Abstract
EmbR, a putative transcriptional regulator from Mycobacterium tuberculosis, is homologous to the OmpR class of transcriptional regulators that possess winged helix-turn-helix DNA binding motifs. In contrast to other OmpR-like response regulators that are usually phosphorylated and controlled by histidine kinases, EmbR was recently shown to be phosphorylated by the cognate mycobacterial serine/threonine kinase PknH. Despite the in vitro evidence of phosphorylation and interaction between the kinase and regulator, the physiological function of the PknH-EmbR pair is still unknown. We identify the embCAB operon encoding arabinosyltransferases in M. tuberculosis as the cellular target of EmbR. Phosphorylation of EmbR enhances its DNA binding activity towards promoter regions of embCAB genes. In vivo studies involving expression of PknH in Mycobacterium smegmatis established its positive regulatory effect on transcription of the embCAB operon via phosphorylation of EmbR. Interestingly, increased transcription of embC, catalyzing arabinosylation of lipomannan (LM) to lipoarabinomannan (LAM), results in a high LAM/LM ratio, which in turn is a crucial factor in mycobacterial virulence. The PknH-mediated increase in the transcription of embAB genes significantly alters resistance to ethambutol, a frontline antituberculosis drug known to target embAB genes. These findings and in vivo upregulation of PknH inside the host macrophages suggest a functionally relevant signaling mechanism involving the PknH-EmbR-embCAB system.
Tuberculosis is a leading cause of human death by an infectious agent, killing approximately 2 million people each year. Mycobacterium tuberculosis, the causative agent of tuberculosis, is one of the most complex pathogens. Recent studies have shown that M. tuberculosis has evolved a number of strategies to survive and persist in the hostile environment of macrophages (13, 28). An important key to the success of pathogenic mycobacteria is the unusual cell wall structure and its interaction with the immune system. The cell wall components are tailored by various complex mechanisms to facilitate the survival of mycobacteria during infection (3, 21). One such mechanism is the arabinosylation of lipomannan (LM) to lipoarabinomannan (LAM), a cell wall lipoglycan that averts the host's proinflammatory response, which can otherwise be evoked by its precursor LM (4). This arabinosylation is catalyzed by an arabinosyltransferase encoded by the embC gene (40). The embC gene is present in an operon with two other arabinosyltransferases, embA and embB, which are involved in the arabinosylation of arabinogalactan (AG), a component of the mycolyl-AG-peptidoglycan cell wall complex (3, 4, 11). Thus, the blocking of arabinosyltransferases and thereby the arabinan synthesis would lead to concomitant disruption of both LAM and AG, making this pathway a potential target for therapeutic intervention.
Initially identified as the major target of ethambutol (an effective antimycobacterial drug) in Mycobacterium avium, the emb genes were found to be ubiquitously present in all mycobacteria (2). In contrast to other mycobacterial species, which contain contiguous embCAB genes, the M. avium emb cluster contains only the embAB genes. Further, a putative transcriptional regulator in M. avium, EmbR, is proposed to modulate the transcription of the downstream emb genes, although this notion lacks direct experimental evidence. A role for embR in modulating the expression of embA and embB in M. avium was originally suggested by the characteristic genetic organization: regulatory protein-promoter region-structural gene (2). However, a similar gene arrangement does not exist in either M. tuberculosis or Mycobacterium smegmatis. In both of these mycobacterial species, an embR homolog is located 2 Mb apart from the embCAB locus (7). Thus, the presence of an additional embC gene and the differential genomic organization of EmbR in M. tuberculosis, with respect to M. avium, prompted us to investigate the role of EmbR in regulating transcription of embCAB genes in M. tuberculosis.
EmbR is a multidomain protein possessing three major domains, namely, DNA binding winged helix-turn-helix (W-HTH) domain, bacterial transcription activation domain, and a forkhead-associated (FHA) domain. The FHA domain present in EmbR is a newly recognized phosphoprotein recognition domain. Recently, this domain was found to play a crucial role in the interaction and phosphorylation of EmbR by PknH, a serine/threonine protein kinase (STPK) of M. tuberculosis (22). PknH and other STPKs have been proposed to mediate signaling between mycobacteria and host cells to establish an environment that is favorable for survival of mycobacteria, and thus kinases have been identified as potential drug targets (1, 17, 31, 36). Interestingly, a mutation in the EmbR FHA domain was shown to be associated with ethambutol (EMB) resistance (26), suggesting that alteration in this “kinase-interacting domain” of EmbR can result in an ethambutol resistance phenotype. This observation provided indirect evidence for the existence of some functional relationship between PknH, EmbR, the embCAB operon, and ethambutol resistance in M. tuberculosis.
In this report, we identify and characterize EmbR-dependent transcriptional regulation of embCAB genes. The regulatory function of EmbR was dependent on its phosphorylation by PknH, which enhances its DNA binding activity. In vivo studies demonstrate that the transcription of embCAB genes is enhanced in response to increased phosphorylation of EmbR. Identification of the regulatory effects of the PknH-EmbR pair on the LAM/LM ratio and ethambutol resistance are the novel aspects of the signal relay involving these proteins.
MATERIALS AND METHODS
Bacterial culture and growth conditions.
Mycobacterial strains were grown in Middlebrook 7H9 broth supplemented with 0.5% glycerol and 10% albumin-dextrose-catalase at 37°C with shaking at 220 rpm for 3 to 4 weeks. The Escherichia coli strains were grown in Luria-Bertani (LB) broth or on LB agar plates at 37°C with shaking at 220 rpm.
Plasmid construction, mutagenesis and protein purification.
Genomic DNA isolated from M. tuberculosis was used as a template for amplification of the gene coding for EmbR (Rv1267c). The 1,167-bp embR gene was amplified using a 5′ primer containing a BamHI site (TATGGATCCATGGCTGGTAGCGCGACAGTGGAGAAGCGG) and a 3′ primer containing a HindIII site (TATAAGCTTCTACGTGCCGCCATG CGTCCCCGCG). This DNA fragment was digested by BamHI and HindIII and ligated into a pET28a vector digested with the same enzymes, thus yielding pET28-embR. To construct an embR gene fragment with deletion of the W-HTH domain, PCR amplification was carried out by using pET28-embR as a template and the following primers: a 5′ primer containing a BamHI site (AAACGCGGATCCATGCTGACCCCACC) and a 3′ primer containing a HindIII site (TATAAGCTTCTACGTGCCGCCATGCGTCCCCGCG). The amplified fragment was then restricted by BamHI and HindIII enzymes and ligated into pET28a vector previously digested with the same enzymes, yielding pET28-embRΔN. The oligonucleotides used to create embR mutants with a substitution of alanine for glycine100 and tyrosine101 were 5′ CGGCTTATCGGCTCAGCAT and 5′ CGGGTGCTCGGCTCAGCAT, respectively (underlined bases mark the mutation to alanine), and the corresponding plasmids were designated pET28-embR-G100A and pET28-embR-Y101A. The sequences of clones were confirmed by DNA sequencing using an automated sequence analyzer (model 3100, ABI). EmbR and its mutants were expressed as His-tagged fusion proteins and purified under denaturing conditions using Ni-nitrilotriacetic acid resin as per the manufacturer's instructions. Polyclonal antiserum against purified EmbR was raised in rabbit. PknH was expressed and purified as in previous studies (30). PknH and its K45M mutant were cloned into pSD5 vector and expressed in M. smegmatis as described previously (9).
Gel mobility shift assay.
For the protein-DNA binding assay, 32P-labeled probe DNA was prepared by end labeling using Polynucleotide kinase (10 U/20-μl reaction). Labeled PCR products representing different promoter regions were incubated with various amounts of EmbR, EmbR mutants, and phosphorylated EmbR at 4°C for 30 min in buffer containing 10 mM Tris-HCl (pH 7.0), 1 mM dithiothreitol, 1 mM EDTA, and 10% glycerol in a total volume of 30 μl. After incubation, complexes and free DNA were resolved by 5% nondenaturing polyacrylamide gels with a running buffer containing 40 mM Tris-HCl (pH 7.8), 20 mM sodium acetate, and 1 mM EDTA. Gels were then dried and subjected to autoradiography. After densitometric analysis, data were analyzed as increases (n-fold) in DNA binding upon EmbR phosphorylation relative to unphosphorylated protein. The statistical significance was tested with Student's t test after repeating the experiment three times. Primer pairs used for amplification of different upstream regions are the following: embR-Rv1268c intergenic region, 5′ ACTTCATGGCCGTCACCACCTGAA 3′ (forward primer) and 5′ TTCTCCACTGTCGCGCTACCAGCCATT 3′ (reverse primer); embC-embA intergenic region, 5′ AGCGGTTGACGCCTTACTACCCCG 3′ (forward primer) and 5′ CCAGAAGATGGTCGCGGTGGTTTGGTT 3′ (reverse primer); embA-embB overlap region, 5′ TTCCTGTTCACCCAGGCGCTGCTGCGCA 3′ (forward primer) and 5′ GCCACCGACAACACAAAGCCAATCA 3′ (reverse primer); and embC upstream region, 5′ GGCAGCCGCCGACCGTCTTCCTCATG 3′ (forward primer) and 5′ GACCCGCCACCACAGCGACGTAC 3′ (reverse primer).
Immunoprecipitation and reverse transcriptase PCR (RT-PCR) analysis.
Whole-cell lysates were prepared from M. smegmatis cells, and EmbR was precipitated for 2 h at 4°C with anti-EmbR antibody raised in rabbits. Immunocomplexes were recovered by adding protein A-Sepharose (SIGMA) as per the manufacturer's instructions and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting using anti-EmbR and antiphosphothreonine antibodies (SIGMA).
RT-PCR analysis was done as described previously (30), and the following primer pairs were used for amplifying transcripts of the emb genes: for embA, 5′ CGCGGGCGCTGATTTTATGGGTAT 3′ and 5′ CGGGCGACGGTCAGAAGGTAGC 3′; for embC, 5′ AGCAAACTACCGGATCGCCCGGTA 3′ and 5′ TGAATGTCAACCGCTGACAGGT 3′; and for embB, 5′ CGCCGCTGCTGCCCGTCGTG 3′ and 5′ TGAGCCCCGCCCGTCCAACTG 3′.
In vitro kinase assay.
The in vitro kinase reactions were carried out routinely as described previously (30). For resin-bound kinase assays, purified EmbR was phosphorylated by PknH in the kinase buffer (25 mM Tris-HCl [pH 7.4], 10 mM MgCl2, 1 mM dithiothreitol) using a modified protocol as described previously (6). The phosphorylated EmbR was eluted from Ni-nitrilotriacetic acid beads using elution buffer (200 mM imidazole in wash buffer). After elution, phosphorylated EmbR was dialyzed against buffer (40 mM Tris, pH 7.6, and 10% glycerol) and stored at −20°C until further use.
Extraction of LAM and LM and silver-PAS.
Lipoglycans were extracted from M. smegmatis expressing PknH (MS-PknH), M. smegmatis expressing the K45M mutant of PknH (MS-PknHK45M), or M. smegmatis carrying vector alone (MS-pSD5) grown to mid-log phase and visualized by silver-periodic acid-Schiff staining (silver-PAS) as described previously (40). Briefly, 50 ml of culture was pelleted and extracted with an equal volume of solvent (10:10:3, chloroform/methanol/water) at 55°C for 30 min. After centrifugation, the residual pellet was extracted with an equal volume of water (15 ml) and phenol. The aqueous layer (containing LAM, LM, and phosphatidyl-myoinositol mannoside) was treated with DNase and RNase at 37°C for 30 min followed by treatment with proteinase K for 2 h at 60°C. The treated sample was microdialyzed and was subjected to lyophilization. After hexose estimation, equal amounts of samples were loaded and resolved by SDS-PAGE. The lipoglycans were visualized by silver-PAS (40), and the relative ratio of LAM to LM was calculated by densitometric analysis. The experiment was repeated three times, and the LAM/LM ratio for different mycobacterial strains was presented as the mean ± standard error of the mean. The statistical analysis was performed using Student's t test. The identity of the lipoglycans was confirmed by immunoblot analysis of LAM and LM preparations from mycobacteria using the monoclonal anti-LAM antibody CS-35. The monoclonal CS-35 antibody was kindly provided by John T. Belisle, Colorado State University, Fort Collins, Colo.
Drug resistance assays.
M. smegmatis transformants carrying PknH, PknHK45M, or pSD5 were plated in duplicate on 7H10 medium with or without ethambutol at various drug concentrations. Isolates were scored as EMB resistant if growth appeared on both types of plates after a 3-day incubation. The MIC, determined by plating on medium containing 0 to 2.5 μg/ml of EMB in increments of 0.25 μg/ml, was defined as the lowest concentration of EMB that completely inhibited growth after 3 to 5 days of incubation. Control strains contained the cloning vector pSD5.
Macrophage infection and quantitative real-time reverse transcription-PCR.
J774A.1 mouse macrophages were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (Gibco Invitrogen Corporation, NY). For each time point, 6 × 107 to 8 × 107 macrophages were infected with a multiplicity of 7 to 10 bacteria per macrophage. Uptake of bacteria by macrophages was analyzed by microscopy and serial dilutions of culture medium on 7H11 agar plates. Extracellular bacteria were removed by washing with cell culture medium prewarmed to 37°C before harvesting of M. tuberculosis. After 24, 48, or 72 h of infection, macrophages were lysed with 0.1% Triton X-100. Cell debris was removed by centrifugation at 200 × g for 5 min at 4°C, and the bacilli in the supernatant were harvested by centrifugation at 2,500 × g for 15 min at 4°C. Bacteria were disrupted by bead beating, RNA was prepared as previously described (10), and cDNA was prepared using gene-specific primers. Real-time PCR was performed using specific TaqMan probes. A reaction lacking reverse transcriptase was performed for every RNA sample. The major housekeeping sigma factor gene sigA was used as an internal control for the normalization of mRNA levels (19). Gene induction ratios were calculated from the comparison with RNA from bacteria grown to mid-log phase in 7H9. RNA was isolated from two independent macrophage infections.
RESULTS
In vitro interaction of EmbR with embCAB genes.
EmbR, a transcriptional regulator suspected of regulating emb genes, is phosphorylated by a cognate serine/threonine kinase of M. tuberculosis, PknH. The embA and embB genes can be expressed from their own individual promoters (11), but the genetic organization and primer extension analysis of the embCAB genes also reveal that, in addition, a polycistronic mRNA encoding the three Emb proteins could also be synthesized in M. tuberculosis (34). Therefore, the questions of whether EmbR-mediated regulation of emb genes is at the operon or individual gene level and whether the genes regulated by EmbR include only the embAB genes or embC is also transcriptionally regulated by EmbR in addition to the other two emb genes remain unanswered.
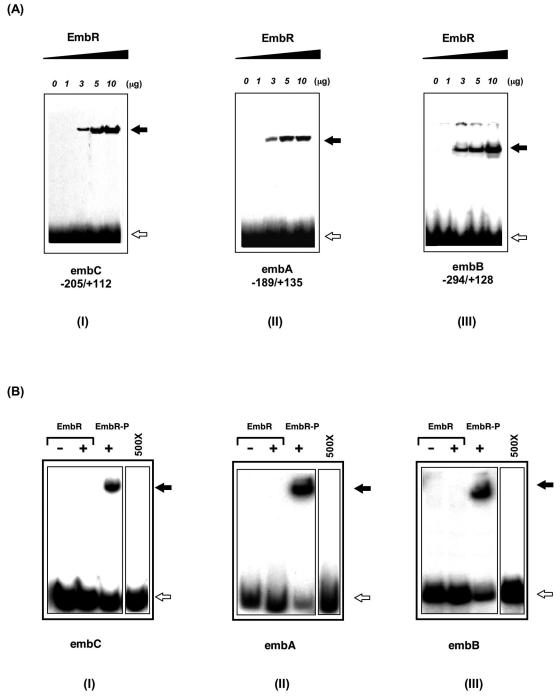
Therefore, as a first step, the possibility of EmbR binding to the upstream region of the embCAB operon in M. tuberculosis as well as the embC-embA and embA-embB intergenic region was evaluated by gel mobility shift assays. Altered electrophoretic mobility, indicative of EmbR binding to DNA, was detected for all three regions with a minimum of 3 μg of EmbR used (Fig. 1A). The binding was found to be specific, as EmbR was unable to bind to the upstream region of the embR gene itself. Further, the addition of the embR upstream region as a nonspecific competitor did not affect binding of EmbR to embCAB upstream regions (data not shown). However, we observed that EmbR binds to embCAB with much lower affinity than other DNA binding proteins. The gel mobility shift assay used here is a standard procedure for the detection of DNA-protein interaction, and under these conditions, for example, only 0.04 μg of AdpA or 0.5 μg of ArpA is sufficient to bind to the respective target DNA sequences (24, 38). In view of the fact that EmbR was recently shown to be phosphorylated by PknH, the effect of such phosphorylation on DNA binding activity of EmbR was examined. The phosphorylated form of EmbR was prepared by in vitro phosphorylation, and a similar gel mobility shift assay with the upstream regions of emb genes was performed. Following its phosphorylation, the strength of DNA binding by EmbR increased manyfold, with only 0.3 μg of protein bringing about a mobility shift similar to that seen for 3 μg of unphosphorylated EmbR (Fig. 1B). The strong retarded signal observed with phosphorylated EmbR for all three regions was lost when competed with a 500× amount of cold probe (data not shown). Quantification of the electrophoretic mobility shift assay blots for EmbR-DNA interaction revealed a phosphorylation-dependent increase in the DNA binding activity of EmbR towards upstream regions of emb genes (increases in DNA binding upon EmbR phosphorylation [n-fold]: embC, 9.4 ± 1.7; embA, 10.7 ± 1.4; and embB, 8.6 ± 1.8 [n = 3, P < 0.01]). The interaction of EmbR with individual upstream regions of embCAB genes suggests that emb genes might serve as in vivo targets of EmbR, and its phosphorylation-dependent DNA binding activity suggests that the PknH-mediated phosphorylation of EmbR is crucial for its interaction with upstream regions of emb genes.
FIG. 1.
Phosphorylation-dependent binding of EmbR to upstream regions of embCAB genes. (A) Gel mobility shift assays showing binding of EmbR to the promoter region of embCAB genes. An increasing amount of EmbR was incubated with 32P-labeled upstream regions of embC, embA, and embB genes at 4°C for 30 min. After incubation, EmbR-DNA complexes and free DNA were separated by nondenaturing polyacrylamide gel electrophoresis and subjected to autoradiography. The positions of EmbR-bound (solid arrows) and free (open arrows) probes are shown. (B) Phosphorylation of EmbR enhances its DNA binding activity. A total of 0.3 μg of unphosphorylated or phosphorylated EmbR (EmbR-P) was subjected to gel mobility shift analysis with 32P-labeled probes of upstream regions of embC, embA, and embB genes as indicated. The positions of EmbR-bound (solid arrow) and free (open arrow) probes are shown. Binding of phosphorylated EmbR with all three regions was examined in the presence of an excess amount (500×) of cold probe.
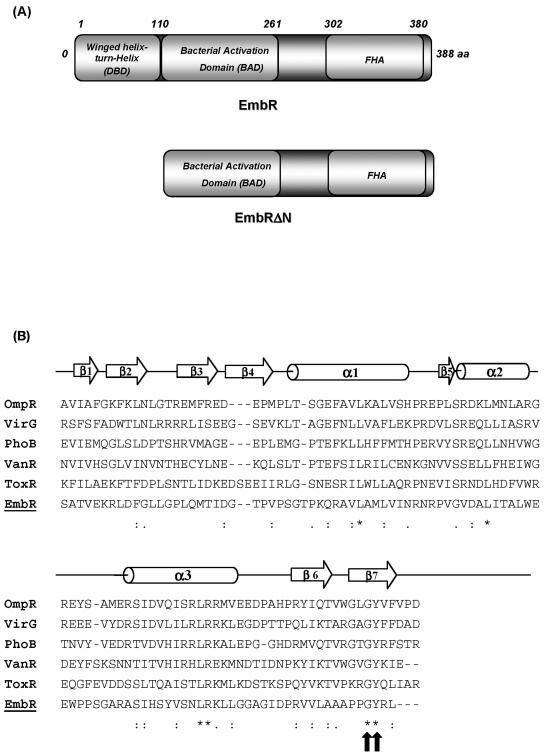
To further characterize its DNA binding activity, we mapped the domain of EmbR involved in DNA recognition and interaction (Fig. 2A). A deletion mutant, EmbRΔN, lacking this DNA binding domain (Fig. 2A) was created and found to be incapable of binding to DNA at concentrations as high as 15 μg (data not shown). The predicted secondary structure of the EmbR DNA binding domain was found to be highly similar to the winged-helix DNA binding domains of the OmpR/PhoB family (Fig. 2B), a large family of transcription factors that bind DNA through the central helix-turn-helix motif (20). To further ascertain that the mapped DNA binding domain of EmbR is involved in DNA interaction and binding, individual alanine substitutions were made at two targeted positions (Gly100 and Tyr101). The resulting mutants were impaired in their DNA binding activity (data not shown). Any mutation in the corresponding Gly and Tyr residues is also known to abrogate DNA binding of other members of the OmpR family. Tyr is one of the crucial amino acid residues that form the hydrophobic core of the OmpR/PhoB DNA binding domain (20).
FIG. 2.
EmbR, a multidomain protein, belongs to the OmpR/PhoB family of transcriptional regulators. (A) Schematic representation of structural components of EmbR and the W-HTH-deleted EmbRΔN mutant. (B) Shown are an alignment of the DNA binding domain of EmbR with winged-helix DNA binding proteins of the OmpR family and a sequence alignment of the DNA binding domains of EmbR and OmpR/PhoB family members. The sequences of E. coli OmpR, Agrobacterium tumefaciens VirG, E. coli PhoB, Enterococcus faecium VanR, Vibrio cholerae ToxR, and M. tuberculosis EmbR DNA binding domains are shown. A schematic diagram of the secondary structure of EmbR is indicated above. The block arrows indicate β-sheets, and cylinders indicate α-helixes. The asterisks (*) indicate identical amino acids, and high similarity is indicated by the double dots (:). The gaps are introduced to optimize the alignment and are indicated by the dashes (-). The block arrows indicate the sites of the G100 and Y101 mutations.
The OmpR/PhoB family represents the DNA binding proteins that are involved in either positive or negative control of transcription of their target genes. The DNA binding activities of most of these OmpR/PhoB family members are controlled in a phosphorylation-dependent manner by the cognate sensor kinase (20). Considering the homology of EmbR with the OmpR/PhoB family and its phosphorylation-dependent DNA binding activity, the concern was then to investigate the kind of control, positive or negative, EmbR phosphorylation exerts on the transcription of embCAB genes.
Positive control of the embCAB operon by EmbR following its phosphorylation by PknH in vivo.
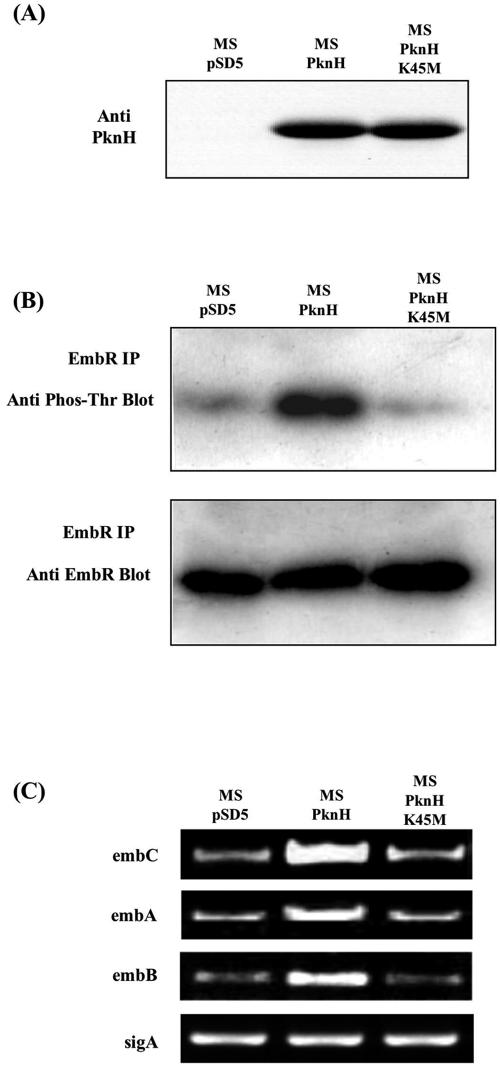
To investigate the possible transcriptional control of the embCAB operon by EmbR through its phosphorylation by PknH, we took advantage of the fact that PknH is absent in M. smegmatis (30) and also that emb genes are ubiquitous in all mycobacteria. Furthermore, the central component of this signal relay, EmbR, was detected in M. smegmatis by immunoprecipitation assays using antisera raised against EmbR of M. tuberculosis. Therefore, it was possible to use M. smegmatis as a model system to explore whether the M. tuberculosis pknH gene when expressed in M. smegmatis can modulate the phosphorylation status of EmbR and thereby regulate the transcriptional status of the embCAB operon. PknH and its inactive K45M mutant (the mutation in the K45 residue in the ATP binding site abrogates the kinase activity of PknH [30]) were expressed in M. smegmatis (Fig. 3A). Since PknH is known to phosphorylate EmbR at its threonine residue(s), the phosphorylation levels of endogenous EmbR in M. smegmatis harboring PknH, PknHK45M, or vector alone were compared after immunoprecipitating EmbR and probing with antiphosphothreonine antibody (Fig. 3B). The phosphorylation of endogenous EmbR was significantly increased in M. smegmatis expressing PknH (MS-PknH) compared with M. smegmatis expressing the K45M mutant of PknH (MS-PknHK45M) or vector alone (MS-pSD5). To elucidate the effect of PknH-mediated phosphorylation of EmbR on the expression level of embCAB genes, transcript levels of embC, embA, and embB genes were compared in the presence and absence of PknH (or the K45M mutant) in M. smegmatis. The transcription of embCAB genes was markedly increased in M. smegmatis upon expression of PknH but not its kinase mutant (Fig. 3C). These results are consistent with a positive regulatory effect of EmbR on transcription of the embCAB operon after its phosphorylation by PknH in vivo.
FIG. 3.
Regulatory effect of PknH on transcription of the embCAB operon through phosphorylation of EmbR. (A) M. smegmatis was electroporated with MS-pSD5 (vector alone), MS-PknH (strain expressing PknH), or MS-PknHK45N (strain expressing inactive kinase mutant). Whole-cell lysates were separated by SDS-PAGE and subjected to immunoblotting with anti-PknH antibody. (B) Whole-cell lysates of MS-pSD5, MS-PknH, and MS-PknHK45N were immunoprecipitated with anti-EmbR antibody. Immunoprecipitates were solubilized and subjected to immunoblotting using antiphosphothreonine and anti-EmbR antibodies. (C) M. smegmatis transformed with vectors encoding PknH/PknHK45M was subjected to RT-PCR analysis for comparison of transcript levels of indicated emb genes. sigA was used as an internal control.
Expression of PknH in M. smegmatis affects the LAM/LM ratio and EMB resistance.
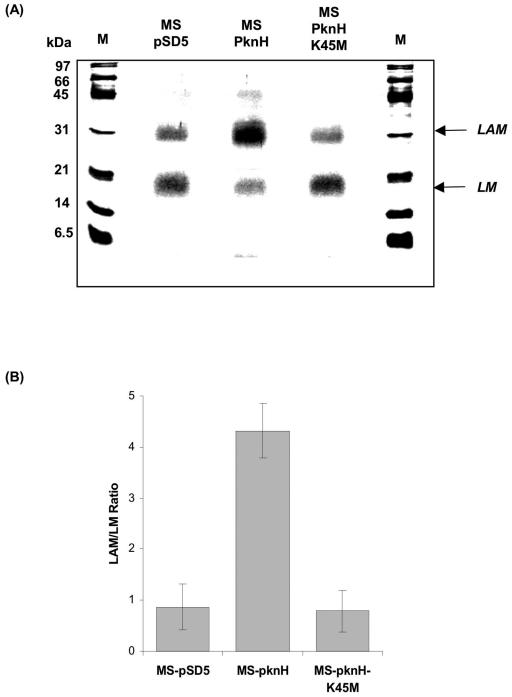
Our results established the effect of PknH expression on the transcript levels of embCAB genes in M. smegmatis. It has been well documented that the arabinosyltransferase encoded by the first gene of the emb operon, embC, catalyzes the arabinosylation of LM to LAM (40). Thus, if PknH expression affects embC transcription levels in vivo, then it may subsequently alter the LAM/LM ratio in the mycobacterial cell wall. Any change in the LAM/LM ratio upon PknH expression will thus imply a signal relay from PknH to EmbC via EmbR. A comparative analysis of the LAM/LM ratio in the cell wall of MS-pSD5, MS-PknH, and MS-PknHK45M was carried out by SDS-PAGE. Interestingly, the expression of PknH in M. smegmatis significantly increased the LAM/LM ratio by approximately fivefold vis-à-vis MS-pSD5.In contrast, expression of the inactive mutant of PknH, MS-PknHK45M, had no such effect on the LAM/LM ratio (Fig. 4). This observation has vital implications, as recent reviews have suggested a direct correlation between mycobacterial virulence and a high LAM/LM ratio (4). The arabinan domain of LAM inhibits the proinflammatory activity of LM on macrophages, presumably by masking the mannan core of LAM. Thus, maintaining a high LAM/LM ratio helps mycobacteria avert the host immune response. Furthermore, the cell wall component LAM from M. tuberculosis is involved in the inhibition of phagosome maturation (12), apoptosis (27), and interferon signaling (14, 15, 25, 35) in macrophages and interleukin-12 cytokine secretion of dendritic cells (16, 29). All these processes are important for the host to mount an efficient immune response. Therefore, our results, for the first time, establish a functionally relevant signaling mechanism involving the PknH-EmbR pair, leading to an altered LAM/LM ratio.
FIG. 4.
Effect of PknH on the LAM/LM ratio. (A) SDS-PAGE analysis of LAM/LM fractions isolated from MS-pSD5, MS-PknH, and MS-PknHK45N grown to mid-log phase by the phenol extraction method as described in Materials and Methods. After total hexose estimation, approximately the same relative proportions of the total yield of lipoglycans were loaded on each lane for all strains, and the lipoglycans were visualized by silver-PAS. A densitometric analysis was performed to calculate the LAM/LM ratio. The SDS-PAGE shown represents the average LAM/LM patterns that were observed in three independent experiments. M represents molecular mass marker. (B) After densitometric analysis, data were expressed as the ratio LAM/LM for different mycobacterial strains. The experiment was repeated three times, and the statistical analysis was performed using Student's t test. LAM/LM ratios were represented as means ± standard errors of the mean (n = 3, P < 0.01) for MS-pSD5 (0.86 ± 0.45), MS-PknH (4.32 ± 0.54), and MS-PknHK45N (0.79 ± 0.41).
Another phenomenon associated with the emb operon is EMB resistance. Several studies have shown that the arabinan biosynthesis of AG is the critical target of EMB (33, 37). EMB specifically inhibits the polymerization of cell wall arabinan by primarily targeting the later two arabinosyltransferases of the emb operon, EmbB and EmbA. The level of EMB resistance depends on the gene copy number of EmbA and EmbB (2). Therefore, it was tempting to examine whether PknH-mediated enhanced transcription of EmbB and EmbA genes in M. smegmatis has any effect on ethambutol resistance. Intriguingly, the susceptibility of MS-PknH to EMB was reduced by threefold (MIC of 0.75 μg/ml) in comparison to MS-PknHK45M and MS-pSD5 (MIC of 0.25 μg/ml for both strains). The above results reveal that the PknH-EmbR pair modulates ethambutol resistance by regulating transcription levels of embB and embA. As mentioned earlier, a mutation in the EmbR FHA domain, the kinase-interacting domain of EmbR, was also shown to be associated with ethambutol resistance (26). It would be interesting to elucidate the mechanism by which such a mutation alters the signal relay between PknH and EmbR, resulting in ethambutol resistance.
Upregulation of M. tuberculosis PknH during macrophage infection.
An important aspect of the life cycle of M. tuberculosis is its ability to survive within host macrophages. Enhanced expression of a bacterial gene during intracellular survival implies a role of its product in circumventing the unfavorable intracellular environment. In order to comprehend the role of PknH during in vivo growth of M. tuberculosis, it was therefore pertinent to investigate the expression of PknH following transition from extracellular to intracellular environment. RNA samples were therefore extracted from exponentially growing cultures and those adapted to the intracellular environment after 24, 48, and 72 h of infection. Real-time PCR was performed using specific TaqMan probes for pknH, sigA, and echA19. The major housekeeping sigma factor gene sigA was used as an internal control for the normalization of mRNA levels, and echA19, a gene known to be upregulated during infection in macrophages, was used as a positive control. As shown in Table 1, exposure to the intracellular environment increased the level of PknH transcription to a significant extent. Therefore, the significance of PknH in M. tuberculosis is apparent from the fact that this kinase is not only absent in nonpathogenic mycobacterial strains (31) but also upregulated inside the host macrophages and involved in the regulation of the LAM/LM ratio, an important determinant for averting the host's immune response.
TABLE 1.
Quantitative real-time PCR analysis of M. tuberculosis PknH regulation after cultivation in macrophages
| Gene | Encoded protein | Gene induction ratioa
|
||
|---|---|---|---|---|
| 24 h | 48 h | 72 h | ||
| sigA | Sigma factor A | −1.1 | 1.0 | 1.3 |
| pknH | Serine/threonine kinase | 5.0 | 6.1 | 6.8 |
| echA19 | Enoyl-coenzyme A hydratase | 2.9 | 3.2 | 1.9 |
Gene induction ratios were calculated from the comparison with RNA from bacteria grown to mid-log phase in 7H9. Data are means from at least three independent measurements analyzing RNA from at least two macrophage infections.
DISCUSSION
Cell growth and development demand the cell wall to have a dynamic structure. In fact, the cell wall changes continuously in the course of cellular growth and developmental processes, such as sporulation, and in response to change in the environment. Besides, morphological adaptations like cell wall thickening in anaerobiosis could be crucial for survival of the mycobacteria (8, 39). Changes in the cell wall in response to various environmental stimuli are central to mycobacterial adaptation during infection (3, 21, 32). Many such stimuli are communicated within the cell by sensor kinases present on the mycobacterial membrane. PknH, a kinase unique to pathogenic mycobacterial strains, was induced on infection of macrophages with M. tuberculosis, underscoring the importance of the encoded gene product in processes that are important during the mycobacterial residence in the host environment. By analogy with eukaryotic signal transduction, we hypothesize that PknH undergoes autophosphorylation on sensing certain external stimuli and transfers the signal to EmbR by means of phosphorylation. Our results show that EmbR is physically and functionally engaged as a mediator of embCAB activation by PknH in vivo. The consequent enhanced transcription of embC results in a higher LAM/LM ratio, an important determinant in mycobacterial virulence. These complex molecules are believed to play important roles in the physiology of the bacterium as well as in the modulation of the host response during infection. LAM is an important modulator of the immune response in the course of tuberculosis and leprosy (5, 23) and a key ligand in the interaction between M. tuberculosis, macrophages, and dendritic cells (18, 29). Furthermore, it was also found that PknH expression modestly but reproducibly alters EMB resistance by virtue of its ability to increase the transcription of embA and embB genes.
These observations raise a number of additional questions. The demonstration that embCAB activation is controlled by EmbR phosphorylation connects the LAM/LM ratio with the phosphorylation status of PknH. In the future, signals/ligands that activate PknH during mycobacterial infection can be identified.
An unresolved question raised by this model is the fate of phosphorylated PknH and EmbR at the end of the signaling cycle. Returning back to the inactive/resting state would require either the synthesis of new proteins or the dephosphorylation of the existing phosphorylated species. In addition, though it has been shown that EmbR is phosphorylated by PknH, we do not rule out the possibility that EmbR may also serve as a substrate of other STPKs in mycobacteria. Experiments to evaluate these possibilities are in progress.
The identification of PknH and EmbR as a pair that regulates arabinan metabolism in M. tuberculosis suggests that it could be targeted therapeutically. Future efforts will address, in addition to molecular details, the role of the PknH-EmbR signaling system in mycobacterial pathogenesis.
Acknowledgments
Financial support was provided by CSIR (SMM 0003).
Many thanks go to John T. Belisle (Colorado State University) for providing monoclonal antibody CS-35 through NIH contract N01-AI-40091 (Tuberculosis Research Materials and Vaccine Testing). N.G. and S.S. were the recipients of “Catch Them Young” fellowships of CSIR given to ACBR, Delhi University. Thanks to Beena Pillai for editing the manuscript.
REFERENCES
- 1.Av-Gay, Y., and V. Deretic. 2005. Two-component systems, protein kinases, and signal transduction in Mycobacterium tuberculosis, p. 359-368. In S. T. Cole, K. D. Eisenach, D. N. McMurray, and W. R. Jacobs, Jr. (ed.), Tuberculosis and the tubercle bacillus. ASM Press, Washington, D.C.
- 2.Belanger, A. E., G. S. Besra, M. E. Ford, K. Mikusova, J. T. Belisle, P. J. Brennan, and J. M. Inamine. 1996. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc. Natl. Acad. Sci. USA 93:11919-11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan, P. J. 2003. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 83:91-97. [DOI] [PubMed] [Google Scholar]
- 4.Briken, V., S. A. Porcelli, G. S. Besra, and L. Kremer. 2004. Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response. Mol. Microbiol. 53:391-403. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee, D., and K. H. Khoo. 1998. Mycobacterial lipoarabinomannan: an extraordinary lipoheteroglycan with profound physiological effects. Glycobiology 8:113-120. [DOI] [PubMed] [Google Scholar]
- 6.Chopra, P., B. Singh, R. Singh, R. Vohra, A. Koul, L. S. Meena, H. Koduri, M. Ghildiyal, P. Deol, T. K. Das, A. K. Tyagi, and Y. Singh. 2003. Phosphoprotein phosphatase of Mycobacterium tuberculosis dephosphorylates serine-threonine kinases PknA and PknB. Biochem. Biophys. Res. Commun. 311:112-120. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham, A. F., and C. L. Spreadbury. 1998. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton α-crystallin homolog. J. Bacteriol. 180:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhar, N., V. Rao, and A. K. Tyagi. 2000. Recombinant BCG approach for development of vaccines: cloning and expression of immunodominant antigens of M. tuberculosis. FEMS Microbiol. Lett. 190:309-316. [DOI] [PubMed] [Google Scholar]
- 10.Ehrt, S., M. Voskuil, G. K. Schoolnik, and D. Schnappinger. 2002. Genome-wide expression profiling of intracellular bacteria: the interaction of Mycobacterium tuberculosis with macrophages, p. 169-180. In S. H. E. Kaufmann and D. Kabelitz (ed.), Immunology of infection. Academic Press, London, United Kingdom.
- 11.Escuyer, V. E., M. A. Lety, J. B. Torrelles, K. H. Khoo, J. B. Tang, C. D. Rithner, C. Frehel, M. R. McNeil, P. J. Brennan, and D. Chatterjee. 2001. The role of the embA and embB gene products in the biosynthesis of the terminal hexaarabinofuranosyl motif of Mycobacterium smegmatis arabinogalactan. J. Biol. Chem. 52:48854-48862. [DOI] [PubMed] [Google Scholar]
- 12.Fratti, R. A., J. Chua, I. Vergne, and V. Deretic. 2003. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc. Natl. Acad. Sci. USA 100:5437-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hingley-Wilson, S. M., V. K. Sambandamurthy, and W. R. Jacobs, Jr. 2003. Survival perspectives from the world's most successful pathogen, Mycobacterium tuberculosis. Nat. Immunol. 4:949-955. [DOI] [PubMed] [Google Scholar]
- 14.Hmama, Z., R. Gabathuler, W. A. Jefferies, G. de Jong, and N. E. Reiner. 1998. Attenuation of HLA-DR expression by mononuclear phagocytes infected with Mycobacterium tuberculosis is related to intracellular sequestration of immature class II heterodimers. J. Immunol. 161:4882-4893. [PubMed] [Google Scholar]
- 15.Hussain, S., B. S. Zwilling, and W. P. Lafuse. 1999. Mycobacterium avium infection of mouse macrophages inhibits IFN-gamma Janus kinase-STAT signaling and gene induction by down-regulation of the IFN-gamma receptor. J. Immunol. 163:2041-2048. [PubMed] [Google Scholar]
- 16.Keane, J., H. G. Remold, and H. Kornfeld. 2000. Virulent Mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J. Immunol. 164:2016-2020. [DOI] [PubMed] [Google Scholar]
- 17.Koul, A., T. Herget, B. Klebl, and A. Ullrich. 2004. Interplay between mycobacteria and host signalling pathways. Nat. Rev. Microbiol. 2:189-202. [DOI] [PubMed] [Google Scholar]
- 18.Maeda, N., J. Nigou, J. L. Herrmann, M. Jackson, A. Amara, P. H. Lagrange, G. Puzo, B. Gicquel, and O. Neyrolles. 2003. The cell surface receptor DC-SIGN discriminates between Mycobacterium species through selective recognition of the mannose caps on lipoarabinomannan. J. Biol. Chem. 278:5513-5516. [DOI] [PubMed] [Google Scholar]
- 19.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Hackert, E., and A. M. Stock. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269:301-312. [DOI] [PubMed] [Google Scholar]
- 21.McNeil, M. R., and P. J. Brennan. 1991. Structure, function and biogenesis of the cell envelope of mycobacteria in relation to bacterial physiology, pathogenesis and drug resistance: some thoughts and possibilities arising from recent structural information. Res. Microbiol. 142:451-463. [DOI] [PubMed] [Google Scholar]
- 22.Molle, V., L. Kremer, C. Girard-Blanc, G. S. Besra, A. J. Cozzone, and J. F. Prost. 2003. An FHA phosphoprotein recognition domain mediates protein EmbR phosphorylation by PknH, a Ser/Thr protein kinase from Mycobacterium tuberculosis. Biochemistry 42:15300-15309. [DOI] [PubMed] [Google Scholar]
- 23.Nigou, J., M. Gilleron, M. Rojas, L. F. Garcia, M. Thurnher, and G. Puzo. 2002. Mycobacterial lipoarabinomannans: modulators of dendritic cell function and the apoptotic response. Microbes Infect. 4:945-953. [DOI] [PubMed] [Google Scholar]
- 24.Onaka, H., M. Sugiyama, and S. Horinouchi. 1997. A mutation at proline-115 in the A-factor receptor protein of Streptomyces griseus abolishes DNA-binding ability but not ligand-binding ability. J. Bacteriol. 179:2748-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pai, R. K., M. Convery, T. A. Hamilton, W. H. Boom, and C. V. Harding. 2003. Inhibition of IFN-gamma-induced class II transactivator expression by a 19-kDa lipoprotein from Mycobacterium tuberculosis: a potential mechanism for immune evasion. J. Immunol. 171:175-184. [DOI] [PubMed] [Google Scholar]
- 26.Ramaswamy, S. V., A. G. Amin, S. Goksel, C. E. Stager, S. J. Dou, H. El Sahly, S. L. Moghazeh, B. N. Kreiswirth, and J. M. Musser. 2000. Molecular genetic analysis of nucleotide polymorphisms associated with ethambutol resistance in human isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:326-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rojas, M., L. F. Garcia, J. Nigou, G. Puzo, and M. Olivier. 2000. Mannosylated lipoarabinomannan antagonizes Mycobacterium tuberculosis-induced macrophage apoptosis by altering Ca+2-dependent cell signaling. J. Infect. Dis. 182:240-251. [DOI] [PubMed] [Google Scholar]
- 28.Russell, D. G. 2001. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell. Biol. 2:569-577. [DOI] [PubMed] [Google Scholar]
- 29.Schlesinger, L. S., S. R. Hull, and T. M. Kaufman. 1994. Binding of the terminal mannosyl units of lipoarabinomannan from a virulent strain of Mycobacterium tuberculosis to human macrophages. J. Immunol. 152:4070-4079. [PubMed] [Google Scholar]
- 30.Sharma, K., H. Chandra, P. K. Gupta, M. Pathak, A. Narayan, L. S. Meena, R. C. D'Souza, P. Chopra, S. Ramachandran, and Y. Singh. 2004. PknH, a transmembrane Hank's type serine/threonine kinase from Mycobacterium tuberculosis is differentially expressed under stress conditions. FEMS Microbiol. Lett. 233:107-113. [DOI] [PubMed] [Google Scholar]
- 31.Sharma, K., P. Chopra, and Y. Singh. 2004. Recent advances towards identification of new drug targets for Mycobacterium tuberculosis. Expert Opin. Ther. Targets 8:79-93. [DOI] [PubMed] [Google Scholar]
- 32.Strohmeier, G. R., and M. J. Fenton. 1999. Roles of lipoarabinomannan in the pathogenesis of tuberculosis. Microbes Infect. 1:709-717. [DOI] [PubMed] [Google Scholar]
- 33.Takayama, K., and J. O. Kilburn. 1989. Inhibition of synthesis of arabinogalactan by ethambutol in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 33:1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Telenti, A., W. J. Philipp, S. Sreevatsan, C. Bernasconi, K. E. Stockbauer, B. Wieles, J. M. Musser, and W. R. Jacobs, Jr. 1997. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat. Med. 3:567-570. [DOI] [PubMed] [Google Scholar]
- 35.Ting, L. M., A. C. Kim, A. Cattamanchi, and J. D. Ernst. 1999. Mycobacterium tuberculosis inhibits IFN-gamma transcriptional responses without inhibiting activation of STAT1. J. Immunol. 163:3898-3906. [PubMed] [Google Scholar]
- 36.Walburger, A., A. Koul, G. Ferrari, L. Nguyen, C. Prescianotto-Baschong, K. Huygen, B. Klebl, C. Thompson, G. Bacher, and J. Pieters. 2004. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 304:1800-1804. [DOI] [PubMed] [Google Scholar]
- 37.Wolucka, B. A., M. R. McNeil, E. de Hoffmann, T. Chojnacki, and P. J. Brennan. 1994. Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J. Biol. Chem. 269:23328-23335. [PubMed] [Google Scholar]
- 38.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2000. An A factor-dependent extracytoplasmic function sigma factor (sAdsA) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan, Y., D. D. Crane, and C. E. Barry III. 1996. Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial alpha-crystallin homolog. J. Bacteriol. 178:4484-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, N., J. B. Torrelles, M. R. McNeil, V. E. Escuyer, K. H. Khoo, P. J. Brennan, and D. Chatterjee. 2003. The Emb proteins of mycobacteria direct arabinosylation of lipoarabinomannan and arabinogalactan via an N-terminal recognition region and a C-terminal synthetic region. Mol. Microbiol. 1:69-76. [DOI] [PubMed] [Google Scholar]