Abstract
Biosynthesis of NAD(P) cofactors is of special importance for cyanobacteria due to their role in photosynthesis and respiration. Despite significant progress in understanding NAD(P) biosynthetic machinery in some model organisms, relatively little is known about its implementation in cyanobacteria. We addressed this problem by a combination of comparative genome analysis with verification experiments in the model system of Synechocystis sp. strain PCC 6803. A detailed reconstruction of the NAD(P) metabolic subsystem using the SEED genomic platform (http://theseed.uchicago.edu/FIG/index.cgi) helped us accurately annotate respective genes in the entire set of 13 cyanobacterial species with completely sequenced genomes available at the time. Comparative analysis of operational variants implemented in this divergent group allowed us to elucidate both conserved (de novo and universal pathways) and variable (recycling and salvage pathways) aspects of this subsystem. Focused genetic and biochemical experiments confirmed several conjectures about the key aspects of this subsystem. (i) The product of the slr1691 gene, a homolog of Escherichia coli gene nadE containing an additional nitrilase-like N-terminal domain, is a NAD synthetase capable of utilizing glutamine as an amide donor in vitro. (ii) The product of the sll1916 gene, a homolog of E. coli gene nadD, is a nicotinic acid mononucleotide-preferring adenylyltransferase. This gene is essential for survival and cannot be compensated for by an alternative nicotinamide mononucleotide (NMN)-preferring adenylyltransferase (slr0787 gene). (iii) The product of the slr0788 gene is a nicotinamide-preferring phosphoribosyltransferase involved in the first step of the two-step nondeamidating utilization of nicotinamide (NMN shunt). (iv) The physiological role of this pathway encoded by a conserved gene cluster, slr0787-slr0788, is likely in the recycling of endogenously generated nicotinamide, as supported by the inability of this organism to utilize exogenously provided niacin. Positional clustering and the cooccurrence profile of the respective genes across a diverse collection of cellular organisms provide evidence of horizontal transfer events in the evolutionary history of this pathway.
Cyanobacteria are among the oldest life forms on earth (64). Their unique role in biogeochemical cycles such as photosynthesis, nitrogen fixation (6, 20) and generation, and homeostasis of oxygenic atmosphere (14) makes this diverse group of bacteria essential for all terrestrial life. Not surprisingly, cyanobacteria attract growing attention in various areas of basic and applied research (11). Recent breakthroughs in genome sequencing opened new opportunities for a fundamental understanding of cyanobacterial physiology and evolution. For example, a comparative genome analysis provided new insights into the photosynthetic machinery of cyanobacteria and related species (12, 54, 59).
Biosynthesis of NAD(P) cofactors is of special importance for cyanobacteria due to their role in photosynthesis and respiration. In addition to its role in innumerable redox reactions, NAD participates as a cosubstrate in a number of metabolic and regulatory processes. Some of these processes, such as NAD-dependent protein deacetylation catalyzed by the CobB/SIR2 enzyme family (68), are leading to the depletion of the NAD pool. Among other enzymes consuming NAD are a NAD-dependent DNA ligase characteristic of bacteria (73), a eukaryotic DNA damage-induced poly(ADP-ribosyl) polymerase (82), and a variety of ADP-ribosyltransferases, including many bacterial toxins (25). Salvage and recycling pathways play an important role in the homeostasis of the NAD pool (composed of four interconverting species, NAD+, NADH, NADP+, and NADPH).
Although most of the biochemical transformations involved in NAD biosynthesis, salvage, and recycling were analyzed in great detail (for reviews, see references 3, 33, 35, and 48), those studies were historically focused on a few model species. NAD-mediated links between metabolic and signaling networks recently revealed in eukaryotic cells (16, 69, 78) triggered a new wave of research in yeast and mammals (for recent reviews, see references 31 and 60). At the same time, relatively little is known about NAD biosynthesis in cyanobacteria. Although many NAD biosynthetic genes are annotated in public archives (e.g., Cyanobase [http://www.kazusa.or.jp/cyanobase/] and KEGG [http://www.genome.ad.jp/kegg/]), some of these annotations are imprecise and have not been consistently projected across all the sequenced cyanobacteria. Moreover, we are not aware of any attempt at metabolic reconstruction of respective pathways in any of the divergent cyanobacterial species. When we began this study, only one of the NAD biosynthetic enzymes, archaea-like nicotinamide mononucleotide (NMN) adenylyltransferase from Synechocystis sp. strain PCC 6803 (encoded by the slr0787 gene), was experimentally characterized (50), although its actual physiological role remained unclear.
We used genome comparative analysis and subsystem annotation tools implemented in the SEED genomic platform (http://theseed.uchicago.edu/FIG/index.cgi) to elucidate NAD biosynthetic pathways in 13 cyanobacterial species with completely sequenced genomes (http://yersinia.uchicago.edu/public/FIG/organisms.cgi?show=cyano). Previously, we applied a similar approach to the analysis of possible drug targets in the NAD biosynthesis of bacterial pathogens (23). A detailed reconstruction of NAD metabolism in cyanobacteria has led us to a number of functional inferences. Some of them were further validated by focused experiments in the model system of Synechocystis sp. strain PCC 6803.
MATERIALS AND METHODS
Bioinformatics analysis.
Most of the bioinformatics analysis in this study was performed using a collection of approximately 300 complete or almost complete genomes integrated in the SEED database with a set of specialized tools for genome comparative analysis and annotation (http://theseed.uchicago.edu/FIG/index.cgi). The SEED database is an open-source genomic platform provided by the Fellowship for Interpretation of Genomes, which supports the encoding and projection of metabolic subsystems across the entire collection of integrated genomes (43). Other bioinformatics tools and resources used in this study are as follows: GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html), PSI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST/), Pfam (http://pfam.wustl.edu/), Cyanobase (http://www.kazusa.or.jp/cyanobase/), KEGG (http://www.genome.ad.jp/kegg/), JGI (http://genome.jgi-psf.org/mic_home.html), and the ERGO genomic database (Integrated Genomics, Inc.) (45). A public version, ERGO Light (http://www.ergo-light.com/), provides annotations and elements of metabolic reconstruction for nine genomes, including Synechocystis sp. strain PCC 6803.
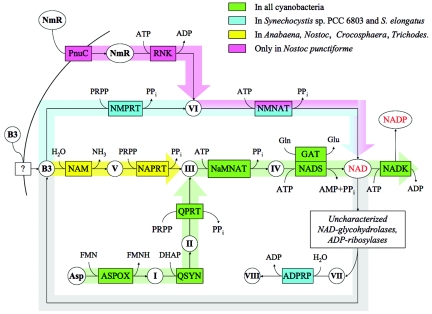
We used established principles of metabolic reconstruction technology (10, 65) to infer individual reactions and pathways based on the presence of respective gene orthologs. Although this approach does not account for quantitative aspects of cellular metabolism (such as stoichiometry or gene expression), it provides an important first approximation of an organism's phenotype (also referred to as genome property in reference 24). The SEED implementation of this technology is based on annotated subsystems, collections of functional roles capturing the knowledge of a biological process in a variety of species (43). The NAD subsystem, which was the focus of this study, included approximately 20 protein families (mostly enzymes) known to be immediately associated with the homeostasis of the NAD pool. A subset of enzymes and biochemical transformations inferred for cyanobacteria is illustrated in Table 1 and Fig. 1.
TABLE 1.
Functional reconstruction of NAD(P) metabolic pathways from genomic dataa
| Organism | De novo pathway |
Universal pathway |
Niacin recycling |
Niacin salvage |
Nicotinamide ribose salvage |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASPOX | QSYN | QPRT | NaMNAT | NADS | GATf | NADK | NMPRTb | NMNATc | NAM | NAPRT | PnuC | RNK | NMNATd | |
| Synechocystis sp. strain PCC 6803 | sll0631 | sll0622 | slr0936 | sll1916 | slr1691 | slr1691 | sll1415 | slr0788 | slr0787 | |||||
| slr0400 | ||||||||||||||
| Synechococcus elongatus PCC 7942 | 1195 | 777 | 294 | 2414 | 2415 | 2415 | 1179 | 2328 | 2329 | |||||
| 1605 | ||||||||||||||
| Synechococcus sp. strain WH 8102 | 2185 | 1311 | 2311 | 500 | 499 | 499 | 1594 | |||||||
| 2261 | ||||||||||||||
| Prochlorococcus marinus MIT 9313 | 177 | 661 | 2064 | 1454 | 1455 | 1455 | 2008 | |||||||
| 368 | ||||||||||||||
| Prochlorococcus marinus CCMP1375 | 119 | 1011 | 213 | 1595 | 1596 | 1596 | 1340 | |||||||
| 180 | ||||||||||||||
| Prochlorococcus marinus CCMP1986 | 100 | 675 | 188 | 1441 | 1442 | 1442 | 1265 | |||||||
| 156 | ||||||||||||||
| Gloeobacter violaceus PCC 7421 | 846 | 192 | 888 | 4016 | 2587 | 2587 | 3525 | 1660 | ||||||
| 473 | ||||||||||||||
| Trichodesmium erythraeum IMS101 | 1272 | 1282 | 3078 | 2493 | 772 | 772 | 141 | 2494 | ||||||
| 1273 | 457 | |||||||||||||
| Thermosynechococcus elongatus BP-1 | 2407 | 231 | 1712 | 1177 | 1179 | 1179 | 483 | 545e | 1176 | |||||
| 857 | ||||||||||||||
| Crocosphaera watsonii WH 8501 | 2684 | 4205 | 654 | 3610 | 3608 | 3608 | 161 | 358 | 3611 | |||||
| 42 | ||||||||||||||
| Nostoc sp. strain PCC 7120 | 1527 | 4980 | 2398 | 2790 | 2792 | 2792 | 5058 | 5469 | 2789 | |||||
| 5370 | 538 | |||||||||||||
| Anabaena variabilis ATCC 29413 | 80 | 2778 | 4661 | 4802 | 4800 | 4800 | 5436 | 1339 | 4803 | |||||
| 4373 | 652 | |||||||||||||
| Nostoc punctiforme | 4768 | 6260 | 6375 | 5605 | 5607 | 5607 | 981 | 1255 | 5604 | 3159 | 3160 | 3160 | ||
| 5019 | 5859 | |||||||||||||
| Escherichia coli K-12 | nadB | nadA | nadC | nadD | nadE | ? | nadK | pncA | pncB | pncC | nadR | nadR | ||
| Staphylococcus epidermidis | 1280 | 1596 | ? | 696 | 1601 | 1597 | ||||||||
| Haemophilus influenzae R2846 | 1197 | 82 | 378 | 378 | ||||||||||
| Rhodospirillum rubrum | 2934 | 2933 | 2935 | 3129 | 1657 | 1657 | 1821 | 642 | ||||||
The presence of genes assigned with respective functional roles are shown by CyanoBase identification numbers for Synechocystis sp. strain PCC 6803, by common gene names in E. coli, and by abbreviated SEED identification numbers for all other genomes. Underlining reflects the proximity of the genes on the chromosome: in each row, numbers with matching underlining mark genes that occur in the same chromosomal cluster (illustrated in Fig. 2). Multifunctional (fused) proteins with more than one functional domain are shown in boldface type. Enzyme abbreviations are as described in the legend of Fig. 1. This table was generated using Subsystem encoding tools in SEED; the complete “NAD and NADP cofactor biosynthesis global” subsystem spreadsheet containg over 200 genomes can be viewed at http://theseed.uchicago.edu/FIG/organisms.cgi?show=cyano (select “all organisms” option).
Members of NadV family, identified in Haemophilus ducreyi (34), present in V-factor-independent Pasteurellaceae but not in E. coli.
NMNAT of NadM family (as in archaea) containing an additional ADPRP domain.
NMNAT of NadR family (as in H. influenzae) containing an additional RNK domain.
Gene is apparently interrupted by a transposon insertion.
?, the mechanism of NaAD amidation by single-domain NADS (lacking a GAT component) remains unknown.
FIG. 1.
Biochemical transformations (arrows) directly involved in the biosynthesis, salvage, and recycling of NAD(P) in cyanobacteria. Functional roles in this subsystem (mostly enzymes) are shown by abbreviations in boxes. ADPRP, ADP-ribose pyrophosphatase (EC 3.6.1.13); ASPOX, l-aspartate oxidase (EC 1.4.3.16); GAT, glutamine amidotransferase chain of NAD synthetase; NADK, NAD kinase (EC 2.7.1.23); NADS, NAD synthetase (EC 6.3.1.5); NAM, nicotinamidase (EC 3.5.1.19); NaMNAT, nicotinate-nucleotide adenylyltransferase (EC 2.7.7.18); NAPRT, nicotinate phosphoribosyltransferase (EC 2.4.2.11); NMNAT, nicotinamide-nucleotide adenylyltransferase (EC 2.7.7.1); NMPRT, nicotinamide phosphoribosyltransferase (EC 2.4.2.12); PnuC, ribosyl nicotinamide transporter, PnuC-like; QPRT, quinolinate phosphoribosyltransferase (decarboxylating) (EC 2.4.2.19); QSYN, quinolinate synthetase (EC 4.1.99.-); RNK, ribosylnicotinamide kinase (EC 2.7.1.22). Key metabolites (precursors and products) and intermediates are shown by abbreviations or roman numerals in circles. Asp, l-aspartate; B3, nicotinamide, vitamin B3; NADP, NAD phosphate; NmR, N-ribosylnicotinamide; I, iminoaspartate; II, quinolinic acid; III, nicotinate mononucleotide (NaMN); IV, deamido-NAD; V, nicotinic acid; VI, nicotinamide mononucleotide (NMN); VII, ADP-ribose; VIII, ribose phosphate. Other metabolites are shown by standard symbols and abbreviations: DHAP, dihydroxyacetone phosphate; FMN, flavin mononucleotide; FMNH, flavin mononucleotide, reduced; PRPP, 5-phosphoribosyl-1-pyrophosphate; PPi, pyrophosphate. Thick bars with arrows outline individual pathways; the coloring scheme reflects the occurrence of respective genes and corresponding pathways in various cyanobacteria, as shown in the inset. For example, the green color indicates genes and pathways (de novo and universal) present in all compared cyanobacteria.
A primary set of gene annotations was generated by conventional homology-based techniques. Refinement of annotations was performed using genome context analysis techniques, most notably clustering of functionally related genes on the prokaryotic chromosome (44). Further analysis of variations in subsystem topology allowed us to accurately access which of the possible scenarios (functional variants) (76) are implemented in different species. Such functional context analysis within the framework of an encoded subsystem usually leads to significant improvement of the quality of gene assignments (43) and helps reveal previously uncharacterized variations, formulate open problems, and generate testable functional predictions.
Genomic sequences of most relevance for this study were as follows: Anabaena variabilis ATCC 29413 (GenBank accession no. NZ_AAEA00000000), Nostoc punctiforme PCC 73102 (GenBank accession no. NZ_AAAY00000000), Nostoc sp. strain PCC 7120 (27) (GenBank accession no. BA000019), Synechocystis sp. strain PCC 6803 (28) (GenBank accession no. BA000022), Synechococcus elongatus PCC 7942 (GenBank accession no. AP008231), Prochlorococcus marinus MIT9313 (59) (GenBank accession no. BX548175), Prochlorococcus marinus subsp. marinus strain CCMP1375 (17) (GenBank accession no. AE017126), Prochlorococcus marinus subsp. pastoris strain CCMP1986 (59) (GenBank accession no. BX548174), Synechococcus sp. strain WH8102 (47) (GenBank accession no. BX548020), Trichodesmium erythraeum IMS101 (GenBank accession no. NZ_AABK00000000), Thermosynechococcus elongatus BP-1 (38) (GenBank accession no. BA000039), Crocosphaera watsonii WH 8501 (GenBank accession no. NZ_AADV00000000), Gloeobacter violaceus PCC 7421 (39) (GenBank accession no. BA000045), Escherichia coli K-12 MG1655 (9) (GenBank accession no. U00096), Haemophilus influenzae Rd KW20 (21) (GenBank accession no. L42023), Rhodospirillum rubrum (GenBank accession no. CP000230), and Staphylococcus epidermidis ATCC 12228 (80) (GenBank accession no. AE015929).
Bacterial strains, medium components, plasmids, and reagents.
E. coli strains DH5α (Invitrogen, Carlsbad, CA), BL21, and BL21(DE3) (Stratagene, La Jolla, CA) were used for cloning and protein overexpression. E. coli cells were grown in LB (Luria-Bertani) broth medium. Synechocystis sp. strain PCC 6803 strains were grown at 30°C in BG-11 medium (56) under continuous white light (50 μmol photons m−2 s−1). Liquid medium was perfused with sterile air. Solid medium was supplemented with 1.5% agar (Difco Laboratories), 0.3% sodium thiosulfate, and 10 mM TES [N-tris(hydroxymethyl)-methyl-2-aminoethanesulfonic acid] buffer, pH 8.3. A pET-derived vector, pODC29, containing a T7 promoter, an N-terminal His6 tag, and a TEV (tobacco etch virus) protease cleavage site described previously (41) or a similar vector, pProEX HTa (Invitrogen), with a trc promoter was used for cloning and protein expression. Primers were obtained from Sigma-Genosys (The Woodlands, TX) and MWG Biotech Inc. (High Point, NC). PCR reagents and enzymes for DNA manipulations were obtained from New England Biolabs (Beverly, MA) and Fermentas (Vilnius, Lithuania). Plasmid purification kits and Ni-nitrilotriacetic acid (NTA) resin were purchased from QIAGEN (Valencia, CA). Reagents for DNA sequencing were obtained from PE Biosystems (Foster City, CA). Antibiotics, buffer components, and all reagents for enzymatic assays were obtained from Sigma (St. Louis, MO).
Cloning of Synechocystis sp. strain PCC 6803 genes selected for validation experiments.
The basic molecular biology procedures for plasmid preparation, restriction analysis, and transformation of competent cells were performed as described previously (63). To perform focused validation experiments, four Synechocystis sp. strain PCC 6803 genes were cloned: (i) slr0787, a genomic fusion of the archaeal nadM homolog with the mutT-like Nudix hydrolase domain, previously shown to possess dual, NMN-specific adenylyltransferase (NMNAT) and ADP-ribose pyrophosphatase (ADPRP) activity; (ii) sll1916, a homolog of bacterial nicotinic acid mononucleotide (NaMN)-specific adenylyltransferase (NaMNAT) (nadD gene in E. coli); (iii) slr1691, a homolog of the nadE gene of E. coli (ATP-dependent amidotransferase component of NAD synthase [NADS]), containing an additional N-terminal domain, tentatively assigned as a glutamine amidotransferase (GAT) component of NADS based on homology with recently analyzed eukaryotic (7) and bacterial (4) enzymes; and (iv) slr0788, a homolog of the nicotinamide phosphoribosyltransferase (NMPRT) gene, recently characterized in Haemophilus ducreyi and termed nadV (34). The DNA fragments corresponding to the coding sequences of these genes (designated here sy_nadM, sy_nadD, sy_nadE, and sy_nadV) located at the following positions in the Synechocystis sp. strain PCC 6803 genome were amplified by PCR: bp 3044833 to 3045852, 770890 to 770291, 1983982 to 1985655, and 3045876 to 3047261, respectively (http://www.kazusa.or.jp/cyanobase/Synechocystis/) (28). The following oligonucleotide primer pairs were used: sy_nadM5′ (gggccATGgAAACTAAATATCAATACGGGATCTACATTGGTC) and sy_nadM3′ (ggggtcgacCTAAACTTTGCTGACAAAATGCTGAATAATC), sy_nadD5′ (gggtcaTGAAAATAGCCCTTTTTGGCACCAGC) and sy_nadD3′ (ggggtcgacTCACTGGCGATAAAGTTG), sy_nadE5′ (gggtcATGaTTACCATTGCCCTTGCCCAGCTTAATC) and sy_nadE3′ (ggggtcgacTTAACTGCCTTGGGGATGGAAAGCTTG), and sy_nadV5′ (ggggtCATGAATACTAATCTCATTCTGGATGTGG) and sy_nadV3′ (ggggtcgaCTAGCTTGCGGGAACATTGACCCTG).
Chromosomal DNA of wild-type Synechocystis sp. strain PCC 6803, isolated as described previously (19), was used as a template. Primers were designed to incorporate an NcoI or BspHI restriction site at the 5′ end and a SalI site at the 3′ end of each gene (underlined in the primer sequences; mutations and added nucleotides are in lowercase letters). PCR fragments were purified, digested with respective enzymes, and cloned into an expression vector linearized with NcoI and SalI. The nucleotide sequences of the inserts were confirmed by DNA sequencing. The resulting constructs were used to transform E. coli DH5α for plasmid preparation and E. coli BL21 or BL21(DE3) for protein expression.
Gene inactivation in Synechocystis sp. strain PCC 6803.
The DNA construct containing sy_nadM (slr0787) cloned into pProEx HTa was digested with StuI and SmaI to excise a 120-bp-long fragment of the coding sequence (12%). It was replaced with the 1.3-kb-long SmaI-HindIII DNA fragment from pUC119Cm (S. Gerdes, unpublished data) containing the Tn9 chloramphenicol acetyltransferase gene. Likewise, a recombinant plasmid containing sy_nadD (sll1916) was digested with ApaI and MunI to introduce a 104-bp-long deletion into nadD. A 3.9-kb SacB/kanamycin resistance cassette from pRL250 (13) was inserted in place of the deleted region. The resultant inactivated variants of sy_nadD and sy_nadM were used to transform wild-type Synechocystis sp. strain PCC 6803 as described previously (74), selecting for resistance to kanamycin (20 μg/ml) or chloramphenicol (5 μg/ml), respectively. Antibiotic-resistant transformants were passaged repeatedly on BG-11 plates with progressively increasing concentrations of kanamycin (up to 120 μg/ml) or chloramphenicol (up to 30 μg/ml) in attempts to achieve complete segregation of the introduced mutations. Additional segregation experiments with the sll1916 mutant were performed in the presence of 100 μM nicotinamide. The vitamin concentration in the medium aliquots was monitored by high-performance liquid chromatography (HPLC) as follows.
Nicotinamide uptake by Synechocystis sp. strain PCC 6803.
Nicotinamide was added to the BG-11 medium to a final concentration of 100 μM, and wild-type Synechocystis sp. strain PCC 6803 and sll1916 mutant strains were propagated on the resultant medium for 72 h. In the case of the sll1916 mutant, 20 μg/ml chloramphenicol was present in the growth medium. Aliquots were withdrawn every 6 h, cells and debris were removed by centrifugation and consecutive microultrafiltration using Microcon YM-10 centrifugal filters (Amicon, Bedford, MA), and the filtrates were analyzed on 50- by 4.6-mm C18 columns (Supelco, Bellerofonte, PA). HPLC separation was carried out isocratically in 50 mM sodium phosphate, pH 6.2, containing 5% methanol at a flow rate of 1 ml/min. Samples were monitored at 254 nm using a Gilson (Middleton, WI) HPLC system. The sensitivity threshold of this procedure was ∼1 μM nicotinamide.
Recombinant protein expression and purification.
All recombinant proteins analyzed in this study (sy_NadD, sy_NadE, and sy_NadV) were expressed as N-terminal His6 tag fusions with a TEV protease cleavage site. The cultures were grown in LB medium with 100 μg/ml Amp at 37°C in shaking flasks (200 rpm) to an optical density at 600 nm of ∼0.8. The temperature was adjusted to 20°C, and expression was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to 0.8 mM. Cells were harvested after ∼12 h of shaking at 20°C.
A rapid partial purification of recombinant proteins on Ni-NTA agarose minicolumns was performed as described previously (42). In particular, cells harvested from a 50-ml culture were resuspended and lysed in 10 mM HEPES-NaOH, pH 7.2, containing 100 mM NaCl, 2 mM β-mercaptoethanol, 0.03% Brij 35, 2 mM phenylmethylsulfonyl fluoride, and 1 mg/ml lysozyme. After lysis by freeze-thawing and sonication, the lysate was centrifuged, and pellets and supernatants were processed separately. Supernatants (adjusted to pH 8 using 50 mM Tris-HCl) were loaded onto 200-μl Ni-NTA agarose minicolumns and equilibrated with the same buffer. After washing with the starting buffer containing 1 M NaCl and 0.3% Brij 35, bound proteins were eluted with 300 μl starting buffer containing 250 mM imidazole. This quick procedure usually allows us to obtain samples of sufficient quality and quantity for activity verification. For a more detailed enzymatic analysis, one of the proteins (sy_NadE) was purified from a larger (typically 6-liter) culture using a two-step procedure using an AKTA-FPLC system similar to methods described previously (15, 29). Briefly, the cell lysate was centrifuged (2 h at 20,000 × g) and applied to a 10-ml Ni-NTA agarose column. The column was washed with the same buffer as described above, followed by washing with 0.5 M NaCl and elution of the target protein with an imidazole gradient (0 to 250 mM in 30 column volumes) with monitoring at 280 nm. Collected peak fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), pooled, treated with 10 mM EDTA, concentrated (Amicon ultrafiltration cell, YM-10 membranes), and loaded onto a gel filtration column, Superdex 200 (16/60; Pharmacia), equilibrated in a solution containing 20 mM HEPES-NaOH, pH 7.2, 100 mM NaCl, 1 mM dithiothreitol, and 0.03% Brij 35. Collected fractions were analyzed by SDS-PAGE and enzymatic activity. Purified recombinant proteins used in this study retained full activity upon storage at −80°C after rapid freezing in liquid nitrogen (as small aliquots).
Enzyme activity assays. (i) NaMNAT and NMNAT activities.
NaMNAT and NMNAT activities were tested using coupled spectrophotometric assays as previously described (29). The NMNAT assay is based on the coupling of NAD formation to alcohol dehydrogenase-catalyzed conversion of NAD to NADH monitored by UV absorbance at 340 nm, as originally developed by Balducci et al. (2). The reaction was started by adding NMN to 1 mM and monitored at 340 nm over a 20-min period. To measure the NaMN-specific activity, the procedure was modified by introducing an additional enzymatic step, a conversion of deamido-NAD (NaAD) to NAD by an added excess of pure recombinant NADS as described previously (29).
(ii) NADS activity.
The continuous coupled spectrophotometric assay for NADS activity was a modification of a method described previously (77). Reaction mixtures contained 1 mM NaAD, 2 mM ATP, 10 mM MgCl2, 7 U/ml alcohol dehydrogenase (Sigma), 46 mM ethanol, 16 mM semicarbazide (or 2 mM NaHSO3), and 4 mM NH4Cl (or 2 mM glutamine) in 100 mM HEPES (pH 8.5). The reactions were carried out at 37°C and monitored by the change in UV absorbance at 340 nm using a Beckman DU-640 spectrophotometer or, for kinetic studies, in 96-well plates using a Tecan-Plus reader as described previously (29).
(iii) NMPRT activity.
An original continuous spectrophotometric assay was developed that coupled the NMPRT activity to NADH formation via two additional enzymatic steps: (a) conversion of NMN to NAD by NMNAT (a recombinant human enzyme PNAT-3 with dual NMN/NaMN specificity overexpressed and purified as described previously [79]) and (b) alcohol dehydrogenase-catalyzed conversion of NAD to NADH. The assay was performed as described above for the NMNAT assay, except that the reaction mixture contained 2.0 mM nicotinamide instead of NMN, 5 mM ATP, and 0.15 U of human NMNAT. The reaction was initiated by the addition of phosphoribosyl pyrophosphate (PRPP) to 2 mM.
A direct HPLC-based assay was used to assess substrate specificity of sy_NadV. The reaction mixture contained 100 mM HEPES-KOH, pH 7.5, 10 mM MgCl2, 1 mM ATP, 5 mM PRPP, and 2 mM nicotinamide or nicotinic acid. After 30 min of incubation with the enzyme at 37°C, protein was removed by microultrafiltration using Microcon YM-10 centrifugal filters (Amicon, Bedford, MA), and the filtrates were analyzed on 50- by 4.6-mm C18 columns (Supelco, Bellerofonte, PA) after appropriate dilution. Ion-pair separation was carried out isocratically in 50 mM sodium phosphate, pH 5.5, containing 8 mM tetrabutylammonium bromide and 8% methanol at 1 ml/min. Samples were monitored at 254 nm using a Gilson HPLC system.
RESULTS
(i) Reconstruction of NAD metabolism from genomic data.
Metabolic reconstruction of NAD biosynthesis in Synechocystis sp. strain PCC 6803 and other cyanobacteria for which genome sequence data are currently available is illustrated in Table 1 and Fig. 1. It is based on the projection of individual functional roles and pathways from model species (such as E. coli), where they were experimentally characterized (for recent overviews, see references 3, 23, 31, and 33). The initial encoding of the NAD biosynthesis subsystem and tentative annotation of respective genes were performed using the SEED software tools and included the entire collection of ∼300 integrated genomes (http://theseed.uchicago.edu/FIG/index.cgi) (43). In this study, we have focused on a more detailed reconstruction of pathways characteristic of cyanobacteria (also reflected in the CyanoSEED website [http://theseed.uchicago.edu/FIG/organisms.cgi?show=cyano]). This analysis allowed us to infer a complete set of biochemical transformations and respective genes that implement the biogenesis of NAD(P) cofactors in cyanobacteria. Some of the inferences related to Synechocystis sp. strain PCC 6803 were validated by a series of focused experiments in the second part of this study.
De novo pathway: from aspartate to NaMN.
Many prokaryotes are capable of synthesizing NAD de novo from aspartate, and they do not require the presence of niacin (vitamin B3) in growth media. A conversion of aspartate to NaMN, as originally established in Salmonella enterica serovar Typhimurium (and E. coli), is implemented via three consecutive reactions catalyzed by l-aspartate oxidase (EC 1.4.3.16; nadB), quinolinate synthetase (EC 4.1.99.-; nadA), and quinolinate phosphoribosyltransferase (decarboxylating) (EC 2.4.2.19; nadC). Reliable orthologs of all three genes are present in all analyzed genomes of cyanobacteria (for example, sll0631, sll0622, and slr0936 in Synechocystis sp. strain PCC 6803). Although the respective genes have a strong tendency to form an operon-like cluster in many prokaryotic genomes (for example, see R. rubrum in Table 1), in all cyanobacteria, they occur in scattered chromosomal loci. Nevertheless, a straightforward orthology provides us with unambiguous evidence of the de novo pathway in all compared cyanobacteria. This conclusion is consistent with annotations by other groups as well as with an autotrophic lifestyle characteristic of these organisms.
Universal pathway: from NaMN to NADP.
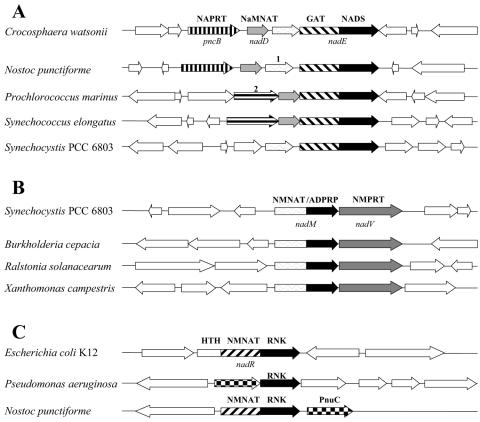
Three enzymatic functions forming this pathway, NaMNAT (EC 2.7.7.18; E. coli nadD gene), NADS (EC 6.3.1.5; nadE gene), and NAD kinase (EC 2.7.1.23; yfjB, now renamed nadK), appear to be present in the vast majority of bacteria, including those that lack a de novo pathway and produce NaMN via niacin salvage (see below). Among a few exceptions are some intracellular pathogens and Pasteurellaceae (as previously discussed) (23, 29). All cyanobacteria contain readily detectable homologs of all three genes of the canonical universal pathway. Moreover, nadD and nadE orthologs form a chromosomal cluster (Fig. 2A) conserved in almost all cyanobacteria. In contrast to that finding, in Synechocystis sp. strain PCC 6803, which was the main focus of our study, nadD (sll1916) and nadE (slr1691) homologs occur in distal chromosomal loci. Moreover, Synechocystis sp. strain PCC 6803 is one of the few cyanobacterial species that also contains an alternative NMN-specific adenylyltransferase encoded within a fusion protein (slr0787), NMNAT (EC 2.7.7.1)/ADP-ribose pyrophosphatase (EC 3.6.1.13) (50). The NMNAT activity is apparently confined to the N-terminal domain of this protein, based on homology with the previously characterized archaeal NMNAT (termed NadM) (52). The simultaneous presence of two alternative adenylyltransferases pointed to a potential ambiguity of the functional assignment of the nadD homolog, a member of a functionally divergent HIGH superfamily of nucleotidyl transferases (1, 81). That prompted us to experimentally test whether sll1916 is the key housekeeping NaMNAT (as suggested by homology, chromosomal clustering, and functional context analysis) or whether this function in Synechocystis sp. strain PCC 6803 could be fulfilled by slr0787 as well.
FIG. 2.
Clustering of the genes controlling NAD(P) metabolism on the chromosome. Shown is the alignment of chromosomal contigs of respective genomes around the homologs of nadE (A), nadM (B), and nadR (C) generated in SEED (display was modified for publication). Homologous genes are shown by arrows with matching patterns. Genes assigned specific functional roles are identified by the same abbreviations defined in the legend of Fig. 1; hypothetical genes are identified by numbers (1, predicted MutT/Nudix family protein slr1690; 2, putative membrane-associated GTPase PMM1444) (A). Genes not conserved within the clusters are shown by open arrows. For multidomain nadE, nadM, and nadR homologs, the domain structure is outlined. Only a few representative cyanobacterial genomes out of the 13 analyzed are shown in each case.
Unlike Synechocystis sp. strain PCC 6803 and other unicellular cyanobacteria in our study, the three closely related filamentous heterocystous species harbor two copies of the nadD gene (Table 1). The first copy, located in a conserved cluster with NAPRT and NADS (alr2483 in Nostoc sp. strain PCC 7120), is similar to NadD homologs in all analyzed cyanobacteria, while the other copy (all5063) is highly divergent and is not functionally coupled to any other genes of NAD metabolism in these organisms. However, the fact that a close homolog of all5063 apparently serves as the only NadD in the cyanobacterium Gloeobacter violaceus, as well as in several gram-positive microbial species (e.g., bsu2560 in Bacillus subtilis), supports the functional assignment of this protein family as nicotinate-nucleotide adenylyltransferase (EC 2.7.7.18). Although its origin in filamentous cyanobacteria is unclear, a horizontal transfer event seems likely.
A remarkable feature that differentiates cyanobacterial NADS from most of the well-characterized bacterial homologs (e.g., NadE proteins of E. coli [46] and B. subtilis [57]) is the presence of an additional nitrilase-like N-terminal domain (Fig. 2A). It was recently confirmed that a similar N-terminal domain of eukaryotic NADS performs a functional role of GAT, which is required to utilize glutamine as a source of an amido group for a consecutive ATP-dependent amidation reaction catalyzed by the C-terminal (NadE-like) domain (7). Although the genes encoding two-domain proteins similar to eukaryotic NADS can be detected in many bacterial genomes, the data on their enzymatic characterization are relatively limited and contradictory. For example, the glutaminase role of the N-terminal (nitrilase-like) domain in the NadE homolog from Mycobacterium tuberculosis was suggested (5) and recently verified (4). On the other hand, an analogous protein from Rhodobacter capsulatus was described as strictly ammonia-dependent NADS (75). This insufficient knowledge is reflected in highly inconsistent annotations of NadE homologs in most public archives. For example, compare annotations of slr1691: “NH(3)-dependent NAD(+) synthetase (EC:6.3.5.1)” in KEGG and “glutamine-dependent NAD(+) synthetase” in Cyanobase. We have decided to experimentally verify the ability of cyanobacterial enzymes with a suggested annotation, NADS/GAT, to utilize glutamine for ATP-dependent amidation of NaAD.
Salvage/recycling pathway I: from niacin to NaMN.
Vitamin B3 naturally occurs and may be utilized by many species in either of two forms, nicotinamide and nicotinic acid, which are often collectively referred to as niacin. The most common salvage pathway is a two-step conversion of nicotinamide to NaMN via nicotinamidase (NAM) (EC 3.5.1.19; gene pncA) followed by NAPRT (EC 2.4.2.11; gene pncB). This pathway is present in many species either in addition to (as in E. coli) or instead of (as in Staphylococcus epidermidis) a de novo pathway (Table 1). Within analyzed cyanobacteria, this type of salvage appears to be largely confined to filamentous species, in keeping with their symbiotic association with fungi and plants (53). Annotations of cyanobacterial NAPRT genes are based on homology supported by chromosomal clustering with other genes of a downstream universal pathway (Fig. 2A). Homology-based NAM annotations are less reliable, since they belong to a large family of paralogs with different enzymatic activities (for example, isochorismatase). Although such paralogs can be disambiguated based on chromosomal arrangement (e.g., pncA-pncB operon in B. subtilis) in some species, this is not the case in cyanobacterial genomes. An additional complexity arises due to the fact that the mechanism of niacin uptake (passive or mediated by a yet-uncharacterized transporter) remains unclear. Since both nicotinamide and nicotinic acid may be generated inside the cell due to nonredox utilization of NAD, it is often impossible to a priori discriminate between the salvage of exogenous and the recycling of endogenous niacin.
Salvage/recycling pathway II: from niacin to NMN.
A similar salvage-versus-recycling ambiguity applies to an alternative (nondeamidating) pathway of nicotinamide utilization via direct conversion to NMN by NMPRT (EC 2.4.2.12). This enzyme, encoded by gene nadV, originally identified in Haemophilus ducreyi (34), occurs in a limited number of diverse species (Table 2). A mammalian ortholog of NadV, previously described as a cytokine-like pre-B-cell colony-enhancing factor (62), was recently shown to be a fully functional NMPRT, both in vitro (61) and in vivo (55). Of all analyzed cyanobacteria, nadV homologs are present only in Synechocystis sp. strain PCC 6803 (gene slr0788) and Synechococcus elongatus. Functional assignment of these genes, as well as a tentative assertion of the respective pathway converting nicotinamide to NAD by consecutive reactions of NMPRT and NMNAT, bypassing NADS (a version of NMN shunt), is strongly supported by genome context analysis: (i) a strong tendency of genes encoding NMPRT and NMNAT/ADPRP to cluster on the chromosome of diverse bacteria (Fig. 2B) and (ii) an almost perfect cooccurrence profile of these genes over the entire collection of ∼300 genomes integrated in the SEED database (Table 2). Finally, the existence of this pathway would provide a rationale for the otherwise puzzling occurrence of two alternative adenylyltransferases (of the NadD and NadM type) in Synechocystis sp. strain PCC 6803 and Synechococcus elongatus. The physiological role of this pathway in cyanobacteria appeared to be less clear than in V-factor-independent Pasteurellaceae, where it was unambiguously implicated with the salvage of exogenous nicotinamide (34). At least two considerations, an autotrophic lifestyle of Synechocystis sp. strain PCC 6803 and the presence of the ADPRP domain fused with one of the enzymes of the pathway, pointed to its possible involvement in the recycling of endogenous nicotinamide. The latter may be generated by a yet-uncharacterized (but previously documented) (66) enzyme(s) cleaving NAD to nicotinamide and ADP-ribose, providing a rationale for the existence of a bifunctional NMNAT/ADPRP enzyme. Experimental verification of the NMPRT activity of the Synechocystis sp. strain PCC 6803 NadV homolog (gene slr0788) and genetic experiments with nadD and nadM knockout mutants accomplished in this study support this interpretation (see below). At the same time, the actual source of free nicotinamide in Synechocystis sp. strain PCC 6803 remains unknown, since its genome does not contain homologs of NAD-dependent protein deacetylase of the SIR2 family (cobB gene in E. coli), the only known candidate for this role, which is broadly conserved in many other bacteria.
TABLE 2.
Phylogenetic distribution of the nadV-nadM gene cassettea
| Organism | NMPRT (nadV) | NMNAT/ ADPRP (nadM) |
|---|---|---|
| Synechocystis sp. strain PCC 6803 | slr0788 | slr0787 |
| Synechococcus elongatus PCC 7942 | 2328 | 2329 |
| Gloeobacter violaceus PCC 7421 | − | 1660 |
| Deinococcus geothermalisb | − | 1173, 1257 |
| Deinococcus radioduransb | 476 | 2607, 3086 |
| Desulfitobacterium hafniense | − | 4279 |
| Burkholderia cenocepacia | 1675 | 1674 |
| Burkholderia cepacia R1808 | 3138 | 3137 |
| Burkholderia cepacia R18194 | 651 | 650 |
| Ralstonia metallidurans | 92 | 93 |
| Ralstonia solanacearum | 4276 | 4275 |
| Psychrobacter sp. strain 273-4 | 1584 | 1583 |
| Francisella tularensis | 620 | 482 |
| Stenotrophomonas maltophilia | 1582 | 1580 |
| Xanthomonas axonopodis | 671 | 670 |
| Xanthomonas campestris | 3396 | 3397 |
Only microbial species for which complete or nearly complete genome sequences are currently available are listed. Synechocystis sp. strain 6803 genes are shown by CyanoBase identification numbers, and the presence of orthologous genes in other genomes is indicated by abbreviated SEED identification numbers. Underlining reflects the proximity of genes on the chromosome.
Two copies of the bifunctional nicotinamide-nucleotide adenylyltransferase/ADP-ribose pyrophosphatase are present in a genome.
Salvage/recycling pathway III: from nicotinamide ribose.
Salvage/recycling pathway III includes a PnuC-like transporter-mediated uptake of nicotinamide ribose, followed by its conversion to NMN by ribosylnicotinamide kinase (RNK) (EC 2.7.1.22) and then to NAD by NMNAT, also bypassing the need for NADS. We have previously shown that in E. coli and H. influenzae, both reactions are performed by another fusion protein, NadR (29, 67). Although this pathway rarely occurs outside the γ-Proteobacteria group, all its components appear to be conserved and clustered on the chromosome of a single cyanobacterial species, Nostoc punctiforme (Fig. 2C).
(ii) Experimental validation. (a) NaMNAT, essential enzyme in the universal pathway of NAD biosynthesis. (I) Biochemical characterization of NadD homolog.
To experimentally assess a tentative assignment of cyanobacterial NadD homologs, we have cloned Synechocystis sp. strain PCC 6803 gene sll1916, overexpressed it in E. coli, and verified the predicted NAMNAT activity of the recombinant protein partially purified (enriched) on a Ni-NTA minicolumn. Due to a relatively low yield of a soluble form of the enzyme, we have also performed a large-scale (6-liter) expression procedure followed by a two-step purification procedure (as described in Materials and Methods for NADS/GAT). The resulting (>90% pure) protein with a predicted molecular mass of 25 kDa was enzymatically characterized using previously established colorimetric coupled assays with both possible substrates, NaMN and NMN (Table 3). The sy_NaMNAT exhibited a more-than-100-fold preference toward NaMN over NMN based on the comparison of catalytic activity at a 1 mM concentration of either substrate. This substrate preference was consistent with the proposed role of NaMNAT in the universal NAD biosynthesis pathway (Fig. 1). Although the observed specific activity (∼1 U/mg of pure enzyme) at a saturating (1 mM) concentration of NaMN is low compared to that of E. coli NaMNAT (∼120 U/mg of pure enzyme), it is comparable to the activity of NaMNAT from Helicobacter pylori (1.2 U/mg of pure enzyme), which was previously characterized by our group (O. V. Kurnasov and A. L. Osterman, unpublished data). This observation may be rationalized in terms of pathway/enzyme adaptation to the vastly different growth rates of these organisms. Indeed, both Synechocystis sp. strain PCC 6803 (doubling rate of 12 to 20 h) and H. pylori (doubling rate of ∼6 h) grow much slower than E. coli (doubling rate of 0.3 h).
TABLE 3.
Biochemical characterization of enzymes in NAD metbolism of Synechocystis sp. strain PCC 6803
| Locus tag | Functional role | Abbreviation | Protein sample | kDae | Enzymatic activity (U/mg)f |
Source or reference | ||
|---|---|---|---|---|---|---|---|---|
| Substrate 1 | Substrate 2 | Ratio | ||||||
| NaMN | NMN | |||||||
| slr0787 | Nicotinamide mononucleotide adenylyltransferase | NMNAT | Recombinant, 3-step purification | 39 | Not reported | 3.4 | ∼1/20 | 50a |
| sll1916 | Nicotinate mononucleotide adenylyltransferase | NaMNAT | Recombinant, 2-step purification | 25 | 1.0 | <0.01 | >100/1 | This studyb |
| Gln | NH3 | |||||||
| slr1691 | NAD synthetase | NADS | Recombinant, 2-step purification | 63 | 0.03 | 0.08 | ∼1/2.5 | This studyc |
| Nam | NA | |||||||
| slr0788 | Nicotinamide phosphoribosyltransferase | NMPRT | Recombinant, 1-step purification | 54 | 0.5 | 0.003 | >150/1 | This studyd |
NMNAT activity determined at 0.2 mM NMN, 0.2 mM ATP, pH 8.2; details of NaMNAT activity were not reported.
Activity at 1 mM NMN or NaMN with 2 mM ATP, pH 7.5.
Activity at 5 mM l-glutamine or 2 mM NH4Cl with 1 mM NaAD, 2 mM ATP, pH 7.5.
Activity at 0.2 mM nicotinamide or nicotinic acid with 1 mM PRPP, pH 7.5.
As determined by SDS-PAGE.
NA, nicotinic acid; Nam, nicotinamide.
(II) Genetic analysis of NadM and NadD homologs.
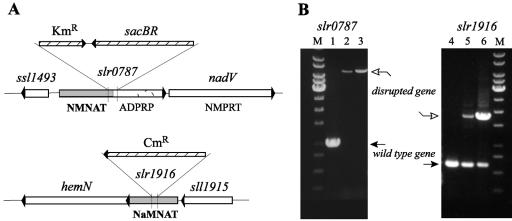
To address possible biological roles of the two alternative adenylyltransferases in Synechocystis sp. strain PCC 6803, NMNAT (50) and NaMNAT, we have attempted to inactivate the respective genes slr0787 and sll1916. Merodiploid antibiotic-resistant transformants (due to the well-known genome multiplicity of Synechocystis sp. strain PCC 6803 [8, 74]) were readily obtained using either of the two knockout plasmids and verified by PCR analysis (Fig. 3). To attain segregation of mutant and wild-type genome copies, continued culturing of the original transformants in the presence of increasing levels of an antibiotic (kanamycin or chloramphenicol, respectively) was performed (71, 74). As previously shown in similar experiments, full segregation cannot be reached in the case of the essential genes, and both wild-type and mutant alleles remain detectable in the culture (22, 40). The segregation of mutant and wild-type genotypes was monitored by PCR (Fig. 3). No wild-type band could be detected in the Synechocystis sp. strain PCC 6803 slr0787 mutant strain after two consecutive passages in BG-11 with kanamycin (about 2 weeks). On the contrary, the presence of the wild-type copy of the sll1916 gene remained evident in addition to the knockout allele (Fig. 3), even after 3 months of continuous culturing of the Synechocystis sp. strain PCC 6803 sll1916 mutant strain in the presence of high concentrations of chloramphenicol. This implied that the inactivation of the NaMNAT-encoding gene (sll1916) in Synechocystis sp. strain PCC 6803 was lethal, while the NMNAT gene (slr0787) was dispensable under the growth conditions tested. No phenotypic differences were detected between the Δslr0787 strain and the wild type under standard growth conditions.
FIG. 3.
Genetic analysis of the two adenylyltransferases in Synechocystis sp. strain PCC 6803. (A) Maps of the Synechocystis sp. strain PCC 6803 chromosome loci containing slr0787 (encoding NMNAT/ADPRP) and sll1916 (encoding NaMNAT). Positions of antibiotic resistance cassette insertions in the corresponding mutant strains are shown. (B) PCR amplification of the slr0787 and sll1916 genes using total chromosomal DNA from wild-type Synechocystis sp. strain PCC 6803 (lanes 1 and 4) and slr0787 (lane 2) and sll1916 (lane 5) mutant strains as templates after two consecutive passages on BG-11 with kanamycin or chloramphenicol, respectively, and after six consecutive passages (lanes 3 and 6). Primer pairs used for PCR amplification are those used for cloning of slr0787 and sll1916 (described in Materials and Methods). The absence of the wild-type-size PCR product in lanes 2 and 3 indicates that in the genome of the slr0787 mutant strain, all copies of slr0787 were successfully substituted with the knockout construct. On the contrary, note the persistent presence of the wild-type sll1916 gene in the sll1916 mutant strain even after multiple passages with an increasing concentration of antibiotic (lanes 5 and 6). Lane M, molecular size markers.
The addition of up to 100 μM nicotinamide to the growth medium did not improve the viability of the sll1916 mutant and did not lead to its segregation. The monitoring of the nicotinamide concentration in growth medium by HPLC failed to detect any decrease in the amount of this compound in the medium, indicating the absence of active uptake of nicotinamide by Synechocystis sp. strain PCC 6803. Altogether, these results suggest that in Synechocystis sp. strain PCC 6803, (i) NaMNAT is an essential component of the main universal NAD biosynthetic pathway, which cannot be compensated for by the alternative NMN-preferring enzyme, and (ii) the role of the NMPRT/NMNAT pathway is likely in the recycling of endogenous nicotinamide rather than in the salvage of exogenously provided vitamins.
(b) NADS/GAT: glutamine-utilizing enzyme in the universal pathway of NAD biosynthesis. Biochemical characterization of the NadE homolog.
The goal of this verification experiment was to establish whether the Synechocystis sp. strain PCC 6803 NadE homolog is capable of utilizing glutamine as an amide donor in the ATP-dependent conversion of NaAD to NAD cofactor. The Synechocystis sp. strain PCC 6803 slr1691 gene was cloned and overexpressed in E. coli. The recombinant enzyme was purified to near homogeneity, as determined by SDS-PAGE (>95%; single major band with an observed molecular mass of 63 kDa). This protein eluted as a single peak in gel filtration with a retention volume corresponding to a monomeric form of the expected Mr. Optimal conditions for the coupled colorimetric assay were established, and the enzyme activity was determined and compared using two alternative amide donors, ammonia (2 mM) and glutamine (5 mM), at saturating concentrations of other substrates, ATP (2 mM) and NaAD (1 mM), as shown in Table 3. The specific activity of the Synechocystis sp. strain PCC 6803 enzyme with glutamine as a substrate was found to be only 2.5-fold lower than that with ammonia (0.03 and 0.08 U/mg, respectively). A comparable activity level as well as similar catalytic rates and Km values with both amide donors were reported for the two-domain form of NADS from M. tuberculosis (5). It is important that although two-domain NADS do not seem to display a strong preference (if any) towards glutamine compared to ammonia, their ability to utilize glutamine at near-physiological concentrations is of crucial importance. In contrast to this, single-domain NADS (e.g., NadE of E. coli) are only active in the presence of ammonia, which, unlike glutamine, is not expected to be abundant in a free form in most living cells. Therefore, the actual mechanism and the immediate source of nitrogen for amidation of NaAD by single-domain NADS present in many bacteria and archaea remain unknown.
A bona fide utilization of glutamine by the Synechocystis sp. strain PCC 6803 enzyme was additionally confirmed by HPLC analysis of reaction products (data not shown). Overall, these results support the functional assignment of NADS/GAT suggested for cyanobacterial NadE homologs and provide additional evidence in favor of projecting this assignment to a large number of bacterial NadE homologs containing a nitrilase-like domain.
(c) NMPRT: the first enzyme in the nondeamidating nicotinamide recycling pathway. Biochemical characterization of the NadV homolog.
To confirm the possible role of the first gene of the nadV-nadM gene cluster in Synechocystis sp. strain PCC 6803, the slr0788 gene was cloned and overexpressed in E. coli. The high overall expression level and the favorable distribution between the soluble (>80%) and insoluble forms allowed us to obtain a highly enriched sample by a one-step purification process using a Ni-NTA minicolumn (∼90% major band on SDS-PAGE with an expected molecular mass of ∼55 kDa). Coupled kinetic assays were developed and used to evaluate enzymatic properties of the purified recombinant protein. In agreement with our expectations, this enzyme displayed a robust NMPRT activity, utilizing nicotinamide at least 100-fold more efficiently than nicotinate, at a 1 mM concentration of either substrate (Table 3). This substrate preference appears to be largely manifested at the level of binding affinity, as reflected in the difference of apparent Km values, ∼0.08 mM for nicotinamide compared to ∼30 mM for nicotinate, at a saturating (1 mM) concentration of PRPP cosubstrate. These values represent only a rough estimate of both kinetic parameters. In the case of a specific substrate, NAM, an apparent saturation at ∼0.5 mM was followed by a pronounced substrate inhibition at NAM concentrations above 2 mM. This is indicative of a rather complex kinetic behavior, which was not further investigated in this study. In contrast to that, saturation was barely detectable within a feasible range of nicotinic acid concentrations (up to 100 mM). Notwithstanding the technical difficulties, these experimental data directly support both the suggested annotation and the two-step pathway (NMN shunt) inferred for Synechocystis sp. strain PCC 6803. As discussed above, this pathway in Synechocystis sp. strain PCC 6803, S. elongatus, and possibly several other divergent species containing the same gene cluster (Table 2) is likely involved in the recycling of nicotinamide generated by the glycohydrolytic cleavage of endogenous NAD.
DISCUSSION
Cyanobacteria uniquely combine oxygenic photosynthesis and aerobic respiration in one membrane system (49). The redox state of the interconnected NAD and quinone pools is believed to be the key regulatory mechanism for these complex bioenergetics as well as numerous other metabolic processes in the cyanobacterial cell (26, 70, 72). In a number of organisms, the NAD pool was shown to be a subject of tight homeostasis and rapid turnover, which includes a variety of catabolic and recycling pathways. For example, NAD consumption by ADP-ribosylating enzymes (detected in Synechocystis sp. strain PCC 6803 [66]) by NAD-dependent DNA ligase and other NAD-hydrolyzing enzymes has to be counterbalanced by its resynthesis and/or recycling. Despite significant progress in the knowledge of NAD biosynthetic machinery in a number of model organisms, relatively little information was available for cyanobacteria. We applied a comparative genome analysis supported by subsystem annotation tools available in the SEED genomic platform (43) for a detailed reconstruction of NAD biosynthetic, salvage, and recycling pathways in all cyanobacterial species for which complete or nearly complete genome sequence data are currently available in public databases. This analysis led to a number of conjectures, some of which were experimentally validated in the model system of Synechocystis sp. strain PCC 6803.
All analyzed cyanobacteria invariably include two aspects of NAD(P) biosynthesis: (i) the de novo biosynthetic pathway (from aspartate to NaMN), feeding into (ii) the universal pathway (from NaMN to NADP) (Fig. 1 and Table 1), which is consistent with the autotrophic lifestyle of these organisms. On the other hand, the routes for salvage of exogenous precursors and/or recycling of intracellular products of NAD degradation vary significantly between representatives of this group.
Two out of three invariable enzymes in the universal pathway represented by products of Synechocystis sp. strain PCC 6803 genes sll1916 and slr1691, tentatively assigned as (i) NaMNAT and (ii) GAT/NADS, were selected for biochemical characterization. The choice of these enzymes was dictated by their essential nature and by a certain level of ambiguity associated with their assignment (as discussed above). Although the product of Synechocystis sp. strain PCC 6803 gene slr0787 was historically the first characterized adenylyltransferase related to NAD metabolism in bacteria, our biochemical and genetic data confirmed that an alternative NaMNAT (gene sll1916) is required and sufficient for NAD biosynthesis. Enzymatic characterization of the pure recombinant protein encoded by the Synechocystis sp. strain PCC 6803 gene slr1691 confirmed that, unlike many well-characterized bacterial single-domain NADS, this enzyme is capable of utilizing glutamine in vitro. Our data, along with those previously reported for the M. tuberculosis enzyme (4), provide sufficient experimental evidence to project the proposed GAT/NADS functional assignment to a large number of bacterial NadE homologs containing a nitrilase-like N-terminal domain.
Although only half of the analyzed cyanobacteria contain recognizable salvage genes, all three known recycling/salvage routes are represented within the rest of the group (Table 1), reflecting their adaptation to distinct ecological niches rather than the accepted phylogeny. For example, while the majority of filamentous cyanobacteria harbor a single (and the most common) niacin utilization pathway (via NAPRT to NaMN), a second pathway, nicotinamide ribose salvage, appears to be present in Nostoc punctiforme (Table 1). This observation is consistent with an expected availability of various NAD precursors in a unique ecological niche of N. punctiforme, which exists for most of its life cycle as a microsymbiont of fungal endocyanosis Geosiphon pyriforme (36).
The existence of an alternative, nondeamidating pathway of nicotinamide utilization was inferred in Synechocystis sp. strain PCC 6803 and Synechococcus elongatus based on the presence of a conserved genomic cluster composed of two genes encoding enzymes for the two-step transformation of nicotinamide to NAD via NMN (Fig. 1). Our experimental data provided additional verification of this assertion: (i) enzymatic activity and substrate specificity of the recombinant NMPRT encoded by the slr0788 gene are consistent with its proposed role in the pathway, and (ii) previously established NMNAT activity of the second gene, slr0787 (50), along with our finding that this gene is dispensable and incapable of compensating for inactivation of the NaMNAT gene sll1916, is in agreement with its proposed role of catalyzing the second step in this pathway. Finally, the inability of Synechocystis sp. strain PCC 6803 to uptake and utilize exogenous niacin points to the likely role of this pathway in the recycling of endogenous nicotinamide. This interpretation provides a rationale for the existence of the previously characterized ADPRP domain in the slr0787 gene product. This domain may be involved in the utilization of ADP-ribose, a by-product generated along with nicotinamide during the glycohydrolytic cleavage of NAD.
The nondeamidating pathway of nicotinamide utilization (NMN shunt) appears to play an important role in mammalian cells (60), but it is rare in bacteria and seems to be absent in archaea. While the first enzyme of the pathway, NadV-like NMPRT, is highly conserved between bacteria and eukaryotes, the second enzyme, NMNAT, is represented by at least three divergent families (32): (i) an NMNAT/NaMNAT family with dual substrate specificity as in humans (18), (ii) a NadR-like NMNAT/RNK family as in Pasteurellaceae (37, 51), and (iii) a NadM-like NMNAT/ADPRP family as in Synechocystis sp. strain PCC 6803 (50). An observed tight functional coupling (positional clustering and cooccurrence profile of NMNAT/ADPRP- and NMPRT-encoding genes) as well as a peculiar phylogenetic distribution of this gene cluster (Table 2 and Fig. 2B) may reflect relatively recent horizontal transfer events.
In summary, the detailed subsystem analysis allowed us to accurately annotate respective genes and elucidate both conserved (de novo and universal pathways) and variable (recycling/salvage pathways) aspects of NAD(P) metabolism in the entire set of 13 cyanobacterial species with completely sequenced genomes. Key conjectures about important aspects of this subsystem were validated by focused genetic and biochemical experiments in the model system of Synechocystis sp. strain PCC 6803. Of special interest is the experimentally supported assertion of the nicotinamide recycling pathway implemented in Synechocystis sp. strain PCC 6803. Positional clustering and a cooccurrence profile of the respective genes across the diverse collection of cellular organisms provide evidence for a possible role of horizontal transfer in the evolutionary history of this interesting pathway.
Acknowledgments
We are grateful to all members of the SEED development team for their invaluable help with bioinformatics aspects of this study and to Rob Edwards for productive discussions.
This work was supported by U.S. Department of Energy grants DE-FG02-02ER15349 to Integrated Genomics, Inc., and DE-FG02-04ER15499 to the Fellowship for Interpretation of Genomes. This work was also partially supported by National Institute of Allergy and Infectious Diseases grant 1-R01-AI059146-01A2 to A.L.O.
REFERENCES
- 1.Aravind, L., V. Anantharaman, and E. V. Koonin. 2002. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA. Proteins 48:1-14. [DOI] [PubMed] [Google Scholar]
- 2.Balducci, E., M. Emanuelli, N. Raffaelli, S. Ruggieri, A. Amici, G. Magni, G. Orsomando, V. Polzonetti, and P. Natalini. 1995. Assay methods for nicotinamide mononucleotide adenylyltransferase of wide applicability. Anal. Biochem. 228:64-68. [DOI] [PubMed] [Google Scholar]
- 3.Begley, T. P., C. Kinsland, R. A. Mehl, A. Osterman, and P. Dorrestein. 2001. The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam. Horm. 61:103-119. [DOI] [PubMed] [Google Scholar]
- 4.Bellinzoni, M., S. Buroni, M. R. Pasca, P. Guglierame, F. Arcesi, E. De Rossi, and G. Riccardi. 2005. Glutamine amidotransferase activity of NAD+ synthetase from Mycobacterium tuberculosis depends on an amino-terminal nitrilase domain. Res. Microbiol. 156:173-177. [DOI] [PubMed] [Google Scholar]
- 5.Bellinzoni, M., E. De Rossi, M. Branzoni, A. Milano, F. A. Peverali, M. Rizzi, and G. Riccardi. 2002. Heterologous expression, purification, and enzymatic activity of Mycobacterium tuberculosis NAD(+) synthetase. Protein Expr. Purif. 25:547-557. [DOI] [PubMed] [Google Scholar]
- 6.Berman-Frank, I., P. Lundgren, and P. Falkowski. 2003. Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res. Microbiol. 154:157-164. [DOI] [PubMed] [Google Scholar]
- 7.Bieganowski, P., H. C. Pace, and C. Brenner. 2003. Eukaryotic NAD+ synthetase Qns1 contains an essential, obligate intramolecular thiol glutamine amidotransferase domain related to nitrilase. J. Biol. Chem. 278:33049-33055. [DOI] [PubMed] [Google Scholar]
- 8.Binder, B. J., and S. W. Chisholm. 1990. Relationship between DNA cycle and growth rate in Synechococcus sp. strain PCC 6301. J. Bacteriol. 172:2313-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 10.Bono, H., H. Ogata, S. Goto, and M. Kanehisa. 1998. Reconstruction of amino acid biosynthesis pathways from the complete genome sequence. Genome Res. 8:203-210. [DOI] [PubMed] [Google Scholar]
- 11.Burja, A. M., B. Banaigs, E. Abou-Mansour, J. G. Burgess, and P. C. Wright. 2001. Marine cyanobacteria—a prolific source of natural products. Tetrahedron 57:9347-9377. [Google Scholar]
- 12.Burja, A. M., S. Dhamwichukorn, and P. C. Wright. 2003. Cyanobacterial postgenomic research and systems biology. Trends Biotechnol. 21:504-511. [DOI] [PubMed] [Google Scholar]
- 13.Cai, Y. P., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canfield, D. E. 1999. Palaeoecology—a breath of fresh air. Nature 400:503. [Google Scholar]
- 15.Daugherty, M., V. Vonstein, R. Overbeek, and A. Osterman. 2001. Archaeal shikimate kinase, a new member of the GHMP-kinase family. J. Bacteriol. 183:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denu, J. M. 2003. Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem. Sci. 28:41-48. [DOI] [PubMed] [Google Scholar]
- 17.Dufresne, A., M. Salanoubat, F. Partensky, F. Artiguenave, I. M. Axmann, V. Barbe, S. Duprat, M. Y. Galperin, E. V. Koonin, F. Le Gall, K. S. Makarova, M. Ostrowski, S. Oztas, C. Robert, I. B. Rogozin, D. J. Scanlan, N. T. de Marsac, J. Weissenbach, P. Wincker, Y. I. Wolf, and W. R. Hess. 2003. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. USA 100:10020-10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emanuelli, M., F. Carnevali, F. Saccucci, F. Pierella, A. Amici, N. Raffaelli, and G. Magni. 2000. Molecular cloning, chromosomal localization, tissue mRNA levels, bacterial expression, and enzymatic properties of human NMN adenylyltransferase. J. Biol. Chem. 276:406-412. [DOI] [PubMed] [Google Scholar]
- 19.Ermakova-Gerdes, S., and W. Vermaas. 1999. Inactivation of the open reading frame slr0399 in Synechocystis sp. PCC 6803 functionally complements mutations near the QA niche of photosystem II: a possible role of Slr0399 as a chaperone for quinone binding. J. Biol. Chem. 274:30540-30549. [DOI] [PubMed] [Google Scholar]
- 20.Field, C. B., M. J. Behrenfeld, J. T. Randerson, and P. Falkowski. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281:237-240. [DOI] [PubMed] [Google Scholar]
- 21.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. Fitzhugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L. I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole genome random sequencing and assembly of Haemophilus Influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 22.Fulda, S., B. Norling, A. Schoor, and M. Hagemann. 2002. The Slr0924 protein of Synechocystis sp. strain PCC 6803 resembles a subunit of the chloroplast protein import complex and is mainly localized in the thylakoid lumen. Plant Mol. Biol. 49:107-118. [DOI] [PubMed] [Google Scholar]
- 23.Gerdes, S. Y., M. D. Scholle, M. D'Souza, A. Bernal, M. V. Baev, M. Farrell, O. V. Kurnasov, M. D. Daugherty, F. Mseeh, B. M. Polanuyer, J. W. Campbell, S. Anantha, K. Y. Shatalin, S. A. Chowdhury, M. Y. Fonstein, and A. L. Osterman. 2002. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 184:4555-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haft, D. H., J. D. Selengut, L. M. Brinkac, N. Zafar, and O. White. 2005. Genome Properties: a system for the investigation of prokaryotic genetic content for microbiology, genome annotation and comparative genomics. Bioinformatics 21:293-306. [DOI] [PubMed] [Google Scholar]
- 25.Han, S., and J. A. Tainer. 2002. The ARTT motif and a unified structural understanding of substrate recognition in ADP-ribosylating bacterial toxins and eukaryotic ADP-ribosyltransferases. Int. J. Med. Microbiol. 291:523-529. [DOI] [PubMed] [Google Scholar]
- 26.Howitt, C. A., P. K. Udall, and W. F. Vermaas. 1999. Type 2 NADH dehydrogenases in the cyanobacterium Synechocystis sp. strain PCC 6803 are involved in regulation rather than respiration. J. Bacteriol. 181:3994-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 29.Kurnasov, O. V., B. M. Polanuyer, S. Ananta, R. Sloutsky, A. Tam, S. Y. Gerdes, and A. L. Osterman. 2002. Ribosylnicotinamide kinase domain of NadR protein: identification and implications in NAD biosynthesis. J. Bacteriol. 184:6906-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeder, D. L., R. B. Weiss, D. M. Dunn, J. L. Cherry, J. M. Gonzalez, J. DiRuggiero, and F. T. Robb. 1999. Divergence of the hyperthermophilic archaea Pyrococcus furiosus and P. horikoshii inferred from complete genomic sequences. Genetics 152:1299-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magni, G., A. Amici, M. Emanuelli, G. Orsomando, N. Raffaelli, and S. Ruggieri. 2004. Enzymology of NAD+ homeostasis in man. Cell. Mol. Life Sci. 61:19-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magni, G., A. Amici, M. Emanuelli, G. Orsomando, N. Raffaelli, and S. Ruggieri. 2004. Structure and function of nicotinamide mononucleotide adenylyltransferase. Curr. Med. Chem. 11:873-885. [DOI] [PubMed] [Google Scholar]
- 33.Magni, G., A. Amici, M. Emanuelli, N. Raffaelli, and S. Ruggieri. 1999. Enzymology of NAD+ synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 73:135-182. [DOI] [PubMed] [Google Scholar]
- 34.Martin, P. R., R. J. Shea, and M. H. Mulks. 2001. Identification of a plasmid-encoded gene from Haemophilus ducreyi, which confers NAD independence. J. Bacteriol. 183:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moat, A. G., and J. W. Foster. 1987. Biosynthesis and salvage pathways of pyridine nucleotides, p. 1-20. In P. R. Dolphin and O. Avramovich (ed.), Pyridine nucleotide coenzymes—part B. John Wiley & Sons, New York, N.Y.
- 36.Mollenhauer, D., and M. Kluge. 1994. Geosiphon pyriforme. Endocytobiosis Cell Res. 10:29-34. [Google Scholar]
- 37.Mushegian, A. 1999. The purloined letter: bacterial orthologs of archaeal NMN adenylyltransferase are domains within multifunctional transcription regulator NadR. J. Mol. Microbiol. Biotechnol. 1:127-128. [PubMed] [Google Scholar]
- 38.Nakamura, Y., T. Kaneko, S. Sato, M. Ikeuchi, H. Katoh, S. Sasamoto, A. Watanabe, M. Iriguchi, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2002. Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res. 9:123-130. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura, Y., T. Kaneko, S. Sato, M. Mimuro, H. Miyashita, T. Tsuchiya, S. Sasamoto, A. Watanabe, K. Kawashima, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, N. Nakazaki, S. Shimpo, C. Takeuchi, M. Yamada, and S. Tabata. 2003. Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res. 10:137-145. [DOI] [PubMed] [Google Scholar]
- 40.Osiewacz, H. D. 1992. Construction of insertion mutants of Synechocystis sp. PCC 6803: evidence for an essential function of subunit IV of the cytochrome b6/f complex. Arch. Microbiol. 157:336-342. [DOI] [PubMed] [Google Scholar]
- 41.Osterman, A., N. V. Grishin, L. N. Kinch, and M. A. Phillips. 1994. Formation of functional cross-species heterodimers of ornithine decarboxylase. Biochemistry 33:13662-13667. [DOI] [PubMed] [Google Scholar]
- 42.Osterman, A. L., D. V. Lueder, M. Quick, D. Myers, B. J. Canagarajah, and M. A. Phillips. 1995. Domain organization and a protease-sensitive loop in eukaryotic ornithine decarboxylase. Biochemistry 34:13431-13436. [DOI] [PubMed] [Google Scholar]
- 43.Overbeek, R., T. Begley, R. M. Butler, J. V. Choudhuri, H.-Y. Chuang, M. Cohoon, V. de Crécy-Lagard, N. Diaz, T. Disz, R. Edwards, M. Fonstein, E. Frank, S. Gerdes, E. Glass, A. Goesmann, A. Hanson, D. Iwata-Reuyl, R. Jensen, N. Jamshidi, L. Krause, M. Kubal, N. Larsen, B. Linke, A. C. McHardy, F. Meyer, H. Neuweger, G. Olsen, R. Olson, A. Osterman, V. Portnoy, G. D. Pusch, D. A. Rodionov, C. Ruckert, J. Steiner, R. Stevens, I. Thiele, O. Vassieva, Y. Ye, O. Zagnitko, and V. Vonstein. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Overbeek, R., M. Fonstein, M. D'Souza, G. D. Pusch, and N. Maltsev. 1999. Use of contiguity on the chromosome to predict functional coupling. In Silico Biol. 1:93-108. [PubMed] [Google Scholar]
- 45.Overbeek, R., N. Larsen, T. Walunas, M. D'Souza, G. Pusch, E. Selkov, Jr., K. Liolios, V. Joukov, D. Kaznadzey, I. Anderson, A. Bhattacharyya, H. Burd, W. Gardner, P. Hanke, V. Kapatral, N. Mikhailova, O. Vasieva, A. Osterman, V. Vonstein, M. Fonstein, N. Ivanova, and N. Kyrpides. 2003. The ERGO genome analysis and discovery system. Nucleic Acids Res. 31:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ozment, C., J. Barchue, L. J. DeLucas, and D. Chattopadhyay. 1999. Structural study of Escherichia coli NAD synthetase: overexpression, purification, crystallization, and preliminary crystallographic analysis. J. Struct. Biol. 127:279-282. [DOI] [PubMed] [Google Scholar]
- 47.Palenik, B., B. Brahamsha, F. W. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb, and J. Waterbury. 2003. The genome of a motile marine Synechococcus. Nature 424:1037-1042. [DOI] [PubMed] [Google Scholar]
- 48.Penfound, T., and J. W. Foster. 1996. Biosynthesis and recycling of NAD, p. 721-730. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 49.Peschek, G. A. 1996. Structure-function relationships in the dual-function photosynthetic-respiratory electron-transport assembly of cyanobacteria (blue-green algae). Biochem. Soc. Trans. 24:729-733. [DOI] [PubMed] [Google Scholar]
- 50.Raffaelli, N., T. Lorenzi, A. Amici, M. Emanuelli, S. Ruggieri, and G. Magni. 1999. Synechocystis sp. Slr0787 protein is a novel bifunctional enzyme endowed with both nicotinamide mononucleotide adenylyltransferase and ‘Nudix’ hydrolase activities. FEBS Lett. 444:222-226. [DOI] [PubMed] [Google Scholar]
- 51.Raffaelli, N., T. Lorenzi, P. L. Mariani, M. Emanuelli, A. Amici, S. Ruggieri, and G. Magni. 1999. The Escherichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J. Bacteriol. 181:5509-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raffaelli, N., F. M. Pisani, T. Lorenzi, M. Emanuelli, A. Amici, S. Ruggieri, and G. Magni. 1997. Characterization of nicotinamide mononucleotide adenylyltransferase from thermophilic archaea. J. Bacteriol. 179:7718-7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rai, A. N., E. Soderback, and B. Bergman. 2000. Cyanobacterium-plant symbioses. New Phytologist 147:449-481. [DOI] [PubMed] [Google Scholar]
- 54.Raymond, J., O. Zhaxybayeva, J. P. Gogarten, S. Y. Gerdes, and R. E. Blankenship. 2002. Whole-genome analysis of photosynthetic prokaryotes. Science 298:1616-1620. [DOI] [PubMed] [Google Scholar]
- 55.Revollo, J. R., A. A. Grimm, and S. Imai. 2004. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 279:50754-50763. [DOI] [PubMed] [Google Scholar]
- 56.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 57.Rizzi, M., C. Nessi, A. Mattevi, A. Coda, M. Bolognesi, and A. Galizzi. 1996. Crystal structure of NH3-dependent NAD+ synthetase from Bacillus subtilis. EMBO J. 15:5125-5134. [PMC free article] [PubMed] [Google Scholar]
- 58.Robb, F. T., D. L. Maeder, J. R. Brown, J. DiRuggiero, M. D. Stump, R. K. Yeh, R. B. Weiss, and D. M. Dunn. 2001. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 330:134-157. [DOI] [PubMed] [Google Scholar]
- 59.Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042-1047. [DOI] [PubMed] [Google Scholar]
- 60.Rongvaux, A., F. Andris, F. Van Gool, and O. Leo. 2003. Reconstructing eukaryotic NAD metabolism. Bioessays 25:683-690. [DOI] [PubMed] [Google Scholar]
- 61.Rongvaux, A., R. J. Shea, M. H. Mulks, D. Gigot, J. Urbain, O. Leo, and F. Andris. 2002. Pre-B-cell colony-enhancing factor, whose expression is upregulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur. J. Immunol. 32:3225-3234. [DOI] [PubMed] [Google Scholar]
- 62.Samal, B., Y. Sun, G. Stearns, C. Xie, S. Suggs, and I. McNiece. 1994. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol. Cell. Biol. 14:1431-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 64.Schopf, J. W. 2000. The fossil record: tracing the roots of the cyanobacterial lineage, p. 13-35. In B. A. Whitton and M. Potts (ed.), Ecology of cyanobacteria: their diversity in time and space. Kluwer Academic Publishers, New York, N.Y.
- 65.Selkov, E., N. Maltsev, G. J. Olsen, R. Overbeek, and W. B. Whitman. 1997. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene 197:GC11-GC26. [DOI] [PubMed] [Google Scholar]
- 66.Silman, N. J., N. G. Carr, and N. H. Mann. 1995. ADP-ribosylation of glutamine synthetase in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 177:3527-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh, S. K., O. V. Kurnasov, B. Chen, H. Robinson, N. V. Grishin, A. L. Osterman, and H. Zhang. 2002. Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities. J. Biol. Chem. 277:33291-33299. [DOI] [PubMed] [Google Scholar]
- 68.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Starai, V. J., H. Takahashi, J. D. Boeke, and J. C. Escalante-Semerena. 2004. A link between transcription and intermediary metabolism: a role for Sir2 in the control of acetyl-coenzyme A synthetase. Curr. Opin. Microbiol. 7:115-119. [DOI] [PubMed] [Google Scholar]
- 70.Vener, A. V., I. Ohad, and B. Andersson. 1998. Protein phosphorylation and redox sensing in chloroplast thylakoids. Curr. Opin. Plant Biol. 1:217-223. [DOI] [PubMed] [Google Scholar]
- 71.Vermaas, W. 1996. Molecular genetics of the cyanobacterium Synechocystis sp. PCC 6803: principles and possible biotechnology applications. J. Appl. Phycol. 8:263-273. [Google Scholar]
- 72.Wedel, N., and J. Soll. 1998. Evolutionary conserved light regulation of Calvin cycle activity by NADPH-mediated reversible phosphoribulokinase/CP12/glyceraldehyde-3-phosphate dehydrogenase complex dissociation. Proc. Natl. Acad. Sci. USA 95:9699-9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilkinson, A., J. Day, and R. Bowater. 2001. Bacterial DNA ligases. Mol. Microbiol. 40:1241-1248. [DOI] [PubMed] [Google Scholar]
- 74.Williams, J. G. K. 1988. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 167:766-778. [Google Scholar]
- 75.Willison, J. C., and G. Tissot. 1994. The Escherichia coli efg gene and the Rhodobacter capsulatus adgA gene code for NH3-dependent NAD synthetase. J. Bacteriol. 176:3400-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ye, Y., A. Osterman, R. Overbeek, and A. Godzik. 2005. Automatic detection of subsystem/pathway variants in genome analysis. Bioinformatics 21:i1-i9. [DOI] [PubMed] [Google Scholar]
- 77.Zalkin, H. 1985. NAD synthetase. Methods Enzymol. 113:297-302. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, Q., D. W. Piston, and R. H. Goodman. 2002. Regulation of corepressor function by nuclear NADH. Science 295:1895-1897. [DOI] [PubMed] [Google Scholar]
- 79.Zhang, X., O. V. Kurnasov, S. Karthikeyan, N. V. Grishin, A. L. Osterman, and H. Zhang. 2003. Structural characterization of a human cytosolic NMN/NaMN adenylyltransferase and implication in human NAD biosynthesis. J. Biol. Chem. 278:13503-13511. [DOI] [PubMed] [Google Scholar]
- 80.Zhang, Y. Q., S. X. Ren, H. L. Li, Y. X. Wang, G. Fu, J. Yang, Z. Q. Qin, Y. G. Miao, W. Y. Wang, R. S. Chen, Y. Shen, Z. Chen, Z. H. Yuan, G. P. Zhao, D. Qu, A. Danchin, and Y. M. Wen. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577-1593. [DOI] [PubMed] [Google Scholar]
- 81.Zhou, T., O. Kurnasov, D. R. Tomchick, D. D. Binns, N. V. Grishin, V. E. Marquez, A. L. Osterman, and H. Zhang. 2002. Structure of human nicotinamide/nicotinic acid mononucleotide adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin. J. Biol. Chem. 277:13148-13154. [DOI] [PubMed] [Google Scholar]
- 82.Ziegler, M. 2000. New functions of a long-known molecule: emerging roles of NAD in cellular signaling. Eur. J. Biochem. 267:1550-1564. [DOI] [PubMed] [Google Scholar]