Abstract
The natural plasmid pBM19 carries the key mdh gene needed for the oxidation of methanol into formaldehyde by Bacillus methanolicus. Five more genes, glpX, fba, tkt, pfk, and rpe, with deduced roles in the cell primary metabolism, are also located on this plasmid. By using real-time PCR, we show that they are transcriptionally upregulated (6- to 40-fold) in cells utilizing methanol; a similar induction was shown for two chromosomal genes, hps and phi. These seven genes are involved in the fructose bisphosphate aldolase/sedoheptulose bisphosphatase variant of the ribulose monophosphate (RuMP) pathway for formaldehyde assimilation. Curing of pBM19 causes higher methanol tolerance and reduced formaldehyde tolerance, and the methanol tolerance is reversed to wild-type levels by reintroducing mdh. Thus, the RuMP pathway is needed to detoxify the formaldehyde produced by the methanol dehydrogenase-mediated conversion of methanol, and the in vivo transcription levels of mdh and the RuMP pathway genes reflect the methanol tolerance level of the cells. The transcriptional inducer of hps and phi genes is formaldehyde, and not methanol, and introduction of multiple copies of these two genes into B. methanolicus made the cells more tolerant of growth on high methanol concentrations. The recombinant strain also had a significantly higher specific growth rate on methanol than the wild type. While pBM19 is critical for growth on methanol and important for formaldehyde detoxification, the maintenance of this plasmid represents a burden for B. methanolicus when growing on mannitol. Our data contribute to a new and fundamental understanding of the regulation of B. methanolicus methylotrophy.
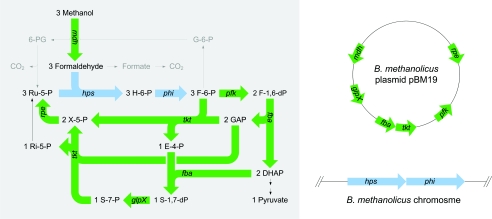
Aerobic methylotrophs are bacteria capable of utilizing reduced one-carbon (C1) compounds as the sole carbon source for growth and energy (2), and the majority of research on these bacteria has focused on their biochemical novelty and commercial viability. A number of gram-positive and thermotolerant Bacillus strains have been isolated and designated Bacillus methanolicus (4, 26). This methylotrophic bacterium has a novel NAD-dependent methanol dehydrogenase (MDH), which contains bound NAD, to oxidize methanol into formaldehyde (11). The enzyme has a remarkably high affinity for methanol, and an activator protein called ACT modulates its in vivo activity. ACT activates MDH by hydrolysis, and the expression of these two proteins is reported to be under coordinate (and methanol-induced) control in B. methanolicus (16). Formaldehyde is the key intermediate in C1 metabolism and can be assimilated via the ribulose monophosphate (RuMP) pathway (Fig. 1). Many RuMP pathway variants are described in the literature, and thermotolerant Bacillus strains are reported to use the fructose-bisphosphate aldolase/transaldolase variant (3, 4, 12).
FIG. 1.
Graphical map of biochemical reactions and genes involved in methanol oxidation and assimilation by B. methanolicus (8) and the physical locations of the genes involved. Pathways for linear and cyclic formaldehyde dissimilation are indicated in gray. pBM19 genes: mdh, methanol dehydrogenase; pfk, phosphofructokinase; fba, fructose bisphosphate aldolase; tkt, transketolase; glpX, fructose/sedoheptulose biphosphatase; rpe, ribulose phosphate epimerase. Chromosomal genes: hps, hexulose phosphate synthase; phi, phosphohexuloisomerase. Metabolites: H-6-P, hexulose-6-phosphate; F-6-P, fructose-6-phosphate; F-1,6-dP, fructose-1,6-bisphosphate; DHAP, dihydroxy acetone phosphate; GAP, glyceraldehyde-3-phosphate; E-4-P, erythrose-4-phosphate; S-7-P, sedoheptulose-7-phosphate; S-1,7-dP, sedoheptulose-1,7-bisphosphate; X-5-P, xylulose-5-phosphate; Ri-5-P, ribose-5-phosphate; Ru-5-P, ribulose-5-phosphate; G-6-P, glucose-6-phosphate; 6-PG, 6-phosphogluconate.
Steady-state cultures of B. methanolicus MGA3 limited by methanol in the feed display sensitivity to minor methanol pulses and respond by a transient decline in biomass concentration. By using 13C nuclear magnetic resonance analysis, substantial intracellular accumulation of formaldehyde has been detected in response to the pulses, and the latter compound was proposed to be the major cell-toxic agent under these conditions (22). A similar conclusion has been described for the facultative methylotroph Methylobacterium extorquens AM1 metabolizing C1 substrates (19). Interestingly, RuMP pathways have recently also been found in certain nonmethylotrophic bacteria, and it has been speculated that they function to detoxify intracellular formaldehyde (20, 21, 23, 28). In the thermotolerant Bacillus sp. strain C1, it was shown that a high MDH activity together with a low hexulose phosphate synthase (HPS) activity may lead to accumulation of intracellular formaldehyde (5). In B. methanolicus a linear pathway for dissimilation of formaldehyde into the formation of CO2 was proposed based upon 13C nuclear magnetic resonance data. In addition to generating reducing power, this pathway was assumed to be important in detoxifying formaldehyde and supplying reducing equivalents (22). Based on enzyme activity measurements in crude extracts, B. methanolicus has been reported to possess a cyclic dissimilatory pathway (3). This route includes three biochemical steps (catalyzed by glucose phosphate isomerase, glucose phosphate dehydrogenase, and phosphogluconate dehydrogenase), converting fructose-6-phosphate into CO2 (Fig. 1). To date, no dissimilatory-pathway genes have been cloned from any B. methanolicus strains. Therefore, understanding the regulation of carbon flux to the RuMP pathway is important to be able to genetically alter the cell yield on methanol, as well as to reduce methanol toxicity.
Methylotrophy does not correlate well with traditional methods of bacterial classification, and it has been postulated that there has been an overemphasis on methylotrophy as a novel taxonomic trait (6, 13). One emerging theory is that such complex metabolic traits can be transferred between bacteria by lateral gene transfer, and thus natural plasmids as carriers of the genes should be of great significance (6, 13). The existence of methylotrophy inherited on plasmids was predicted decades ago (18), and the first evidence of plasmid-dependent methylotrophy in gram-positive bacteria was recently documented by us. We discovered that the only copy of the mdh gene, which is indispensable for growth on methanol by B. methanolicus, is carried by a 19.1-kb natural plasmid called pBM19 (8). To our knowledge, no similar findings have been reported in the literature for gram-negative methylotrophs. In the present report, we explore new biological functions of plasmid pBM19. By using real-time PCR, we show that the transcription of mdh and five more pBM19 genes (glpX, fructose/sedoheptulose bisphosphatase; fba, fructose bisphosphate aldolase; tkt, transketolase; pfk, phosphofructokinase (PFK); and rpe, ribulose phosphate epimerase) is induced upon growth in methanol. Two chromosomal genes (hps, hexulose phosphate synthase, and phi, phosphohexuloisomerase) are under coordinate control with the pBM19 genes, and together all these genes are involved in the fructose bisphosphate aldolase/sedoheptulose bisphosphatase variant of the RuMP pathway for methanol assimilation. The finding that the RuMP pathway is also important for methanol and formaldehyde detoxification should expand the fundamental role of plasmid pBM19. This knowledge could also be used to improve the methylotrophic properties of B. methanolicus, including both methanol assimilation and the methanol tolerance level, by metabolic engineering.
MATERIALS AND METHODS
Biological materials, growth conditions, and DNA manipulations.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5α was used as a standard cloning host, and recombinant cells were grown at 37°C in liquid or solid Luria-Bertani medium supplemented with ampicillin (100 μg/ml) or chloramphenicol (15 μg/ml) when appropriate. Recombinant E. coli procedures were performed as described elsewhere (25). B. methanolicus strains were grown at 50°C in the following media. SOBsuc medium is SOB medium (Difco) supplemented with 0.25 M sucrose. Solid medium is regeneration plates as described elsewhere (10). Mannitol growth of B. methanolicus was performed in Mann10 medium containing salt buffer, 1 mM MgSO4, vitamins, trace metals, 0.025% yeast extract (Difco), and mannitol (10 g/liter; Sigma), pH 7.2. Salt buffer, vitamins, and trace metals were essentially as for MV medium, described elsewhere (26). Mann10-Y medium is Mann10 without yeast extract, pH 7.0. Methanol growth of B. methanolicus was performed in MeOH200 medium, which is similar to Mann10, except that the mannitol is replaced with methanol (200 mM). Mixed medium is MeOH200 with 10 g/liter mannitol. A small amount of yeast extract was used in the defined media to avoid lag phases after inoculation. The yeast extract supports a limited biomass production (optical density at 600 nm [OD600] ≈ 0.2), and beyond this level, cell growth is not affected. Growth in shake flasks (500 ml) was performed in 100 ml medium at 200 rpm, and silicone sponge closures (Bellco) were used to decrease evaporation. Growth in 96-well format plates was performed in 100 μl medium at 950 rpm and 75% relative humidity. Specific growth rates were calculated by linear regression of semilogarithmic plots of optical density versus time over an 8-hour period from an OD600 of ∼0.5 to an OD600 of ∼4. Neomycin (25 μg/ml in SOBsuc medium; 5 μg/ml in defined media) and chloramphenicol (5 μg/ml) were supplied when appropriate. Plasmid and genomic DNAs from B. methanolicus were isolated by using QIAGEN Midi prep and DNeasy tissue kits (QIAGEN Gmbh, Hilden, Germany), respectively, according to the manufacturer's instructions. Bacterial growth was monitored by measuring the OD600. DNA sequencing was performed by using the BigDye kit from Applied Biosystems.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| B. methanolicus | ||
| MGA3 | Wild-type strain | 26 |
| MGA3C-A6 | MGA3 cured of natural plasmid pBM19 | 8 |
| E. coli | ||
| DH5α | General cloning host | BRL |
| Plasmids | ||
| pGEM-11zf | E. coli cloning vector; Apr | Promega |
| pTB1.9 | E. coli-B. subtilis shuttle vector; Apr Neor | 8 |
| pTB1.9mdhL | pTB1.9 carrying the mdh gene | 8 |
| pHP13 | E. coli-B. subtilis shuttle vector; Clmr | 14 |
| pHPhps+phib | pHP13 harboring hps and phi genes | This study |
Apr, ampicillin resistance; Neor, neomycin resistance; Clmr, chloramphenicol resistance.
This plasmid is described in Materials and Methods.
Isolation of total RNA and synthesis of cDNA.
B. methanolicus cells were grown in shake flasks in Mann10 or MeOH200 medium to exponential phase (OD600 = 1.0), and 6 ml cells was harvested by centrifugation (6,000 × g; 5 min). The pellets were resuspended in 1 ml fresh medium, and 2 ml RNA Protect (QIAGEN) was added, followed by incubation at room temperature for 5 min. The samples were centrifuged (6,000 × g; 5 min), and the resulting pellets were resuspended in 0.2 ml Tris-EDTA buffer (pH 8.0) supplemented with lysozyme (15 mg/ml) and mutanolysine (25 U/ml) (Sigma). Cell lysis was achieved by incubation at room temperature for 30 min. Total RNA was isolated by using the RNAqueous kit (Ambion) according to the manufacturer's descriptions. The material obtained was analyzed spectrophotometrically (OD260 and OD280), and 3 μg was treated with the DNA-free kit (Ambion) to degrade any contaminating DNA as described in the manufacturer's descriptions. The RNA materials were tested by PCR with primers rmp-F5 and rmp-R4, which correspond to the amplification of a 551-bp region of the B. methanolicus hps gene (8). In all cases, no product was obtained, confirming that the material was DNA free. The total RNA was then used as a template for cDNA synthesis by using a first-strand cDNA synthesis kit (Amersham) according to the instructions of the manufacturer. The resulting cDNAs were then used as templates for the real-time (RT)-PCR experiments.
Real-time PCR.
To optimize conditions for RT-PCR, a series of standard PCRs were run with different primer concentrations (1 to 5 pmol per 20 μl reaction mixture) and with different annealing temperatures (48°C to 65°C). B. methanolicus genomic DNA was used as a template, and the product yields were analyzed by gel electrophoresis (not shown). The optimized RT-PCR profile was as follows: segment 1 (1 cycle), 95°C for 10 min; segment 2 (40 cycles), 95°C for 30 s, 55°C for 60 s, and 68°C for 30 s; segment 3 (1 cycle), 95°C for 60 s, 55°C for 30 s, and slowly up to 95°C (dissociation curve). For each gene and intergenic region to be analyzed, a pair of RT-PCR primers was designed (Table 2). Care was taken to ensure that all primers had similar melting temperatures (around 60°C), lengths (18 to 20 nucleotides), and GC contents (50 to 55%). For each gene, the primers were designed for the amplification of 220 to 360 bp of the 5′-terminal coding regions, and for analysis of the cotranscription of individual genes, primers were designed for the amplification of 320- to 540-bp fragments representing intergenic regions. We used the iTaq SYBR Green Supermix with Rox (Bio-Rad), and amplification reaction mixtures contained 5 μl of diluted cDNA templates and 3 pmol of each primer in a final volume of 20 μl. RT-PCR products were detected by monitoring the increase in fluorescence using the Mx3000P cycler system (Stratagene). Cycle thresholds that intersected the amplification curves in the linear regions of the semilogarithmic plots were set for each specific primer pair and cDNA template. For each primer pair, a standard curve of cycle threshold values as a function of at least four different template dilutions (40- to 640-fold) was made, and from the resulting plots we calculated primer efficiencies, which were found to be above 90% in all cases. Inspection of the dissociation curves confirmed negligible levels of primer self-hybridizations. Amplification of 16S rRNA and the pBM19 replication initiator gene repB were both used for sample normalization, and the results of the quantitative RT-PCRs are expressed as stimulation indices (ratios of the amounts of RNA obtained from cells growing on methanol to the amounts from those growing on mannitol as a sole C source). All experiments were run in triplicate, and the mean values were calculated.
TABLE 2.
Deoxyoligonucleotide primers used for rt-PCR
| Primer name | Sequence (5′-3′) | Orientation and position in target |
|---|---|---|
| pBM19-primers | ||
| rpe-CF | CGGAGCCGATTACATTCA | Sense, 4432 in pBM19 |
| rpe-CR | GCCAGGATTAACCGTCAT | Antisense, 4778 in pBM19 |
| pfk-CF | AAGTGCCATCTCCACCAATC | Sense, 8142 in pBM19 |
| pfk-CR | CCAGGAATGAACGCTGCTAT | Antisense, 8419 in pBM19 |
| tkt-TR | AACGACCGCACGAATAGC | Sense, 8386 in pBM19 |
| tkt-TF | ATGGCCTGTGCCAATTCT | Antisense, 8916 in pBM19 |
| tkt-CF | CGATATCATTGCCGTCCTCA | Sense, 10436 in pBM19 |
| tkt-CR | ATGCCACAACAGGTCCGTTA | Antisense, 10772 in pBM19 |
| fbaT-rt1 | GGATGGCCTGATCCAACCTT | Sense, 11020 in pBM19 |
| fbaT-rt2 | CTCCACGGTGGTACAGGTAT | Antisense, 11382 in pBM19 |
| fba-RTB | CTTCTTGTCCACCGATACGA | Sense, 11564 in pBM19 |
| fba-RTA | CGTAACGGTACCAGTTGCTA | Antisense, 11776 in pBM19 |
| glpXT-R | CTGTCCAACAGCATATCC | Sense, 11934 in pBM19 |
| glpXT-F | GTCGCAGTAACGATTAGC | Antisense, 12476 in pBM19 |
| glpX-RTA | GTTCCGAGTTCCTCTCCAAT | Sense, 13311 in pBM19 |
| glpX-RTB | AAGCGTGTTCAGTCGTTAGC | Antisense, 13535 in pBM19 |
| mdhT-RT | ACGGTACGAGACATCATTCC | Sense, 13832 in pBM19 |
| mdhT-RT | CACGCGTTCCTACTGTTCAA | Antisense, 14156 in pBM19 |
| mdh-RTA | GCTGTATCGTGAGAGCTACC | Sense, 14944 in pBM19 |
| mdh-RTB | TCCACCAGCCAGCGTAATTG | Antisense, 15232 in pBM19 |
| parA-rt1 | TCTTCGGCACTGTTGAAGGA | Sense, 16916 in pBM19 |
| parA-rt2 | TCCAGCCTGAAGGATATAGC | Antisense, 17080 in pBM19 |
| repB-rt1 | GTCTGTTCGTCCTTGGACTT | Sense, 18220 in pBM19 |
| repB-rt2 | AATGCTTGGAGCCGAGGATA | Antisense, 18587 in pBM19 |
| Chromosomal primers | ||
| hps-RTA | GTAGCTGAGGTTCAGGAGTA | Sense, 54 in hps |
| hps-RTB | CTGCGATCATGTCAACAAGG | Antisense, 336 in hps |
| hpsT-rtF | TCATTGTTGGCGGCGGTATT | Sense, 549 in hps |
| hpsT-rtR | AGTGCTTCGGCTTCTTCATC | Antisense, 85 in phi |
| phi-rt1 | GAAGAAGCCGAAGCACTGGT | Sense, 68 in phi |
| phi-rt2 | TGATCGTTACAGCCGCGATG | Antisense, 339 in phi |
| act-RTA | CCTGGTGCTGTAGCTGTAAT | Sense, 128 in act |
| act-RTB | GCTTCGTCAAGTGTCAGTTC | Antisense, 463 in act |
| rRNA-1F | AAGATGGCTTCGGCTATCAC | Sense, 179 in 16SrRNA |
| rRNA-1R | CGAACGGTACTTGTTCTTCC | Antisense, 443 in 16SrRNA |
Construction of expression vector pHP13hps+phi.
The 1,700-bp DNA fragment including the hps and phi coding regions and 500-bp upstream sequences was PCR amplified from total DNA of MGA3 by using the following primers: rump-f10 (5′-TTTTCGGCCGCGCCAAACAAGACAAAGG-3′) and rump-r11 (5′-TTTTCGGCCGCTACTCGAGATTGGCATGTC-3′). PCRs were performed as described previously (8). Underlined in both primers are EagI restriction sites used for the cloning of the resulting PCR fragment into the NotI site of plasmid pGEM11-zf. From the resulting construct, the 1,756-bp EcoRI/HindIII insert was isolated and ligated into the corresponding sites of plasmid pHP13, yielding the vector pHP13hps+phi. This vector was verified by DNA sequencing. To simplify future cloning experiments, the complete pHP13 DNA sequence was determined.
C1 induction of B. methanolicus cultures.
Methanol or formaldehyde was gradually added to cultures growing exponentially in Mann10 medium. The formaldehyde concentrations used were the highest levels that did not cause significant growth perturbations of the cells. After inoculation, the OD600 was typically 0.15. Methanol was added to MGA3 and MGA3C-A6 cell cultures as follows: 7.5 mM at OD600s of 0.4 and 0.5, 15 mM at an OD600 of 0.6, and 22.5 mM at OD600s of 0.75 and 0.9. Formaldehyde was added to MGA3 as follows: 0.5 mM at OD600s of 0.4 and 0.5, 0.67 mM at an OD600 of 0.6, 1.0 mM at an OD600 of 0.75, and 1.5 mM at an OD600 of 0.9. Formaldehyde was added to MGA3C-A6 as follows: 0.5 mM at OD600s of 0.4 and 0.5 and 0.6 mM at OD600s of 0.6, 0.75, and 0.9. The cells were harvested at an OD600 of 1.0 for measurements of HPS plus phosphohexuloisomerase (PHI) activities as described below.
Preparation of crude extracts and measurements of HPS plus PHI activities.
B. methanolicus cells growing exponentially (OD600 = 1.0) in defined media were harvested and washed twice in ice-cold potassium phosphate buffer (50 mM; pH 7.6). The cells were disrupted by sonication as previously described (7). Coupled HPS-PHI activities were assayed by monitoring the fixation of formaldehyde with ribulose-5-phosphate (28). Protein concentrations were determined by the method of Bradford (Bio-Rad), using bovine serum albumin as a standard. All experiments were done in triplicate.
Growth rate analyses of B. methanolicus strains.
Phenotypic analyses were performed by monitoring the growth of B. methanolicus strains in liquid media. Ampoules of B. methanolicus cells were prepared from exponentially growing cultures (OD600 = 1.0 to 1.5) and stored at −80°C after 15% (vol/vol) glycerol was added. The ampoules were thawed, 250 μl of cell material was used to inoculate 100 ml Mann10 or MeOH200 medium, and the cells were grown to an OD600 of 0.5 to 1.5. From these cultures, 5 to 10% was inoculated into fresh and prewarmed media for growth rate analyses. At the time of inoculation, the methanol concentration in the MeOH200 inoculum was typically 150 mM. For the methanol sensitivity experiments, the cultures were grown in shake flasks in MeOH200 or Mann10 medium to an OD600 of 1.0 (about three generations after inoculation). Methanol was added to the cell cultures to increase the methanol concentrations up to 2,880 mM, and growth was monitored. Growth rates after methanol pulses were calculated based on at least five separate data points collected over a 5-hour period from the time of pulsing. For the formaldehyde sensitivity experiments, cultures were grown in well plates in Mann10-Y medium to an OD600 of 0.05 to 0.06 (two to three generations after inoculation). Ten microliters of various stock solutions of formaldehyde in Mann10-Y was added to bring the B. methanolicus cultures to final formaldehyde concentrations of 0.2 mM to 5.4 mM. Eight parallel cultures were run for each formaldehyde concentration.
Electroporation of B. methanolicus.
A method was developed for the transformation of plasmid pHP13 and its derivatives into B. methanolicus MGA3 using electroporation. Competent cells were prepared as follows: 250 μl culture from a frozen ampoule was used to inoculate 100 ml SOBsuc medium, and the cells were grown for 16 h. From this culture, 2 ml was transferred to shake flasks with 100 ml prewarmed SOBsuc medium, and cell growth was continued to an OD600 of 0.25. Cells (35 ml) were harvested by centrifugation (3,000 × g; 5 min), washed twice in 3.5 ml EP buffer (1 mM HEPES, 25% polyethylene glycol 8000, pH 7.0), and resuspended in 0.2 ml EP buffer. Competent cells were stored at −80°C. Competent cells (100 μl) were mixed with about 1 μg of plasmid DNA and incubated on ice for 30 min. The mixture was transferred to an ice-cold electroporation cuvette (0.2-cm electrode gap; Bio-Rad Laboratories), and the cells were exposed to a single electrical pulse using the Bio-Rad Gene-Pulser (200 Ω; 25 μF; 2.5 kV). The cells were grown in 5 ml of SOBsuc medium for 16 h, and 4 ml of the cell culture was transferred to a shake flask containing 100 ml of prewarmed SOBsuc medium supplemented with chloramphenicol. Growth was continued for 6 h, and the culture was plated on solid media with chloramphenicol and incubated overnight. From the plates, one single colony was picked and transferred to a shake flask containing 100 ml MeOH30 medium (similar to MeOH200 medium, but with 30 mM MeOH) supplemented with chloramphenicol. After 4 h of growth, an additional 170 mM methanol was added to the cell culture. The culture was grown to an OD600 of 1.0 to 1.5, and the ampoules were stored at −80°C after 15% (vol/vol) glycerol was added.
High-performance liquid chromatography analysis of methanol and mannitol.
To measure methanol and mannitol concentrations in the culture media, samples (0.8 ml) were collected from the cell cultures and added to cold perchloric acid (0.6 M; 0.2 ml), centrifuged (10,000 × g; 5 min), and filtered through 0.2-μm syringe filters before high-performance liquid chromatography analyses were performed. A Shimadzu chromatograph was used, equipped with an autoinjector (SIL-9A; Shimadzu, Japan), an Aminex HPX-87-H (Bio-Rad Laboratories) column at 45°C, and a refractive index detector (RID 6A; Shimadzu, Japan). A 5 mM H2SO4 solution was used as the eluent (0.6 ml/min). Standards of mannitol (5.0 g/liter) and methanol (300 mM) were used for calibration.
Nucleotide sequence accession number.
The complete pHP13 DNA sequence was deposited in the GenBank nucleotide sequence database under accession number DQ 297764.
RESULTS
Maintenance of pBM19 represents a burden for B. methanolicus growing on mannitol.
B. methanolicus is a restricted facultative methylotroph, and besides mannitol, few multicarbon sources can be consumed by this bacterium. We previously demonstrated that MGA3 can be cured of pBM19 by introducing the pBM19 derivative pTB1.9 and that the resulting strain, MGA3C-A6, had lost the ability to grow on methanol (8). Besides encoding mdh, the biological significance of pBM19 for maintaining the viability of B. methanolicus in the environment remains unknown. The pBM19 genes glpX, fba, tkt, pfk, and rpe encode proteins with proposed functions in the RuMP pathway (Fig. 1), but enzymes may be shared between several pathways, and it was of interest to test if pBM19 is also important for nonmethylotrophic growth. The specific growth rates of wild-type MGA3 in shake flask cultures on methanol and mannitol were found to be similar (0.32 h−1 ± 0.02 and 0.30 h−1 ± 0.02, respectively). Interestingly, the specific growth rate of the pBM19-cured strain MGA3C-A6 on mannitol is higher (0.37 h−1 ± 0.02) than that of its parental wild-type strain. We next exposed the wild-type MGA3 cells for prolonged growth in Mann10 medium, and after 70 and 140 generations, samples were collected from the growing culture and plated on solid medium, and single colonies were picked and tested for the ability to grow on methanol medium (MeOH200). After 70 generations, 2 of 10 colonies (20%) tested had lost the ability to grow on methanol, while after 140 generations, 16 of 20 colonies (80%) tested could not grow on methanol. All colonies tested grew well on mannitol medium (Mann10). To verify that the observed phenotypes were due to loss of pBM19, we isolated total DNA from selected colonies and used this material as templates for PCR analyses using primers directed toward both pBM19 and chromosomal targets (8). The results of these experiments confirmed that all colonies that had lost the ability to grow on methanol were B. methanolicus cured of pBM19 (data not shown). Total DNA isolated from the wild-type strain, MGA3, was used as a positive control template in these PCR experiments. Together, these data demonstrate that the maintenance of the natural plasmid pBM19 represents a metabolic burden for the B. methanolicus cells when growing on mannitol.
Six pBM19 genes, together with two chromosomal genes, are transcriptionally induced upon growth of B. methanolicus in methanol.
It has been shown that expression of genes may be selectively induced upon C1 growth in autotrophic (27) and methylotrophic (1, 3, 15, 17, 29) bacteria. To fully explore the biological function of pBM19 for methylotrophic growth, we analyzed the transcriptional regulation of a selection of its genes by RT-PCR. MGA3 cells were grown exponentially in defined mannitol (Mann10) and methanol (MeOH200) media, and samples were harvested for isolation of total RNA and concomitant cDNA synthesis (see Materials and Methods). The latter material served as a template for the RT-PCR analyses. Based on the growth rate data presented above, we expected that pBM19 replication should not be affected by the C source. This assumption was confirmed by RT-PCR analysis showing that the transcript levels of the pBM19 replication initiator gene repB are similar in cells utilizing mannitol and methanol (data not shown). To avoid the high template dilutions needed when analyzing 16S rRNA transcription levels, we chose to use repB as the internal standard in all further RT-PCR experiments. The results of these experiments (Table 3) show that mdh transcription is induced about threefold in cells growing on methanol medium compared to those growing on mannitol medium, which is in agreement with MDH enzyme activity data presented by others (4, 11). The transcript levels of glpX, fba, tkt, pfk, and rpe are also induced (between 6- and 40-fold) upon methanol growth, supporting the roles of these genes for methanol assimilation by B. methanolicus. The methanol-mediated induction differs considerably between the genes tested. The pfk transcription is very low upon growth in mannitol and strongly induced (40-fold) upon growth in methanol. This may suggest that the deduced pfk gene product, an ATP-dependent PFK protein (8), is needed only during C1 growth. This is similar to Amycolatopsis methanolica, in which the ATP-dependent (irreversible) PFK activity is induced by methanol, whereas pyrophosphate (PPi)-dependent PFK enzyme (reversible) is expressed upon growth on glucose (1).
TABLE 3.
Upregulation of transcription levels of selected genes in cells growing on methanol (methylotrophic) relative to cells growing on mannitol (nonmethylotrophic)a
| Gene | Localization | Biological function | Upregulationb |
|---|---|---|---|
| mdh | pBM19 | Methanol oxidation | 2.9× ± 0.4× |
| glpX | pBM19 | RuMP pathway | 9.0× ± 0.8× |
| fba | pBM19 | RuMP pathway | 17× ± 1.2× |
| tkt | pBM19 | RuMP pathway | 15× ± 1.5× |
| pfk | pBM19 | RuMP pathway | 40× ± 1.5× |
| rpe | pBM19 | RuMP pathway | 6.0× ± 0.2× |
| hps | Chromosome | RuMP pathway | 6.8× ± 0.8× |
| phi | Chromosome | RuMP pathway | 6.0× ± 1.0× |
| act | Chromosome | Methanol oxidation | 0.8× ± 0.05× |
| parA | pBM19 | pBM19 replication | 0.9× ± 0.1× |
Total RNA was isolated from cells growing in methanol (MeOH200) or mannitol (Mann10) medium and was used as templates for cDNA synthesis. The cDNA templates were used for RT-PCR experiments by using the primers listed in Table 2.
Upregulation, given as transcription level in methanol medium compared to in mannitol medium, was for each gene determined by using comparative quantification as described in the Materials and Methods section. Mean values from three independent experiments are given and standard errors are indicated.
Plasmid pBM19 does not encode a complete RuMP pathway in B. methanolicus, and we have cloned two chromosomal genes, hps and phi, with deduced roles in this metabolic route (8). The MDH activator gene act has also been described (16), and we proceeded to analyze the regulation of these three chromosomal genes by RT-PCR as described above. The results of these experiments (Table 3) show that transcription of hps and phi is induced approximately sixfold upon methanol growth. act transcription is not affected by the C source, which is contradictory to previous reports stating that mdh and act expression is under coordinated and methanol-induced control in B. methanolicus strain C1 (16). The reason for this discrepancy is unknown. As a negative control in the RT-PCR experiments, we chose the pBM19 plasmid partition gene parA (8), and as with repB, we expected its transcription not to be regulated by the C source. The result obtained confirmed that parA transcript levels are similar in cells growing on both media tested (Table 3). These data demonstrate that there is a coordinated up-regulation of two chromosomal and six pBM19 genes during growth of B. methanolicus on methanol.
Cotranscription of methylotrophy genes in B. methanolicus.
The pBM19 genes mdh, glpX, fba, tkt, and pfk are oriented as an operon (8), but the different transcript levels of these genes determined here suggest that they are expressed individually. The fba and tkt coding sequences are separated by 32 nucleotides, while the remaining intergenic regions are long (547 to 653 nucleotides). Sequence analysis identified a putative strong stem-and-loop structure in the intergenic region between the mdh and glpX coding sequences (calculated binding energy, −19.3 kcal) that may function as a transcription termination signal. No such structures were found in the remaining intergenic regions tested, and due to the high AT content (64 to 72%), it is difficult to predict promoter elements. To experimentally analyze for cotranscription of these genes, we designed primer pairs for the amplification of relevant intergenic regions and used them for RT-PCR analyses as described above. A similar strategy has been used to predict promoters for methanol oxidation genes in M. extorquens AM1 (30). The level of transcript covering the mdh/glpX intergenic region was below 0.5% of the level representing the glpX coding sequence, indicating that transcription is efficiently terminated downstream of the mdh coding region. The transcript levels detected for the tkt/pfk and the glpX/fba intergenic regions contribute to 7% and 23% of the total transcript levels of the pfk and fba coding sequences, respectively. The transcript level detected for the fba/tkt intergenic region was the same as that of the fba coding sequence and more than twice the level detected for the tkt coding sequence. Together, these results indicate that mdh, glpX, and pfk are mainly transcribed from endogenous promoters into single mRNAs, while fba and tkt are cotranscribed into one polycistronic mRNA. By using a similar strategy, we also demonstrated that the chromosomal hps and phi genes are cotranscribed into one polycistronic mRNA, similar to what has been reported for other methylotrophic bacteria (24, 29). The rpe and act genes were not included in these analyses due to their distant locations compared to the other methylotrophy genes.
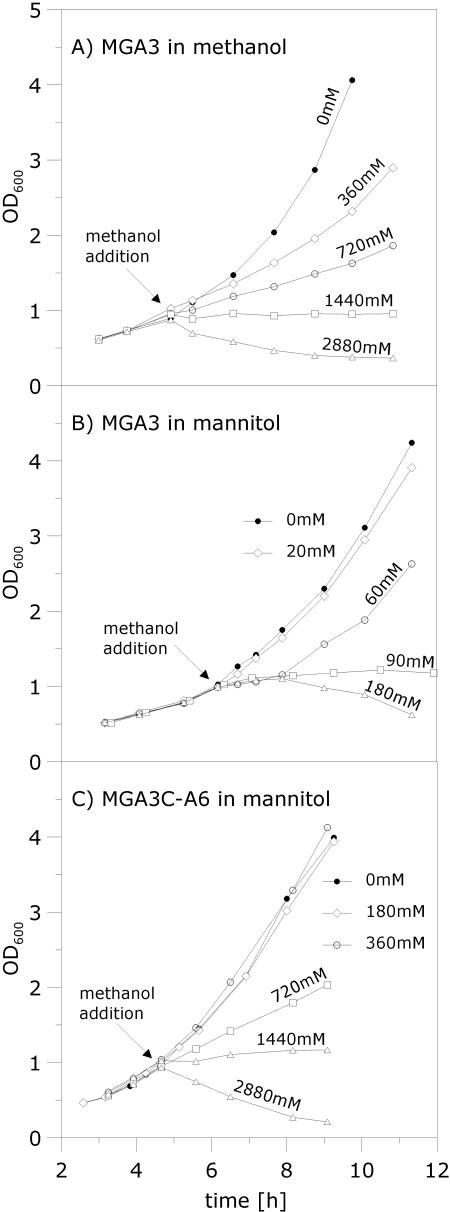
The methanol tolerance of the cells is modulated by transcription levels of mdh and the RuMP pathway genes.
For large-scale and high-cell-density fed-batch fermentations, methanol concentrations may not be uniform, and understanding the physiological response of B. methanolicus to fluctuating methanol levels is therefore of great importance. Previous studies have indicated that continuous B. methanolicus MGA3 cultures limited by methanol are sensitive to small (10 to 20 mM) methanol pulses (22) before cell growth is inhibited. B. methanolicus is reported to have a linear and a cyclic pathway (3, 22) for the dissimilation of formaldehyde into CO2, and the biological impact of the RuMP pathway on methanol tolerance is unknown. We hypothesized that evidence for a role of the RuMP pathway in methanol detoxification should be detected by comparing the methanol tolerance of the cells under conditions of induced and noninduced transcription of the RuMP pathway genes. To investigate this, we compared the effects of different methanol pulses on the growth rate of B. methanolicus MGA3 growing exponentially in shake flasks on either methanol or mannitol. In these phenotypic analyses, any toxic effects of the compounds added will have an effect on the growth rates of the cultures (19, 21, 28). The results of these experiments (Fig. 2A and B) show that cells already utilizing methanol can tolerate about 10-fold-higher methanol concentrations than cells growing on mannitol. Taken together with the RT-PCR results above, these data suggest that the induced state of the RuMP pathway genes is important for the tolerance of B. methanolicus for methanol. Possibly the expression of the cyclic dissimilatory pathway enzymes is also upregulated under these conditions and thus may contribute to the higher methanol tolerance. It was of interest to test if B. methanolicus MGA3 can discriminate between methanol and mannitol used for growth, and we therefore cultivated cells in mixed medium (see Materials and Methods) containing both these C sources. Cell growth, as well as depletion of methanol and mannitol in the medium, was monitored and used to calculate the consumption of each of the two C sources. The result of this experiment showed that approximately 75% of the total moles of carbon consumed was derived from the methanol, which implies that B. methanolicus utilizes both C sources when they are available. Interestingly, this result also indicates that methanol is consumed more effectively than mannitol under these conditions. Pulsing of methanol to this culture revealed that the methanol tolerance was similar to that of cells exclusively consuming methanol (data not shown). This implies that B. methanolicus can induce its RuMP pathway genes when growing on mannitol, as long as methanol is present, indicating that methanol utilization is not subject to catabolite control by this sugar.
FIG. 2.
Analysis of methanol tolerances of B. methanolicus strains. Growth perturbations caused by the adding of different methanol concentrations, between 0 mM (control) and 2,880 mM, to exponentially growing cell cultures were investigated. (A) Wild-type MGA3 growing in MeOH200 medium. (B) Wild-type MGA3 growing in Mann10 medium. (C) pBM19-cured MGA3C-A6 growing in Mann10 medium. The methanol was added at an OD600 of ∼1.0, as indicated by the arrows, and cell growth was continued and monitored for 6 to 8 h.
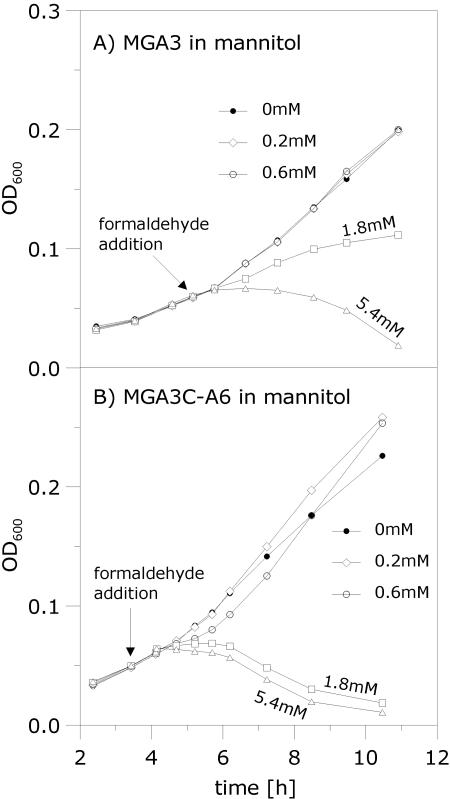
The pBM19-cured strain MGA3C-A6 has a higher methanol tolerance and a lower formaldehyde tolerance than the wild-type strain upon growth on mannitol.
The pBM19-cured strain MGA3C-A6 lacks mdh needed to convert methanol into formaldehyde (8), and it also lacks a functional RuMP pathway. Interestingly, during growth on mannitol, MGA3C-A6 cells can tolerate the addition of about a 10-fold-higher methanol concentration than wild-type MGA3 cells under similar conditions (Fig. 2B and C). This result suggests that the toxic effect observed in response to methanol pulses for the wild-type cells is largely due to accumulation of formaldehyde produced by the MDH-mediated conversion of methanol (Fig. 1). We analyzed the phenotypic responses of pulsing formaldehyde instead of methanol to the cell cultures upon growth in mannitol. The results show that the tolerance of MGA3C-A6 for formaldehyde is significantly lower than that of the wild-type strain under these conditions (Fig. 3), which supports our assumption that the RuMP pathway is important for formaldehyde detoxification. Together, these results demonstrate that both mdh and the RuMP pathway genes play major roles in methanol and formaldehyde tolerance by B. methanolicus.
FIG. 3.
Analysis of formaldehyde tolerances of B. methanolicus strains. Growth perturbations caused by the adding of different formaldehyde concentrations, between 0 mM (control) and 5.4 mM, to exponentially growing cell cultures were investigated. (A) Wild-type MGA3 growing in Mann10-Y medium. (B) pBM19-cured MGA3C-A6 growing in Mann10-Y medium. The formaldehyde was added at an OD600 of ∼0.05, as indicated by the arrows, and cell growth was continued and monitored for 6 to 8 h. The use of well plates in these experiments give rise to relatively low measured OD600 values, but the cell densities are comparable to those in Fig. 2 (data not shown).
Reintroduction of the homologous mdh gene in MGA3C-A6 reduces methanol tolerance to the wild-type level.
Inspection of the RT-PCR data indicated that mdh, in contrast to the RuMP pathway genes tested, is also transcribed at relatively high levels during mannitol growth, and our data imply that this causes a rapid accumulation of formaldehyde to toxic levels when these cells are pulsed with methanol. To further investigate this, we used plasmid pTB1.9mdhL (Table 1), which is a pBM19 derivative that carries mdh and no RuMP pathway genes. We have shown that this plasmid is not sufficient to restore methanol growth when introduced into the pBM19-cured strain MGA3C-A6 (8). To rule out the possibility that this observation is due to poor expression of mdh, hps, and phi, we analyzed strain MGA3C-A6(pTB1.9mdhL) upon growth in mannitol (μ = 0.26 h−1 ± 0.02) by RT-PCR as described above. The results showed that the transcription levels of all three genes are similar to those in the wild-type strain under these conditions (data not shown). We next performed phenotypic characterization of this strain upon growth in mannitol, and the results show that the reconstitution of the homologous mdh gene caused a decrease in the methanol tolerance level of MGA3C-A6 down to wild-type levels. As observed for the wild-type strain (Fig. 2B), the pulsing of 180 mM methanol inhibited the growth of MGA3C-A6(pTB1.9mdhL), while this level had no effect on the growth of the control strain, MGA3C-A6(pTB1.9), which lacks the mdh gene (data not shown). These results support our theory that the high expression of mdh causes the formation of formaldehyde when methanol is pulsed into the growth medium, and in the absence of a functional RuMP pathway, the formaldehyde rapidly accumulates to toxic levels in the cells.
Overexpression of homologous hps and phi genes in wild-type MGA3 causes a higher growth rate on methanol and improved methanol tolerance.
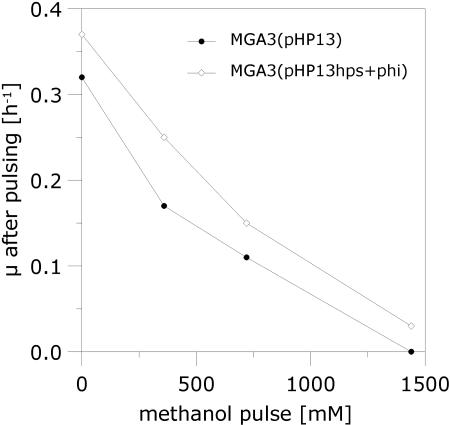
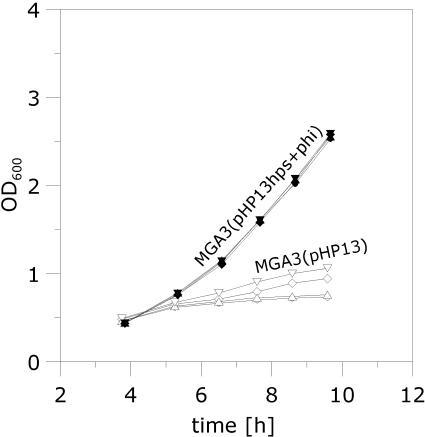
Our data suggest that the methanol tolerance of B. methanolicus is dependent on the coordinated expression of mdh and the RuMP pathway genes, in addition to the proposed linear and cyclic pathways of methanol dissimilation (3, 22). The pBM19 copy number is 15 per genome, while the hps and phi genes are present as single chromosomal copies (8). The last two genes are needed in the initial fixation phase of the RuMP pathway (Fig. 1). We hypothesized that increasing the number of copies of these two genes should elevate the expression levels of HPS and PHI activities in the cells, leading to a higher formaldehyde assimilation rate and hence an improved tolerance for methanol. Plasmid vectors and a protoplast transformation method for B. methanolicus have been described (10), but recombinant work in this bacterium has so far been severely limited by poor efficiency of gene delivery and also by the incompatibility of plasmid vectors with the natural plasmid pBM19 (8). To our knowledge, no recombinant B. methanolicus strains that maintain the ability to grow on methanol have been reported in the literature. We therefore developed a new method for electroporation of vector pHP13 (Table 1) and its derivatives into B. methanolicus (see Materials and Methods section), yielding recombinant strains which retain the ability to grow on methanol. The DNA fragment that includes the hps and phi operon was PCR amplified from MGA3 total DNA and cloned into pHP13 (see Materials and Methods). The resulting plasmid, pHP13hps+phi, expresses recombinant HPS-PHI activities in E. coli (data not shown), confirming the deduced biochemical functions of the hps and phi gene products. Next, we established MGA3(pHP13hps+phi) and MGA3(pHP13) and compared the HPS-PHI enzyme activities in crude extracts prepared from these two strains when grown on methanol or mannitol. The results (Table 4) show that MGA3(pHP13hps+phi) expresses two- to threefold-higher HPS-PHI activities than the control strain during growth on either of the two C sources tested. Thus, the elevated copy numbers of hps and phi result in elevated enzyme activities of the corresponding gene products. Interestingly, the specific growth rate on methanol (Table 4) of strain MGA3(pHP13hps+phi) is significantly higher (0.37 h−1) than that of the control strain MGA3(pHP13) (0.32 h−1), and it is also higher than that of the wild-type strain, MGA3 (0.32 h−1), under the conditions tested. We noticed that introduction of pHP13 causes growth inhibition to the cell exclusively when growing on mannitol, and the biological reason for this observation is unknown. We proceeded to monitor the cell growth of the recombinant strains in response to different methanol pulses (up to 1,440 mM). The results confirmed that MGA3(pHP13hps+phi) displays a higher specific growth rate than the control strain, MGA3(pHP13), under all conditions tested (Fig. 4). We extended the experiment by transferring cells growing exponentially in MeOH200 medium into fresh MeOH500 medium (containing 500 mM methanol) while monitoring cell growth (Fig. 5). The results show that while the cells harboring empty plasmid pHP13 can grow poorly, the recombinant cells maintained a high growth rate (0.28 h−1) under these conditions. These data indicate that methanol assimilation efficiency and methanol tolerance are closely connected traits of B. methanolicus that can be altered by increasing the gene dosage of the RuMP pathway genes hps and phi.
TABLE 4.
Recombinant expression of coupled HPS+PHI activity in B. methanolicus MGA3 cells growing exponentially in methanol (MeOH200) or mannitol (Mann10) medium
| Strain | C source | Specific growth rate (h−1)a | HPS+PHI activityb |
|---|---|---|---|
| MGA3(pHP13) | Mannitol | 0.26 ± 0.02 | 1,850 ± 250 |
| MGA3(pHP13) | Methanol | 0.32 ± 0.02 | 3,600 ± 600 |
| MGA3(pHP13hps+phi) | Mannitol | 0.25 ± 0.02 | 3,700 ± 300 |
| MGA3(pHP13hps+phi) | Methanol | 0.37 ± 0.02 | 11,000 ± 3,400 |
The corresponding values of the wild-type MGA3 cells are 0.32 h−1 (methanol) and 0.30 h−1 (mannitol); see Results.
The numbers given are nmol of substrate consumed per minute per mg of total protein in crude enzyme extracts, as described in Materials and Methods.
FIG. 4.
Phenotypic characterization of recombinant MGA3(pHP13hps+phi). Methanol was added to cells growing in MeOH200 medium to increase the methanol concentration to up to 1,440 mM, and cell growth was monitored. The methanol additions were performed at an OD600 of ∼1.0. Specific growth rates were calculated based on OD600 measurements over a 4- to 6-h period from the time of methanol addition. MGA3(pHP13) was included as a reference strain.
FIG. 5.
Growth of recombinant MGA3(pHP13hps+phi) under conditions of high methanol concentration. Cells growing exponentially in MeOH200 medium were transferred to MeOH500 medium, and cell growth was monitored. MGA3(pHP13) was included as a reference strain, and four parallels of each strain are plotted.
Formaldehyde, and not methanol, is the inducer of RuMP pathway genes in B. methanolicus.
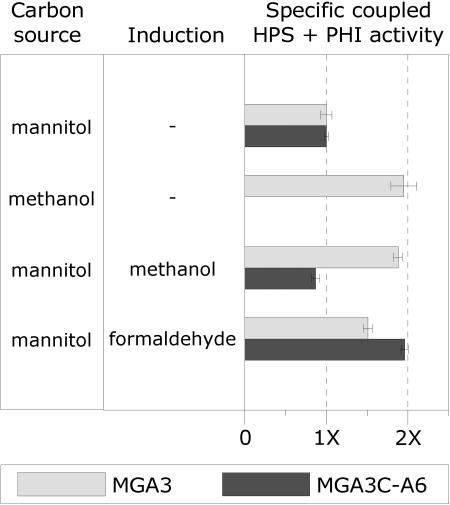
Two important questions remain regarding regulation of the RuMP pathway genes in B. methanolicus: (i) which C1 compound, methanol or formaldehyde, is the true transcriptional inducer and (ii) does this induction rely on any unknown pBM19 function? In the nonmethylotrophic bacteria Bacillus subtilis (28) and Burkholderia cepacia TM1 (21), the expression of HPS-PHI activities is induced when a small amount of formaldehyde (below 1.0 mM) is added to the growth medium, and it has been proposed that formaldehyde can induce HPS activity in B. methanolicus C1 (5). To answer the questions above, MGA3 and MGA3C-A6 cells were cultivated in mannitol medium, and the C1 compounds formaldehyde and methanol were then added to the growing cell cultures (see Materials and Methods). Both strains have the hps-phi operon, and the corresponding HPS-PHI activities were measured and used as parameters to determine induction of RuMP pathway genes under the different conditions. We have shown (Table 4) that the coupled specific HPS-PHI activities are approximately two times higher (defined as induced) in B. methanolicus cells growing on methanol than when they are growing on mannitol (noninducing conditions). The HPS-PHI activities of the two strains growing on mannitol and under noninduced growth conditions were similar. Interestingly, methanol addition induced HPS-PHI activities in the MGA3 cells upon growth in mannitol, while it had no such effect for MGA3C-A6 (Fig. 6). We hypothesized that the formaldehyde formed by the MDH-mediated conversion of methanol causes the induced phenotype observed exclusively in the wild-type strain. We performed similar experiments with formaldehyde as an inducer instead of methanol, and the results show that under these conditions HPS-PHI activity was also induced in the MGA3C-A6 strain. We noticed that MGA3 is not fully induced under these conditions, which may be due to assimilation of the formaldehyde by these cells. Together, our data confirm that formaldehyde is the inducer of the RuMP pathway genes hps and phi and that this induction is not dependent on any pBM19 function. In B. subtilis, the yckH gene is required for expression of the yckG and yckF genes encoding HPS-PHI activities. Plasmid pBM19 has no genes with similarity to yckH, and the presence of such a function on the genome of B. methanolicus is unknown.
FIG. 6.
Effect of induction (methanol and formaldehyde) of B. methanolicus cultures on the expression of HPS-PHI activities. The reported specific coupled HPS-PHI activities for each strain are relative to the value obtained during growth on mannitol (defined as 1×). Standard deviations based on two to four parallel experiments are indicated as error bars.
DISCUSSION
Our previous discovery of plasmid pBM19 represented the first documentation of plasmid-dependent methylotrophy in any microorganism (8). In the present report, we unravel new biological functions of this plasmid for B. methanolicus. By using in vivo quantitative PCR, together with phenotypic analyses of mutant and recombinant strains, we demonstrated that at least six pBM19 genes for methanol oxidation and assimilation via the RuMP pathway are transcriptionally upregulated during growth on methanol by this organism. Moreover, our data show that the RuMP pathway is important for the methanol and formaldehyde tolerance of the cells and that these phenotypes are largely determined by the induced state of the corresponding genes. The growth rates of B. methanolicus on methanol and mannitol are similar, and when both C sources are available, the majority of the total carbon consumed derives from methanol. Under such mixed growth, both mdh and the RuMP pathway genes are transcriptionally induced, indicating that methanol is a preferred C source for the bacterium. Proteome profiling of the facultative methylotroph M. extorquens AM1 indicated that expression of 68 of its proteins was induced upon methylotrophic growth, including proteins involved in the serine pathway for C1 assimilation (17). It was later shown that its serine cycle genes are under the transcriptional control of the positive regulator QscR (15). Whether induction of the B. methanolicus methylotrophy genes is mediated by a transcriptional regulator is not known. One potential candidate could be the pBM19 orf-1 gene product, which has a strong helix-turn-helix motif and also shows similarity to the transcriptional regulator DeoR (accession number ZP_00237682) of Bacillus cereus. Interestingly, despite the documented fundamental role of pBM19 for B. methanolicus, this plasmid represents a metabolic burden for the cells when they are growing nonmethylotrophically. Prolonged cell growth on mannitol selected for pBM19-cured cells with lost ability to grow on methanol and an improved growth rate on mannitol.
The biological advantage of carrying mdh and some RuMP pathway genes on a high-copy-number plasmid while keeping other RuMP pathway genes at single chromosomal copies is puzzling. Plasmid pBM19 is presumably capable of carrying methylotrophy to bacteria that can already to some extent detoxify formaldehyde, because in the absence of such a property, the uptake of pBM19 could be lethal to the cells if methanol is present in the environment. We assumed that this genetic organization, together with the transcription profiles of mdh versus the initial RuMP pathway genes hps and phi in particular, could be critical for the methanol assimilation rate of B. methanolicus. This assumption was verified by introducing multiple copies of the hps-phi operon, resulting in a recombinant B. methanolicus strain with improved growth rates on a broad range of methanol concentrations. To our knowledge, this represents the first example of an improved specific growth rate on methanol of any methylotroph by metabolic engineering. An analogous approach of the nonmethylotrophic bacterium Burkholderia cepacia TM1 has been reported. When this organism consumes vanillic acid, formaldehyde is formed as a by-product, and a recombinant B. cepacia strain overexpressing heterologous HPS-PHI activities displayed improved degradation of vanillic acid (21).
It is plausible to assume that the dissimilatory pathways (3, 22) could also play important roles for the C1 tolerance level and detoxification of formaldehyde by B. methanolicus. In B. methanolicus strain C1, it has been shown that the specific glucose phosphate dehydrogenase activity of the cyclic dissimilatory pathway is three- to fourfold higher in cells growing on methanol than in cells growing on glucose (3). At least four different pathways for bacterial formaldehyde detoxification, including oxidation and fixation, have been described (9, 19, 20), and it has been shown that some bacteria possess several of these pathways. Interestingly, B. subtilis mutants with yckG or yckF (encoding HPS and PHI activities, respectively) disrupted display only a minor decrease in formaldehyde tolerance, suggesting that the bacterium has alternative biochemical routes for detoxification of formaldehyde (28). We are at present on the way to cloning the formaldehyde dehydrogenase and formate dehydrogenase genes of B. methanolicus MGA3, in order to investigate their regulation and their impacts on formaldehyde detoxification. In conclusion, our study contributes new and valuable basic knowledge of the genes and their regulation involved in the methanol assimilation and detoxification of the methylotrophic bacterium B. methanolicus. In addition, we have demonstrated that this basic knowledge can be utilized to alter and improve the methylotrophic properties of B. methanolicus by metabolic engineering, and it should therefore be valuable in using this organism in large-scale bioprocesses.
Acknowledgments
We are grateful to Per Bruheim for assistance during development of the electroporation protocol for B. methanolicus. We thank Cornelia Schlee for help during growth inhibition experiments.
This work was supported by a grant from the Norwegian Research Council.
REFERENCES
- 1.Alves, A. M. C. R., G. J. W. Euverink, H. Santos, and L. Dijkhuizen. 2001. Different physiological roles of ATP- and PPi-dependent phosphofructokinase isoenzymes in the methylotrophic actinomycete Amycolaptis methanolica. J. Bacteriol. 183:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, C. 1991. Assimilation of carbon by methylotrophs, p. 79-109. In I. Goldberg and J. S. Rokem (ed.), Biology of methylotrophs. Butterworth-Heinemann, Boston, Mass. [DOI] [PubMed]
- 3.Arfman, N., E. M. Watling, W. Clement, R. J. van Oosterwijk, G. E. De Vries, W. Harder, M. M. Attwood, and L. Dijkhuizen. 1989. Methanol metabolism in thermotolerant methylotrophic Bacillus strains involving a novel catabolic NAD-dependent methanol dehydrogenase as a key enzyme. Arch. Microbiol. 152:280-288. [DOI] [PubMed] [Google Scholar]
- 4.Arfman, N., L. Dijkhuizen, G. Kirchof, W. Ludwig, K.-H. Schleifer, E. S. Bulygina, K. M. Chumakov, N. I. Govorhukhina, Y. A. Trotsenko, D. White, and R. J. Sharp. 1992. Bacillus methanolicus sp. nov., a new species of thermotolerant, methanol utilizing, endospore-forming bacteria. Int. J. Syst. Bacteriol. 42:439-445. [DOI] [PubMed] [Google Scholar]
- 5.Arfman, N., K. J. de Vries, H. R. Moezelaar, N. M. Arrwood, G. K. Robinson, M. van Geel, and L. Dijkhuizen. 1992. Environmental regulation of alcohol metabolism in thermotolerant methylotrophic Bacillus strains. Arch. Microbiol. 157:372-378. [DOI] [PubMed] [Google Scholar]
- 6.Boucher, Y., C. J. Douady, R. T. Papke, D. A. Walsh, M. E. Boudreau, C. L. Nesbo, R. J. Case, and W. F. Doolittle. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37:283-328. [DOI] [PubMed] [Google Scholar]
- 7.Brautaset, T., M. D. Williams, R. D. Dillingham, C. Kaufmann, A. Bennaars, E. Crabbe, and M. C. Flickinger. 2003. The role of the Bacillus methanolicus citrate synthase II gene, citY, in regulating the secretion of glutamate in lysine-secreting mutants. Appl. Environ. Microbiol. 69:3986-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brautaset, T., Ø. M. Jakobsen, M. C. Flickinger, S. Valla, and T. E. Ellingsen. 2004. Plasmid-dependent methylotrophy in thermotolerant Bacillus methanolicus. J. Bacteriol. 186:1229-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chistosterdova, L., L. Gomelsky, J. A. Vorholt, M. Gomelsky, Y. D. Tsygankov, and M. E. Lidstrom. 2000. Analysis of two formaldehyde oxidation pathways in Methylobacillus flagellatus KT, a ribulose monophosphate cycle methylotroph. Microbiology 146:233-238. [DOI] [PubMed] [Google Scholar]
- 10.Cue, D., H. Lam, R. L. Dillingham, R. S. Hanson, and M. C. Flickinger. 1997. Genetic manipulation of Bacillus methanolicus, a gram-positive thermotolerant methylotroph. Appl. Environ. Microbiol. 63:1406-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Vries, G. E., N. Arfman, P. Terpstra, and L. Dijkhuizen. 1992. Cloning, expression, and sequence analysis of the Bacillus methanolicus C1 methanol dehydrogenase gene. J. Bacteriol. 174:5346-5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dijkhuizen, L., P. R. Levering, and G. E. De Vries. 1992. The physiology and biochemistry of aerobic methanol-utilizing gram negative and gram positive bacteria, p. 149-181 In J. C. Murrell and H. Dalton (ed.), Methane and methanol utilizers. Plenum Press, New York, N.Y.
- 13.Gogarten, J. P., W. F. Doolittle, and J. G. Lawrence. 2002. Procaryotic evolution in light of gene transfer. Mol. Biol. Evol. 19:2226-2238. [DOI] [PubMed] [Google Scholar]
- 14.Haima, P., S. Bron, and G. Venema. 1987. The effect of restriction and shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol. Gen. Genet. 209:335-342. [DOI] [PubMed] [Google Scholar]
- 15.Kalyuzhnaya, M. G., and M. E. Lidstrom. 2005. QscR-mediated transcriptional activation of serine cycle genes in Methylobacterium extorquens AM1. J. Bacteriol. 187:7511-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloosterman, H., J. W. Vrijbloed, and L. Dijkhuizen. 2002. Molecular, biochemical, and functional characterizatiuon of a nudix hydrolase protein that stimulates the activity of a nicotinoprotein alcohol dehydrogenase. J. Biol. Chem. 277:34785-34792. [DOI] [PubMed] [Google Scholar]
- 17.Laukel, M., M. Rossignol, G. Borderies, U. Völker, and J. A. Vorholt. 2004. Comparison of the proteome of Methylobacterium extorquens AM1 grown under methylotrophic and nonmethylotrophic conditions. Proteomics 4:1247-1264. [DOI] [PubMed] [Google Scholar]
- 18.Lidstrom, M. E., and A. E. Wopat. 1984. Plasmids in methylotrophic bacteria: isolation, characterization, and hybridization analysis. Arch. Microbiol. 140:27-33. [DOI] [PubMed] [Google Scholar]
- 19.Marx, C. J., L. Chistosterdova, and M. E. Lidstrom. 2003. Formaldehyde-detoxifying role of the tetrahydromethanopterin-linked pathway in Methylobacterium extorquens AM1. J. Bacteriol. 185:7160-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marx, C. J., J. A. Miller, L. Chistoserdova, and M. E. Lidstrom. 2004. Multiple formaldehyde oxidation/detoxification pathways in Burkholderia fungorum LB400. J. Bacteriol. 186:2173-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitsui, R., Y. Kusano, H. Yurimoto, Y. Sakai, N. Kato, and M. Tanaka. 2003. Formaldehyde fixation contributes to detoxification for growth of a nonmethylotroph, Burkholderia cepacia TM1, on vanillic acid. Appl. Environ. Microbiol. 69:6128-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pluschkell, S. B., and M. C. Flickinger. 2002. Dissimilation of [13C]methanol by continuous cultures of Bacillus methanolicus MGA3 at 50°C studied by 13C NMR and isotope-ratio mass spectrometry. Microbiology 148:3223-3233. [DOI] [PubMed] [Google Scholar]
- 23.Reizer, J., A. Reizer, and M. H. Saier, Jr. 1997. Is the ribulose monophosphate pathway widely distributed in bacteria? Microbiology 143:2519-2520. [DOI] [PubMed] [Google Scholar]
- 24.Sakai, Y., R. Mitsui, Y. Katayama, H. Yanase, and N. Kato. 1999. Organization of the genes involved in the ribulose monophosphate pathway in an obligate methylotrophic bacterium, Methylomonas aminofaciens 77a. FEMS Microbiol. Lett. 176:125-130. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Schendel, F. J., C. E. Bremmon, M. C. Flickinger, M. Guettler, and R. S. Hanson. 1990. l-Lysine production at 50°C by mutants of a newly isolated and characterized Bacillus sp. Appl. Environ. Microbiol. 56:963-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van den Bergh, E. R. E., S. C. Baker, R. J. Taggers, P. Terpstra, E. C. Woudstra, L. Dijkhuizen, and W. G. Meijer. 1996. Primary structure and phylogeny of the Calvin cycle enzymes transketolase and fructose-bisphosphate aldolase of Xanthobacter flavus. J. Bacteriol. 178:888-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yasueda, H., Y. Kawahara, and S. I. Sugimoto. 1999. Bacillus subtilis yckG and yckF encode two key enzymes of the ribulose monophosphate pathway used by methylotrophs, and yckH is required for their expression. J. Bacteriol. 181:7154-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yurimoto, H., R. Hirai, H. Yasueda, R. Mitsui, Y. Sakai, and N. Kato. 2002. The ribulose monophosphate pathway operon encoding formaldehyde fixation in a thermotolerant methylotroph, Bacillus brevis S1. FEMS Microbiol. Lett. 214:189-193. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, M., and M. E. Lidstrom. 2003. Promoters and transcripts for genes involved in methanol oxidation in Methylobacterium extorquens AM1. Microbiology 149:1033-1040. [DOI] [PubMed] [Google Scholar]