 1998. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 12:692-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piruat, J. I., S. Chavez, and A. Aguilera. 1997. The yeast HRS1 gene is involved in positive and negative regulation of transcription and shows genetic characteristics similar to SIN4 and GAL11. Genetics 147:1585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799-809. [DOI] [PubMed] [Google Scholar]
- 52.Qiu, H., C. Hu, F. Zhang, G. J. Hwang, M. J. Swanson, C. Boonchird, and A. G. Hinnebusch. 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol. Cell. Biol. 25:3461-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rouillon, A., R. Barbey, E. E. Patton, M. Tyers, and D. Thomas. 2000. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30)complex. EMBO J. 19:282-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki, Y., Y. Nogi, A. Abe, and T. Fukasawa. 1988. GAL11 protein, an auxiliary transcription activator for genes encoding galactose-metabolizing enzymes in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:4991-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swanson, M. J., H. Qiu, L. Sumibcay, A. Krueger, S. J. Kim, K. Natarajan, S. Yoon, and A. G. Hinnebusch. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 23:2800-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas, D., I. Jacquemin, and Y. Surdin-Kerjan. 1992. MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:1719-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas, D., and Y. Surdin-Kerjan. 1997. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61:503-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson, C. M., A. J. Koleske, D. M. Chao, and R. A. Young. 1993. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73:1361-1375. [DOI] [PubMed] [Google Scholar]
- 60.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, F., L. Sumibcay, A. G. Hinnebusch, and M. J. Swanson. 2004. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol. Cell. Biol. 24:6871-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.