Abstract
Previous studies with Corynebacterium diphtheriae and Mycobacterium species revealed that the transcriptional regulator DtxR and its ortholog IdeR play a central role in the control of iron metabolism. In the present work, we used genome-based approaches to determine the DtxR regulon of Corynebacterium glutamicum, a nonpathogenic relative of C. diphtheriae. First, global gene expression of a dtxR deletion mutant was compared with that of the wild type using DNA microarrays. Second, we used a computer-based approach to identify 117 putative DtxR binding sites in the C. glutamicum genome. In the third step, 74 of the corresponding genome regions were amplified by PCR, 51 of which were shifted by the DtxR protein. Finally, we analyzed which of the genes preceded by a functional DtxR binding site showed altered mRNA levels in the transcriptome comparison. Fifty-one genes organized in 27 putative operons displayed an increased mRNA level in the ΔdtxR mutant and thus are presumably repressed by DtxR. The majority of these genes are obviously involved in iron acquisition, three encode transcriptional regulators, e.g., the recently identified repressor of iron proteins RipA, and the others encode proteins of diverse or unknown functions. Thirteen genes showed a decreased mRNA level in the ΔdtxR mutant and thus might be activated by DtxR. This group included the suf operon, whose products are involved in the formation and repair of iron-sulfur clusters, and several genes for transcriptional regulators. Our results clearly establish DtxR as the master regulator of iron-dependent gene expression in C. glutamicum.
Corynebacterium glutamicum is a nonpathogenic, aerobic, gram-positive soil bacterium used for the large-scale biotechnological production of amino acids, mainly l-glutamate (1.5 million tons/year) and l-lysine (0.7 million tons/year). In addition, this species has gained interest as a model organism for the Corynebacterineae, a suborder of the Actinomycetales, which also includes the genus Mycobacterium (34). An overview on the current knowledge on C. glutamicum can be found in a recent monograph (9).
Our group has initiated studies on the regulation of C. glutamicum genes and enzymes involved in the citric acid cycle, which is of central importance for metabolism in general and for amino acid production in particular because it provides the precursors of the aspartate and glutamate family of amino acids. We identified and characterized AcnR, a member of the TetR family of transcriptional regulators, which functions as a repressor of the aconitase gene acn (24). In the course of these studies, it became evident that acn expression is controlled by iron in an AcnR-independent manner, being reduced under iron limitation. Subsequently, we were able to show that the iron-dependent transcriptional regulation of aconitase is exerted by the AraC-type regulator RipA, which represses aconitase under iron limitation but not under iron excess (40). RipA stands for “regulator of iron proteins A,” and this name was given because RipA represses not only aconitase but also succinate dehydrogenase (sdhCAB operon), isopropylmalate dehydratase (leuCD operon), nitrate reductase (narKGHJI operon), catechol 1,2-dioxygenase (catA), catalase (katA), and phosphotransacetylase and acetate kinase (pta-ackA operon). Except for the latter two enzymes and the nitrate/nitrite transporter NarK, all other enzymes contain iron, and RipA thus functions to reduce the cellular iron demand under iron limitation by reducing the synthesis of prominent iron proteins.
The observation that RipA functions only under iron limitation is due to the fact that expression of ripA itself is repressed under iron excess (24). It thus behaves like typical iron starvation genes. In many high-GC gram-positive species, e.g., Corynebacterium diphtheriae, Mycobacterium smegmatis, Mycobacterium tuberculosis, or Streptomyces coelicolor, proteins of the DtxR family function as global iron regulators. In complex with iron, these proteins are active as transcriptional regulators, usually as repressors of genes coping with iron starvation, e.g., genes encoding high-affinity iron uptake systems or siderophore biosynthesis enzymes. In some cases, DtxR proteins can also function as transcriptional activators, as shown for the bacterioferritin gene bfrA of M. tuberculosis (14). When not complexed with iron, the DtxR proteins appear to be inactive. Since the DNA-binding site of DtxR and its homologs is known (14, 36), we were able to identify a well-conserved DtxR operator upstream of the C. glutamicum ripA gene and we could show that the DtxR protein from C. glutamicum (encoded by NCgl1845) binds in vitro to the ripA promoter/operator region (40). Thus, we proposed that ripA expression is controlled by DtxR, being repressed under iron excess.
In this work, we performed a genome-wide search for the DtxR target genes in C. glutamicum. To this end, we constructed a dtxR deletion mutant and compared its global gene expression with that of the parent wild type using DNA microarrays. In parallel, we searched the genome for putative DtxR-binding sites and tested these in vitro using band shift assays with purified DtxR. By combining the results of these two approaches, we were able to identify 27 operons with 51 genes that are presumably repressed by DtxR and 7 operons with 13 genes that might be activated by DtxR.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All strains and plasmids used in this work are listed in Table 1. The C. glutamicum type strain ATCC 13032 (22) was used as the wild type. The ΔdtxR strain is a derivative containing an in-frame deletion within the dtxR gene. For growth experiments, 5 ml of brain heart infusion medium (Difco Laboratories, Detroit, MI) was inoculated with colonies from a fresh LB agar plate (31) and incubated for 6 h at 30°C and at 170 rpm. After washing with 5 ml 0.9% NaCl, the cells of this first preculture were used to inoculate a 500-ml shake flask containing 50 ml CGXII minimal medium (21) with 4% (wt/vol) glucose and either 1 μM FeSO4 (iron starvation) or 100 μM FeSO4 (iron excess). This second preculture was cultivated overnight at 30°C and then used to inoculate the main culture to an optical density at 600 nm of ∼1. The main culture contained the same iron concentration as the second preculture. The trace element solution with iron salts omitted and the FeSO4 solution were always added after autoclaving. For all cloning purposes, Escherichia coli DH5α (Invitrogen) was used as the host, for overproduction of DtxR E. coli BL21(DE3) (35). The E. coli strains were cultivated aerobically in LB medium at 37°C (DH5α) or at 30°C [BL21(DE3)]. When appropriate, kanamycin was added to a concentration of 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| C. glutamicum | ||
| ATCC 13032 | Biotin-auxotrophic wild type | 22 |
| 13032ΔdtxR | In-frame deletion of the dtxR gene | This work |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| BL21(DE3) | ompT hsdSB(rB−mB−) gal dcm (DE3) | 35 |
| Plasmids | ||
| pK19mobsacB | Kanr; vector for allelic exchange in C. glutamicum; (pK18 oriVE.c.sacB lacZα) | 32 |
| pK19mobsacB-ΔdtxR | Kmr; pK19mobsacB derivative containing a crossover PCR product covering the up- and downstream regions of dtxR | This work |
| pET24b | Kanr; vector for overexpression of genes in E. coli, adding a C-terminal hexahistidine affinity tag to the synthesized protein (pBR322 oriVE.c.PT7lacI) | Novagen |
| pET24b-dtxR-C | Kanr; pET24b derivative for overproduction of DtxR with a C-terminal decahistidine tag—the four additional histidines were attached to the dtxR fragment | 40 |
Recombinant DNA work.
The enzymes for recombinant DNA work were obtained from Roche Diagnostics (Mannheim, Germany) or New England Biolabs (Frankfurt, Germany). The oligonucleotides used in this study were obtained from Operon (Cologne, Germany) and are listed in Table S1 in the supplemental material. Routine methods like PCR, restriction, or ligation were carried out according to standard protocols (31). Chromosomal DNA from C. glutamicum was prepared as described previously (10). Plasmids from E. coli were isolated with the QIAprep spin miniprep kit (QIAGEN, Hilden, Germany). E. coli was transformed by the RbCl method (15), C. glutamicum by electroporation (37). DNA sequencing was performed with a 3100-Avant genetic analyzer (Applied Biosystems, Darmstadt, Germany). Sequencing reactions were carried out with the Thermo Sequenase primer cycle sequencing kit (Amersham Biosciences, Freiburg, Germany).
An in-frame dtxR deletion mutant of C. glutamicum was constructed via a two-step homologous recombination procedure as described previously (27). The dtxR up- and downstream regions (approximately 450 bp each) were amplified using the oligonucleotide pairs Delta-dtxR-1 with Delta-dtxR-2 and Delta-dtxR-3 with Delta-dtxR-4, respectively, and the products served as a template for crossover PCR with oligonucleotides Delta-dtxR-1 and Delta-dtxR-4. The resulting PCR product of ∼0.9 kb was digested with EcoRI and HindIII and cloned into pK19mobsacB (32). DNA sequence analysis confirmed that the cloned PCR product did not contain spurious mutations. Transfer of the resulting plasmid, pK19mobsacB-ΔdtxR, into C. glutamicum and selection for the first and second recombination event were performed as described previously (27). Kanamycin-sensitive and saccharose-resistant clones were tested by PCR analysis of chromosomal DNA with the primer pair Delta-dtxR-for and Delta-dtxR-rev (see Table S1 in the supplemental material). Of 20 clones tested, 11 showed the wild-type situation (1.6-kb fragment) and 9 had the desired in-frame deletion of the dtxR gene (0.9-kb fragment), in which all nucleotides except for the first 6 codons and the last 12 codons were replaced by a 21-bp tag.
Preparation of total RNA.
Cultures of the wild type and the ΔdtxR mutant were grown in CGXII minimal medium containing 4% (wt/vol) glucose under iron limitation (1 μM FeSO4) or iron excess (100 μM FeSO4). In the exponential growth phase at an optical density at 600 nm of 4 to 6, 25 ml of the cultures was used for the preparation of total RNA as described previously (26). Isolated RNA samples were analyzed for quantity and quality by UV spectrophotometry and denaturing formaldehyde agarose gel electrophoresis (31), respectively, and stored at −70°C until use.
DNA microarray analyses.
The generation of whole-genome DNA microarrays (39), synthesis of fluorescently labeled cDNA from total RNA, microarray hybridization, washing, and data analysis were performed as described previously (18, 25, 29). Genes that exhibited significantly changed mRNA levels (P < 0.05 in Student's t test) by at least a factor of 2 were determined in two series of DNA microarray experiments: (i) four comparisons of the wild type and the ΔdtxR mutant cultivated in CGXII minimal medium with 4% (wt/vol) glucose under iron excess (100 μM FeSO4); (ii) three comparisons of the wild type and the ΔdtxR mutant cultivated in CGXII-glucose medium under iron limitation (1 μM FeSO4).
Overproduction and purification of DtxR.
The C. glutamicum DtxR protein containing 12 additional amino acid at the carboxyl terminus (HHHHLEHHHHHH) was overproduced in E. coli BL21(DE3) using the expression plasmid pET24b-dtxR and purified by Ni2+-chelate affinity chromatography as described previously (40).
Gel shift assays.
For testing the binding of DtxR to putative target promoters, purified DtxR protein (0 to 4 μM dimeric form) was mixed with DNA fragments (200 to 450 bp; final concentration, 8 to 20 nM) in a total volume of 20 μl. The binding buffer contained 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 40 mM KCl, 5% (vol/vol) glycerol, 1 mM dithiothreitol (DTT), 150 μM MnCl2. Approximately 20 nM of a promoter fragment lacking a DtxR binding site (acn, pta, katA, or porB) was used as negative control. The reaction mixture was incubated at room temperature for 30 min and then loaded onto a 15% native polyacrylamide gel containing 1 mM DTT and 150 μM MnCl2. Electrophoresis was performed at room temperature and 170 V using 1× TB (89 mM Tris base, 89 mM boric acid) supplemented with 1 mM DTT and 150 μM MnCl2 as an electrophoresis buffer. The gels were subsequently stained with Sybr Green I according to the instructions of the supplier (Sigma-Aldrich, Taufkirchen, Germany) and photographed. All PCR products used in the gel shift assays were purified with the PCR purification kit (QIAGEN, Hilden, Germany) and eluted in EB buffer (10 mM Tris-HCl, pH 8.5).
RESULTS
Construction and growth properties of a C. glutamicum dtxR deletion mutant.
The genome of the C. glutamicum type strain ATCC 13032 (20) contains a gene (NCgl1845) encoding a protein of 228 amino acid residues (25.484 kDa) with 72% sequence identity to C. diphtheriae DtxR. The corresponding protein from C. glutamicum strain ATCC 13869 (previously designated “Brevibacterium lactofermentum”), which differs in only two positions from the ATCC 13032 homolog, was shown to repress the tox promoter from C. diphtheriae in an iron-dependent manner (28). Therefore, the DtxR protein from C. glutamicum might have the same regulatory function as its ortholog from C. diphtheriae, i.e., mainly transcriptional control of genes involved in iron metabolism.
In order to identify the target genes of DtxR in C. glutamicum, an in-frame dtxR deletion mutant was constructed by two-step homologous recombination and verified by PCR analysis (see Materials and Methods). In order to exclude the possibility that large genomic alterations (duplication or deletion) had occurred in the course of the deletion of the dtxR gene, the chromosomal DNA of the wild type and the ΔdtxR mutant was compared with DNA microarrays. The only significant difference observed was the loss of the dtxR gene in the mutant DNA.
In a first set of experiments, the growth behavior of the mutant was compared with that of the wild type (Fig. 1). Under iron limitation (1 μM FeSO4), the ΔdtxR mutant showed a slightly decreased growth rate (μ = 0.32 ± 0.02 h−1) compared to the wild type (μ = 0.35 ± 0.03 h−1) but reached a higher final cell density (10.6 ± 1.4 g dry weight/liter versus 8.1 ± 0.2 g dry weight/liter). Under iron excess (100 μM FeSO4), the ΔdtxR mutant grew significantly slower (μ = 0.27 ± 0.03 h−1) than the wild type (μ = 0.41 ± 0.01 h−1) and reached a final cell density (14.6 ± 0.8 g dry weight/liter) similar to that of the wild type (15.6 ± 2.0 g dry weight/liter). Thus, deletion of the dtxR gene in C. glutamicum has a negative influence on the growth rate under iron excess and a positive influence on the growth yield under iron limitation.
FIG. 1.
Growth of C. glutamicum wild type (squares) and the ΔdtxR mutant (triangles). The cells were cultivated in CGXII minimal medium with 4% (wt/vol) glucose and either 100 μM FeSO4 (filled symbols) or 1 μM FeSO4 (unfilled symbols).
Comparison of the expression profiles of ΔdtxR mutant and wild type with DNA microarrays.
To investigate the effect of the dtxR deletion on global gene expression and to identify target genes of DtxR, the transcriptomes of the ΔdtxR mutant and the C. glutamicum wild type were compared using DNA microarray analysis (39). Two series of experiments were performed: (i) four comparisons of the wild type and the ΔdtxR mutant cultivated in glucose minimal medium under iron excess (100 μM FeSO4); (ii) three comparisons of the wild type and the ΔdtxR mutant cultivated in glucose minimal medium under iron limitation (1 μM FeSO4). RNA was always isolated from cells in the exponential growth phase. Table S2 in the supplemental material shows all genes whose mRNA level was changed ≥2-fold under iron excess or under iron limitation in at least two independent experiments. Under the chosen conditions, 164 genes fell under these criteria, of which 118 showed a higher mRNA level and 46 a lower mRNA level in the ΔdtxR mutant. Seventy-three of the genes encode hypothetical proteins.
Remarkably, more than 50 genes within the NCgl numbers 1611 and 1816 showed more than twofold-increased mRNA levels in the ΔdtxR mutant under iron excess and to a lesser extent also under iron limitation. This region, which spans about 187 kb, was identified as the prophage region CGP3 (19). Its G+C content is significantly lower than the overall G+C content of the C. glutamicum genome (53.8%), the insertion site is a tRNA-Val gene indicated by the presence of a direct repeat flanking the CGP3 element, and it contains a phage-type integrase gene at the left border, which appears to be disrupted by a frameshift mutation. With few exceptions, e.g., NCgl1646, encoding a secretory serine protease, and NCgl1703 to NCgl1705, encoding a type II restriction-modification system, the genes within CGP3 encode hypothetical proteins.
Table 2 lists a subset of the genes with ≥2-fold-altered mRNA level in the ΔdtxR mutant, i.e., those presumably involved in iron acquistion, storage, or metabolism (group I), those known to be RipA targets (group II), those encoding transcriptional regulators (group III), and those further genes showing a ≥10-fold-increased mRNA level in the ΔdtxR mutant (group IV).
TABLE 2.
Selection of genes whose average mRNA ratio was altered ≥2-fold (P value, ≤0.05) under iron excess or under iron limitation in at least two independent experimentsa
| Category and NCgl gene no. | Description of product and gene designation | mRNA ratio
|
|
|---|---|---|---|
| Fe excess | Fe limitation | ||
| Genes presumably involved in iron acquisition, storage, or metabolism | |||
| NCgl0377 | Put. secreted heme transport associated protein, C-term. TMH | 16.82 | 5.00 |
| NCgl0378 | Heme ABC transporter, secreted lipoprotein, hmuT | 3.54 | 2.64* |
| NCgl0379 | Heme ABC transporter, permease, hmuU | 9.33 | 3.98 |
| NCgl0381 | Put. secreted heme transport associated protein, C-term. TMH | 17.66 | 5.99 |
| NCgl0382 | Put. secreted heme transport associated protein, C-term. TMH | 13.99 | 6.51* |
| NCgl0482 | Put. siderophore ABC transporter, ATPase | 5.72 | 1.73 |
| NCgl0483 | Put. siderophore ABC transporter, permease, FecCD family | 6.43 | 1.55* |
| NCgl0635 | Put. soluble, cytoplasmic siderophore-interacting protein | 6.53 | 1.70 |
| NCgl0636 | Put. siderophore ABC transporter, ATPase | 6.78 | 1.62 |
| NCgl0637 | Put. siderophore ABC transporter, permease, FecCD family | 5.70 | 1.74 |
| NCgl0638 | Put. siderophore ABC transporter, permease, FecCD family | 2.35 | 1.42 |
| NCgl0639 | Put. siderophore ABC transporter, secreted lipoprotein | 7.85 | 1.72 |
| NCgl0773 | Put. soluble, cytoplasmic siderophore-interacting protein | 11.63 | 2.38 |
| NCgl0774 | Put. siderophore ABC transporter, secreted lipoprotein | 8.43 | 2.51 |
| NCgl0776 | Put. siderophore ABC transporter, secreted lipoprotein | 4.16 | 1.17* |
| NCgl0779 | Put. siderophore ABC transporter, ATPase | 4.21 | 1.27* |
| NCgl1200 | Put. soluble, cytoplasmic siderophore-interacting protein | 4.76 | 2.42* |
| NCgl1209 | Put. siderophore ABC transporter, secreted lipoprotein | 3.31 | 1.94* |
| NCgl1502 | Fe-S cluster assembly protein, sufD | 0.47 | 0.43 |
| NCgl1503 | Fe-S cluster assembly protein, sufB | 0.48 | 0.43 |
| NCgl1959 | Put. secreted siderophore binding lipoprotein | 8.48 | 3.26 |
| NCgl2146 | Heme oxygenase, hmuO | 4.02 | 3.17 |
| NCgl2439 | Ferritin, ftn | 1.07* | 2.54 |
| NCgl2897 | Starvation-inducible DNA-binding protein, dps | 1.28** | 6.39** |
| NCgl2969 | Transporter of major facilitator superfamily | 4.35 | 1.43 |
| NCgl2970 | Put. secreted siderophore binding lipoprotein | 4.94 | 1.37 |
| Genes known to be regulated by RipA | |||
| NCgl0251 | Catalase, katA | 0.27 | 3.06 |
| NCgl0359 | Succinate dehydrogenase, sdhC | 0.16 | 0.46* |
| NCgl0360 | Succinate dehydrogenase, sdhA | 0.08 | 0.28* |
| NCgl0361 | Succinate dehydrogenase, sdhB | 0.18 | 0.46 |
| NCgl1141 | Nitrate reductase, β subunit, narH | 0.21 | 0.24* |
| NCgl1142 | Nitrate reductase, α subunit, narG | 0.10 | 0.41 |
| NCgl1143 | Nitrate/nitrite transporter, narK | 0.34 | 0.60* |
| NCgl1262 | 3-Isopropylmalate dehydratase, large subunit, leuC | 0.28 | 0.50* |
| NCgl1263 | 3-Isopropylmalate dehydratase small subunit, leuD | 0.28 | 0.59* |
| NCgl1482 | Aconitase, acn | 0.29 | 0.63* |
| NCgl2319 | Catechol 1,2-dioxygenase, catA | 0.05 | 0.14 |
| NCgl2657 | Phosphotransacetylase, pta | 0.26 | 1.10* |
| Genes encoding transcriptional regulators | |||
| NCgl0358 | Transcriptional regulator, ramB | 0.39 | 0.67* |
| NCgl0430 | Transcriptional regulator, ArsR family | 6.68 | 7.39 |
| NCgl0943 | Transcriptional regulator, AraC family, ripA | 12.16 | 4.13 |
| Further genes with a >10-fold increased mRNA level in the ΔdtxR mutant | |||
| NCgl0122 | Hypothetical protein | 31.33 | 21.49* |
| NCgl0123 | Hypothetical protein | 20.93 | 10.76 |
| NCgl1618 | Hypothetical protein | 5.88 | 17.48 |
| NCgl1635 | Hypothetical protein | 24.00 | 20.55 |
| NCgl1646 | Secretory serine protease | 33.09 | 11.92 |
| NCgl1651 | Hypothetical exported protein | 13.72 | 11.53 |
| NCgl1677 | Hypothetical protein | 7.47 | 14.78 |
| NCgl1678 | Hypothetical protein | 23.04 | 10.11 |
| NCgl1679 | Hypothetical protein | 16.00 | 9.27* |
| NCgl1680 | Hypothetical protein | 14.99 | 9.97 |
| NCgl1682 | Hypothetical protein | 10.40 | 8.04 |
| NCgl1685 | Hypothetical protein | 12.54 | 9.10 |
| NCgl2450 | Put. 2-methylcitrate dehydratase | 10.10 | 11.52 |
The mRNA ratios (ΔdtxR mutant/wild type) represent average values obtained from four (iron excess) or three (iron limitation) DNA microarray experiments performed with RNA isolated from four or three independent cultures in CGXII minimal medium containing either 100 μM FeSO4 or 1 μM FeSO4. Ratios labeled with a single asterisk could be analyzed in only a single experiment; for the ratios labeled with two asterisks, the P value was above 0.05. A list of all genes showing twofold-altered mRNA levels in the ΔdtxR mutant is provided in Table S2 in the supplemental material. Put., putative; TMH, transmembrane helix.
Group I includes 26 genes, 22 of which encode proteins presumably involved in heme uptake, in heme degradation (hmuO), or in iron acquisition via siderophores. All of these 22 genes showed increased expression in the ΔdtxR mutant under iron excess, whereas under iron limitation their mRNA levels are unaltered or increased to a lower extent. The latter situation indicates that even under our conditions of iron limitation there is still some active DtxR protein present in the wild type. Overall the expression pattern corresponds to the expectation that these genes are repressed by DtxR under iron excess but not, or more weakly, under iron limitation. Two genes of group I (sufB and sufD) encode components involved in the formation of iron-sulfur clusters. Their mRNA levels were decreased under both iron excess and iron starvation. Similar to the situation with M. tuberculosis (12, 17), the genes encoded by the suf operon may constitute the only iron-sulfur cluster assembly machinery in C. glutamicum. Finally, group I included the two genes of the C. glutamicum genome that are involved in iron storage, i.e., the ftn gene encoding ferritin and the dps gene encoding a protein that protects DNA from oxidative damage by nonspecific binding to DNA and catalyzing the oxidation of ferrous iron and its mineralization as a ferric core inside the protein (1). Both the ftn and dps mRNA levels were unaltered under iron excess but increased in the ΔdtxR mutant under iron limitation.
Group II includes 12 genes previously shown to be repressed by the RipA protein under iron limitation (40). Most of them encode iron-containing proteins (catalase, succinate dehydrogenase, nitrate reductase, isopropylmalate dehydratase, aconitase, and catechol 1,2-dioxygenase), but some also noniron proteins (nitrate/nitrite transporter, phosphotransacetylase). The mRNA level of all these genes was 3- to 20-fold decreased in the ΔdtxR mutant under iron excess, as expected when ripA expression is no longer repressed by DtxR. Under iron limitation, most of the RipA targets still showed decreased mRNA levels, but not as strongly as under iron limitation, again indicating that there is still some active DtxR protein present in the wild type that represses ripA. The only exceptions were the mRNA ratios for pta and katA, whose mRNA levels were unaltered or even increased in the ΔdtxR mutant under iron limitation, respectively, presumably because expression of these genes is controlled by additional regulatory proteins or mechanisms.
Group III includes three genes which encode transcriptional regulators. In our previous work, ripA was proposed to be a target of DtxR, since its expression is induced under iron limitation (24) and DtxR binds to the ripA promoter region (40). In agreement with this model, the ripA gene showed a strongly (>10-fold) increased mRNA level in the ΔdtxR mutant under iron excess, confirming its control by DtxR. Under iron limitation, the ripA mRNA level was still increased but to a significantly lesser extent than under iron excess. The ripA expression pattern fits perfectly with that of its target genes (see above). The ramB gene encodes a regulator which is involved in the control of carbon metabolism in C. glutamicum (13). Its mRNA level was decreased in the ΔdtxR mutant under iron excess and to a lesser extent also under iron limitation, indicating that its expression in the wild type is positively influenced by DtxR. The NCgl0430 gene encodes a transcriptional regulator of the ArsR family, whose mRNA level was strongly increased in the ΔdtxR mutant under both iron excess and iron limitation. The function of the regulator encoded by NCgl0430 and its target genes are not yet known.
Group IV includes 11 further genes whose mRNA levels were more than 10-fold increased in the ΔdtxR mutant under iron excess and which do not obviously belong to group I, II, or III. With three exceptions, all these genes are located in the CGP3 prophage region (see above). Nine of the genes encode hypothetical proteins, one encodes a putative secreted serine protease, and one a putative 2-methylcitrate dehydratase.
Identification and testing of putative DtxR boxes in C. glutamicum.
Besides the transcriptome comparison of the wild type and the ΔdtxR mutant, we used a second, completely independent approach for the identification of putative DtxR target genes. We searched for potential DtxR operators in the C. glutamicum genome, amplified the corresponding DNA regions by PCR, and tested them for their ability to bind purified DtxR. A 19-bp consensus binding site of the C. diphtheriae DtxR protein has been defined as TWAGGTWAGSCTWACCTWA (36) and is probably also correct for DtxR from C. glutamicum, since C. glutamicum DtxR was shown to repress the C. diphtheriae tox promoter in an iron-dependent manner (28). Using the ERGO software suite (Integrated Genomics), we searched the C. glutamicum genome for sequences deviating in maximally five positions from the consensus and allowing neither insertions nor deletions. As shown in Table S3 in the supplemental material, 117 putative DtxR binding sites could be identified that fulfilled the given criteria. Seventy of these sites were located maximally 500 bp upstream or maximally 100 bp downstream of the start codon of the neighboring gene and were thought to be physiologically the most relevant.
To test whether DtxR binds in vitro to the potential binding sites, DNA fragments covering 74 of the corresponding sequences were amplified by PCR and used for gel shift assays with the purified C. glutamicum DtxR protein. For this purpose, the DtxR protein was overproduced in E. coli and isolated by means of a carboxy-terminal histidine tag as described previously. For the gel shift assays, the DNA fragments (8 to 20 nM) were mixed with various concentrations of the DtxR protein (0 to 4 μM dimeric form) and then separated on 15% native polyacrylamide gels. Mn2+ was included in all reactions instead of Fe2+ on account of its redox stability. As a negative control, all reactions included a promoter fragment of acn, pta, katA, or porB which was not shifted by DtxR. As shown in Fig. 2, the DtxR protein shifted 51 of the 74 tested DNA fragments containing a putative DtxR box, albeit with different affinities. In Table 3, the DtxR boxes present within the shifted fragments, the neighboring genes, the distances of the DtxR boxes to the neighboring genes, and the relative affinities are summarized. According to the affinity, three classes of DtxR binding sites were distinguished: those requiring a 25-fold molar excess of dimeric DtxR for a complete shift (indicated by “++” in Table 3), those requiring a 50-fold excess for a complete shift (“+”), and those requiring a 200-fold molar excess for a complete or a partial shift [“(+)”]. There was no correlation between the binding affinity and individual positions in the DtxR binding site; however, there was a correlation between binding affinity and the number of deviations from the consensus binding site: 3.9 on average for those with high affinity, 4.5 on average for those with intermediate affinity, and always 5 for those with low affinity. The genes located adjacent to the DtxR binding sites were ordered into four groups. The first group includes 17 genes coding for proteins involved in iron acquistion, e.g., siderophore binding proteins, siderophore ABC uptake systems, and heme uptake ABC transporters. Most of the corresponding binding sites showed a high affinity for DtxR, with DtxR-DNA complex formation being observed at a 15- to 25-fold molar excess of dimeric DtxR (data not shown). The second group includes the genes for the iron storage proteins ferritin and Dps. Whereas the DtxR binding site in front of the dps gene showed a high affinity similar to that of the iron uptake genes, the DNA fragment covering the ferritin promoter region showed a lower affinity, requiring a 200-fold molar excess of DtxR for a complete shift. The third group consists of eight genes coding for transcriptional regulators, including ramB, ripA, sufR, and cgtR11. The corresponding DNA fragments showed various affinities for DtxR: in the cases of ramB, NCgl0430, ripA, sufR, cgtR11 and NCgl2877, a 50-fold molar excess of dimeric DtxR was sufficient for a complete shift, whereas in the cases of NCgl0120 and NCgl1127, even a 200-fold molar excess of dimeric DtxR was not sufficient for a complete shift. The fourth group, designated as “others,” includes genes encoding proteins of various functions, e.g., two cation-transporting ATPases, a fatty acid synthase, two secreted serine proteases, the SecD subunit of the Sec protein translocase, a glycogen phosphorylase, and others. The corresponding DNA fragments showed various affinities for DtxR, requiring a 50- to 200-fold molar excess of dimeric DtxR for a complete shift. Binding of DtxR to all of these fragments was strictly dependent on the presence of Mn2+ ions, since in the absence of Mn2+ no shift was observed even at a 200-fold molar excess of dimeric DtxR (data not shown).
FIG.2.
Binding of DtxR to the predicted DtxR boxes. DNA fragments (200 to 400 bp in size; final concentration, 10 to 20 nM) covering promoter regions with putative DtxR binding sites were incubated for 30 min at room temperature without DtxR (left lanes) or with a 50-fold or a 200-fold (labeled with an asterisk) molar excess of purified DtxR protein (dimeric form) (right lanes) before separation by native polyacrylamide (15%) gel electrophoresis and staining with SybrGreen I. DNA fragments (100 to 200 bp) covering the promoter regions of acn, pta, and katA, which do not contain putative DtxR binding sites, served as negative controls.
TABLE 3.
DtxR binding sites in the C. glutamicum genome verified by bandshift analysisa
| NCgl gene no. or description | DtxR binding site | Category of neighboring gene | Annotated function of neighboring gene | Shift | Location |
|---|---|---|---|---|---|
| Consensus site of C. diphtheriae | TWAGGTWAGSCTWACCTWA | ||||
| NCgl0027 | TTTGCGCAGGCTAACCTTT | Iron acquisition | ABC transporter, permease | ++ | +125.5 |
| NCgl0329 | TAAGGATAACCTTGCCTTA | Iron acquisition | Secreted siderophore binding lipoprotein | ++ | −42.5 |
| NCgl0377 | TTAAGTTAGCATAGCCTTA | Iron acquisition | Heme transport-associated protein | ++ | −150.5 |
| NCgl0381 | TAAGGTTACCCTACCCTCT | Iron acquisition | Heme transport-associated protein | ++ | −90.5 |
| NCgl0484 | TTAGTAAAGGCTCACCTAA | Iron acquisition | Siderophore ABC transporter, permease | + | −100.5 |
| NCgl0618 | ATAGGATAGGTTAACCTGA | Iron acquisition | Secreted siderophore binding lipoprotein | ++ | −34.5 |
| NCgl0639 | GTCGGGCAGCCTAACCTAA | Iron acquisition | Siderophore ABC transporter, secreted lipoprotein | ++ | −49.5 |
| NCgl0774 | TAAGGTTTGCCTAATGTTT | Iron acquisition | Secreted siderophore binding lipoprotein | + | −39.5 |
| NCgl0776 | TTTAGGTAACCTAACCTCA | Iron acquisition | Secreted siderophore binding lipoprotein | ++ | −73.5 |
| NCgl0777 | TTAGGTTAGGCTCTAATAT | Iron acquisition | Siderophore ABC transporter, permease | ++ | −182.5 |
| NCgl0914 | TTAAGTCAGTGTTACCTAA | Iron acquisition | Siderophore export ABC transporter, permease | ++ | −92.5 |
| NCgl1200 | TTTTGTTAGGCTTGCCTAG | Iron acquisition | Siderophore-interacting protein | + | −42.5 |
| NCgl1209 | TTAGGTAAGGTTTGCATAC | Iron acquisition | Secreted siderophore binding lipoprotein | + | −40.5 |
| NCgl1395 | TTAGGTTAGGCAAGCCATA | Iron acquisition | Cytoplasmic siderophore-interacting protein | ++ | −48.5 |
| NCgl1959 | TTAGGCAAGGCTACCTTTT | Iron acquisition | Secreted siderophore binding lipoprotein | ++ | −19.5 |
| NCgl2146 | GTAGGTGTGGGTAACCTAA | Iron acquisition | Heme oxygenase | + | −129.5 |
| NCgl2752 | TAAGGCAAGCCTAAATTAG | Iron acquisition | Hyp. exported protein, heme transport | ++ | −106.5 |
| NCgl2970 | TTGCGTTAGGATAGCCTAA | Iron acquisition | Secreted siderophore binding lipoprotein | ++ | −8.5 |
| NCgl2439 | TTATGCTGCGCTAACCTAT | Iron storage | Ferritin, ftn | (+) | −46.5 |
| NCgl2897 | TCAGGATAGGACAACCTAA | Iron storage | DNA protection during starvation protein, dps | ++ | −70.5 |
| NCgl1504 | TTGGCTTAGGGTTCGCTTA | Iron-sulfur cluster assembly | Fe-S cluster assembly, suf operon | ++ | −141.5 |
| NCgl0120 | TTGGCTATGGTTTACCTAT | Transcriptional regulator | Transcriptional repressor | (+) | +13.5 |
| NCgl0358 | TTAGGATAGCCTTACTTTA | Transcriptional regulator | Transcriptional regulator, ramB | ++ | −390.5 |
| NCgl0430 | TTAGGCTTGCCATACCTAT | Transcriptional regulator | ArsR-type transcriptional regulator | ++ | −25.5 |
| NCgl0943 | TGAGGTTAGCGTAACCTAC | Transcriptional regulator | AraC-type transcriptional regulator, ripA | ++ | −42.5 |
| NCgl1127 | TAAGGGAATTGTAATCTAA | Transcriptional regulator | Transcriptional regulator, Crp family | (+) | −353.5 |
| NCgl1444 | TTAGGGACGCTTTACCTGC | Transcriptional regulator | Put. transcriptional regulator | ++ | −87.5 |
| NCgl2834 | ATGAGTAAGGCTAGACTAA | Transcriptional regulator | Response regulator, cgtR11 | + | −95.5 |
| NCgl2877 | TTTGGCAAGACTTACCGAC | Transcriptional regulator | Transcriptional regulator, PadR-like family | ++ | −110.5 |
| NCgl0123 | AATGGTTAGGCTAACCTTA | Other | Hypothetical protein | ++ | +10.4 |
| NCgl0376 | TAAGGCTATGCTAACTTAA | Other | Copper-transporting P-type ATPase | ++ | −79.5 |
| NCgl0465 | TAGGGAAAGCCCATCCTTA | Other | Copper-transporting P-type ATPase | ++ | −31.5 |
| NCgl0486 | GTTGGTAAGGCAAACATGA | Other | Ribosomal protein S10, rpsJ | + | −378.5 |
| NCgl0594 | TTTGGTCTGGCCTACCTAT | Other | Lycopene elongase, crtEb | + | +78.5 |
| NCgl0802 | TTCGGCTACGCTCACGTAA | Other | Fatty acid synthase, fasA | + | −73.5 |
| NCgl0891 | TGAGGTACGCGTTACCTGT | Other | Hypothetical protein | + | −186.5 |
| NCgl1145 | CGTGGGAAGCCTAACTTAA | Other | Putative secreted serine protease | + | −51.5 |
| NCgl1339 | TTAGGGAAGGAAAACATAT | Other | Putative secreted lipoprotein | (+) | +7.5 |
| NCgl1569 | TTCCGTACGGCTATGCTTA | Other | Putative Holliday junction resolvase | (+) | −130.5 |
| NCgl1594 | TATGGGAAGGCAAAACTAC | Other | Protein translocase, secD | (+) | −160.5 |
| NCgl1645 | TAACTTAAGCCTCACATAC | Other | Putative resolvase, res | (+) | −759.5 |
| NCgl1646 | TTAGGTAAAGCTTGCCTAT | Other | Secretory serine protease | ++ | −183.5 |
| NCgl1677 | TTAGGTTATGTCAAAGTTA | Other | Hypothetical protein | + | −563.5 |
| NCgl1955 | TTAGATAAGCCTGACATCA | Other | Predicted endonuclease | (+) | −342.5 |
| NCgl2006 | CAATCTTAGGCTTAGTTTA | Other | Glycogen phosphorylase, glgP2 | + | −14.5 |
| NCgl2027 | TCAAGTAAGGTTTACCTTA | Other | SAM-dependent methyltransferase | ++ | −8.5 |
| NCgl2282 | TTAGGTCAAGCTTGCATTT | Other | Hypothetical membrane-spanning protein | + | −29.5 |
| NCgl2414 | TAATGTATGCCTTGACTTG | Other | Xanthosine triphosphate pyrophosphatase | + | −227.5 |
| NCgl2719 | TTAGGTTAGGTTCACCGTG | Other | Putative sulfite reductase, cysJ | + | −213.5 |
| NCgl2750 | ATTGGTACGGGTTACCTTG | Other | Predicted UDP-glucose 6-dehydrogenase, udgA2 | ++ | +24.5 |
| NCgl2766 | TTAACTTTGCCCTACCTAA | Other | Putative permease | + | −199.5 |
| NCgl2971 | TTGCATTAGGCTATCCTAA | Other | Putative Zn-dependent oxidoreductase | ++ | −173.5 |
The DtxR binding sites shown in this table were identified by a motif search of the C. glutamicum genome using the consensus sequence TWAGGTWAGSCTWACCTWA from C. diphtheriae and allowing up to five mismatches, but no insertions or deletions. All binding sites fulfilling the applied criteria are listed in Table S3 in the supplemental material. This table includes only those sites that were successfully shifted by purified DtxR protein. The column labelled “Shift” indicates whether DtxR showed a high (++), medium (+), or low [(+)] affinity to the corresponding DNA fragment. The position of the center of the binding sites relative to the predicted translation start site of the neighboring gene is given by the numbers in the “Location” column. Boldface indicates consensus.
Combining gel shift and DNA microarray data for the identification of the DtxR target genes.
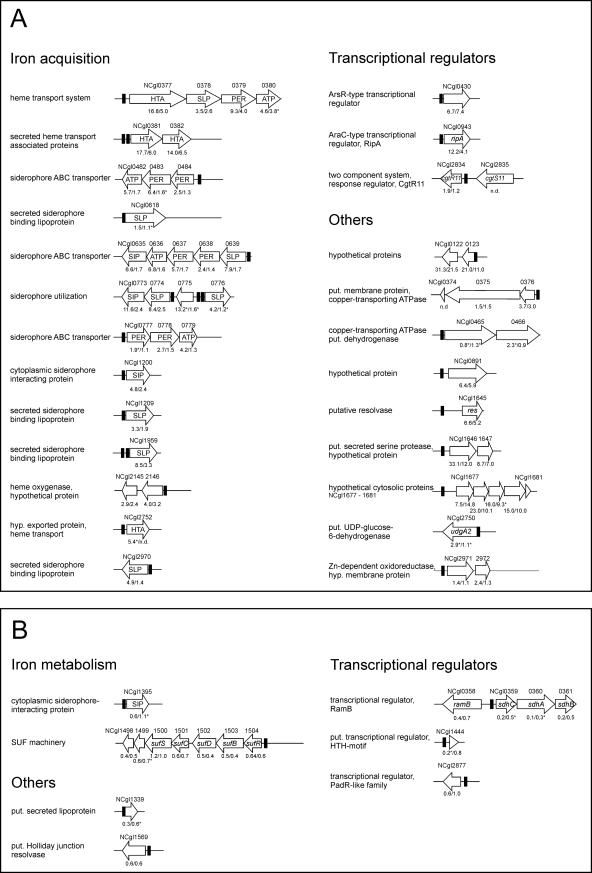
The transcriptome comparison of the wild type and the ΔdtxR mutant identified genes and operons whose mRNA levels were altered as a consequence of the dtxR deletion; however, this approach was not able to distinguish between genes that are directly controlled by DtxR and those whose expression is indirectly influenced by the absence of DtxR. In contrast, the gel shift assay with purified DtxR and DNA regions containing potential DtxR binding sites was able to identify putative direct target genes of DtxR, but this approach gave no information on the influence of DtxR on the expression of these genes in vivo. To identify genes or operons that are probably directly regulated by DtxR in vivo, we analyzed which of the genes/operons containing a functional DtxR binding site in their promoter region showed more than 1.5-fold-altered mRNA levels in the ΔdtxR mutant and which did not. Based on this analysis, four groups of genes were identified, which are shown in Fig. 3.
FIG. 3.
Overview of C. glutamicum operons preceded by a functional DtxR binding site. The black boxes indicate the positions of the identified DtxR boxes. The mRNA ratios (ΔdtxR mutant versus wild type) obtained from the DNA microarray experiments are shown below the corresponding gene. The first value gives the ratio under iron excess, and the second value gives the ratio under limitation. It has to be taken into account that for the majority of genes the indicated function was derived from the genome annotation rather than from experimental data. The genes/operons were separated in four groups. Group A contains genes whose mRNA level was at least 1.5-fold increased in the ΔdtxR mutant under iron excess. These genes are presumably repressed by DtxR. Group B comprises genes whose mRNA level was at least 1.5-fold decreased in the ΔdtxR mutant under iron excess. These genes might be activated by DtxR. Group C contains two genes whose mRNA levels were almost unaltered in the ΔdtxR mutant under iron excess but were increased under iron limitation. How DtxR controls expression of these genes is not clear yet. Group D contains all genes whose mRNA ratios could not be determined and those whose mRNA levels were altered less than 1.5-fold in the ΔdtxR mutant. Whether these genes are indeed regulated by DtxR requires further studies.
The first group (Fig. 3A) includes 51 genes organized in 27 putative operons which are presumably repressed by DtxR, since they show an increased mRNA level in the ΔdtxR mutant under iron excess. The majority of these genes are obviously involved in iron acquisition, three encode transcriptional regulators, i.e., the AraC-type regulator RipA (40), the response regulator CgtR11 (23), and an ArsR-type regulator, and the others encode proteins of diverse or unknown functions. Three DtxR-repressed operons are located within the CGP3 prophage region.
The second group (Fig. 3B) comprises 12 genes that show a decreased mRNA level in the ΔdtxR mutant under iron excess. These genes could be activated by DtxR, as previously reported for the bfrA gene of M. tuberculosis (14). Interestingly, this group included the suf operon (NCgl1504 to NCgl1498), whose products are involved in formation and repair of iron-sulfur clusters, and several genes for transcriptional regulators, including RamB, which is involved in the regulation of acetate metabolism (13). In the case of ramB, the situation is exceptional, since this gene is located upstream and divergent to the RipA-controlled sdhCAB operon encoding succinate dehydrogenase (Fig. 3B). The DtxR binding site dedicated to ramB is located 390 bp upstream of the translation start of ramB and 105 bp upstream of the transcription start site of sdhC, which we recently identified by primer extension to be located 15 bp upstream of the sdhC start codon (S. Degraf and M. Bott, unpublished data). Although sdhCAB expression followed the same pattern as that of the other RipA target genes, the possibility that DtxR directly activates sdhCAB expression under iron excess cannot be excluded from our data.
The third group (Fig. 3C) contains two genes, ftn and dps, which are of particular importance for iron homoeostasis, since they encode proteins capable of iron storage, namely ferritin and Dps (for a review see reference 1). The mRNA levels of these genes were almost unaltered in the ΔdtxR mutant under iron excess but increased under iron limitation, a behavior not yet understood.
The fourth group (Fig. 3D) includes 32 genes organized in 18 putative operons that contain a functional DtxR binding site but whose mRNA levels could not be determined or were changed less than 1.5-fold in the ΔdtxR mutant. With few exceptions, most of the corresponding genes show no obvious connection to iron metabolism. The influence of DtxR on the expression of these genes in vivo is unclear, since small changes in expression might remain undetected with the DNA microarray method used.
On average, high mRNA ratios (ΔdtxR mutant/wild type) under iron excess correlated with high in vitro binding affinities of the respective DtxR binding sites and low mRNA ratios with low in vitro binding affinities.
DISCUSSION
The transcriptional regulator DtxR was first identified in C. diphtheriae, where it controls the expression of the diphtheria toxin gene carried by corynebacteriophage β in an iron-dependent manner (4, 33). Subsequent studies with DtxR and its ortholog IdeR from Mycobacterium species revealed that DtxR plays a key role in the control of iron metabolism and thus is a functional equivalent of the Fur protein from gram-negative and low-G+C gram-positive bacteria (2, 11, 16). In this work, we performed a genome-wide search for DtxR-regulated genes in C. glutamicum by comparing the gene expression profiles of the wild type and a ΔdtxR mutant and by testing the functionality of 74 putative DtxR binding sites with electrophoretic mobility shift assays with purified DtxR protein from C. glutamicum. Genes which are preceded by a functional DtxR binding site and which show an at least 1.5-fold-altered mRNA level in the ΔdtxR mutant are considered probable direct target genes of DtxR.
As expected, the majority of the target genes are repressed by DtxR (increased mRNA level in the ΔdtxR mutant) and are involved in iron acquisition (Fig. 3A). One operon (NCgl0377 to NCgl0380) is homologous to the hmu operon of C. diphtheriae (7) and encodes an ABC transporter for heme uptake. Three other genes are predicted to encode proteins that are also involved in heme transport, all of which contain a signal peptide and a putative C-terminal transmembrane helix (NCgl0381-NCgl0382 and NCgl2752). A further gene repressed by DtxR and probably also required for heme utilization is hmuO, encoding heme oxygenase (NCgl2146), an enzyme converting protoheme to biliverdin IXα and free iron. Nineteen genes repressed by DtxR encode proteins presumably involved in the acquisition of iron from siderophores. They include three ABC transporters (permeases and ATPases), seven secreted siderophore-binding lipoproteins, and three cytoplasmic siderophore-interacting proteins. In contrast to the high number of genes involved in siderophore utilization, none of the genes repressed by DtxR was annotated to be involved in siderophore biosynthesis, in contrast, for example, to M. smegmatis or M. tuberculosis (8, 14, 30). Inspection of the C. glutamicum ATCC 13032 genome sequence (20) failed to reveal genes that are obviously involved in siderophore biosynthesis. Thus, the strategy of C. glutamicum ATCC 13032 for coping with iron starvation is to make use of a variety of siderophores produced by other microbes and perhaps also of heme compounds. However, since another strain of C. glutamicum, ATCC 14067, produces the cyclic catecholate siderophore corynebactin (5), strain-specific differences exist with respect to siderophore production.
In addition to its function in repressing iron starvation genes, we recently provided evidence for another role of DtxR, namely, its influence on the expression of several prominent iron-containing proteins via the AraC-type regulator RipA (40). We proposed that ripA expression is controlled by DtxR, since this protein binds to a well-conserved binding site upstream of ripA (40). This proposal was clearly confirmed in this work. The ripA mRNA level was more than 10-fold increased in the ΔdtxR mutant, whereas the mRNA level of the RipA target genes was strongly reduced in the ΔdtxR mutant (Table 2). The fact that the RipA protein synthesized in the ΔdtxR mutant repressed its target genes in the presence of excess iron indicates that RipA repressor activity is not directly controlled by iron, in contrast to the case with DtxR.
Besides ripA, two further genes for transcriptional regulators are likely to be repressed by DtxR, i.e., NCgl0430 and cgtR11. The mRNA level of NCgl0430, encoding an ArsR-type regulator, was sevenfold increased in the ΔdtxR mutant under both iron excess and iron starvation. Members of the SmtB/ArsR family usually repress the expression of genes required to cope with stress-inducing concentrations of di- and multivalent heavy metal ions. Depression results from direct binding of metal ions to the homodimeric repressors (6). The metal ion that might be sensed by the NCgl0430 protein as well as its target genes are not yet known. The mRNA level of cgtR11 was about twofold increased in the ΔdtxR mutant under iron excess. This gene encodes the response regulator of the CgtSR11 two-component system of C. glutamicum (23). Since the DtxR binding site is located upstream of cgtR11 (Fig. 3A), only this gene, but not the upstream cgtS11 gene, appears to be under direct control of DtxR. The CgtS11 and CgtR11 proteins show high sequence identity to the C. diphtheriae proteins encoded by the genes DIP2268 (56% identity) and DIP2267 (86%), respectively, and weaker similarity to the sensor kinase ChrS (30%) and the response regulator ChrA (50%) from C. diphtheriae. Since the ChrAS two-component system was recently shown to control the expression of the hmuO gene encoding heme oxygenase (3), the CgtSR11 two-component system might also be involved in the control of genes required for heme utilization.
Studies with M. tuberculosis indicate that DtxR can function not only as a transcriptional repressor but also as a transcriptional activator (14). In the course of our studies, a number of genes were identified that were preceded by a functional DtxR binding site and which showed a decreased mRNA level in the ΔdtxR mutant (Fig. 3B). Thus, expression of these genes appears to be activated by DtxR. One interesting operon belonging to this group was the suf operon, which includes seven genes (NCgl1504 to NCgl1498). A DtxR box upstream of NCgl1504, designated here as sufR, was able to bind purified DtxR (Fig. 2), whereas a second putative DtxR box localized upstream of NCgl1501 was not. With the exception of the promoter-proximal sufR, the genes of this operon encode proteins involved in the assembly and repair of iron-sulfur clusters, which form the so-called SUF machinery. As in M. tuberculosis (17), the SUF machinery appears to be the exclusive system for this function in C. glutamicum. The SufR protein of C. glutamicum, which is a DeoR-type transcriptional regulator, might have the same function as the SufR protein from Synechocystis sp. strain PCC6803 (38). In Synechocystis, SufR functions as a repressor of the sufBCDS operon, and it was proposed that SufR senses the levels of iron-sulfur clusters in the cell through its own unstable iron-sulfur cluster: if the cluster is present, SufR binds to its operator and represses the suf operon; if it is oxidatively damaged or absent, repression is relieved. If the C. glutamicum SufR protein functions in a similar fashion, the C. glutamicum suf operon is under dual transcriptional control, i.e., positive control by DtxR and negative control by SufR.
Genes of particular importance for iron homeostasis are those encoding iron storage proteins. The genome of C. glutamicum harbors two such genes, i.e., ftn (NCgl2439), encoding a ferritin (NCgl2439), and dps (NCgl2897), encoding a protein of the Dps family. Both genes contain a functional DtxR binding site in the 5′ noncoding region and showed almost unchanged mRNA levels in the ΔdtxR mutant under iron excess but increased mRNA levels under iron limitation (Fig. 3C). The simplest explanation would assume that DtxR represses ftn and dps under iron limitation, which is physiologically meaningful, since iron storage proteins are probably required in only minute amounts under these conditions. However, purified DtxR bound with high affinity to the dps promoter and with low affinity to the ftn promoter only in the presence of Mn2+ but not in its absence. Thus, further studies are required to understand how DtxR attenuates ftn and dps expression under iron limitation.
In summary, the results presented here indicate that about 60 genes are likely to be directly controlled by DtxR and thus form the DtxR regulon. Since several of these themselves encode transcriptional regulators, such as ripA, a multitude of further genes is controlled indirectly by DtxR. Thus, DtxR was shown to be the master regulator of iron-dependent gene expression in C. glutamicum.
ADDENDUM IN PROOF
After this article was accepted for publication, another study on the identification of DtxR target genes in Corynebacterium glutamicum was published (I. Brune, H. Werner, A. T. Hüser, J. Kalinowski, A. Pühler, and A. Tauch, BMC Genomics 7:21, 2006).
Supplementary Material
Acknowledgments
We thank Hermann Sahm for continuous support, Volker Wendisch for establishing the DNA chip technique in our institute, and Melanie Brocker for help with DNA microarray analyses.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 2.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 3.Bibb, L. A., N. D. King, C. A. Kunkle, and M. P. Schmitt. 2005. Analysis of a heme-dependent signal transduction system in Corynebacterium diphtheriae: deletion of the chrAS genes results in heme sensitivity and diminished heme-dependent activation of the hmuO promoter. Infect. Immun. 73:7406-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, J., M. N. Oza, and J. R. Murphy. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. USA 87:5968-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budzikiewicz, H., A. Bossenkamp, K. Taraz, A. Pandey, and J. M. Meyer. 1997. Corynebactin, a cyclic catecholate siderophore from Corynebacterium glutamicum ATCC 14067 (Brevibacterium sp. DSM 20411). Z. Naturforsch. 52c:551-554.
- 6.Busenlehner, L. S., M. A. Pennella, and D. P. Giedroc. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27:131-143. [DOI] [PubMed] [Google Scholar]
- 7.Drazek, E. S., C. A. Hammack, and M. P. Schmitt. 2000. Corynebacterium diphtheriae genes required for acquisition of iron from haemin and haemoglobin are homologous to ABC haemin transporters. Mol. Microbiol. 36:68-84. [DOI] [PubMed] [Google Scholar]
- 8.Dussurget, O., M. Rodriguez, and I. Smith. 1996. An ideR mutant of Mycobacterium smegmatis has derepressed siderophore production and an altered oxidative-stress response. Mol. Microbiol. 22:535-544. [DOI] [PubMed] [Google Scholar]
- 9.Eggeling, L., and M. Bott. 2005. Handbook of Corynebacterium glutamicum. CRC Press, Inc., Boca Raton, Fla.
- 10.Eikmanns, B. J., N. Thum-Schmitz, L. Eggeling, K. U. Lüdtke, and H. Sahm. 1994. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140:1817-1828. [DOI] [PubMed] [Google Scholar]
- 11.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontecave, M., S. O. Choudens, B. Py, and F. Barras. 2005. Mechanisms of iron-sulfur cluster assembly: the SUF machinery. J. Biol. Inorg. Chem. 10:713-721. [DOI] [PubMed] [Google Scholar]
- 13.Gerstmeir, R., A. Cramer, P. Dangel, S. Schaffer, and B. J. Eikmanns. 2004. RamB, a novel transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 186:2798-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gold, B., G. M. Rodriguez, S. A. Marras, M. Pentecost, and I. Smith. 2001. The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol. Microbiol. 42:851-865. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning: a practical approach, vol. 1. IRL-Press, Oxford, United Kingdom. [Google Scholar]
- 16.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 17.Huet, G., M. Daffe, and I. Saves. 2005. Identification of the Mycobacterium tuberculosis SUF machinery as the exclusive mycobacterial system of [Fe-S] cluster assembly: evidence for its implication in the pathogen's survival. J. Bacteriol. 187:6137-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishige, T., M. Krause, M. Bott, V. F. Wendisch, and H. Sahm. 2003. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 185:4519-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalinowski, J. 2005. The genomes of amino acid-producing corynebacteria, p. 37-56. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, Fla.
- 20.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Kramer, B. Linke, A. C. McHardy, F. Meyer, B. Mockel, W. Pfefferle, A. Puhler, D. A. Rey, C. Ruckert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegrabe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 21.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinoshita, S., S. Udaka, and M. Shimono. 1957. Studies on the amino acid fermentation. I. Production of L-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 3:193-205. [PubMed] [Google Scholar]
- 23.Kocan, M., S. Schaffer, T. Ishige, U. Sorger-Herrmann, V. F. Wendisch, and M. Bott. 2006. Two-component systems of Corynebacterium glutamicum: deletion analysis and involvement of the PhoS-PhoR system in the phosphate starvation response. J. Bacteriol. 188:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krug, A., V. F. Wendisch, and M. Bott. 2005. Identification of AcnR, a TetR-type repressor of the aconitase gene acn in Corynebacterium glutamicum. J. Biol. Chem. 280:585-595. [DOI] [PubMed] [Google Scholar]
- 25.Lange, C., D. Rittmann, V. F. Wendisch, M. Bott, and H. Sahm. 2003. Global expression profiling and physiological characterization of Corynebacterium glutamicum grown in the presence of L-valine. Appl. Environ. Microbiol. 69:2521-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Möker, N., M. Brocker, S. Schaffer, R. Kramer, S. Morbach, and M. Bott. 2004. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 54:420-438. [DOI] [PubMed] [Google Scholar]
- 27.Niebisch, A., and M. Bott. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175:282-294. [DOI] [PubMed] [Google Scholar]
- 28.Oguiza, J. A., X. Tao, A. T. Marcos, J. F. Martin, and J. R. Murphy. 1995. Molecular cloning, DNA sequence analysis, and characterization of the Corynebacterium diphtheriae dtxR homolog from Brevibacterium lactofermentum. J. Bacteriol. 177:465-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polen, T., D. Rittmann, V. F. Wendisch, and H. Sahm. 2003. DNA microarray analyses of the long-term adaptive response of Escherichia coli to acetate and propionate. Appl. Environ. Microbiol. 69:1759-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning. a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 33.Schmitt, M. P., and R. K. Holmes. 1991. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect. Immun. 59:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stackebrandt, E., F. A. Rainey, and N. L. WardRainey. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47:479-491. [Google Scholar]
- 35.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 36.Tao, X., and J. R. Murphy. 1994. Determination of the minimal essential nucleotide sequence for diphtheria tox repressor binding by in vitro affinity selection. Proc. Natl. Acad. Sci. USA 91:9646-9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Rest, M. E., C. Lange, and D. Molenaar. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52:541-545. [DOI] [PubMed] [Google Scholar]
- 38.Wang, T., G. Shen, R. Balasubramanian, L. McIntosh, D. A. Bryant, and J. H. Golbeck. 2004. The sufR gene (sll0088 in Synechocystis sp. strain PCC 6803) functions as a repressor of the sufBCDS operon in iron-sulfur cluster biogenesis in cyanobacteria. J. Bacteriol. 186:956-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wendisch, V. F. 2003. Genome-wide expression analysis in Corynebacterium glutamicum using DNA microarrays. J. Biotechnol. 104:273-285. [DOI] [PubMed] [Google Scholar]
- 40.Wennerhold, J., A. Krug, and M. Bott. 2005. The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J. Biol. Chem. 280:40500-40508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.