Abstract
The expression of the phase 2 detoxification enzymes and antioxidant proteins is induced at the transcriptional level by Nrf2 and negatively regulated at the posttranslational level by Keap1 through protein-protein interactions with and subsequent proteolysis of Nrf2. We found that the Neh2 domain of Nrf2 is an intrinsically disordered but biologically active regulatory domain containing a 33-residue central α-helix followed by a mini antiparallel β-sheet. Isothermal calorimetry analysis indicated that one Neh2 molecule interacts with two molecules of Keap1 via two binding sites, the stronger binding ETGE motif and the weaker binding DLG motif. Nuclear magnetic resonance titration study showed that these two motifs of the Neh2 domain bind to an overlapping site on the bottom surface of the β-propeller structure of Keap1. In contrast, the central α-helix of the Neh2 domain does not have any observable affinity to Keap1, suggesting that this region may serve as a bridge connecting the two motifs for the association with the two β-propeller structures of a dimer of Keap1. Based on these observations, we propose that Keap1 recruits Nrf2 by the ETGE motif and that the DLG motif of the Neh2 domain locks its lysine-rich central α-helix in a correct position to benefit ubiquitin signaling.
Bodies are inevitably and constantly insulted by different environmental stresses caused by toxic chemicals, free radicals, carcinogens, and xenobiotic metabolites, which can induce the pathogenesis of many diseases, including cancer, diabetes, atherosclerosis, Alzheimer's disease, and arthritis (3, 29, 51, 53). The expression of an array of antioxidant and phase 2 drug-metabolizing enzymes protects cells from various oxidative stresses (2, 5, 12, 16, 17, 43, 44). The induction of these cytoprotective genes is regulated at the transcriptional level by a specific cis-acting element, the antioxidant/electrophile response element (ARE/EpRE) found in the promoter regions of these genes (21, 39, 40). Nrf2 (nuclear factor E2-related factor 2) has been found to be the central transcription factor that interacts with the ARE/EpRE to transactivate cytoprotective gene expression constitutively or to induce the expression in response to oxidative stress signals (19, 21, 42).
Nrf2 belongs to the Cap'n'Collar (CNC) family of transcription factors that contain a conserved basic region-leucine zipper structure (19, 38). By comparing the human and chicken Nrf2 amino acid sequences, six highly homologous regions have been defined in Nrf2 (Neh1 to Neh6 domains) (20). Of these Neh domains, Neh1, Neh3, and Neh6 locate in the C-terminal half of Nrf2. Neh1 contains a CNC-type basic-leucine zipper DNA binding motif, and Neh6 contains a serine-rich conserved region. In the N-terminal half, there are two acidic transactivation domains, Neh4 and Neh5, which have been shown to interact with the KIX and CH3 domains of CBP for transactivation (24). Neh2 locates at the N terminus of Nrf2 and acts as the regulatory domain for cellular stress response. Neh2 interacts with a cytoplasmic protein Keap1 (Kelch-like ECH-associated protein 1) (20). Keap1 possesses four functional domains: BTB (Broad complex, Tramtrack, and Bric-a-Brac) (4), IVR (intervening region), DGR (double glycine repeat or Kelch repeat) (1, 60), and CTR (C-terminal region). The BTB domain, like some of its structural homologs (54), has been shown to serve as a dimerization domain, and dimerization of Keap1 appears to be important for effective function of Keap1 (64).
Being a substrate adaptor of the Cul3-based E3 ligase machinery (6, 14, 30, 61) and an oxidative stress sensor (8, 57), Keap1 utilizes the DGR and CTR domains to interact with the Neh2 domain of Nrf2. This intermolecular interaction allows Keap1 to regulate the rate of Nrf2 protein turnover through ubiquitin signaling and proteasomal proteolysis. Oxidative/electrophilic stress signals are transduced by modification of the sulfhydryl groups of reactive cysteines within the IVR domain (8, 57), which attenuates both polyubiquitination and proteasomal degradation of Nrf2 (30) and results in an enhanced nuclear accumulation of Nrf2 for transactivating ARE-dependent cellular protective enzymes (17), such as heme oxygenase 1 and NAD(P)H-quinone oxidoreductase 1.
On the other hand, there are two evolutionarily conserved motifs within the Neh2 domain among the CNC protein family. The DLG motif, which locates at the N-terminal region, has been reported to be important for ubiquitination and degradation of Nrf2 (25, 36), while the ETGE motif is essential for interacting with Keap1 (32). In addition, seven lysine residues of the Neh2 domain, which reside upstream of the ETGE motif, have been shown to be indispensable for Keap1-dependent polyubiquitination and degradation of Nrf2 (61).
Recently, X-ray crystal structures of human and mouse Keap1 including the DGR and CTR domains were reported (35, 47, 47a). The DGR and CTR domains of Keap1 (hereafter Keap1-DC) form a six-bladed β-propeller conformation with high symmetry. Analogous to the substrate recognition mode reported for the Skp-Cdc4-CPD complex (45), a short ETGE peptide (76LDEETGEFL84) containing the conserved ETGE motif binds to Keap1-DC at the entrance of the central cavity on the bottom side of the β-propeller (47a). Although this crystal structure has provided the molecular basis of substrate recognition for this protein adaptor of the E3 ligase holoenzyme, further analysis of the entire Neh2 domain and its interaction with Keap1 may provide insights into the biological switch for mediating ubiquitin signaling and the cellular response against oxidative stress.
In this paper, we present biophysical studies using nuclear magnetic resonance (NMR) and isothermal calorimetry (ITC) on the mouse Neh2 domain and its interaction with the Keap1-DC. Based on the results of these analyses, we propose that Keap1 recruits Nrf2 by recognizing the ETGE and DLG motifs within the Neh2 domain and positions the lysine residues of the Neh2 domain in a correct orientation to facilitate ubiquitin signaling for protein turnover.
MATERIALS AND METHODS
Plasmids.
A gene fragment encoding mouse Neh2 (Met-1 to Gly-98) or Neh2[Δ1-33] (Arg-34 to Gly-98) was inserted into pET15b prokaryotic expression vector (Novagen) via the NdeI and BamHI sites, resulting in pET15b-Neh2 or pET15b-Neh2[Δ1-33], respectively. The 79ETGE82 deletion (ΔETGE) and the double deletion (Δ1-33, ΔETGE) mutants of Neh2 were constructed by amplifying the corresponding DNA fragments from pCMVNrf2ΔETGE (30) and inserting them into pET15b via NdeI and BamHI sites. The construct that expresses mouse Keap1-DC (Thr-309 to Cys-624) as the Mxe intein/chitin binding domain fusion protein was made as described previously (47). Constructs expressing arginine point mutants of Keap1-DC were prepared by the PCR overlap extension method, and resulting DNA fragments were inserted into the pET21a expression vector via NdeI and XhoHI sites. pET21a-Keap1 encoding Met-1 to Arg-614 of mouse Keap1 was constructed by inserting the corresponding DNA fragment via NdeI and XhoHI into the expression vector.
Protein expression and purification.
All Neh2 or Keap1-DC constructs were expressed in Escherichia coli BL21 Codon Plus (DE3)-RIL (Stratagene). Unlabeled or uniformly 15N- or 15N/13C-labeled protein samples of mouse Neh2 and its mutants were prepared by expressing the proteins in modified minimal M9 medium supplemented with [15N]ammonium chloride or [15N]ammonium chloride/13C6-d-glucose (Cambridge Isotope Laboratory and Isotec, Inc.) as sole nitrogen and carbon sources. Preparation of the growth medium and expression protocols for preparing proteins isotopically labeled with different selective amino acids, 15N- or 15N/13C-Gln, Arg, Lys, or Leu (Cambridge Isotope Laboratory), are described elsewhere (K. I. Tong, M. Yamamoto, and T. Tanaka, submitted for publication). His-tagged Neh2, Neh2 mutants, Keap1-DC arginine mutants, and the full-length Keap1-DC were purified using Ni-nitrilotriacetic acid beads (QIAGEN). N-terminal His tags of Neh2 and Neh2 mutants were removed by thrombin (Calbiochem). All proteins were further purified with MonoQ and Superdex S75 16/60PG or S200 26/60PG column chromatography (Pharmacia). Mouse Keap1-DC was expressed and purified as previously described (47). All proteins were near homogeneity, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. NMR samples were prepared in the range of 0.25 to 1.2 mM in 20 mM sodium phosphate, 100 mM NaCl, 10 mM [2H10]dithiothreitol (Cambridge Isotope Laboratory), 1 mM 4-(2-aminoethyl)-benzenesulfonyl-fluoride (Roche), and 90% H2O-10% 2H2O (Isotec Inc.), pH 8.0, or in 20 mM [2H11]Tris(hydroxymethyl)methylamine (Cambridge Isotope Laboratory) at pH 8.2 in place of the sodium phosphate buffer. The DLG peptide (Met-17 to Val-36, 82% in purity) containing the DLG motif of the Neh2 domain was purchased from Genosys, Sigma.
NMR data acquisition.
All NMR spectra were acquired at 25°C on a Varian UNITY INOVA 500 or a Bruker AVANCE DRX 800 spectrometer equipped with a triple resonance pulse field gradient probe. Two-dimensional (2D) 1H, 15N heteronuclear single-quantum correlation (HSQC) spectra (27) were recorded with the following numbers of complex points and acquisition times: 15N(F1) 256, 128 ms; 1H(F2) 1,024, 102 ms (64 transients) on a 500 MHz spectrometer; 15N(F1) 256, 102 ms; 1H(F2) 1,024, 92 ms (80 transients) on an 800 MHz spectrometer. All transverse relaxation optimized spectroscopy-type 2D 1H, 15N HSQC spectra (hereafter TROSY-HSQC) (48) were recorded on an 800 MHz spectrometer. The following numbers of complex points and acquisition times were used: 15N(F1) 256, 102 ms; 1H(F2) 1,024, 92 ms (80 transients).
The 15N-{1H} nuclear Overhauser effect (NOE) was measured as described previously (11) using the uniformly 15N-labeled Neh2. Two sets of spectra with and without the NOE were recorded on a 500 MHz spectrometer with the following numbers of complex points and acquisition times: 15N(F1) 256, 128 ms; 1H(F2) 1,024, 102 ms (32 transients). Resonance intensities were used to determine the NOE values, and the average values of results from two experiments were calculated. Homonuclear 2D or three-dimensional (3D) 15N-edited nuclear Overhauser effect spectroscopy (NOESY) experiments (22, 62) were performed on unlabeled or uniformly 15N-labeled Neh2, respectively, with an NOE mixing time of 100 ms or 200 ms and using an 800 MHz spectrometer to detect protons within a distance of 5 Å. The following numbers of complex points and acquisition times were used: 1H(F1) 256, 22 ms; 1H(F3) 1,024, 88 ms (64 transients) for homonuclear 2D NOESY; 1H(F1) 256, 22 ms; 15N(F2) 32, 13 ms; 1H(F3) 1,024, 88 ms (16 transients) for 3D 15N-edited NOESY. Triple-resonance 3D spectra of the uniformly or selective amino acid 15N/13C-labeled Neh2 were recorded with the following numbers of complex points and acquisition times: HNCO, 13CO (F1) 64, 43 ms; 15N (F2) 32, 16 ms; 1H (F3) 1,024, 102 ms (8 transients); CBCA(CO)NH, 13Cα/β (F1) 44, 6 ms; 15N (F2) 32, 16 ms; 1H (F3) 1,024, 102 ms (16 transients); HNCACB, 13Cα/β (F1) 96, 12 ms; 15N (F2) 32, 16 ms; 1H (F3) 1,024, 102 ms (16 transients) (41).
NMR data processing and analysis.
All spectra were processed and displayed using NMRPipe and NMRDraw (7) and analyzed by Pipp (15). Sequential resonance assignments for backbone 1HN, 13Cα, 13Cβ, 13CO, and 15N nuclei of Neh2 were generated from 3D HNCO, CBCA(CO)NH, and HNCACB data sets (52). The secondary structure of Neh2 was characterized using chemical shift indexes (CSI) based on Cα and Cβ chemical shifts (37, 58) and NOE connectivity patterns.
Mouse Keap1-DC titration.
Neh2 or Neh2 mutants (0.25 mM) uniformly labeled with 15N or [15N]Leu were titrated with an increasing molar ratio (1:0.25 to 1:2) of unlabeled Keap1-DC. [15N]Arg-labeled Keap1-DC (0.3 mM) was titrated with unlabeled Neh2, Neh2[Δ1-33], Neh2[ΔETGE], Neh2[Δ1-33, ΔETGE], or the DLG peptide (0.1 mM to 0.6 mM for deletion mutants of the Neh2 domain and 0.1 mM to 1.5 mM for the DLG peptide).
Analytical ultracentrifugation.
All analyses were carried out at 25°C using a Beckman Optima XL-I analytical ultracentrifuge monitoring at 280 nm. Data sets were collected at a rotor speed of 60,000 rpm and a protein concentration of 155 μM in the sedimentation velocity experiments. Results were analyzed by the van Holde-Weischet method (56). Data sets of sedimentation equilibrium analyses for the Neh2 domain were collected at a rotor speed of 32,000 rpm and three protein concentrations of 77.5, 124, and 155 μM. Those for the full-length Keap1 were collected at a rotor speed of 12,000 rpm and concentrations of 3, 4.3, and 8.5 μM. The resulting data were subjected to global fitting, and the molecular weight was calculated using a nonlinear least-squares technique (23).
ITC titration experiments.
ITC titration experiments were performed at 25°C on a VP-ITC system (MicroCal, Inc.). All proteins were extensively dialyzed against 60 mM sodium phosphate buffer containing 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (Sigma) at pH 8.1. All samples and buffers were filtered and degassed before use. In a typical experiment, 5-μl aliquots of 0.326 mM Neh2 or Neh2 mutants were injected 56 times at 4-min intervals from a stirring syringe (300 rpm) into the sample cell containing 1.40 ml of 0.046 mM Keap1-DC. For the titration of Keap1-DC or its arginine mutants against a 9-residue ETGE peptide (Leu-76 to Leu-84; Promega) of the Neh2 domain, 10-μl aliquots of 0.400 mM peptide solution were injected 28 times into 1.40 ml of 0.040 mM of Keap1-DC or its mutants. The heat changes in binding of the last 5 injections after saturation were averaged and used to correct the heat of protein dilution. All runs were done in triplicate. Binding data were analyzed using the computer program Origin, version 5.0, supplied by MicroCal, Inc. The binding stoichiometry (n), the enthalpy change (ΔH) and binding constant (Ka) upon protein-protein interaction can be obtained directly from the experimental titration curve (see lower panels of Fig. 3 and 5). Gibbs energy change (ΔG = −RTlnKa) and the entropy change [ΔS = (−ΔG +ΔH)/T] of the association were calculated from the Ka and ΔH with T (the absolute temperature) and R (the gas constant).
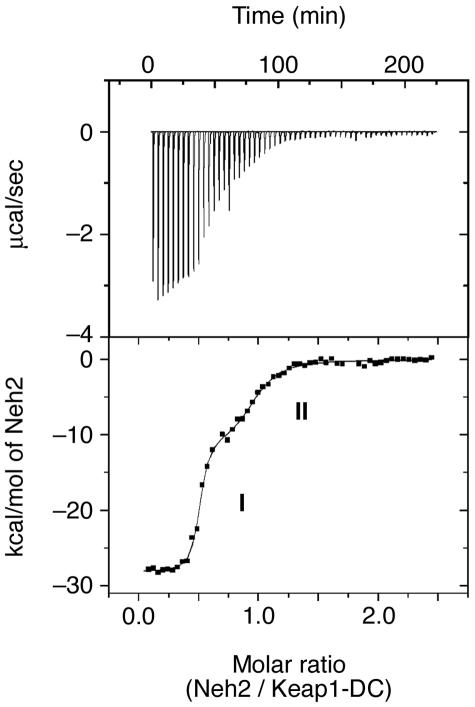
FIG. 3.
Two binding sites for Keap1 reside in Neh2 that differ in their affinities by 2 orders of magnitude. A representative ITC profile of the titration of Keap1-DC with Neh2 is shown. The upper panel shows the raw ITC thermograms, and the lower panel shows the fitted binding isotherms. The two phases on the curve represent the higher (I) and lower (II) binding affinities in the two binding sites of Neh2.
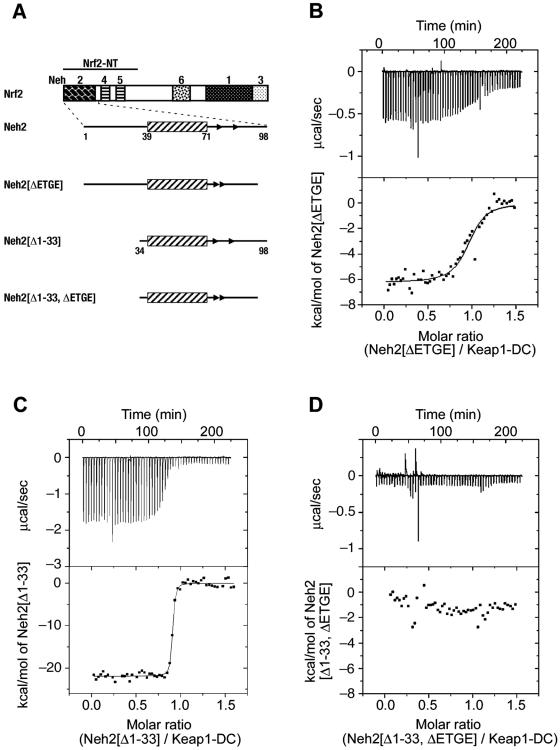
FIG. 5.
The ETGE motif and DLG motifs constitute the stronger and weaker binding sites, respectively, to Keap1. (A) Schematic presentation of expression constructs for the Neh2 domain and its deletion mutants for ITC and NMR studies. (B to D) Representative ITC titration profiles for the titration of Keap1-DC with Neh2[ΔETGE] (B), Neh2[Δ1-33] (C), and Neh2[Δ1-33, ΔETGE] (D). The upper panels represent the raw ITC thermograms, and the lower panels represent the fitted binding isotherms. The integrated binding isotherms are plotted against the molar ratio of different mutants of Neh2. A single-site binding model fits best the single-phase curve for the titration with Neh2[ΔETGE] and Neh2[Δ1-33]. Neh2[Δ1-33, ΔETGE] shows heat change similar to that of the heat of dilution of the proteins.
RESULTS
The Neh2 domain is intrinsically disordered with local elements of secondary structure.
Keap1 negatively regulates transactivation activity of Nrf2 through physical intermolecular interaction, followed by ubiquitination and proteasomal degradation of Nrf2 (21, 30). Since Keap1 represses the Nrf2 activity through binding to the Neh2 domain, in this study, we examined the structure of the Neh2 domain and its interaction with Keap1-DC using solution NMR spectroscopy and ITC analysis. The TROSY-HSQC spectrum of unbound Neh2 exhibited limited chemical shift dispersion (Fig. 1A) (backbone amide protons resonate between 7.7 to 8.7 ppm around the random coil region of 8.0 to 8.5 ppm) (58), a feature typical of intrinsically disordered polypeptides (10, 55). As the N-terminal half of Nrf2 (Nrf2-NT), comprised of Neh2 and two transactivation domains, Neh4 and Neh5 (Fig. 2A), has been shown to be physiologically active in luciferase reporter assays (24), we tested whether the disordered nature of Neh2 is intrinsic or due to immature truncation. The TROSY-HSQC spectrum of the Nrf2-NT (data not shown) showed resonances of limited dispersion in the amide proton dimension similar to that of Neh2. In addition, we noticed in the spectrum of the Nrf2-NT construct the same chemical shift values as those present in the spectrum of the Neh2 domain (Fig. 1A). These results suggest that the disordered nature of Neh2 is intrinsic.
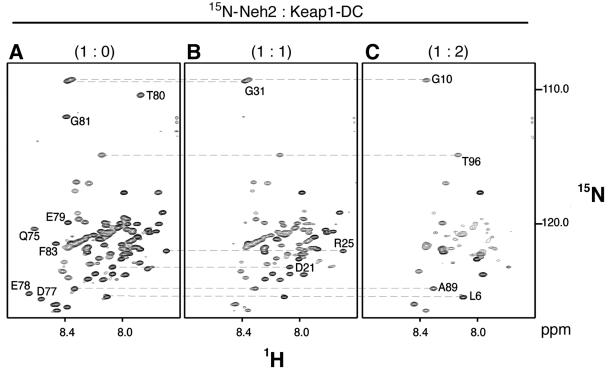
FIG. 1.
Neh2 is an intrinsically disordered but biologically active domain. 1H, 15N-TROSY-HSQC spectra of uniformly 15N-labeled Neh2 (A) and its complex with a one (B) or two (C) molar ratio of unlabeled Keap1-DC. Some of the isolated resonance signals representing residues in the β-sheet (Gln-75, Asp-77, Glu-78, Glu-79, Thr-80, Gly-81, and Phe-83) (A), residues close to the DLG motif (Asp-21, Arg-25 and Gly-31) (B), and residues at both flexible terminal ends of Neh2 (Leu-6, Gly-10, Ala-89, and Thr-96) (C) are labeled with a 1-letter amino acid code and a residue number. The relatively poor dispersion of amide proton (1H) chemical shifts indicates that Neh2 is disordered in solution (A). The disappearance of resonance signals upon the addition of Keap1-DC suggests the formation of a Neh2-Keap1 complex (B and C). The lost resonance is due to either an increase in molecular size through complex formation or an intermediate chemical exchange between free and bound conformations of Neh2.
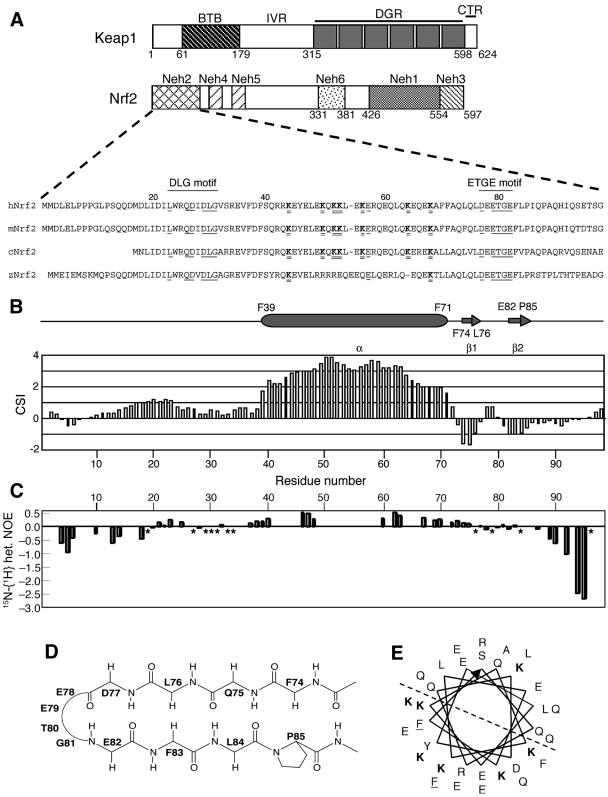
FIG. 2.
The secondary structure and low backbone rigidity exhibited in the Neh2 domain. (A) Schematic diagram showing the protein architecture of Keap1 and Nrf2 and sequence alignment of the Neh2 domain from different species (h, human; m, mouse; c, chicken; z, zebra fish). Numbers corresponding to the residues of mouse Nrf2 are shown above the sequences. Amino acids that are conserved within a subgroup of the CNC protein family, including Nrf2, are underlined in the sequences, and regions containing the DLG and ETGE motifs are indicated by solid lines. Seven lysine residues (K) within the α-helix are in boldface type and double underlined. (B) 13Cα-13Cβ CSI of mouse Neh2 plotted against residue number. Consecutive positive CSI values (>1) indicate an α-helix, whereas negative values (<−0.5) indicate a β-strand. Based on the CSI values, the secondary structural elements, the starts and ends of which have been labeled by residue number, are shown above the plot. (C) A chart showing the steady-state 1H-{15N} heteronuclear NOEs of Neh2 and indicating the backbone dynamics of the protein. Asterisks denote values of zero. Overall heteronuclear NOE values are equal or less than 0.5. (D) Schematic diagram of the mini antiparallel β-sheet (Phe-74 to Pro-85), where the ETGE motif (Glu-79 to Glu-82) forms a hydrophilic hairpin loop. (E) A helical wheel representation of the central α-helix from Phe-39 to Phe-71. Following the direction of the arrow, the helical wheel starts at Phe-39 (underlined) in the inner circle and ends at Phe-71 (underlined) in the outer circle. All seven lysines (K) are shown in boldface type. The dashed line divides the helical wheel into two equal parts, highlighting the concentration of the six lysines lying on one half of the helical surface.
Since some of the unstructured but biologically active polypeptides retain, albeit partially, secondary structure (33), we examined whether Neh2 falls into this category of polypeptides. Hence, we determined the sequence-specific backbone assignments of Neh2. As depicted in Fig. 2B, the CSI values show that Neh2 is composed of a single central α-helix (Phe-39 to Phe-71) and two very short β-strands (β1, Phe-74 to Leu-76; β2, Glu-82 to Pro-85). Several sequential and medium-range 1H, 1H NOEs indicative of helical structure (59) were observed within residues from Phe-39 to Phe-71. In addition, weak-to-moderate, but evident, 1H, 1H NOEs were observed between residues within the two β-strands (Table 1), suggesting that the two β-strands shape into a mini antiparallel β-sheet with a hydrophilic hairpin loop (Glu-78 to Gly-81) formed by the conserved ETGE motif (Fig. 2D). In addition, hydrophobic interaction among the hydrophobic residues (Phe-74, Leu-76, Phe-83, and Pro-85) in the β-strands may be attributed to the stabilizing force for this antiparallel β-sheet structure.
TABLE 1.
Sets of protons within the β-loop-β region that show 1H, 1H NOEs
| Region | Amino acid | Proton | Amino acid | Proton |
|---|---|---|---|---|
| β-Sheet | Phe-74 | Hδ | Pro-85 | Hα |
| Leu-84 | HN | |||
| Hɛ | Pro-85 | Hα | ||
| Gln-75 | HN | Hα | ||
| Leu-84 | HN | |||
| Hβ | HN | |||
| Hγ | HN | |||
| Leu-76 | HN | Phe-83 | Hδ, Hɛ | |
| Leu-84 | HN | |||
| Hδ | Phe-83 | HN, Hδ, Hɛ | ||
| Glu-82 | HN | |||
| Gly-81 | HN | |||
| Asp-77 | HN | Leu-84 | Hδ | |
| Glu-82 | HN | |||
| Hβ | HN | |||
| Loop | Glu-78 | HN | Glu-79 | HN |
| Hα | Thr-80 | HN | ||
| Glu-79 | HN | HN | ||
| Thr-80 | HN | Gly-81 | HN | |
| Gly-81 | HN | Glu-82 | HN |
To draw a relation between the secondary structure and backbone dynamics, we measured 15N-{1H} NOEs for the Neh2 domain. Due to significant resonance overlaps, we could determine approximately 50% of the heteronuclear NOEs (Fig. 2C). Both the N terminus and C terminus of Neh2 display negative heteronuclear NOE values less than −0.2, which indicate significant backbone motions on a subnanosecond timescale (9, 28). However, there are positive heteronuclear NOEs observed for the regions with secondary structure. NOE values vary from +0.2 to +0.5 for the central α-helix and −0.1 to +0.1 for the β-sheet region with slight negative values primarily found within the β-hairpin loop. Although these regions with secondary structure exhibit positive or close to zero heteronuclear NOEs, the overall values fall below the average of +0.8 for rigid proteins of similar size tumbling isotropically in solutions (26). These suggest that there is low degree of motional restriction within these regions. Moreover, given that 15N-{1H} heteronuclear NOE value of +0.5 corresponds to an overall rotational correlation time of less than 2 ns for a rigid globular molecule (26) and that the overall rotational correlation time for globular proteins of similar size is much slower, approximately 7 ns for 100 residues and 4 ns for a ubiquitin of only 76 residues on 500 MHz (34, 46), we conclude that Neh2 is, in general, flexible and the two individual secondary structural domains are not likely to pack into any folded tertiary structure.
The absence of 1H, 1H NOEs between α and β secondary structural elements is consistent with the 15N-{1H} NOE data and suggests the lack of a stable or rigid tertiary fold formed between the α-helix and the β-strands. An extended conformation of Neh2 is also implied by the slower than expected migration rate of the protein on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, as well as its faster than expected elution time from a size exclusion column (data not shown). These observations thus indicate that the Neh2 domain is a nonglobular protein with some native-like secondary structure and high backbone flexibility.
Neh2 contains two binding sites for Keap1-DC.
To examine how this intrinsically disordered but biologically active Neh2 domain binds to its repressor Keap1, ITC was used to characterize the interaction between the Neh2 and Keap1-DC by probing the binding stoichiometry (n), binding constant (Ka), enthalpy change (ΔH), and entropy change (ΔS) of binding. The integrated heats of reaction are shown in Fig. 3 as a function of the Neh2/Keap1-DC molar ratio. The integrated isotherms exhibit a biphasic curve, which is fitted best with a two-site binding model using a nonlinear least-squares algorithm provided by the instrument. As shown in Table 2, the binding stoichiometry (n) obtained for the interaction between Neh2 and Keap1-DC is close to 0.5 for both site 1 (0.49) and site 2 (0.46), where n = Neh2/Keap1-DC upon titrating Neh2 into Keap1-DC, indicating that one Neh2 domain interacts with two molecules of Keap1-DC.
TABLE 2.
Thermodynamic parameters for the binding of the wild-type Neh2 or Neh2 mutants to Keap1-DC at 25°Ca
| Neh2 | nc | Kab (107 M−1) | ΔHb (kcal/mol) | TΔSb (kcal/mol) | ΔGb (kcal/mol) |
|---|---|---|---|---|---|
| Wild type | |||||
| Site 1 | 0.49 ± 0.00 | 19.0 ± 4.0 | −28.4 ± 0.1 | −17.1 ± 0.1 | −11.3 ± 0.2 |
| Site 2 | 0.46 ± 0.00 | 0.1 ± 0.0 | −11.2 ± 0.3 | −3.0 ± 0.3 | −8.1 ± 0.0 |
| Neh2[Δ1-33] | 1.01 ± 0.01 | 12.4 ± 0.9 | −21.2 ± 0.3 | −10.2 ± 0.2 | −11.0 ± 0.1 |
| Neh2[ΔETGE] | 0.94 ± 0.02 | 0.2 ± 0.0 | −6.9 ± 0.3 | 1.8 ± 0.3 | −8.7 ± 0.1 |
All standard deviations are derived from triplicate runs.
Ka is the binding constant. ΔH, ΔS, and ΔG are the change in binding enthalpy, entropy, and Gibbs energy, respectively. −RTlnKa = ΔG = ΔH − TΔS, where T and R are the absolute temperature and the gas constant, respectively.
n, binding stoichiometry.
However, the binding constants (Ka) are quite different between these two sites, (1.9 ± 0.4) × 108 M−1 for site 1 and (1.0 ± 0.0) × 106 M−1 for site 2. The binding events of the two sites are also accompanied by different contributions of enthalpy and entropy changes. The protein association is enthalpy driven, and site 1 exhibits a more negative enthalpy of −28.4 ± 0.1 kcal/mol than that of site 2 (−11.2 ± 0.3 kcal/mol). These results support the contention that site 1 has a stronger driving power than site 2 for protein association.
The ETGE motif in the hydrophilic loop docks with Keap1-DC as site 1.
The ITC study suggests that there are two binding sites on Neh2 of different affinity for interacting with Keap1-DC. To map these two binding sites in Neh2, NMR titration analyses were performed. Upon forming complex with Keap1-DC, there was increasing disappearance of resonance signals in the TROSY-HSQC spectra (Fig. 1B and C). The loss of resonance signals is most likely to be due to an increase in overall molecular size caused by the protein complex formation. In addition, an intermediate chemical exchange between the free and bound states of Neh2 may also contribute, to certain extent, to the line broadening and reduction of the signal-to-noise ratio. This observation, together with the problem of resonance overlaps, led us to employ [15N]Leu-labeled Neh2 samples to reduce spectral complexity for further NMR chemical shift perturbation analysis.
We selected leucine residues for isotopic labeling because of their relatively even distribution in the α-helix, the β-sheet, and in the rest of the flexible regions, including the conserved DLG motif of the Neh2 domain (25) (Fig. 2A). Based on the assumption that resonance signals would disappear when protein complex formation was taking place, our strategy is to monitor the loss of resonance signals of the [15N]leucines. We kept in mind that this approach can only be taken as a qualitative analysis, since the loss of resonance signals can be due to chemical exchange effect, increase of overall molecular mass, or nonspecific aggregation. Of the possibilities, the nonspecific aggregation seems less likely, as the results of both the ITC and [15N]Arg-labeled Keap1-DC NMR titration experiments performed under similar solution conditions showed that there are two distinct binding sites in the Neh2 domain (see below).
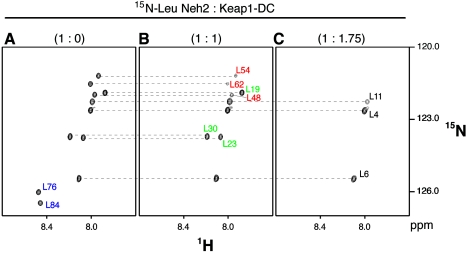
In the TROSY-HSQC spectrum of [15N]Leu-labeled Neh2, only 11 labeled leucine residues were visible (Fig. 4A). Nonetheless, the resonance signals of [15N]Leu-labeled Neh2 were well resolved and easily traced during the titration experiment. Upon the addition of an increasing molar ratio of unlabeled Keap1-DC, the resonances corresponding to Leu-76 and Leu-84 located within the two β-strands disappeared at a molar ratio of Neh2/Keap1-DC of 1:1 (Fig. 4B). When the system is close to saturation at anapproximately 1:1.75 molar ratio (Neh2/Keap1-DC), leucines located in the α-helix (Leu-48, Leu-54. and Leu-62) and less rigid region immediately upstream of the α-helix (Leu-19, Leu-23, and Leu-30) also disappeared from the spectrum, leaving three resonance signals from the most N-terminal leucines (Leu-4, Leu-6, and Leu-11) in the very flexible region of the polypeptide (Fig. 4C). Since ITC data have implied the presence of two Keap1 binding sites of higher and lower association affinities in Neh2, the disappearance of Leu-76 and Leu-84 at the lower molar ratio of approximately 1:1 suggests that the C terminus of the Neh2 domain containing a β-sheet possesses the higher affinity binding site. Indeed, the conserved ETGE motif in the hairpin loop of this β-sheet has been demonstrated to be important in the association with Keap1 (32), which corroborates the present data.
FIG. 4.
The ETGE motif marks the initiation of binding between Neh2 and Keap1. 1H, 15N-TROSY-HSQC spectra of [15N]Leu-labeled Neh2 (A) and its complex with a 1 (B) or 1.75 (C) molar ratio of unlabeled Keap1-DC. Only 1H, 15N cross peaks for leucine residues are visible on the spectra. (A) Leucine peaks representing the β-sheet of Neh2 are labeled in blue (Leu-76 and Leu-84). (B) Leucine peaks representing the putative second binding site upstream of the central α-helix (Leu-19, Leu-23, and Leu-30, labeled in green) and the α-helix (Leu-48, Leu-54, and Leu-62, labeled in red) of Neh2 are shown. (C) Leucine peaks representing the very flexible N-terminal ends are labeled in black (Leu-4, Leu-6, and Leu-11).
To verify that the ETGE motif is a part of the high-affinity binding site in Neh2, we prepared a deletion mutant (Neh2[ΔETGE]) of Neh2 (Fig. 5A), in which the residues of 79ETGE82 were removed, and examined its effect on the interaction with Keap1-DC by the ITC method. Mutations in the ETGE motif did not impose structural changes to the α-helix nor the N-terminal region (data not shown). Unlike the wild-type Neh2, the ETGE deletion mutant gave rise to a binding curve that is best fitted for a one-site binding model with a stoichiometry of 0.94 ± 0.02 (Fig. 5B and Table 2). The binding affinity of this mutant (2.0 × 106 M−1) resembles to that of site 2 of the wild-type Neh2 (1.0 × 106 M−1). This similarity suggests that, by removing the ETGE motif, the higher affinity Keap1 binding site is abolished in Neh2. The residual binding affinity in Neh2[ΔETGE] reflects that of the weaker binding site in the Neh2 domain, which is plausibly upstream of the β-sheet as implied by the NMR titration data.
The weaker Keap1 binding site lies upstream of the central α-helix of Neh2.
Since data from NMR and ITC analyses revealed that there is another weaker Keap1 binding site N-terminal to the short antiparallel β-sheet of Neh2, we prepared an N-terminal deletion mutant of Neh2 that lacks the first 33 residues, including the DLG motif (Neh2[Δ1-33]) (Fig. 5A). The TROSY-HSQC spectrum for this mutant showed that there is almost no chemical shift changes to all of the residual [15N]Leu signals (data not shown), suggesting that deletion of the first 33 residues did not induce structural change to the rest of the Neh2 molecule. Binding isotherms of Neh2[Δ1-33] against Keap1-DC also display a curve that is best fitted for a one-site binding model with a binding stoichiometry of 1.01 ± 0.01 and a Ka value of (1.24 ± 0.09) × 108 M−1 (Fig. 5C and Table 2). In this case, both Neh2[Δ1-33] and site 1 of the wild-type Neh2 domain show the same order of binding affinity (108 M−1). These results indicate that the weaker binding site was indeed eliminated in this N-terminal deletion mutant. In summary, these ITC analyses suggest that the stronger and weaker Keap1 binding sites locate in the conserved ETGE and DLG motifs of the Neh2 domain.
There is no independent binding affinity in the central α-helix.
A question remains as to the role the central α-helix plays in the intermolecular interaction between Keap1 and Nrf2. Since the Neh2[Δ1-33] mutant exhibits a high association affinity to Keap1-DC, resembling the stronger binding site in the wild-type Neh2 domain, it is interesting to examine whether further deletion of the ETGE sequence from the mutant diminishes the entire binding capacity. To our expectation, the ITC isotherms of this double deletion mutant (Neh2[Δ1-33, ΔETGE]) titrating against Keap1-DC essentially display a magnitude of heat change similar to that of the heat of dilution of these proteins (Fig. 5D). The binding capacity of Neh2[Δ1-33, ΔETGE] is undetectable, suggesting that the central α-helix of the Neh2 domain does not contribute to its molecular interaction with Keap1-DC.
Neh2 interacts with Keap1-DC via the ETGE and DLG motifs, utilizing an overlapping binding site.
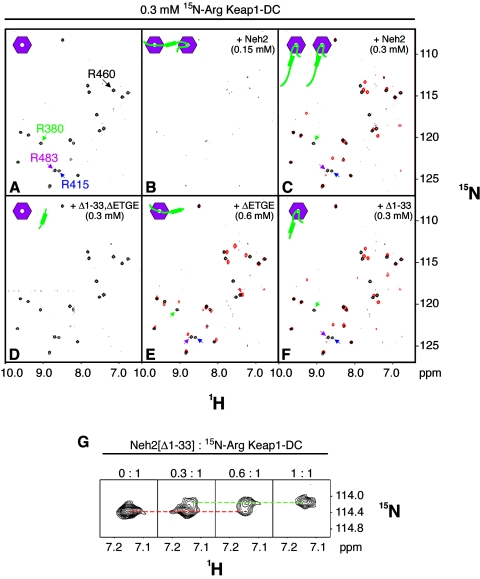
A molecular surface rich in positively charged residues in Keap1-DC is expected to serve as the site of interaction with Neh2, since the hydrophilic loop containing the ETGE motif is negatively charged and there are also multiple aspartates within the DLG motif (Fig. 2A and B). Our recent crystal structure of mouse Keap1-DC complexed with a 9-residue ETGE peptide (Leu-76 to Leu-84) provides evidence that there are multiple bonding interactions between the two acidic glutamates of the ETGE motif and conserved arginines (Arg-380, Arg-415, and Arg-483) at the entrance of the central cavity on the bottom side of the β-propeller structure of Keap1-DC (47a). The present NMR and ITC data also suggest the importance of the ETGE motif for the intermolecular interaction. To illustrate the binding interface of the DLG motif to Keap1-DC, we employed NMR titration experiments using [15N]Arg-labeled Keap1-DC and deletion mutants of the Neh2 domain. [15N]Arg-labeled Keap1-DC shows 20 well-resolved arginine signals, but 3 signals from 23 arginine residues of Keap1-DC are missing (Fig. 6A). Exploiting a combination of [15N]Arg or [13C, 15N]Arg labeling along with three arginine substitution mutants (R380G, R415K, and R483G) of Keap1-DC, we assigned the resonances of four arginine residues, Arg-380, Arg-415, Arg-460, and Arg-483 (Fig. 6A), which locate on the bottom side of Keap1-DC.
FIG. 6.
Neh2 binds two molecules of Keap1-DC through both the ETGE and DLG motifs. (A to F) 1H, 15N-TROSY-HSQC spectra of [15N]Arg-labeled Keap1-DC (0.3 mM) (A) titrated with 0.15 mM Neh2 (B), 0.3 mM Neh2 (C), 0.3 mM Neh2[Δ1-33, ΔETGE] (D), 0.6 mM Neh2[ΔETGE] (E), and 0.3 mM Neh2[Δ1-33] (F). The assigned resonances for Arg-380, Arg-415, Arg-460, and Arg-483 of Keap1-DC are as indicated in panel A. Panels C, E, and F show overlaid spectra of the unbound (black) and bound (red) states of the [15N]Arg-labeled Keap1-DC with the respective wild-type Neh2 or mutants. The resonances for Arg-380, Arg-415, and Arg-483 in their unbound states (black signals) are as indicated by arrows in green, blue, and magenta, respectively, on panels A, C, E, and F. Miniature cartoons at the top left corners of each spectrum show respective protein-protein interaction status, with hexagons in magenta representing Keap1-DC and green hockey sticks representing the Neh2 domain. (G) A zoomed spectral view showing Arg-460 resonance of [15N]Arg-labeled Keap1-DC in the course of titration with unlabeled Neh2[Δ1-33]. The red dotted line indicates the diminishing Arg-460 resonance in its unbound state, and the green dotted line indicates the appearing Arg-460 resonance in its bound state.
Addition of equimolar unlabeled Neh2 or Neh2[Δ1-33], both of which contain the stronger Keap1 binding motif, leads to significant shifts of the three surface arginine resonances (Arg-380, Arg-415, and Arg-483), indicating involvement of these arginines in the molecular interaction with Neh2 (Fig. 6C and F). On the other hand, titrating the [15N]Arg-labeled Keap1-DC with Neh2[ΔETGE], which contains the DLG motif, incurs chemical shift changes distinct from these three arginine resonances (Fig. 6E). Since deletion of the ETGE motif did not impose any structural changes to the α-helix nor the N-terminal region of the Neh2 domain, the spectral change observed in this case is likely due to binding upstream of the ETGE motif.
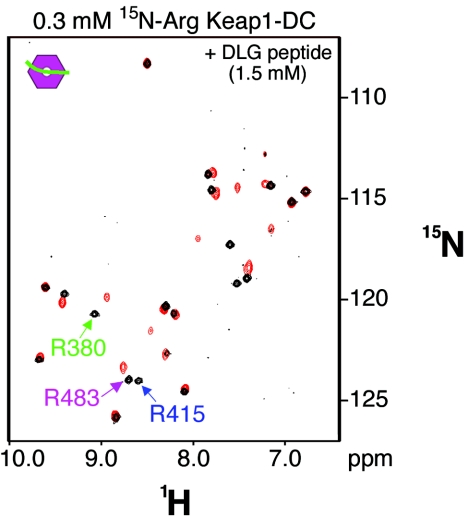
To clarify the region of this alternate binding site on Neh2, we prepared a peptide that contains only the DLG motif (DLG peptide; Met-17 to Val-36) of the Neh2 domain and studied its effects on the arginine signals. This 20-amino-acid-long peptide causes spectral changes to all three of these arginine signals (Fig. 7). This observation suggests that the weaker binding site falls between Met-17 and Val-36 of the Neh2 domain and strengthens the conclusion that the DLG motif is site 2. Moreover, both the deletion mutants and the DLG peptide introduce spectral changes to a total of 9 common arginine signals, including the three assigned residues in Keap1-DC (Fig. 6E and F and 7). These spectral changes suggest that both the ETGE and DLG motifs of the Neh2 domain are interacting with an overlapping site on Keap1-DC.
FIG. 7.
The DLG motif is the weaker Keap1 binding site within Met-17 to Val-36. 1H, 15N-TROSY-HSQC spectra of [15N]Arg-labeled Keap1-DC (0.3 mM) titrated with the DLG peptide of the Neh2 domain incurs spectral changes similar to those provoked by the Neh2[ΔETGE] mutant. An overlaid spectrum shows arginine resonance signals of unbound (black) and bound (red) states of the [15N]Arg-labeled Keap1-DC; the latter is in complex with a 20-amino-acid DLG peptide (Met-17 to Val-36) of the Neh2 domain. The color code for the arrows of Arg-380, Arg-415, and Arg-483 is the same as described for Fig. 6.
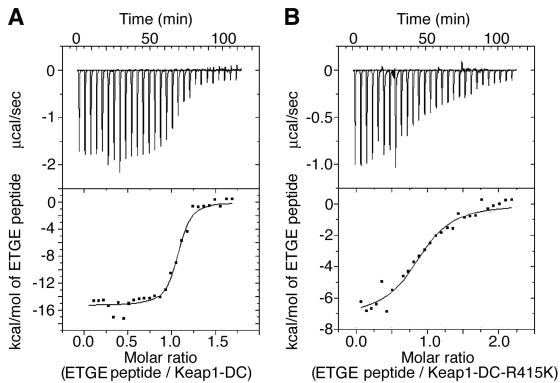
On the other hand, we have tested the effect of a point mutation of Arg-415 to lysine in Keap1-DC on its binding affinity with the peptide containing the ETGE motif. There is one order of magnitude lower in binding affinity when titrating the ETGE peptide against this arginine mutant of the Keap1-DC in ITC analyses compared with that against wild-type Keap1-DC (Fig. 8 and Table 3). Similarly, a R380G mutant of Keap1-DC was also 1 order of magnitude lower in binding affinity (data not shown). These data further support the notion that these arginines are located in the binding interface, and the observed spectral changes of these three arginines upon addition of Neh2 mutants or peptide containing either the ETGE or DLG motif alone are due to intermolecular interaction. Taken together, these experimental observations suggest that both the ETGE and DLG motifs are involved in binding with Keap1-DC via an overlapping binding site of conserved arginine residues at the entrance of the central cavity on the bottom side of the β-propeller structure.
FIG. 8.
Single-arginine substitution mutation in Keap1-DC attenuates the binding affinity of Keap1-DC to the ETGE peptide. (A, B) ITC titration profiles showing the titration of Keap1-DC (A) and Keap1-DC-R415K mutant (B) with the ETGE peptide (Leu-76 to Leu-84) of the Neh2 domain. Note that the integrated binding isotherms fit best for a one-site binding model.
TABLE 3.
Thermodynamic parameters for the binding of the wild-type Keap1-DC and Keap1-DC R415K to the ETGE peptide (Leu-76 to Leu-84) of the Neh2 domain at 25°Ca
| Keap1-DC | nc | Kab (106 M−1) | ΔHb (kcal/mol) | TΔSb (kcal/mol) | ΔGb (kcal/mol) |
|---|---|---|---|---|---|
| Wild type | 0.98 ± 0.04 | 5.5 ± 0.3 | −14.8 ± 0.3 | −5.6 ± 0.3 | −9.2 ± 0.0 |
| R415K | 0.92 ± 0.00 | 0.5 ± 0.1 | −6.4 ± 0.3 | 1.4 ± 0.3 | −7.8 ± 0.1 |
All standard deviations are derived from triplicate runs.
Ka is the binding constant. ΔH, ΔS, and ΔG are the change in binding enthalpy, entropy, and Gibbs energy, respectively. −RTlnKa = ΔG = ΔH − TΔS, where T and R are the absolute temperature and the gas constant, respectively.
n, binding stoichiometry.
One Neh2 molecule binds two molecules of Keap1-DC of a Keap1 dimer.
Data so far described have revealed that one Neh2 molecule binds two molecules of Keap1-DC and there is no apparent binding affinity involved in the central α-helix. NMR titration experiments of [15N]Arg-labeled Keap1-DC with the Neh2 domain further support this contention. Intriguingly, while a very poor signal-to-noise ratio was observed in the TROSY-HSQC spectrum for [15N]Arg-labeled Keap1-DC and unlabeled full-length Neh2 at a ratio of 0.5:1 (Neh2:Keap1-DC) (Fig. 6B), all 20 arginine resonances showed up with the same chemical shift changes as those observed in the Neh2[Δ1-33] and Keap1-DC complex at a 1:1 molar ratio when the amount of Neh2 was increased to the same molar concentration as Keap1-DC (Fig. 6C).
One interpretation for this disappearance and appearance of resonance signals is that each Neh2, via both ETGE and DLG motifs, is able to bind with two molecules of Keap1-DC at 0.5:1 molar ratio (Fig. 6B). The complex formation increased the overall molecular mass beyond 80 kDa and might slow down its molecular tumbling time in solution, resulting in line broadening and signal-to-noise ratio reduction. On the contrary, when the amount of Neh2 is further increased to an equimolar concentration, more ETGE motif, with 2 orders of magnitude stronger binding affinity than that of the DLG motif, is available to compete for the interaction, so that eventually every Keap1-DC molecule was occupied by the Neh2 domain through its ETGE motif (Fig. 6C). Therefore, the TROSY-HSQC spectrum of [15N]Arg-labeled Keap1-DC and unlabeled full-length Neh2 at a ratio of 1:1 resembles that of Keap1-DC and the Neh2[Δ1-33] complex, which contains only the ETGE Keap1 binding site (Fig. 6F). These observations further illustrate that the binding stoichiometry between Keap1-DC and the Neh2 domain is 2:1, and the stronger binding capacity resides in the ETGE motif.
An alternative explanation for the disappearance of resonance signals at a 0.5:1 molar ratio is that it may be due to broadening by chemical exchange effects of the bound and unbound forms of Keap1-DC. However, we did not observe a similar loss of resonance signals during titration of the [15N]Arg-labeled Keap1-DC with either Neh2[Δ1-33] or Neh2[ΔETGE]. Arginine signals of both the bound and unbound forms could be traced during the course of the titration in either case of the two mutants (Fig. 6G). In addition, arginine signals that are not affected upon complex formation remain detectable throughout the titration process with the deletion mutants (data not shown). Therefore, we think it is less likely that the loss of signals seen in Fig. 6B is derived solely from the effects of chemical exchanges.
Although there are multiple arginine chemical shift changes found in Keap1-DC when associating with Neh2[Δ1-33] at a 1:1 molar ratio, the TROSY-HSQC spectrum for [15N]Arg-labeled Keap1-DC remains unchanged when Neh2[Δ1-33, ΔETGE] is added at the same molar ratio (Fig. 6D). This result coincides very well with our ITC analysis in which no observable binding affinity was detected between Keap1-DC and this double deletion mutant of Neh2 (Fig. 4D). Since Neh2 interacts with two molecules of Keap1-DC by means of both ETGE and DLG motifs to an overlapping binding site on the same surface plane of Keap1-DC, we surmise that the central α-helix of Neh2 may serve as a bridge connecting the two binding sites (Fig. 9A).
FIG. 9.
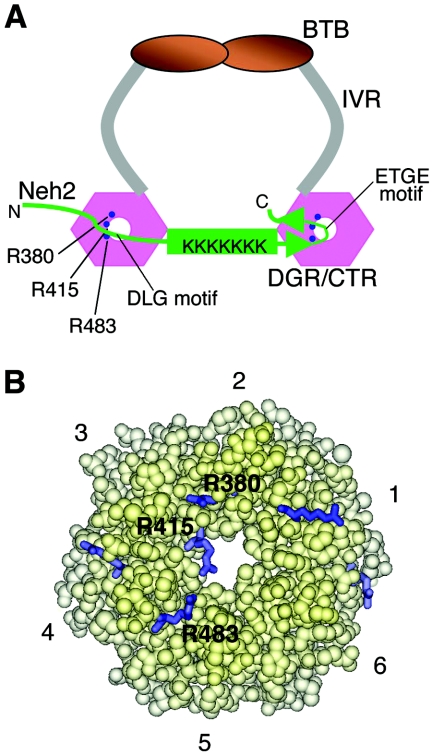
Two-site molecular recognition model for Keap1 and Neh2 interaction. (A) Binding model of Keap1 and Neh2. Keap1 forms a homodimer through intermolecular interaction of its BTB domain. IVR (in gray) connects the BTB domain with the β-propeller structure of the DGR/CTR domain (Keap1-DC, pink hexagons) of Keap1. The Neh2 domain (in green) is recruited to one molecule of the Keap1-DC via the ETGE motif within the antiparallel β-sheet (green arrowheads) and the DLG motif upstream of the central α-helix to an overlapping site on another molecule of Keap1-DC. The central α-helix (green rod) of Neh2 serves as a bridge between the two molecules of Keap1. Lysine residues within the central α-helix are labeled K in the schematic model. Binding of the DLG motif with Keap1 may help to correctly orientate these lysines in space for other biological functions. The arginine triad (Arg-380, Arg-415, and Arg-483) at the entrance of the central channel of Keap1 is shown as solid blue circles. (B) A space-filled model generated by InsightII (Accelrys) and the coordinates of the mouse Keap1-DC (47a) (PDB accession code 1X2J), showing the bottom view of the molecule. The six Kelch repeats are as numbered. Arginine residues located on the bottom surface are shown as blue sticks. The three arginines (Arg-380, Arg-415, and Arg-483) that are involved in protein-protein interaction with the Neh2 domain are labeled.
Dimerization of Keap1 was proposed to be biologically significant for its activity in vivo (64). Indeed, our sedimentation analysis data, acquired under similar buffer conditions as those in NMR studies, show that full-length Keap1 is a homogeneous homodimer (theoretical molecular mass of 69,456 Da versus empirical molecular mass of 162,000 Da), while Neh2 domain exists as a monomer (theoretical molecular mass of 11,700 Da versus empirical molecular mass of 11,922 Da) in solution. In addition, full-length Keap1 exhibits a binding curve, which is best fitted for a two-site binding model, when titrated against full-length Neh2. The binding constant for the stronger site is about 2.2 × 108 M−1, while that for the weaker site is about 3.6 × 105 M−1 from duplicate runs (raw data not shown). The 1 order of magnitude lower affinity for the weaker site, in the case of the full-length Keap1, may be due to some distance or orientation constraint associated with the homodimer in contrast with the freely circulating Keap1-DC monomers in solution.
DISCUSSION
Being a substrate adaptor for the Cul3-based E3 ligase machinery, Keap1 facilitates the ubiquitination of Nrf2, the target substrate, and signals its degradation by the proteasomal pathway (6, 14, 30, 61). Previous studies demonstrated that Neh2 contains the key to this signaling event for protein turnover (25, 36, 61). In the present study, we found that the structural properties of the Neh2 domain render merits for its regulatory role of Nrf2. The ETGE motif in the Neh2 domain of Nrf2 has been shown to be responsible for the interaction with Keap1 (32). In agreement with this contention, we found that the ETGE motif resides in the hydrophilic loop of the β-loop-β structure at the C-terminal region of Neh2 and binds the β-propeller of Keap1-DC with high affinity. A striking feature is that the conserved DLG motif of Neh2, which lies in a flexible region upstream of the central α-helix, also binds Keap1-DC with a comparatively lower affinity. In addition, these two evolutionarily conserved motifs of Neh2 interact with two molecules of Keap1-DC individually via overlapping binding sites on the bottom surface of the β-propeller involving three conserved arginine residues of Keap1.
Since Keap1 forms a dimer in solution by utilizing the BTB domain and since the dimerization of Keap1 has been proposed to be functionally significant (64), we envisage that the DGR domain of Keap1 recruits the substrate Nrf2 molecule through binding to the higher affinity ETGE motif, followed by the docking of the weaker affinity DLG motif in the same Neh2 domain to an adjacent β-propeller structure of a Keap1 dimer (Fig. 9A). Considering the fact that the Neh2 domain possesses intrinsic flexibility, but lacks any rigid tertiary fold, and that these two conserved motifs are separated by a 33-residue-long α-helix, which does not have observable affinity to Keap1, its structural plasticity and the length of the central α-helix (approximately 50 Å) may confer on Neh2 an advantage to configure itself to interact with a dimer of Keap1.
It is intriguing that Keap1 recruits its substrate by means of the two-site binding mechanism. Besides Cul3, higher eukaryotic genomes encode another six closely related Cullin paralogs (Cul1, 2, 4A, 4B, 5, and 7), all of which associate with Roc1 to form ubiquitin ligases (reviewed in references 49 and 50). In spite of their striking similarity in overall composition and structure, there is a high specificity for substrate recognition among these subtypes of Cul-based E3 ligases. For instance, both Cul1 and Cul7 utilize SKP1 and F-box proteins as their adaptor proteins and substrate receptors, respectively, whereas Elongin BC and suppressor of cytokine signaling-box proteins are involved in the Cul2 and Cul5 ligase complexes. Recently, it was found that DNA-damage-binding protein 1 (DDB1) binds to Cul4A and serves as an adaptor protein. However, mechanisms for linking substrate receptors to this subtype of E3 ligase still remain to be clarified (18). In the case of Cul3-based E3 ligases, BTB proteins serve both as the protein adaptor and the substrate-specific receptor. Except for the DDB1, core BTB folds are found in SKP1, Elongin C and other BTB proteins (54). Intriguingly, of these protein adaptors, only Keap1 forms a homodimer via its BTB domain. Therefore, our present study showing the two-site binding mechanism in the Keap1-Neh2 association suggests that this modular organization could be unique within this subtype of Cul3-based E3 ligases for the purpose of both substrate recognition and regulation.
How do these substrate recognition determinants help us to elucidate the ubiquitin targeting mechanism for signaling Nrf2 to proteasomal proteolysis? When evaluating the helical wheel of the central α-helix of the Neh2 domain, we found that six of the seven lysines are located on one side of the helical surface (Fig. 2A and E). Since these lysine residues are targets for Keap1-mediated ubiquitination by E3 ligase (61), this arrangement led us to speculate that these lysine residues are biologically important for the regulation of Nrf2 stability. Since ubiquitin conjugation often requires close proximity of the epsilon amino group of a target lysine residue to the catalytic pocket of an E2 enzyme, a conserved, correct, and rigidly defined substrate spatial orientation may facilitate the presentation of the substrate for the ubiquitin transfer, as has also been proposed by the model of the ubiquitin-E2-SCFCdc4-CPD complex (45, 63).
The importance of the conserved DLG motif within Neh2 for Keap1-dependent ubiquitination and degradation has been pointed out previously (25). Deletion of Met-17 to Val-32, including the DLG motif, significantly hampered the Keap1-dependent polyubiquitination (36). Either deletion of the N-terminal half (Met-1 to Arg-43) of the Neh2 domain or alanine substitution mutation within the DLG motif attenuates the Keap1-mediated proteasomal degradation of Nrf2 (25), and the lysine residues within the Neh2 domain are essential for both Keap1-mediated ubiquitination and proteasomal degradation of Nrf2 (61). Showing very good agreement with our NMR and ITC studies with Neh2[Δ1-33], deletion of Met-17 to Val-32 or mutations of the lysine residues did not interfere the interaction of Neh2 with Keap1. Presumably, the high binding affinity (1.9 × 108 M−1) of the ETGE motif is self-sufficient in securing the interaction between Neh2 and Keap1. Therefore, the physiological function of the conserved DLG motif may be beyond a simple purpose of protein-protein interaction. We surmise that the ETGE motif engages Nrf2 with Keap1 to secure a tight binding and the DLG motif helps to anchor the lysine-rich central α-helix of Nrf2 in a confined spatial arrangement to benefit polyubiquitination of these lysines.
Another notable characteristic of this two-site binding model is the differential binding affinity inherent in these two conserved motifs, while both the ETGE and DLG motifs recognize a similar or overlapping interface in Keap1. It should be noted that Keap1 not only acts as a substrate adaptor but also acts as an oxidative/electrophilic stress sensor (8, 57). It has been postulated that stress signals are transduced by electrophilic or oxidative modification of reactive cysteines within the IVR domain of Keap1 and the modification confers conformational changes to Keap1 and releases the Keap1 repression on Nrf2. Given that one of the important features of the β-propeller structure of Keap1-DC is the structural stability (13, 47a), stress signals sensed by Keap1 seem less likely to provoke conformational alterations in the β-propeller structure. Indeed, treatment with chemical inducers did not discourage the association of Nrf2 to Keap1 but liberated Nrf2 from the Keap1-dependent polyubiquitination and proteasomal degradation (31).
If Keap1-dependent polyubiquitination requires proper positioning of the lysine-rich central α-helix with the help of the DLG motif, there emerges a simple but basic question of whether the distortion of the IVR linker by oxidizing the reactive cysteines within this region may easily dislodge the weaker-binding DLG motif from the Keap1-DC domains or not. We think that this mechanism is plausible, since there are certain constraints on the length adhered to the central α-helix, which is suspended between the two β-propeller structures following substrate recognition. Even a slight distortion in the IVR linker domains of a dimer of Keap1 may introduce a sufficient increase in distance between the two downstream Keap1-DC domains to displace the weaker-binding DLG motif, hence disorienting the arrangement of the central α-helix or the lysine residues in the space, albeit the tight binding of the ETGE motif may not be affected. We therefore propose that the DLG motif may possess the signaling switch for the repressive regulation of Nrf2 via the Keap1-dependent ubiquitination and proteasomal degradation. We are waiting for the detailed studies on the 3D structure of the quaternary Cul3-Roc1-Keap1-Nrf2 complex to reveal their respective spatial arrangements and on the in vivo function of the DLG motif in mice to unravel the biological determinants that govern the regulation of Nrf2 turnover and the oxidative stress response for rendering both cellular protection and homeostasis of the body.
Acknowledgments
We thank Kouhei Tsumoto, Midori Oinuma, and Rieko Ishima for advice on interpreting the ITC data, for instruction on the use of the ITC instrument, and for discussion of the NMR data, respectively. We also thank Balasundaram Padmanabhan for the gift of the ETGE peptide and Akira Kobayashi for discussions. We thank Tania O'Connor for critical reading of the manuscript.
This work is supported in part by grants-in-aid from JST-ERATO, the Ministry of Education, Culture, Sports, Science, and Technology. T.T. is supported by the 21st Century COE Program and the Protein 3000 project.
REFERENCES
- 1.Adams, J., R. Kelso, and L. Cooley. 2000. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 10:17-24. [DOI] [PubMed] [Google Scholar]
- 2.Bakin, A. V., N. V. Stourman, K. R. Sekhar, C. Rinehart, X. Yan, M. J. Meredith, C. L. Arteaga, and M. L. Freeman. 2005. Smad3-ATF3 signaling mediates TGF-beta suppression of genes encoding phase II detoxifying proteins. Free Radic. Biol. Med. 38:375-387. [DOI] [PubMed] [Google Scholar]
- 3.Balaban, R. S., S. Nemoto, and T. Finkel. 2005. Mitochondria, oxidants, and aging. Cell 120:483-495. [DOI] [PubMed] [Google Scholar]
- 4.Bardwell, V. J., and R. Treisman. 1994. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 8:1664-1677. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C., and A. N. Kong. 2004. Dietary chemopreventive compounds and ARE/EpRE signaling. Free Radic. Biol. Med. 36:1505-1516. [DOI] [PubMed] [Google Scholar]
- 6.Cullinan, S. B., J. D. Gordan, J. Jin, J. W. Harper, and J. A. Diehl. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 24:8477-8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 8.Dinkova-Kostova, A. T., W. D. Holtzclaw, R. N. Cole, K. Itoh, N. Wakabayashi, Y. Katoh, M. Yamamoto, and P. Talalay. 2002. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 99:11908-11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duggan, B. M., H. J. Dyson, and P. E. Wright. 1999. Inherent flexibility in a potent inhibitor of blood coagulation, recombinant nematode anticoagulant protein c2. Eur. J. Biochem. 265:539-548. [DOI] [PubMed] [Google Scholar]
- 10.Dyson, H. J., and P. E. Wright. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6:197-208. [DOI] [PubMed] [Google Scholar]
- 11.Farrow, N. A., R. Muhandiram, A. U. Singer, S. M. Pascal, C. M. Kay, G. Gish, S. E. Shoelson, T. Pawson, J. D. Forman-Kay, and L. E. Kay. 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33:5984-6003. [DOI] [PubMed] [Google Scholar]
- 12.Forman, H. J., and D. Ruden. 2004. Introduction to serial reviews on EpRE and its signaling pathway. Free Radic. Biol. Med. 36:1197-1198. [DOI] [PubMed] [Google Scholar]
- 13.Fulop, V., and D. T. Jones. 1999. Beta propellers: structural rigidity and functional diversity. Curr. Opin. Struct. Biol. 9:715-721. [DOI] [PubMed] [Google Scholar]
- 14.Furukawa, M., and Y. Xiong. 2005. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 25:162-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrett, D. S., R. Powers, A. M. Gronenborn, and G. M. Clore. 1991. A common sense approach to peak picking in two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 95:214-220. [DOI] [PubMed] [Google Scholar]
- 16.Hayes, J. D., J. U. Flanagan, and I. R. Jowsey. 2005. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 45:51-88. [DOI] [PubMed] [Google Scholar]
- 17.Holtzclaw, W. D., A. T. Dinkova-Kostova, and P. Talalay. 2004. Protection against electrophile and oxidative stress by induction of phase 2 genes: the quest for the elusive sensor that responds to inducers. Adv. Enzyme Regul. 44:335-367. [DOI] [PubMed] [Google Scholar]
- 18.Hu, J., C. M. McCall, T. Ohta, and Y. Xiong. 2004. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6:1003-1009. [DOI] [PubMed] [Google Scholar]
- 19.Itoh, K., K. Igarashi, N. Hayashi, M. Nishizawa, and M. Yamamoto. 1995. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell Biol. 15:4184-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh, K., K. I. Tong, and M. Yamamoto. 2004. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic. Biol. Med. 36:1208-1213. [DOI] [PubMed] [Google Scholar]
- 22.Jeener, J., B. H. Meier, P. Bachmann, and R. R. Ernst. 1979. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 71:4546-4553. [Google Scholar]
- 23.Johnson, M. L., J. J. Correia, D. A. Yphantis, and H. R. Halvorson. 1981. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys. J. 36:575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh, Y., K. Itoh, E. Yoshida, M. Miyagishi, A. Fukamizu, and M. Yamamoto. 2001. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 6:857-868. [DOI] [PubMed] [Google Scholar]
- 25.Katoh, Y., K. Iida, M. I. Kang, A. Kobayashi, M. Mizukami, K. I. Tong, M. McMahon, J. D. Hayes, K. Itoh, and M. Yamamoto. 2005. Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome. Arch. Biochem. Biophys. 433:342-350. [DOI] [PubMed] [Google Scholar]
- 26.Kay, L. E., D. A. Torchia, and A. Bax. 1989. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28:8972-8979. [DOI] [PubMed] [Google Scholar]
- 27.Kay, L. E., P. Keifer, and T. Saarinen. 1992. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114:10663-10665. [Google Scholar]
- 28.Kay, L. E. 1998. Protein dynamics from NMR. Nat. Struct. Biol. 5(Suppl.):513-517. [DOI] [PubMed] [Google Scholar]
- 29.Kim, D., and F. P. Guengerich. 2005. Cytochrome P450 activation of arylamines and heterocyclic amines. Annu. Rev. Pharmacol. Toxicol. 45:27-49. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi, A., M. I. Kang, H. Okawa, M. Ohtsuji, Y. Zenke, T. Chiba, K. Igarashi, and M. Yamamoto. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24:7130-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi, A., M. I. Kang, Y. Watai, K. I. Tong, T. Shibata, K. Uchida, and M. Yamamoto. 2006. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol. Cell. Biol. 26:221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi, M., K. Itoh, T. Suzuki, H. Osanai, K. Nishikawa, Y. Katoh, Y. Takagi, and M. Yamamoto. 2002. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells 7:807-820. [DOI] [PubMed] [Google Scholar]
- 33.Lacy, E. R., I. Filippov, W. S. Lewis, S. Otieno, L. Xiao, S. Weiss, L. Hengst, and R. W. Kriwacki. 2004. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat. Struct. Mol. Biol. 11:358-364. [DOI] [PubMed] [Google Scholar]
- 34.Lee, A. L., and A. J. Wand. 1999. Assessing potential bias in the determination of rotational correlation times of proteins by NMR relaxation. J. Biomol. NMR 13:101-112. [DOI] [PubMed] [Google Scholar]
- 35.Li, X., D. Zhang, M. Hannink, and L. J. Beamer. 2004. Crystal structure of the Kelch domain of human Keap1. J. Biol. Chem. 279:54750-54758. [DOI] [PubMed] [Google Scholar]
- 36.McMahon, M., N. Thomas, K. Itoh, M. Yamamoto, and J. D. Hayes. 2004. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 279:31556-31567. [DOI] [PubMed] [Google Scholar]
- 37.Metzler, W. J., K. L. Constantine, M. S. Friedrichs, A. J. Bell, E. G. Ernst, T. B. Lavoie, and L. Mueller. 1993. Characterization of the three-dimensional solution structure of human profilin: 1H, 13C, and 15N NMR assignments and global folding pattern. Biochemistry 32:13818-13829. [DOI] [PubMed] [Google Scholar]
- 38.Moi, P., K. Chan, I. Asunis, A. Cao, and Y. W. Kan. 1994. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 91:9926-9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Motohashi, H., T. O'Connor, F. Katsuoka, J. D. Engel, and M. Yamamoto. 2002. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294:1-12. [DOI] [PubMed] [Google Scholar]
- 40.Motohashi, H., and M. Yamamoto. 2004. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 10:549-557. [DOI] [PubMed] [Google Scholar]
- 41.Muhandiram, D. R., and L. E. Kay. 1994. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. B 103:203-216. [Google Scholar]
- 42.Nguyen, T., P. J. Sherratt, and C. B. Pickett. 2003. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 43:233-260. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen, T., C. S. Yang, and C. B. Pickett. 2004. The pathways and molecular mechanisms regulating Nrf2 activation in response to chemical stress. Free Radic. Biol. Med. 37:433-441. [DOI] [PubMed] [Google Scholar]
- 44.Nioi, P., and J. D. Hayes. 2004. Contribution of NAD(P)H:quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutat. Res. 555:149-171. [DOI] [PubMed] [Google Scholar]
- 45.Orlicky, S., X. Tang, A. Willems, M. Tyers, and F. Sicheri. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112:243-256. [DOI] [PubMed] [Google Scholar]
- 46.Ortega, A., and J. Garcia de la Torre. 2005. Efficient, accurate calculation of rotational diffusion and NMR relaxation of globular proteins from atomic-level structures and approximate hydrodynamic calculations. J. Am. Chem. Soc. 127:12764-12765. [DOI] [PubMed] [Google Scholar]
- 47.Padmanabhan, B., M. Scharlock, K. I. Tong, Y. Nakamura, M.-I. Kang, A. Kobayashi, T. Matsumoto, A. Tanaka, M. Yamamoto, and S. Yokoyama. 2005. Purification, crystallization and preliminary X-ray diffraction analysis of the Kelch-like motif region of mouse Keap1. Acta Crystallogr. F 61:153-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Padmanabhan, B., K. I. Tong, T. Ohta, Y. Nakamura, M. Scharlock, M. Ohtsuji, M. I. Kang, A. Kobayashi, S. Yokoyama and M. Yamamoto. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell, in press. [DOI] [PubMed]
- 48.Pervushin, K., R. Riek, G. Wider, and K. Wuthrich. 1997. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA 94:12366-12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6:9-20. [DOI] [PubMed] [Google Scholar]
- 50.Pintard, L., A. Willems, and M. Peter. 2004. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 23:1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pratico, D., J. Rokach, J. Lawson, and G. A. FitzGerald. 2004. F2-isoprostanes as indices of lipid peroxidation in inflammatory diseases. Chem. Phys. Lipids 128:165-171. [DOI] [PubMed] [Google Scholar]
- 52.Sattler, M., J. Schleucher, and C. Griesinger. 1999. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. NMR Spectrosc. 34:93-158. [Google Scholar]
- 53.Schulze, P. C., and R. T. Lee. 2005. Oxidative stress and atherosclerosis. Curr. Atheroscler. Rep. 7:242-248. [DOI] [PubMed] [Google Scholar]
- 54.Stogios, P. J., G. S. Downs, J. J. Jauhal, S. K. Nandra, and G. G. Prive. 2005. Sequence and structural analysis of BTB domain proteins. Genome Biol. 6:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tompa, P. 2005. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 579:3346-3354. [DOI] [PubMed] [Google Scholar]
- 56.van Holde, K. E., and W. O. Weischet. 1978. Boundary analysis of sedimentation velocity experiments with monodisperse and paucidisperse solutes. Biopolymers 17:1387-1403. [Google Scholar]
- 57.Wakabayashi, N., A. T. Dinkova-Kostova, W. D. Holtzclaw, M. I. Kang, A. Kobayashi, M. Yamamoto, T. W. Kensler, and P. Talalay. 2004. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. USA 101:2040-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wishart, D. S., C. G. Bigam, A. Holm, R. S. Hodges, and B. D. Sykes. 1995. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR 5:67-81. [DOI] [PubMed] [Google Scholar]
- 59.Wuthrich, K. 1986. NMR of proteins and nucleic acids. John Wiley, New York, N.Y.
- 60.Xue, F., and L. Cooley. 1993. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72:681-693. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, D. D., S. C. Lo, J. V. Cross, D. J. Templeton, and M. Hannink. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24:10941-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, O., L. E. Kay, J. P. Olivier, and J. D. Forman-Kay. 1994. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR 4:845-858. [DOI] [PubMed] [Google Scholar]
- 63.Zheng, N., B. A. Schulman, L. Song, J. J. Miller, P. D. Jeffrey, P. Wang, C. Chu, D. M. Koepp, S. J. Elledge, M. Pagano, R. C. Conaway, J. W. Conaway, J. W. Harper, and N. P. Pavletich. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416:703-709. [DOI] [PubMed] [Google Scholar]
- 64.Zipper, L. M., and R. T. Mulcahy. 2002. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J. Biol. Chem. 277:36544-36552. [DOI] [PubMed] [Google Scholar]