Abstract
Hepatitis C virus (HCV) NS5B protein is a membrane-associated phosphoprotein that possesses an RNA-dependent RNA polymerase activity. We recently reported that NS5A protein interacts with TRAF2 and modulates tumor necrosis factor alpha (TNF-α)-induced NF-κB and Jun N-terminal protein kinase (JNK). Since NS5A and NS5B are the essential components of the HCV replication complex, we examined whether NS5B could modulate TNF-α-induced NF-κB and JNK activation. In this study, we have demonstrated that TNF-α-induced NF-κB activation is inhibited by NS5B protein in HEK293 and hepatic cells. Furthermore, NS5B protein inhibited both TRAF2- and IKK-induced NF-κB activation. Using coimmunoprecipitation assays, we show that NS5B interacts with IKKα. Most importantly, NS5B protein in HCV subgenomic replicon cells interacted with endogenous IKKα, and then TNF-α-mediated IKKα kinase activation was significantly decreased by NS5B. Using in vitro kinase assay, we have further found that NS5B protein synergistically activated TNF-α-mediated JNK activity in HEK293 and hepatic cells. These data suggest that NS5B protein modulates TNF-α signaling pathways and may contribute to HCV pathogenesis.
Hepatitis C virus (HCV) is the major cause of non-A, non-B hepatitis, which often leads to liver cirrhosis and hepatocellular carcinoma (1, 14). More than 170 million individuals worldwide are infected with HCV. HCV belongs to a member of the Flaviviridae family and contains a single-stranded, positive-sense RNA genome of ∼9,600 nucleotides in length. The HCV genome encodes a single polyprotein precursor of approximately 3,010 amino acids that is cleaved by both cellular signal peptidase and viral protease to generate structural and nonstructural proteins (19, 21, 33, 35). The N-terminally localized core and envelope proteins (E1 and E2) are viral structural proteins, and the remainders of the genome are nonstructural proteins. The nonstructural protein 5B (NS5B) is an RNA-dependent RNA polymerase. The NS5B is the key enzyme that catalyzes the replication of HCV. We have previously demonstrated that NS5B is a phosphoprotein that is predominantly localized in the perinuclear region in the cytoplasm (24). Although RNA-dependent RNA polymerase activities have been demonstrated using both bacterially and baculovirus-expressed NS5B proteins (8, 16, 34, 39, 40, 60), the detailed mechanism of HCV replication is poorly understood by the lack of an efficient cell culture system. The NS5B has been extensively characterized at the biochemical (8, 34) and structural levels (10, 30) and has been a prime target for inhibitors of HCV replication.
The nuclear transcription factor NF-κB plays a critical role in regulating the expression of many cytokines and immunoregulatory proteins (4-6). The NF-κB complex is composed of homodimers or heterodimers of Rel and NF-κB proteins, including NF-κB1, NF-κB2, p65, Rel B, and c-Rel (4). The activity of NF-κB can be elevated by various stimuli, including tumor necrosis factor (TNF), interleukin 1 (IL-1), and phorbol esters (55). In most cells, NF-κB proteins are sequestered in the cytoplasm, where they are complexed with one of three IκB proteins, IκBα, IκBβ, or IκBɛ (26, 36, 57). Stimulation of the cells with TNF or several other activators leads to the phosphorylation of IκBs on the N-terminal serine residues by IκB kinase (IKK) complex. The IκBs are polyubiquitinated and then rapidly degraded by the proteasome (3). Then NF-κB can be translocated to the nucleus and activates target genes by binding with high affinity to κB elements in their promoters (4, 5).
The three proteins, IKKα, IKKβ, and IKKγ (also called NEMO), were identified as the components of the IKK complex (15, 37, 47, 49, 58, 59, 62, 63). IKKα and IKKβ are activated by IL-1 and TNF, specifically phosphorylate Ser32 and Ser36 of IκBα, and are crucial for NF-κB activation (63). IKKα is an 85-kDa protein, while IKKβ is an 87-kDa protein. Both kinases have two related catalytic subunits and contain an N-terminal kinase domain, a leucine zipper motif, and a helix-loop-helix motif (25). IKKα and IKKβ can form either a homodimer or a heterodimer via their leucine zipper motif, but the predominant IKK complex forms heterodimer (49). In IKKα and IKKβ knockout cells, NF-κB activation is completely inhibited (31). However, IKKα and IKKβ knockout mice show different phenotypes (23, 32, 54). IKKγ is the regulatory subunit of the IKK complex, and it binds to IKKα and IKKβ (49). In IKKγ-deficient primary murine embryonic fibroblasts, NF-κB cannot be activated by TNF-α, IL-1, lipopolysaccharide, and other stimuli (50).
In the present study, we asked whether TNF-α-induced NF-κB and Jun N-terminal protein kinase (JNK) activations could be modulated by the HCV NS5B protein. Indeed, NS5B protein inhibited TNF-α-induced NF-κB activation in a dose-dependent manner. This inhibition was mediated by NS5B-IKKα interaction. Interaction between NS5B and IKKα was further confirmed in Huh7 cells harboring the HCV subgenomic replicon. Endogenous IKKα kinase activity was also inhibited by the NS5B protein. Moreover, TNF-α-stimulated JNK activity was synergistically elevated by NS5B protein. These findings thereby provide a potential mechanism for HCV pathogenesis.
MATERIALS AND METHODS
Plasmid construction.
cDNA corresponding to the NS5B coding sequence of HCV was amplified by PCR using the Korean isolate of HCV (genotype 1b) (11) and subcloned into the BamHI site of pcDNA3 (Invitrogen) or pEF6/Myc-His (Invitrogen). TNFR1, TRADD, TRAF2, MEKK-1, the dominant-negative mutant form of MEKK1 (DN MEKK1), Flag-IKKα, and Flag-IKKβ expression vectors were described previously (28, 43, 44, 64). The Flag-IκBα expression vector was a gift from Dean Ballard (Vanderbilt University).
Cell culture and DNA transfection.
HEK293 cells, Huh7 cells, HepG2 cells, and Cos7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 U/ml streptomycin, and 100 U/ml penicillin (Invitrogen). Huh7 cells harboring HCV subgenomic replicon, which was provided by Christoph Seeger (Fox Chase Cancer Center, Philadelphia, Pa.), were cultured in the presence of G418 (0.5 mg/ml). To establish the interferon (IFN)-cured cells, HCV replicon cells were treated with 100 U/ml of IFN-α for 2 weeks. Elimination of HCV replicon RNA was confirmed by reverse transcription-PCR and Western blotting and by the loss of resistance to G418. For the transfection experiment, ∼5 × 105 cells plated on 60-mm dishes were transfected with DNA by using either LipofectAMINE (Invitrogen) or the calcium phosphate method as described previously (44).
Reporter gene assay.
HEK293 cells and Huh7 cells were transfected by the calcium phosphate method with 2 μg of expression plasmid, 0.1 μg of either NF-κB luciferase reporter (29), and 0.1 μg of pCH110 (Amersham Biosciences) reference plasmid. The total DNA concentration in each transfection mixture was kept constant by adjusting with an empty vector. At 24 h after transfection, the cells were stimulated with TNF-α (20 ng/ml; Invitrogen) for 6 h. For the AP-1 luciferase reporter assay, the AP-1 reporter gene (38) was cotransfected into HepG2 cells. Luciferase and β-galactosidase assays were performed as described previously (44).
EMSA.
HEK293 cells, HepG2 cells, and Huh7 cells were transfected with either vector or NS5B expression plasmid by using Lipofectamine (Invitrogen). At 24 h, cells were stimulated with TNF-α (20 ng/ml) for the indicated times and nuclear extracts were prepared as described previously (29). Protein concentrations were determined using the method of Bradford (Bio-Rad). Nuclear extracts (10 μg) were assayed for NF-κB DNA-binding activity by incubation with 1 × 105 cpm of a 32P-end-labeled 22-mer double-stranded NF-κB oligonucleotide (5′-AGTTGAGGGGACTTTCCCAGGC-3′) (Promega) and for SP1 DNA-binding activity with a 22-mer double-stranded SP1 oligonucleotide (5′-ATTCGATCGGGGCGGGGCGAGC-3′) (Promega) in binding buffer [10 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM dithiothreitol, 1 mM EDTA, pH 7.5, 5% glycerol, and 2 μg of poly(dI-dC), 5% Nonidet P-40] for 20 min at room temperature. For AP-1 DNA-binding activity in the electrophoretic mobility shift assay (EMSA), nuclear extracts (10 μg) were incubated with 1 × 105 cpm of a 32P-end-labeled 21-mer double-stranded AP-1 oligonucleotide (Promega) in binding buffer [20% glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM dithiothretol, 250 mM NaCl, 50 mM Tris-HCl, pH 7.5, 0.25 mg/ml of poly(dI-dC)] for 20 min at room temperature. The protein-DNA complexes were separated by electrophoresis on 5% native polyacrylamide gels using 0.25× Tris-borate EDTA and detected by autoradiography. For competition analysis, unlabeled oligonucleotide was incubated with nuclear extract in binding buffer for 15 min before the addition of radiolabeled oligonucleotide.
In vitro binding assay and coimmunoprecipitation.
cDNA corresponding to full-length NS5B was subcloned into the EcoRI and XhoI site of the pGEX-4T expression vector (Novagen). Glutathione S-transferase (GST)-NS5B fusion protein was expressed in Escherichia coli BL21(DE) (Novagen) and purified with glutathione-Sepharose 4B beads (Amersham Biosciences) as specified by the manufacturer. HEK293 cells were lysed in 400 μl of cell lysis buffer A (50 mM HEPES, pH 7.6, 250 mM NaCl, 5 mM EDTA, 0.2% Nonidet P-40, and 1 mM phenylmethylsulfonyl fluoride). The cell lysates were further centrifuged at 14,000 rpm for 10 min at 4°C, and pellets were discarded. The protein concentration was determined using a Bio-Rad protein assay kit.
For in vitro binding assay, each cell lysate of Flag-IKKα, Flag-IKKβ, and Flag-IκBα was incubated with either GST or GST-NS5B fusion protein for 2 h at 4°C in cell lysis buffer A. Samples were washed five times with cell lysis buffer A, and the bound proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a nitrocellulose membrane, and detected by immunoblot analysis using anti-Flag monoclonal antibody (Sigma-Aldrich).
For coimmunoprecipitation assay, Cos7 cells were infected with recombinant vaccinia virus (vTF7-3) expressing T7 RNA polymerase (17). Two hours after infection, cells were transfected with 4 μg of corresponding plasmids. Following incubation at 37°C for 12 h, cells were harvested, washed twice in cold-phosphate-buffered saline, and incubated in 400 μl of cell lysis buffer A. The cell lysates were triturated by 10 passes through a 25-gauge needle on ice and centrifuged at 14,000 rpm for 10 min. The supernatant was incubated with anti-Flag monoclonal antibody for 2 h. The samples were further incubated with protein A beads (Zymed) for 1 h. Beads were washed five times with cell lysis buffer A, and the bound proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and detected by immunoblot analysis using mouse anti-Myc antibody (Santa Cruz Biotechnology). For coimmunoprecipitation assay using HCV subgenomic replicon cells, cell lysates were incubated with anti-IKKα polyclonal antibody (Santa Cruz Biotechnology) for 2 h. Following incubation with protein A-Sepharose beads, bound proteins were immunoblotted with anti-NS5B monoclonal antibody.
For in vivo binding assay between NS5B and endogenous IKK proteins, NS5B-Myc was transfected into HEK293 cells. At 36 h after transfection, cells were immunoprecipitated with either anti-IKKα, anti-IKKβ, or anti-IκBα antibodies and then bound protein was immunoblotted with anti-Myc antibody.
IκBα degradation.
HEK293 cells were transfected with either an empty vector or NS5B expression plasmid. At 24 h after transfection, cells were treated with TNF-α (20 ng/ml) for various times. The cell extracts were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and detected by immunoblotting with anti-IκBα polyclonal antibody (Santa Cruz Biotechnology).
IKK kinase assay.
The IKK kinase assay was performed as described previously (44). Briefly, cells were treated with TNF-α for the indicated times and lysed in 400 μl of cell lysis buffer B (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride). Cell extracts were immunoprecipitated with either anti-IKKα or anti-IKKβ antibodies (Santa Cruz Biotechnology) for 2 h at 4°C, and protein A-Sepharose was added for 1 h at 4°C. Immune complexes were washed twice with lysis buffer B and then washed twice with kinase buffer (20 mM HEPES, pH 7.4, 1 mM MnCl2, 5 mM MgCl2, 10 mM β-glycerophosphate, 0.1 mM Na orthovanadate, 2 mM NaF, and 1 mM dithiothreitol). Samples were incubated for 30 min at 30°C in 20 μl of kinase buffer containing 5 μCi of [γ-32P]ATP (NEN Life Science) and 1 μg of GST-IκBα (amino acids [aa] 1 to 54) as a substrate. The reaction mixtures were analyzed by SDS-PAGE and then detected by autoradiography. Kinase activity was quantified by measuring the radioactivity using a densitometric scanner (Bio-Rad).
JNK assay.
The JNK assay was performed as described previously (43). Briefly, HEK293 cells were transfected with either an empty vector or NS5B expression vector. At 24 h after transfection, cells were stimulated with TNF-α (20 ng/ml) for 15 min. Total cell lysates were prepared, and JNK activity was determined using a JNK assay kit according to the manufacturer's instructions (New England Biolabs).
RESULTS
HCV NS5B protein inhibits TNF-α-induced NF-κB and IKK activations.
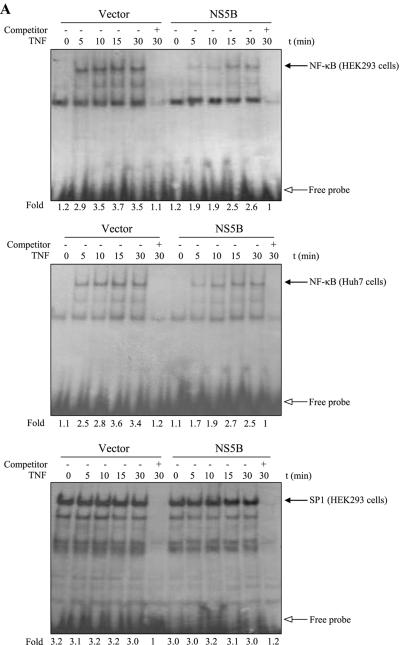
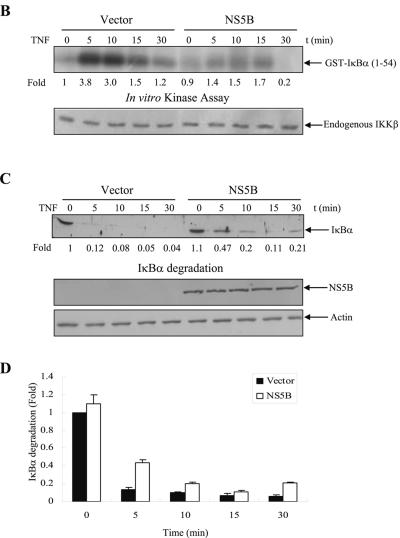
We have previously reported that NS5A protein modulated TNF-α-induced NF-κB and JNK activations in HEK293 cells (44, 45). Since NS5A is physically associated with NS5B (52) and both NS5A and NS5B are essential components of the HCV RNA replication complex (46), we examined whether NS5B could modulate TNF-α-induced NF-κB signaling pathways. For this purpose, HEK293 cells and Huh7 cells transfected with either empty vectors or NS5B expression plasmids were examined to see whether NS5B protein could affect TNF-α-stimulated NF-κB activation, endogenous IKK activity, and IκBα degradation. As demonstrated by EMSA in Fig. 1A, NF-κB was activated by TNF-α as a DNA-protein complex in vector control cells. However, TNF-α-induced NF-κB activation was significantly inhibited by HCV NS5B protein in both HEK293 and Huh7 cells. Furthermore, TNF-α-induced SP1 activation was not affected by NS5B (Fig. 1A), indicating that NS5B specifically inhibits TNF-α-induced NF-κB activation. To examine IKK activity, endogenous IKKβ was immunoprecipitated with IKKβ antibody and immunocomplex kinase activity was determined by using GST-IκBα as a substrate. Similar to NF-κB, endogenous IKKβ activation was also inhibited by NS5B protein (Fig. 1B). We have also found that NS5B protein inhibited the exogenously expressed IKKβ kinase activity (data not shown). IκBα is rapidly phosphorylated on Ser32 and Ser36 by TNF-α stimulation and subsequently degraded by the 26S proteasome (57). Figure 1C and D showed that IκBα was immediately degraded by TNF-α stimulation in vector-transfected cells. However, TNF-α-induced IκBα degradation was also inhibited by NS5B protein.
FIG.1.
The NS5B of hepatitis C virus inhibits TNF-α-induced NF-κB and IKK activations. (A) NS5B inhibits TNF-α-induced NF-κB DNA binding activity. HEK293 cells and Huh7 cells were transfected with either empty vector or NS5B expression plasmid. At 24 h after transfection, cells were treated with human TNF-α (20 ng/ml) for the indicated times. Nuclear extracts were prepared and assessed for activated NF-κB by EMSA with γ-32P-labeled double-stranded NF-κB oligonucleotide (top and middle panels) and SP1 activation with double-stranded SP1 oligonucleotide (bottom panel) as described in Materials and Methods. Arrows indicate retarded DNA-protein complexes. (B) HCV NS5B protein inhibits TNF-α-induced endogenous IKKβ activation. HEK293 cells were transiently transfected with either empty vector or NS5B expression plasmid. At 24 h after transfection, cells were incubated with human TNF-α (20 ng/ml) for the indicated times. Cell lysates were then precipitated with anti-IKKβ antibody, and endogenous IKKβ kinase activity was determined using GST-IκBα (1 to 54 aa) as a substrate (upper panel). Protein levels of endogenous IKKβ in the same cell lysates were determined by immunoblotting (lower panel). (C) NS5B protein inhibits TNF-α-stimulated IκBα degradation. HEK293 cells transfected with either vector or NS5B plasmid DNA were stimulated with human TNF-α (20 ng/ml) for the indicated times. Equal amounts of cell lysates were analyzed by immunoblotting with anti-IκBα antibody (top panel). The results shown are representative of three independent experiments. Protein levels of HCV NS5B (middle panel) and β-actin (bottom panel) in the same cell lysates were determined by immunoblotting. (D) Triplicate experimental data of Fig. 1C were quantified, and each bar represents the average IκBα degradation.
HCV NS5B protein inhibits IKK-mediated NF-κB activation.
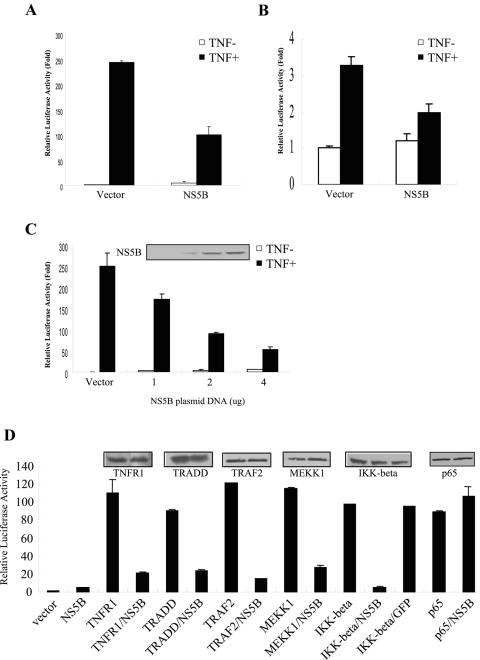
To address the mechanism of the above finding, we further examined the effect of NS5B protein on TNF-α-mediated NF-κB activation using a reporter gene assay system. HEK293 cells were cotransfected with an NF-κB-luciferase reporter plasmid with either empty vectors or NS5B expression plasmids. At 24 h after transfection, cells were either left untreated or treated with human TNF-α (20 ng/ml) for 6 h, and then NF-κB activity was determined. TNF-α induced an approximately 250-fold increase of NF-κB activity compared with the endogenous level (Fig. 2A). On the other hand, overexpression of NS5B protein inhibited TNF-α-induced NF-κB activity by ∼60%. We further investigated the effect of NS5B protein on TNF-α-induced NF-κB activation in Huh7 cells. Although the sensitivity of TNF-α-induced NF-κB activation in Huh7 cells was not comparable to that in HEK293 cells, NS5B protein also inhibited TNF-α-induced NF-κB activity in hepatic cells (Fig. 2B). To further substantiate this finding, we have examined the dosage effect of NS5B on TNF-α-induced NF-κB activation. HEK293 cells were cotransfected with reporter plasmid and increasing amounts of NS5B expression plasmid (1 to 4 μg). As shown in Fig. 2C, NS5B protein significantly inhibited TNF-α-induced NF-κB activation in a dose-dependent manner (∼40 to 80% reduction in reporter gene activity), whereas NS5B itself had little effects on the endogenous NF-κB activity. To determine whether the inhibition of NF-κB activity by HCV NS5B protein is mediated through TNFR1 signaling cascades, we examined the effects of NS5B protein on NF-κB activation induced by TNFR1 signaling transducers, including TNFR1, TRADD, TRAF2, MEKK1, IKKβ, and p65. GFP was used as a control. Overexpression of NS5B significantly inhibited TNFR1-, TRADD-, TRAF2-, MEKK1-, and IKKβ-mediated NF-κB activation (∼80 to 95% reduction in luciferase activity) (Fig. 2D). However, overexpression of NS5B did not inhibit p65-mediated NF-κB activation, indicating that NS5B was acting as an upstream transducer of NF-κB. Furthermore, IKKβ-mediated NF-κB activation was not inhibited by GFP, suggesting that the inhibition of NF-κB activity induced by TNFR1 signaling molecules was NS5B specific. These results suggest that HCV NS5B protein acts as an inhibitor of IKK-mediated NF-κB activation in TNFR1 signaling transducers.
FIG. 2.
NS5B protein inhibits NF-κB activation stimulated by TNF-α signaling molecules. (A, B) Effect of NS5B protein on TNF-mediated NF-κB activation by reporter gene assay. Either HEK293 cells (A) or Huh7 cells (B) were cotransfected with NF-κB-Luc and pCH110 reporter plasmids together with empty vector or NS5B expression plasmid. The total DNA amount was held constant by the addition of an empty vector. At 24 h after transfection, cells were either left untreated or treated with human TNF-α (20 ng/ml) for 6 h, and NF-κB activity was measured as described in Materials and Methods. (C) NS5B protein inhibits NF-κB activation in a dose-dependent manner. Reporter plasmids were cotransfected with selected amounts (1, 2, and 4 μg) of NS5B plasmid. The total DNA amount was held constant by the addition of an empty vector. The results shown are representative of three independent experiments. (D) IKK-induced NF-κB activity is inhibited by NS5B protein. The effects of NS5B protein on NF-κB activity in TNF-α signaling molecules were determined. HEK293 cells were cotransfected with reporter plasmids and TNF signaling molecules (TNFRI, TRADD, TRAF2, MEKK1, IKKβ, and p65) either alone or together with NS5B plasmid. HEK293 cells were also cotransfected with IKKβ and GFP as a control. NF-κB reporter gene activity was determined. The results shown are representative of three independent experiments. Protein expressions of each plasmid are shown in insets.
HCV NS5B protein interacts with both IKKα and IKKβ.
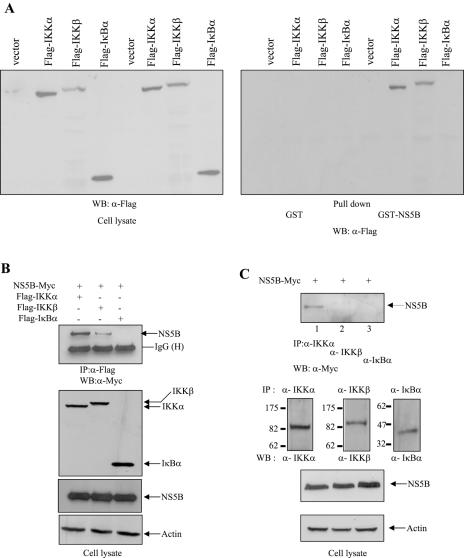
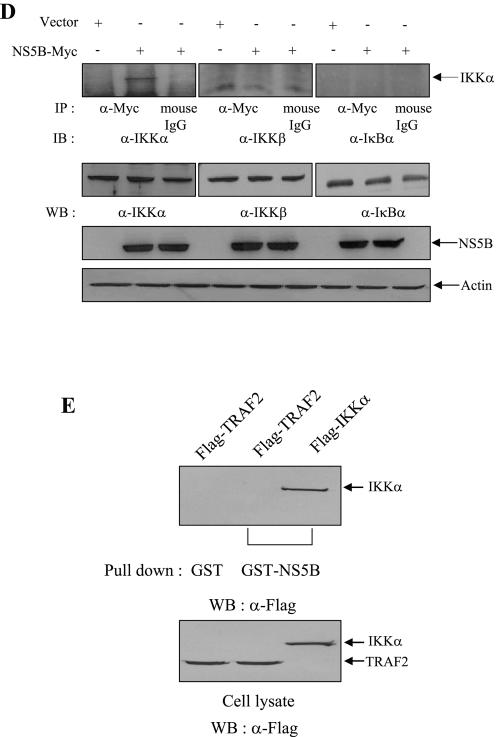
To determine how NS5B protein inhibits IKK-mediated NF-κB activation, we asked whether NS5B could interact with IKK proteins. We first performed a GST pull-down assay using either GST or GST-NS5B fusion protein. In this experiment, both vector and IκΒα were used as controls. HEK293 cells were transiently transfected with empty vector, Flag-IKKα, Flag-IKKβ, and Flag-IκBα expression plasmid, individually. At 36 h after transfection, cell extracts were incubated with either GST or GST-NS5B beads, and the bound proteins were detected by anti-Flag antibody. As shown in Fig. 3A, GST-NS5B protein was selectively bound to IKKα and IKKβ but not to vector or IκBα. However, GST protein failed to retain IKK proteins. To further substantiate this finding, we employed a coimmunoprecipitation assay. For this purpose, NS5B-Myc was coexpressed with either Flag-IKKα, Flag-IKKβ, or Flag-IκBα in Cos7 cells paired with recombinant vaccinia virus (vTF7-3) system (17). Cell lysates were immunoprecipitated with anti-Flag monoclonal antibody, and the coimmunoprecipitated protein was detected by immunoblotting with anti-Myc monoclonal antibody. In Fig. 3B, it can be seen that not only IKKα but also IKKβ is coprecipitated with NS5B. Nevertheless, NS5B bound IKKβ very poorly in this system. To further confirm this finding, we investigated whether NS5B could interact with endogenous IKK proteins by using a coimmunoprecipitation assay. NS5B-Myc was transfected into HEK293 cells, and cell lysates were immunoprecipitated with either anti-IKKα, anti-IKKβ, or IκBα antibody. Prior to in vivo binding assay, the specificity of each antibody was confirmed by immunoprecipitation as shown in Fig. 3C. The coimmunoprecipitated protein was then subsequently detected by immunoblotting with anti-Myc monoclonal antibody. Indeed, NS5B protein interacts with endogenous IKKα (Fig. 3C, lane 1) but not with endogenous IKKβ, although these two proteins are 52% identical in sequence (25). Reciprocally, the same cell lysates were immunoprecipitated with anti-Myc antibody, and then bound protein was detected by immunoblotting with anti-IKKα, IKKβ, and IκBα, respectively. We confirmed that NS5B protein interacted with endogenous IKKα (Fig. 3D). We also found that NS5B protein did not interact with IKKγ (data not shown). Taken together, these data clearly demonstrate that NS5B directly interacts with IKKα both in vitro and in vivo.
FIG.3.
The NS5B protein interacts with both IKKα and IKKβ. (A) HEK293 cells were transiently transfected with empty vector, Flag-IKKα, Flag-IKKβ, and Flag-IκΒα, individually. At 36 h after transfection, total cell lysates were incubated with either GST or purified GST-NS5B protein. Bound proteins were precipitated by glutathione beads and then detected by immunoblot using anti-Flag monoclonal antibody (right panel). Protein expressions of IKKα, IKKβ, and IκΒα were verified using the same cell lysates by immunoblotting with anti-Flag monoclonal antibody (left panel). (B) Cos7 cells were transfected with the indicated combinations of expression plasmids paired with recombinant vaccinia virus expressing T7 RNA polymerase (vTF7-3). At 12 h after transfection, cell lysates were immunoprecipitated with anti-Flag monoclonal antibody and then bound protein was detected by immunoblotting with anti-Myc monoclonal antibody (top panel). Protein levels of IKKα, IKKβ, IκΒα, NS5B, and β-actin in the same cell lysates were determined by immunoblotting (bottom three panels). IP, immunoprecipitation; WB, Western blot. (C) NS5B protein interacts with endogenous IKKα protein in vivo. HEK293 cells were transfected with NS5B expression plasmid. At 36 h after transfection, cells were immunoprecipitated with anti-IKKα, anti-IKKβ, and anti-IκBα antibodies, and then bound protein was immunoblotted with anti-Myc antibody (top panel). The specificity of each antibody was verified by immunoprecipitation with IKKα, IKKβ, and IκBα antibodies in the same cell lysates (second panels from the top). Protein levels of NS5B and β-actin in the same cell lysates were determined by immunoblotting. (D) Reciprocally, cell lysates used in panel C were immunoprecipitated with either anti-Myc antibody or mouse normal immunoglobulin G (IgG), and bound protein was immunoblotted with anti-IKKα, anti-IKKβ, and anti-IκBα antibody, respectively. Protein expressions of each plasmid in the same cell lysates were verified by immunoblotting with corresponding antibodies. (E) The NS5B protein does not interact with TRAF2 protein. Huh7 cells were transiently transfected with either Flag-TRAF2 or Flag-IKKα plasmid. Either GST or GST-NS5B purified from E. coli was incubated with cell lysates containing either TRAF2 or IKKα. Bound proteins were precipitated by glutathione beads and detected by immunoblot using Flag antibody (top panel). Protein expressions of both TRAF2 and IKKα in Huh7 cells were verified by Flag antibody (bottom panel).
HCV NS5B protein does not interact with TRAF2.
In the previous studies, we showed that HCV NS5A protein modulated both NF-κB and JNK activities through interaction with TRAF2 protein (44, 45). Since NS5B is physically associated with NS5A, we examined the potential interaction between NS5B and TRAF2 by GST pull-down assay using E. coli-expressed GST and GST-NS5B fusion protein. Cell extracts containing either TRAF2 or IKKα were incubated with either GST or GST-NS5B beads. The NS5B selectively bound to IKKα, whereas NS5B failed to retain TRAF2 (Fig. 3E), indicating that NS5B modulated TNF signal transduction through effects on IKK protein.
HCV subgenomic replicon inhibits TNF-α-induced IKKα activation through interaction with NS5B and IKK.
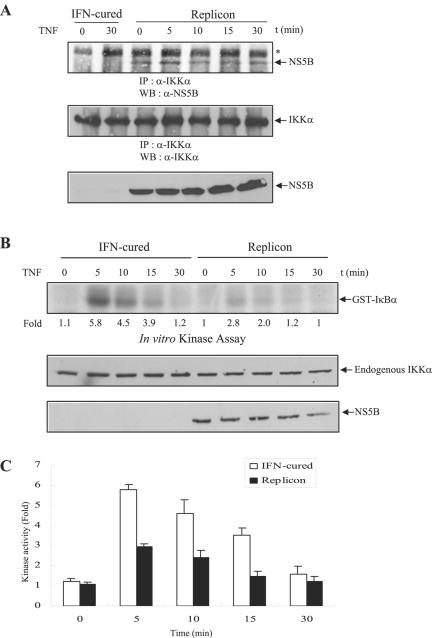
We then asked how the inhibitory effect of NS5B on NF-κB activation might occur in the context of viral infection or even viral RNA replication. For this purpose, both IFN-cured Huh7 cells and HCV subgenomic replicon cells were treated with human TNF-α and cell lysates were immunoprecipitated with anti-IKKα polyclonal antibody, and then bound proteins were immunoblotted with anti-NS5B monoclonal antibody. As shown in Fig. 4A, NS5B protein in HCV subgenomic replicon cells interacts with endogenous IKKα regardless of TNF-α treatment. To further examine how this interaction affects TNF-α-induced endogenous IKKα activation in HCV subgenomic replicon cells, both IFN-cured Huh7 cells and HCV replicon cells were treated with human TNF-α and then endogenous IKKα kinase activity was determined using GST-IκBα as a substrate. Figures 4B and C show that TNF-α-mediated IKKα kinase activation is significantly decreased in HCV replicon cells. Since endogenous IKKα expression level is not altered in replicon cells, this result clearly shows that NS5B inhibits TNF-α-induced IKK kinase activation through effects on IKKα protein.
FIG. 4.
HCV subgenomic replicon cells inhibit TNF-α-induced IKKα activation through interaction with NS5B and IKKα. (A) The NS5B protein of HCV subgenomic replicon cells interacts with endogenous IKKα protein in vivo. Both IFN-cured Huh7 cells and HCV subgenomic replicon cells were treated with human TNF-α (20 ng/ml) for the indicated times. Cell lysates were immunoprecipitated with anti-IKKα, and then bound proteins were immunoblotted with anti-NS5B monoclonal antibody (top panel). Membrane was reprobed with anti-IKKα polyclonal antibody to confirm the expression of endogenous IKKα (middle panel). The same cell lysates were immunoblotted with anti-NS5B monoclonal antibody to confirm the expression of NS5B protein in HCV subgenomic replicon cells (bottom panel). The asterisk indicates a nonspecific band. IP, immunoprecipitation; WB, Western blot. (B) HCV subgenomic replicon cells inhibit TNF-α-induced endogenous IKKα activation. Both IFN-cured Huh7 cells and HCV subgenomic replicon cells were incubated with human TNF-α (20 ng/ml) for the indicated times. Cell lysates were precipitated with anti-IKKα antibody, and then endogenous IKKα kinase activity was determined using GST-IκBα (1 to 54 aa) as a substrate (top panel). Protein levels of endogenous IKKα and NS5B in the same cell lysates were determined by immunoblotting with either IKKα antibody (middle panel) or NS5B antibody (bottom panel). (C) Data from triplicate experiments shown in Fig. 4B were quantified, and each bar represents the average of IKKα kinase activities.
HCV NS5B protein enhances TNF-α-initiated JNK activation.
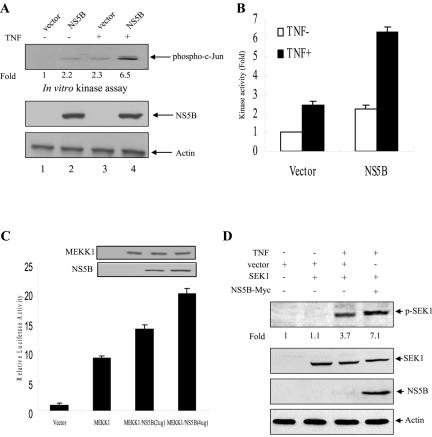
Since TNF-α induces activation of both transcription factor NF-κB (6, 41) and the JNK (9, 28) through TNFR1, we have examined whether NS5B could modulate TNF-α-induced JNK activation. For this purpose, HEK293 cells were transiently transfected with either an empty vector or NS5B expression plasmid. At 24 h after transfection, cells were either untreated or treated with human TNF-α (20 ng/ml) for 15 min, and then JNK activity was determined by using GST-c-Jun (1 to 89) fusion protein as a substrate. NS5B protein itself slightly induced JNK activation (Fig. 5A, lane 2). Moreover, TNF-α-stimulated JNK activation was significantly increased by HCV NS5B protein (Fig. 5A, lane 3 versus lane 4), and this was further confirmed by triplicate experiments (Fig. 5B). Since the protein expression level of NS5B was not affected by TNF (Fig. 5A, lanes 2 and 4), JNK activation was not due to the overexpression of NS5B protein. This result indicated that NS5B protein potentiated TNF-α-induced JNK activation. To investigate if c-Jun phosphorylation is necessarily leading to activity, AP-1 luciferase reporter gene assay was performed by cotransfecting HepG2 cells with AP-1 reporter gene (38) and MEKK1 with increasing amounts of NS5B expression plasmid. MEKK1, MEK kinase 1, is required for the phosphorylation of c-Jun (13). Overexpression of MEKK1 stimulated the AP-1 transcriptional activity, and this MEKK1-mediated AP-1 activity was synergistically elevated by NS5B protein in a dose-dependent manner (Fig. 5C). In a reporter gene assay, we further confirmed that TNF-α-induced AP-1 activity was synergistically elevated by NS5B protein (data not shown). TNF-induced c-Jun phosphorylation is mediated through the MEKK1/SEK1/JNK1 pathway. To investigate whether the increase of JNK activity by NS5B protein is TNF dependent, the effect of NS5B protein on TNF-induced SEK1 phosphorylation was examined. Huh7 cells were cotransfected with SEK1 and NS5B expression plasmids. At 36 h after transfection, cells were treated with TNF (20 ng/ml) for 15 min, and then the phosphorylation level of SEK1 was determined by Western blotting with p-SEK1 antibody (Cell Signaling Technology). Upon TNF treatment, SEK1 was phosphorylated, and this TNF-induced SEK1 phosphorylation was synergistically elevated by NS5B protein, although the protein expression level of SEK1 was not affected by TNF or NS5B (Fig. 5D). This result further confirmed that NS5B enhanced the TNF-induced JNK activity. These data indicate that NS5B protein negatively regulates TNF-α-induced NF-κB activation and acts as a positive regulator of TNF-α-induced JNK signaling cascades.
FIG. 5.
HCV NS5B protein potentiates TNF-α-induced JNK activation. (A) NS5B protein enhances TNF-α-induced JNK activation. HEK293 cells were transiently transfected with either an empty vector or plasmid expressing NS5B. At 24 h after transfection, cells were either left untreated or treated with human TNF-α (20 ng/ml) for 15 min. Using cell lysates, JNK activity was determined (top panel) as described in Materials and Methods. Both NS5B protein and β-actin (middle two panels) levels were determined by immunoblotting. (B) Data from triplicate experiments shown in Fig. 5A were quantified, and each bar represents the average of JNK kinase activity. (C) MEKK1-mediated AP-1 transcriptional activity is synergistically elevated by NS5B protein. HepG2 cells were cotransfected with AP-1 reporter gene and MEKK1 expression plasmid and with selected amounts of NS5B expression plasmid. The total DNA amount was kept constant by adjusting with an empty vector. At 36 h after transfection, luciferase and β-galactosidase assays were performed as described in Materials and Methods. The results shown are representative of three independent experiments. Protein expressions of both MEKK1 and NS5B plasmids are shown in insets. (D) Huh7 cells were transfected with either SEK1 alone or SEK1 and NS5B expression vector together. At 36 h after transfection, cells were treated with human TNF (20 ng/ml) for 15 min, and then the phosphorylation level of SEK1 was determined by Western blotting with p-SEK1 antibody (top panel). Protein expressions of SEK1, NS5B, and β-actin in the same cell lysates were verified by immunoblot analysis (lower three panels).
NS5B protein inhibits MEKK1-mediated IKKα activity but promotes SEK1 activity.
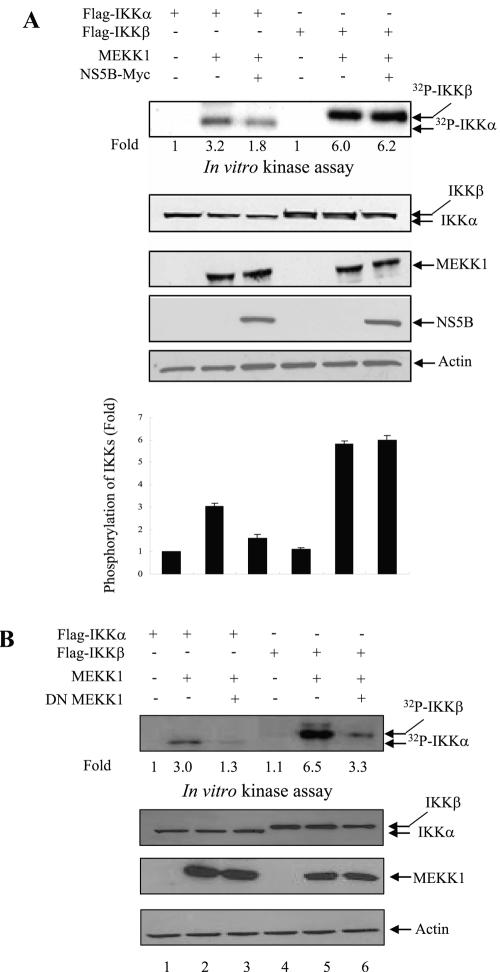
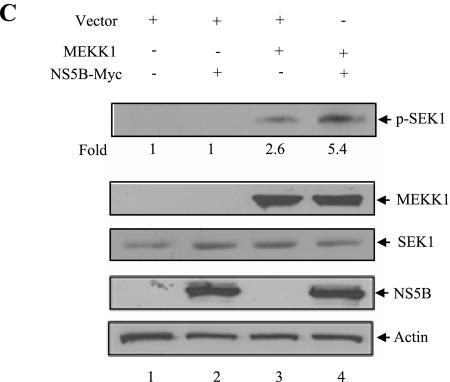
MEKK1 is required for the activation of both transcription factor NF-κB and JNK. NF-κB activation is mediated through IKK protein (27) and JNK activation is mediated through SEK1 protein (42, 61). To investigate the molecular mechanism of JNK activation by NS5B, Huh7 cells were cotransfected with IKK and MEKK1 in the presence or absence of NS5B expression plasmid. At 36 h after transfection, cell lysates were immunoprecipitated with anti-Flag antibody, and kinase assay was performed using an immunocomplex. As shown in Fig. 6A, overexpression of MEKK1 activated both IKKα and IKKβ activities. However, NS5B protein inhibited MEKK1-mediated IKKα activation (Fig. 6A, top panel). This was further confirmed by triplicate experiments as demonstrated in Fig. 6A (bottom graph). Interestingly, MEKK1-mediated IKKβ activation was not affected by NS5B protein. To investigate whether MEKK1 specifically activated both IKKα and IKKβ, we used DN MEKK1 as a control. Indeed, MEKK1-stimulated IKKα and IKKβ activations were significantly inhibited by DN MEKK1 (Fig. 6B, lanes 3 and 6). To examine the effect of NS5B protein on MEKK1-mediated SEK1 activation, Huh7 cells were cotransfected with either MEKK1 and vector or MEKK1 and NS5B expression plasmid. At 36 h after transfection, cell lysates were immunoblotted with phospho-SEK1 antibody to determine SEK1 activity. Figure 6C shows that MEKK1-mediated SEK1 activation is significantly increased by NS5B protein (top panel, lane 4). Since the protein expression level of endogenous SEK1 was not affected by MEKK1 or NS5B protein (Fig. 6C, lane 3 versus lane 4), SEK1 activation was not due to the overexpression of SEK1 protein. This result indicated that NS5B protein enhanced JNK activation through SEK1 activation.
FIG.6.
NS5B protein inhibits MEKK1-mediated IKK activity but promotes SEK1 activity. (A) MEKK1-mediated IKK activity was inhibited by NS5B protein. Huh7 cells were cotransfected with either Flag-IKKα or Flag-IKKβ and MEKK1 in the absence (−) or presence (+) of NS5B as indicated. At 36 h after transfection, cell lysates were immunoprecipitated with Flag antibody and proteins coprecipitated with Flag-IKK were assayed for kinase activity using [γ-32P]ATP. The reaction mixtures were resolved by 10% SDS-PAGE and then detected by autoradiography (top panel). Protein expressions of IKKα, IKKβ, MEKK1, NS5B, and β-actin in the same cell lysates were verified by immunoblot analysis (middle four panels). The average IKK phosphorylation is graphically demonstrated from triplicate experimental data (bottom graph). (B) MEKK1 specifically activates both IKKα and IKKβ. Huh7 cells were cotransfected with either Flag-IKKα or Flag-IKKβ, and MEKK1 in the absence (−) or presence (+) of DN MEKK1, and a kinase assay was performed as described for panel A (top panel). Protein expressions of IKKα, IKKβ, MEKK1, and β-actin in the same cell lysates were verified by immunoblot analysis (lower three panels). Since DN MEKK1 has the same molecular mass as active MEKK1 (13), protein expression of DN MEKK1 was examined separately and not shown here. (C) MEKK1-mediated SEK1 activity was synergistically elevated by NS5B protein. Huh7 cells were cotransfected with MEKK1 and HCV NS5B expression plasmids. At 36 h after transfection, cell lysates were separated by 10% SDS-PAGE and transferred to a nitrocellulose membrane, and SEK1 activity was determined by immunoblotting with rabbit anti-phospho-SEK1 antibody (top panel). Protein levels in the same cell lysates were determined by immunoblot analysis using antibodies against MEKK1, SEK1, NS5B, and β-actin, individually (lower four panels). The results shown are representative of three independent experiments.
DISCUSSION
The most prominent feature of HCV is its ability to persist in virus-infected patients. We speculate that if HCV could modulate the cellular responses to immune stimulatory cytokines, it could be advantageous for the virus to maintain persistent infection in the host. To investigate the molecular mechanism underlying HCV pathogenesis, we have explored the potential involvement of HCV NS5B protein in TNF-α signaling pathways, especially TNFR1-mediated NF-κB and JNK activations. TNF-α has a wide range of actions in inflammation, infection, and immunity (7). Nuclear transcription factor NF-κB plays a key role in regulating expression of many cytokines and immunoregulatory proteins (4-6). It has been well established that TNFR1 interacts with a number of cellular proteins including TRADD, RIP, TRAF2, NIK, and IKKs that are responsible for TNF-α-induced NF-κB signaling cascade. Recently, we reported that NS5A modulated TNF-α-stimulated NF-κB and JNK activations (44, 45). Since NS5A and NS5B are the essential components of the HCV replication complex (46), we asked whether HCV NS5B protein could modulate TNF-α signaling pathways, especially TNFR1-mediated NF-κB and JNK activations. Using both EMSA and reporter gene assays, we have demonstrated that NS5B protein inhibits TNF-α-induced NF-κB activation in HEK293 cells and hepatic cells. Furthermore, NS5B inhibits TNF-α-induced IKK activation and subsequent IκBα degradation. We have further shown that NS5B protein inhibits TNFR1-, TRADD-, TRAF2-, MEKK1-, and IKK-mediated NF-κB activation in HEK293 cells. Since NF-κB activity is immediately controlled by IKK, these results suggest that the inhibition of TNF-α-induced NF-κB activation by HCV NS5B is mediated through effects on IKK proteins.
We utilized a vaccinia virus T7 expression system (vTF7-3) to analyze the physical association between NS5B and IKKs. Indeed, NS5B protein binds to IKKα and IKKβ in both in vitro GST binding and in vivo coimmunoprecipitation assays. The IKK complex consists of two catalytic subunits, IKKα and IKKβ (also called IKK-1 and IKK-2, respectively) (63), and a regulatory subunit termed IKK-γ (also named NEMO) (59). Both IKKα (p85) and IKKβ (p87) have 52% identity in sequence and contain an amino-terminal catalytic domain in addition to carboxyl-terminal helix-loop-helix and leucine zipper domains. In our system, NS5B protein strongly associates with IKKα but binds to IKKβ only weakly. Moreover, NS5B protein interacts with endogenous IKKα but not with endogenous IKKβ, suggesting that HCV may utilize IKKα to evade host immune surveillance or to promote RNA replication. Indeed, a recent study shows that IKKα but not IKKβ regulates mitogenic signaling through transcriptional induction of cyclin D1 (2).
We further asked how the inhibitory effect of NS5B on NF-κB activation might occur in the context of viral infection or even viral RNA replication. For this purpose, we performed a coimmunoprecipitation assay to determine whether NS5B and IKKα could interact in Huh7 cells harboring HCV subgenomic replicons. Indeed, NS5B protein in HCV subgenomic replicon cells interacts with endogenous IKKα regardless of TNF-α stimulation. Furthermore, TNF-α-mediated IKKα kinase activation was significantly decreased in HCV replicon cells. This result further suggests that HCV may utilize cellular IKKα to modulate a specific signaling pathway.
The NF-κB pathway provides an attractive target for viruses to escape immune suppression, leading to persistent infection. Activation of NF-κB is an early event that occurs within minutes after exposure to TNF. Many viruses have evolved their gene products to modulate the NF-κB pathway for viral replication, host cell survival, and evasion of immune responses. For example, latent membrane protein 1 of Epstein-Barr virus, Tat protein of human immunodeficiency virus type 1, and Tax protein of the human T-cell leukemia virus type 1 can activate NF-κB through several distinct mechanisms (18, 22). Meanwhile, the adenovirus E1A protein inhibits TNF-induced NF-κB activation through suppression of IKK activity and IκBα phosphorylation (51). The IκB-like protein encoded by the African swine fever virus inhibits TNF-induced NF-κB activation through the prevention of binding between p50 and p65 (48). It has previously been reported that HCV core protein either enhanced TNF-α-induced NF-κB activation in HEK293 and HeLa cells (12, 20) or suppressed TNF-α-induced NF-κB activity in stable cells expressing HCV core (53, 64). These conflicting results may be due to the difference in the cell types stably or transiently expressing core protein. In HBV, X protein directly interacts with IκBα to prevent the reassociation of IκBα with NF-κB (56).
We previously reported that NS5A inhibited TNF-α-induced NF-κB activation through interaction with TRAF2 (44). To examine whether both NS5A and NS5B proteins would work synergistically in TNF-α-induced NFκB signaling pathway, we performed a luciferase reporter assay by cotransfecting both NS5A and NS5B in Huh7 cells. Indeed, both NS5A and NS5B protein inhibited TNF-α-induced NF-κB activation. However, coexpression of both NS5A and NS5B showed no synergism of inhibitory effect on NF-κB activation (data not shown). Since TRAF2 is the upstream transducer of IKK, most of the TNF-α signaling cascade might be already blocked at TRAF2 by NS5A and, hence, the effect of NS5B on the TNF-α signaling cascade was insignificant at the downstream signaling molecule. We speculate that the combinative effects may not be expected in natural infection because NS5A and NS5B may function at different stages of infection. Therefore, HCV may utilize either NS5A or NS5B protein in a temporally regulated manner to modulate TNF-α signal transduction to evade host immune responses.
In this report, we show that NS5B protein participates as a novel regulator in TNF-α signaling events. NS5B showed divergent effects on NF-κB and JNK activities. By interacting with IKK proteins, NS5B inhibits TNF-α-initiated IKK kinase activity and subsequently inhibits NF-κB activation. NS5B not only inhibited MEKK1-mediated NF-κB activation through interaction with IKK but also enhanced JNK activation by promoting SEK1 activation. The biological function of TNF-α-induced JNK activation is still controversial. Hence, we are not sure why NS5B shows pleiotropic responses to the signaling pathway. HCV may utilize more than one mechanism for survival and inhibition of apoptosis to evade cellular immune responses. It is also possible that the IKK subunit may function as an accessory protein in the HCV replication complex, although there is no evidence showing that IKK is associated with any viral replication complex. Taken together, modulation of TNF-α-induced NF-κB and JNK activations by NS5B protein may play a key role in HCV pathogenesis.
Acknowledgments
We thank Ebrahim Zandi (University of Southern California) for valuable comments.
This study was supported by a grant (0220010-2 to S.B.H.) of the National Cancer Control R&D Program 2002, Ministry of Health and Welfare, Korea, and by Hallym University.
REFERENCES
- 1.Aach, R. D., C. E. Stevens, F. B. Hollinger, J. W. Mosley, D. A. Peterson, P. E. Taylor, R. G. Johnson, L. H. Barbosa, and G. J. Nemo. 1991. Hepatitis C virus infection in post-transfusion hepatitis. An analysis with first- and second-generation assays. N. Engl. J. Med. 325:1325-1329. [DOI] [PubMed] [Google Scholar]
- 2.Albanese, C., K. Wu, M. D'Amico, C. Jarrett, D. Joyce, J. Hughes, J. Hulit, T. Sakamaki, M. Fu, A. Ben-Ze'ev, J. F. Bromberg, C. Lamberti, U. Verma, R. B. Gaynor, S. W. Byers, and R. G. Pestell. 2003. IKKalpha regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Mol. Biol. Cell 14:585-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arenzana-Seisdedos, F., P. Turpin, M. Rodriguez, D. Thomas, R. T. Hay, J. L. Virelizier, and C. Dargemont. 1997. Nuclear localization of IkappaB alpha promotes active transport of NF-kappa B from the nucleus to the cytoplasm. J. Cell Sci. 110:369-378. [DOI] [PubMed] [Google Scholar]
- 4.Baeuerle, P. A., and D. Baltimore. 1996. NF-kappa B: ten years after. Cell 87:13-20. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin, A. S., Jr. 1996. The NF-kappa B and IkappaB proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649-683. [DOI] [PubMed] [Google Scholar]
- 6.Barnes, P. J., and M. Karin. 1997. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 336:1066-1071. [DOI] [PubMed] [Google Scholar]
- 7.Bazzoni, F., and B. Beutler. 1996. The tumor necrosis factor ligand and receptor families. N. Engl. J. Med. 334:1717-1725. [DOI] [PubMed] [Google Scholar]
- 8.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner, D. A., M. O'Hara, P. Angel, M. Chojkier, and M. Karin. 1989. Prolonged activation of jun and collagenase genes by tumour necrosis factor-alpha. Nature 337:661-663. [DOI] [PubMed] [Google Scholar]
- 10.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, Y. G., J. W. Yoon, K. L. Jang, C. M. Kim, and Y. C. Sung. 1993. Full genome cloning and nucleotide sequence analysis of hepatitis C virus from sera of chronic hepatitis patients in Korea. Mol. Cells 3:195-202. [Google Scholar]
- 12.Chung, Y. M., K. J. Park, S. Y. Choi, S. B. Hwang, and S. Y. Lee. 2001. Hepatitis C virus core protein potentiates TNF-α-induced NF-κB activation through TRAF2-IKKβ-dependent pathway. Biochem. Biophys. Res. Commun. 284:15-19. [DOI] [PubMed] [Google Scholar]
- 13.Deak, J. C., and D. J. Templeton. 1997. Regulation of the activity of MEK kinase 1 (MEKK1) by autophosphorylation within the kinase activation domain. Biochem. J. 322:185-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Bisceglie, A., M. L. H. Simpson, M. T. Lotze, and J. H. Hoofnagle. 1994. Development of hepatocellular carcinoma among patients with chronic liver disease due to hepatitis C viral infection. J. Clin. Gastroenterol. 19:222-226. [DOI] [PubMed] [Google Scholar]
- 15.DiDonato, J. A., M. Hayakawa, D. M. Rothwarf, E. Zandi, and M. Karin. 1997. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature 388:548-554. [DOI] [PubMed] [Google Scholar]
- 16.Ferrari, E., J. Wright-Minogue, J. W. Fang, B. M. Baroudy, J. Y. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuerst, T. A., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geleziunas, R., S. Ferrell, X. Lin, Y. Mu, E. T. Cunningham, Jr., M. Grant, M. A. Connelly, J. E. Hambor, K. B. Marcu, and W. C. Greene. 1998. Human T-cell leukemia virus type 1 Tax induction of NF-kappaB involves activation of the IkappaB kinase alpha (IKKalpha) and IKKbeta cellular kinases. Mol. Cell. Biol. 18:5157-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hideo, Y., N. Kato, Y. Shiratori, M. Otsuka, S. Maeda, J. Kato, and M. Omata. 2001. Hepatitis C virus core protein activates nuclear factor kappa B-dependent signaling through tumor necrosis factor receptor-associated factor. J. Biol. Chem. 276:16399-16405. [DOI] [PubMed] [Google Scholar]
- 21.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, and K. Shimotohno. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. USA 88:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiscott, J., H. Kwon, and P. Genin. 2001. Hostile takeovers: viral appropriation of the NF-kappaB pathway. J. Clin. Investig. 107:143-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu, Y., V. Baud, M. Delhase, P. Zhang, T. Deerinck, M. Ellisman, R. Johnson, and M. Karin. 1999. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science 284:316-320. [DOI] [PubMed] [Google Scholar]
- 24.Hwang, S. B., K. J. Park, Y. S. Kim, Y. C. Sung, and M. M. C. Lai. 1997. Hepatitis C virus NS5B protein is a membrane-associated phosphoprotein with a predominantly perinuclear localization. Virology 227:439-446. [DOI] [PubMed] [Google Scholar]
- 25.Karin, M. 1999. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene 18:6867-6874. [DOI] [PubMed] [Google Scholar]
- 26.Krappmann, D., and C. Scheidereit. 1997. Regulation of NF-kappaB activity by IkappaB alpha and IkappaB beta stability. Immunobiology 198:3-13. [DOI] [PubMed] [Google Scholar]
- 27.Lee, F. S., R. T. Peters, L. C. Dang, and T. Maniatis. 1998. MEKK1 activates both IκB kinase α and IκB kinase β. Proc. Natl. Acad. Sci. USA 95:9319-9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, S. Y., A. Reichlin, A. Santana, K. A. Sokol, M. C. Nussenzweig, and Y. Choi. 1997. TRAF2 is essential for JNK but not NF-kappaB activation and regulates lymphocyte proliferation and survival. Immunity 7:703-713. [DOI] [PubMed] [Google Scholar]
- 29.Lee, S. Y., S. Y. Lee, G. Kandala, M.-L. Liou, H. C. Liou, and Y. Choi. 1997. CD30/TNF receptor-associated factor interaction: NF-kappaB activation and binding specificity. Proc. Natl. Acad. Sci. USA 93:9699-9703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 31.Li, Q., G. Estepa, S. Memet, A. Israel, and I. M. Verma. 2000. Complete lack of NF-kappaB activity in IKK1 and IKK2 double-deficient mice: additional defect in neurulation. Genes Dev. 14:1729-1733. [PMC free article] [PubMed] [Google Scholar]
- 32.Li, Q., D. Van Antwerp, F. Mercurio, K. F. Lee, and I. M. Verma. 1999. Severe liver degeneration in mice lacking the IkappaB kinase 2 gene. Science 284:321-325. [DOI] [PubMed] [Google Scholar]
- 33.Lin, C., B. D. Lindenbach, B. M. Pragai, D. W. McCourt, and C. M. Rice. 1994. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 68:5063-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manabe, S., I. Fuke, O. Tanishita, C. Kaji, Y. Gomi, S. Yoshida, C. Mori, A. Takamizawa, I. Yoshida, and H. Okayama. 1994. Production of nonstructural proteins of hepatitis C virus requires a putative viral protease encoded by NS3. Virology 198:636-644. [DOI] [PubMed] [Google Scholar]
- 36.May, M. J., and S. Ghosh. 1997. Rel/NF-kappaB and IkappaB proteins: an overview. Semin. Cancer Biol. 8:63-73. [DOI] [PubMed] [Google Scholar]
- 37.Mercurio, F., H. Zhu, B. W. Murray, A. Shevchenko, B. L. Bennett, J. Li, D. B. Young, M. Barbosa, M. Mann, A. Manning, and A. Rao. 1997. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science 278:860-866. [DOI] [PubMed] [Google Scholar]
- 38.Offringa, R., S. Gebel, H. van Dam, M. Timmers, A. Smits, R. Zwart, B. Stein, J. L. Bos, A. van der Eb, and P. Herrlich. 1990. A novel function of the transforming domain of E1a: repression of AP-1 activity. Cell 62:527-538. [DOI] [PubMed] [Google Scholar]
- 39.Oh, J. W., T. Ito, and M. M. C. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oh, J. W., G. T. Sheu, and M. M. C. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 41.Osborn, L., S. Kunkel, and G. J. Nabel. 1989. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc. Natl. Acad. Sci. USA 86:2336-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park, H. S., M. S. Kim, S. H. Huh, J. Park, J. Chung, S. S. Kang, and E. J. Choi. 2002. Akt (protein kinase B) negatively regulates SEK1 by means of protein phosphorylation. J. Biol. Chem. 277:2573-2578. [DOI] [PubMed] [Google Scholar]
- 43.Park, K. J., S. H. Choi, M. S. Koh, D. J. Kim, S. W. Yie, S. Y. Lee, and S. B. Hwang. 2001. Hepatitis C virus core protein potentiates c-Jun N-terminal kinase activation through a signaling complex involving TRADD and TRAF2. Virus Res. 74:89-98. [DOI] [PubMed] [Google Scholar]
- 44.Park, K. J., S. H. Choi, S. Y. Lee, S. B. Hwang, and M. M. C. Lai. 2002. Nonstructural 5A protein of hepatitis C virus modulates tumor necrosis factor α-stimulated nuclear factor κB activation. J. Biol. Chem. 277:13122-13128. [DOI] [PubMed] [Google Scholar]
- 45.Park, K. J., S. H. Choi, D. H. Choi, J. M. Park, S. W. Yie, S. Y. Lee, and S. B. Hwang. 2003. Hepatitis C virus NS5A protein modulates c-Jun N-terminal kinase through interaction with tumor necrosis factor receptor-associated factor 2. J. Biol. Chem. 278:30711-30718. [DOI] [PubMed] [Google Scholar]
- 46.Penin, F., J. Dubuisson, F. A. Rey, D. Moradpour, and J. M. Pawlotsky. 2004. Structural biology of hepatitis C virus. Hepatology 39:5-19. [DOI] [PubMed] [Google Scholar]
- 47.Regnier, C. H., H. Y. Song, X. Gao, D. V. Goeddel, Z. Cao, and M. Rothe. 1997. Identification and characterization of an IkappaB kinase. Cell 90:373-383. [DOI] [PubMed] [Google Scholar]
- 48.Revilla, Y., M. Callejo, J. M. Rodriguez, E. Culebras, M. L. Nogal, M. L. Salas, E. Vinuela, and M. Fresno. 1998. Inhibition of nuclear factor kappaB activation by a virus-encoded IkappaB-like protein. J. Biol. Chem. 273:5405-5411. [DOI] [PubMed] [Google Scholar]
- 49.Rothwarf, D. M., E. Zandi, G. Natoli, and M. Karin. 1998. IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature 395:297-300. [DOI] [PubMed] [Google Scholar]
- 50.Rudolph, D., W. C. Yeh, A. Wakeham, B. Rudolph, D. Nallainathan, J. Potter, A. J. Elia, and T. W. Mak. 2000. Severe liver degeneration and lack of NF-kappaB activation in NEMO/IKKgamma-deficient mice. Genes Dev. 14:854-862. [PMC free article] [PubMed] [Google Scholar]
- 51.Shao, R., M. C. Hu, B. P. Zhou, S. Y. Lin, P. J. Chiao, R. H. von Lindern, B. Spohn, and M. C. Hung. 1999. E1A sensitizes cells to tumor necrosis factor-induced apoptosis through inhibition of IkappaB kinases and nuclear factor kappaB activities. J. Biol. Chem. 274:21495-21498. [DOI] [PubMed] [Google Scholar]
- 52.Shirota, Y., H. Luo, W. Qin, S. Kaneko, T. Yamashita, K. Kobayashi, and S. Murakami. 2002. Hepatitis C virus NS5A binds RNA-dependent RNA polymerase (RdRp) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277:11149-11155. [DOI] [PubMed] [Google Scholar]
- 53.Shrivastava, A., S. K. Manna, R. Ray, and B. B. Aggarwal. 1998. Etopic expression of hepatitis C virus core protein differentially regulates nuclear transcription factors. J. Virol. 72:9722-9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka, M., M. E. Fuentes, K. Yamaguchi, M. H. Durnin, S. A. Dalrymple, K. L. Hardy, and D. V. Goeddel. 1999. Embryonic lethality, liver degeneration, and impaired NF-kappaB activation in IKK-beta-deficient mice. Immunity 10:421-429. [DOI] [PubMed] [Google Scholar]
- 55.Thanos, D., and T. Maniatis. 1995. NF-kappaB: a lesson in family values. Cell 80:529-532. [DOI] [PubMed] [Google Scholar]
- 56.Weil, R., H. Sirma, C. Giannini, D. Kremsdorf, C. Bessia, C. Dargemont, C. Brechot, and A. Israel. 1999. Direct association and nuclear import of the hepatitis B virus X protein with the NF-κB inhibitor IκBα. Mol. Cell. Biol. 19:6345-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whiteside, S. T., and A. Israel. 1997. IkappaB proteins: structure, function and regulation. Semin. Cancer Biol. 8:75-82. [DOI] [PubMed] [Google Scholar]
- 58.Woronicz, J. D., X. Gao, Z. Cao, M. Rothe, and D. V. Goeddel. 1997. IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science 278:866-869. [DOI] [PubMed] [Google Scholar]
- 59.Yamaoka, S., G. Courtois, C. Bessia, S. T. Whiteside, R. Weil, F. Agou, H. E. Kirk, R. J. Kay, and A. Israel. 1998. Complementation cloning of NEMO, a component of the IkappaB kinase complex essential for NF-kappaB activation. Cell 93:1231-1240. [DOI] [PubMed] [Google Scholar]
- 60.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 61.Yan, M., T. Dai, J. C. Deak, J. M. Kyriakis, L. I. Zon, J. R. Woodgett, and D. J. Templeton. 1994. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature 372:798-800. [DOI] [PubMed] [Google Scholar]
- 62.Zandi, E., D. M. Rothwarf, M. Delhase, M. Hayakawa, and M. Karin. 1997. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell 91:243-252. [DOI] [PubMed] [Google Scholar]
- 63.Zandi, E., Y. Chen, and M. Karin. 1998. Direct phosphorylation of IkappaB by IKKalpha and IKKbeta: discrimination between free and NF-kappaB-bound substrate. Science 281:1360-1363. [DOI] [PubMed] [Google Scholar]
- 64.Zhu, N., A. Khoshnan, R. Schneider, M. Matsumoto, G. Dennert, C. Ware, and M. M. C. Lai. 1998. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J. Virol. 72:3691-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]